Abstract
In the search for common genetic variants that contribute to prevalent human diseases, patterns of linkage disequilibrium (LD) among linked markers should be considered when selecting SNPs. Genotyping efficiency can be increased by choosing tagging SNPs (tagSNPs) in LD with other SNPs. However, it remains to be seen whether tagSNPs defined in one population efficiently capture LD in other populations; that is, how portable tagSNPs are. Indeed, tagSNP portability is a challenge for the applicability of HapMap results. We analyzed 144 SNPs in a 1-Mb region of chromosome 22 in 1055 individuals from 38 worldwide populations, classified into seven continental groups. We measured tagSNP portability by choosing three reference populations (to approximate the three HapMap populations), defining tagSNPs, and applying them to other populations independently on the availability of information on the tagSNPs in the compared population. We found that tagSNPs are highly informative in other populations within each continental group. Moreover, tagSNPs defined in Europeans are often efficient for Middle Eastern and Central/South Asian populations. TagSNPs defined in the three reference populations are also efficient for more distant and differentiated populations (Oceania, Americas), in which the impact of their special demographic history on the genetic structure does not interfere with successfully detecting the most common haplotype variation. This high degree of portability lends promise to the search for disease association in different populations, once tagSNPs are defined in a few reference populations like those analyzed in the HapMap initiative.
It is estimated that the human genome contains >5 million common SNPs with a minor allele frequency of ≥10% (Kruglyak and Nickerson 2001; Carlson et al. 2003; The International HapMap Consortium 2003, 2005). Common SNP analysis lies at the core of current approaches to unravel the genetic bases of complex diseases; it may allow identification of individual risk factors and an understanding of the biological processes that may lead to disease through the implication of specific gene products. The main approach consists of identifying variants (SNPs) that occur at significantly higher (or lower) frequencies in patients compared with controls and, therefore, might predispose to (or protect from) the disease. This association approach is potentially powerful (Risch and Merikangas 1996; Kruglyak 1999; Carlson et al. 2004a), but may require large numbers (500,000–1,000,000) of SNPs to be genotyped in genome-wide association studies.
Efforts are being made to reduce the number of SNPs that may be required for such studies to ∼300,000 (Gabriel et al. 2002; Carlson et al. 2004a) by utilizing the patterns of linkage disequilibrium (LD) present in human populations. Deep understanding of LD structure can allow selection of tagSNPs (Johnson et al. 2001), that is, the SNPs that most efficiently represent the others in a genomic region with high LD, which saves genotyping costs. One of the major goals of the HapMap initiative (http://www.hapmap.org) is to construct an LD map of the whole human genome by determining the genotypes of >1 million SNPs, which is being further developed and completed under HapMap Phase II. The project also aims to characterize the structure of common haplotypes and identify tagSNPs throughout the genome (The International HapMap Consortium 2003, 2005). The SNPs used in HapMap are selected from dbSNP, which is constructed from a wide variety of sources; however, this does not avoid completely ascertainment bias (Ardlie et al. 2002). The HapMap samples comprise 270 individuals from four populations: 30 both-parent-and-adult-child trios from the Yoruba, in Ibadan, Nigeria; 45 unrelated individuals from Tokyo, Japan; 45 unrelated Han Chinese individuals from Beijing (Japanese and Chinese are sometimes considered as a single Asian sample); and 30 trios from the CEPH collection (Utah residents with ancestry from Northern and Western Europe).
Independent efforts are being undertaken to define both common haplotypes and tagSNPs for gene regions, encompassing both the coding and regulatory regions that will be of special interest in a candidate gene approach (Crawford et al. 2004). In such cases, resequencing is preferred over genotyping in order to prevent ascertainment bias of SNPs (Soldevila et al. 2005), but this has the drawback that the number of individuals included is reduced due to the effort required. In most cases, samples from only two or three populations are studied (usually European, Asian, and African), even though previous worldwide surveys of small genomic regions have shown that haplotype composition, LD structure, and LD decay with physical distance are heterogeneous across populations (Pritchard and Przeworski 2001; Mateu et al. 2002; Bertranpetit et al. 2003 and references therein). Nonetheless, a genetic difference measured, for example, in terms of allele or haplotype frequency differences does not necessarily imply differences in the utility of tagSNPs (Ramirez-Soriano et al. 2005). It is not feasible to screen for tagSNPs in all populations of interest, since this involves extensive assessment of large SNP sets at a very high density in order to select those that retain most of the fine-scale structure. Therefore, population heterogeneity in LD patterns and tagSNP portability across populations should be clarified before the tagSNPs defined in current surveys become widespread in the scientific community.
Two main questions require answers. First, how well do tagSNPs defined in one population perform in another population from the same or from a different continent? Second, should haplotype maps of the human genome be developed urgently in other populations for tagSNP selection besides the three main groups included in HapMap? Indeed, portability of tagSNPs among populations and continental groups is fundamental for the future application of HapMap-defined tagSNPs into other populations. To address these questions we have analyzed the LD structure and tagSNP portability across a worldwide set of samples in a region of chromosome 22.
Results
In a well characterized geneless region of chromosome 22, SNPs were selected based on physical distance criteria at a mean distance of 7 kb across 987,872 kb, dense enough to provide a consistent view of general LD patterns in the region (Ke et al. 2004). Selected SNPs were genotyped over 1000 individuals from 38 worldwide populations that contain most of the human genome variation (Table 1) classified in seven geographic regions according to population structure assessments (Rosenberg et al. 2002; see Methods). We then defined tagSNPs in three reference populations, close to those selected by the HapMap project, and analyzed the performance of these tagSNPs in the other populations, both within geographic regions and across geographic regions. We defined tagSNPs for the three reference populations using the r2-based approach of Carlson et al. (2004b), which is independent of haplotype block definition. Three different thresholds of r2 have been used (0.8, 0.64, and 0.5), covering a wide range of possibilities. Results based on the intermediate value (0.64) are presented throughout this paper; results based on the other values are given in the Supplemental material. Table 1 shows the geographic regions, populations, sample size, and number of common polymorphic SNPs within each region.
Table 1.
Descriptive parameters of the studied populations
| Region | Population | Abbreviation | N chromosomesa | Common SNPsb |
|---|---|---|---|---|
| Africa | ||||
| Bantu | BAN | 40 | 92 | |
| Mandenka | MAN | 48 | 92 | |
| Yoruba | YOR | 50 | 92 | |
| San | SAN | 14 | 92 | |
| Mbuti Pygmies | MBU | 30 | 92 | |
| Biaka Pygmies | BIA | 70 | 92 | |
| Europe | ||||
| Orcadian | ORC | 32 | 97 | |
| Adygei | ADY | 34 | 97 | |
| Russian | RUS | 50 | 97 | |
| French Basque | FBAS | 48 | 97 | |
| French France | FRA | 58 | 97 | |
| Continental Italian | CIT | 44 | 97 | |
| Sardinian | SAR | 56 | 97 | |
| Middle East/North Africa | ||||
| Mozabite | MOZ | 60 | 120 | |
| Bedouin | BED | 94 | 120 | |
| Druze | DRU | 88 | 120 | |
| Palestinian | PAL | 98 | 120 | |
| Central/South Asia | ||||
| Balochi | BAL | 50 | 78 | |
| Brahui | BRA | 50 | 78 | |
| Makrani | MAK | 50 | 78 | |
| Sindhi | SIN | 50 | 78 | |
| Pathan | PAT | 50 | 78 | |
| Burusho | BUR | 50 | 78 | |
| Hazara | HAZ | 50 | 78 | |
| Kalash | KAL | 50 | 78 | |
| East Asia | ||||
| Han | HAN | 90 | 89 | |
| North China | NCH | 138 | 89 | |
| South China | SCH | 140 | 89 | |
| Cambodian | CAM | 22 | 89 | |
| Japanese | JAP | 60 | 89 | |
| Yakut | YAK | 50 | 89 | |
| Oceania | ||||
| NAN Melanesian | NAN | 44 | 63 | |
| Papuan | PAP | 34 | 63 | |
| America | ||||
| Karitiana | KAR | 48 | 50 | |
| Surui | SUR | 42 | 50 | |
| Colombian | COL | 28 | 50 | |
| Maya | MAY | 50 | 50 | |
| Pima | PIM | 50 | 50 |
Number of chromosomes analyzed.
Common polymorphic SNPs within regions.
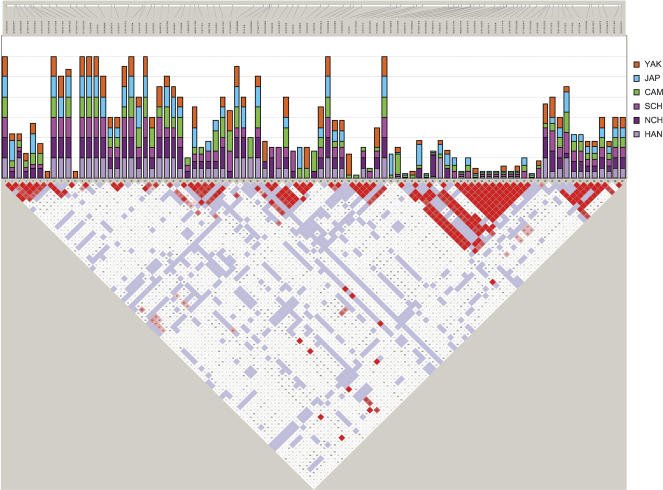
To assess whether tagSNPs are consistent among populations within regions, we calculated, for each SNP and population, the probability of being selected as a tagSNP (see Methods). As an example, results for the six East Asian populations are shown together in Figure 1 (top), along with the LD structure (bottom). As expected, the probabilities are high in regions with low LD and small in those exhibiting high LD. More interesting are the similar probability values found for the various populations within continents, with highly significant coefficients of multiple correlation (seven populations in Europe, R = 0.606; six in East Asia, R = 0.469; six in Africa, R = 0.576; P-values < 10–4), indicating a common pattern of LD. Nonetheless, these correlations within continents do not directly translate in terms of high tagSNP portability across populations; they simply show a common LD pattern among populations within each of the three continental groups.
Figure 1.
Plot of the probability of SNPs being tagSNPs (bar graph, upper middle), added together for the six Asian populations studied; the bar is made up by the sum of the probabilities in the six populations, and thus its maximum value is six. According to the ldSelect algorithm used for tagSNP selection, one or more SNPs within a bin can be specified as a tagSNP, and only one tagSNP need be genotyped per bin. Probability values are from 0 (no new information given by the SNP within the bin) to 1 (unique tagSNP selected in a bin). These values are compared with LD values (D′ parameter), shown in the bottom part of the figure as performed with the Haploview software package. In the D-plot, each diagonal represents a different SNP, with each square representing a pairwise comparison between two SNPs. (Red squares) Statistically significant LD between the pair of SNPs; (dark red) the higher values of D′, up to a maximum of 1. (White squares) Pairwise D′ values <1 with no statistically significant evidence of LD. (Blue squares) Pairwise D′ values of 1 but without statistical significance. (Top) Physical map of the region is shown. Population abbreviations are as in Table 1.
A direct insight into the issue of the portability of tagSNPs defined in specific populations into others of the same geographic region can be reached by focusing, among the populations in our study, those that may be considered as references for the three continents (and closest to the ones used in HapMap). TagSNPs are defined in these populations, those SNPs are applied into other populations within its regional groups as if they were their own tagSNPs, and their validity as tagSNPs is measured. The populations used as reference include: Yoruba (YOR) as African, French (FRA) as European, and Han Chinese (HAN) as Asian; Japanese was also used as a representative of Asia with very similar results as for Han Chinese (results not shown).
For each non-tagSNP in a population being tested, r2 was calculated with every tagSNP selected from a reference population and the maximum value recorded. This maximum r2 value is a measure of the utility of the SNPs that were defined as tags in another population. In the analysis, we considered all the SNPs independently of whether or not they were polymorphic in the compared population. Two approaches have been used: “blind” (in which nothing will be done if a tagSNP has no information in the compared population for being monomorphic or not successfully genotyped) and “ideal” (if a tagSNP has no information in the compared population, a replacement tag is selected in the reference population to ensure all tagSNPs contain information in the compared population); see Methods for more detail.
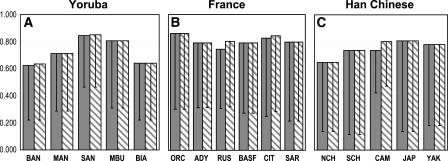
Mean values for those maximum r2 values (both for “blind” and “ideal” analysis) are presented in Figure 2 for the three geographic regions containing one of the three reference populations: Africa, with Yorubas as reference; Europe, with French as reference; and East Asia, with Han Chinese as reference. As expected, the highest r2 value for each non-tagSNP was found with the closest or with a very close SNP to the tagSNPs (defined in the reference population), even for singleton bin tagSNPs. <2% of the non-tagSNPs showing the maximum LD were at a distance further than three SNPs from the tagSNPs defined in the reference population. If those distant SNPs were removed, r2 dropped on average a mere 1.5–2% in various combinations of populations; thus, the signal of the LD measure comes overwhelmingly from the vicinity of the considered SNP.
Figure 2.
Average maximum r2 values of non-tag SNPs in a population with tagSNPs selected in HapMap proxy populations from the same geographic region. Population abbreviations are as in Table 1. An r2 threshold of 0.64 is used for tagSNP selection and evaluation. (A) Values when tagSNPs defined in Yorubas from Africa are used in the rest of African populations, (B) tagSNPs defined in French being used in the rest of European populations, (C) tagSNPs defined in Han Chinese being used in the rest of East Asian populations. For each case results for the “blind” test (opaque bars) and “ideal” test (dashed bars) are shown. Detailed information on number of SNPs and distribution of r2 values can be found in Supplemental Table 1. The 95th percentile values are shown as central bars from each mean value.
Results obtained using all three r2 values (0.8, 0.64, and 0.5) for both tests are provided in Supplemental Table 1, along with parameters including the number of SNPs, number of tagSNPs, tag efficiency, and proportion of values higher than the three threshold values used (0.5, 0.64, and 0.8) to give a more detailed distribution of maximum r2 values. Robustness of portability of tagSNPs was verified by comparing with the results of random SNP sets, which in all cases showed a strong decrease in average r2 values. The increase of average r2 achieved by using tagSNPs rather than random SNPs is in the order of 30%, with variation depending on the populations being used (results for the three reference populations and four compared populations are given in Supplemental Table 2). Results for the SNPs that are in both HapMap and the present study are very similar for the CEPH sample of HapMap and the French population used here (results not shown), as expected given the strong similarity among European populations in LD patterns.
When a 0.64 r2 threshold is used for selecting tagSNPs, the mean values of the maximum r2 of non-tagSNPs in other populations within each continent are very high, >0.60 in all cases and some of them >0.8, but with differences among continents and among populations in some cases (Fig. 2). The average maximum r2 values are highest in Europe (Fig. 2B); that is, on average, tagSNPs selected in the French population will tag SNPs in other European populations with very high r2 values. Thus, a tagSNP selected in one European population behaves as a good tagSNP in another European population, as previously seen in four gene regions in several European populations (Mueller et al. 2005). This is probably a consequence of the small genetic stratification of Europe (Simoni et al. 2000). The average maximum r2 values are slightly lower in East Asian populations (Fig. 2C) and in some African populations (Fig. 2A), while other African populations show lower values <0.8 but >0.60; as discussed below, Africa is the most diverse region for the portability of tagSNPs from one population to another within the continent.
The dispersion of mean r2 values obtained when using tagSNPs of a reference population into the compared one can be measured through the 95th percentile, shown as central bars in Figure 2, defined by the value that leaves only 5% of the r2 values below it (other parameters of the distribution are given in Supplemental Table 1). For Europe, 95th percentiles are mostly ∼0.3, meaning that less than about one in 20 non-tagSNPs will give results worse than r2 = 0.3 by tagSNPs defined in another population. For Asian populations, 95th percentiles are wider and reach smaller values, some <0.2; Africans have heterogeneous intervals, according to the variable r2 values.
Thus, although the portability of tagSNPs defined in the reference samples is reasonably high on average, the variability is such that some tagSNPs may not be informative in other populations from the same region. It is also relevant to global association studies to query on the portability of tagSNPs to populations from continents not covered by the three initial reference populations. Beyond the human populations that are represented by the three continental groups discussed here, an interesting question is to what extent human groups from different continents than the populations of reference could be productively analyzed using the initial three populations.
Although the existence of a unique underlying LD map in the human genome has been qualitatively suggested when comparing data from three or four populations (Ke et al. 2004; de la Vega 2005; The International HapMap Consortium 2005), the extension into other populations in a quantitative manner deserves our attention. To tackle this issue, we analyzed the portability of tagSNPs into populations of the Middle East and North Africa (which have genetic ties to Europeans [Jobling et al. 2004]), Central and South Asia (populations with either European or mixed European–East Asian ancestries [Comas et al. 2004]), and the more distant Oceania and America, where founder events and subsequent genetic drift have created both a clear genetic differentiation from their parental populations in Asia and a high level of interpopulation heterogeneity (Jobling et al. 2004).
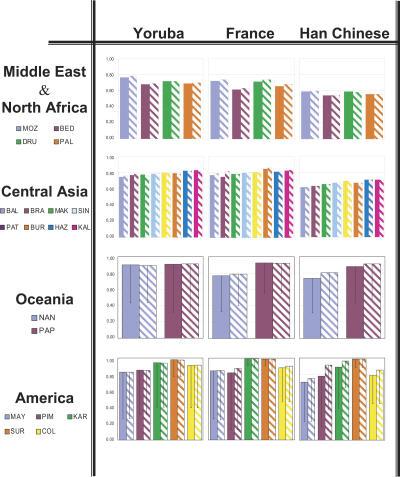
For all populations of these four regional groups, the same approach has been followed, using the tagSNPs defined in all three reference populations and applying them to each population following the same methods (“blind” and “ideal”). Results using a 0.64 r2 threshold are shown in Figure 3, but similar results were obtained applying 0.5 and 0.8 values (see Supplemental Table 1). Surprisingly, the mean r2 value is moderate to high for most populations, and it is rarely <0.6, even between distant groups. TagSNPs defined in the Yoruba are as portable as those defined in the French or the Han Chinese, although, since overall LD is lower in general in Yoruba, more tagSNPs are needed to represent a specific region (24–49 depending on the region as compared with 21–38 in French or 17–29 in Han Chinese; Supplemental Table 1). Therefore, for both Middle Eastern/North African and Central/South Asian populations, the utility of tagSNPs defined in Europeans is promising and much better than those defined in Han Chinese.
Figure 3.
Average maximum r2 obtained for non-tagSNPs when tagSNPs selected in the three reference populations are applied to populations of other geographic regions: Middle East, Central Asia, Oceania, and America. Population abbreviations are as in Table 1. For each case, results for the “blind” and “ideal” analysis are shown. Detailed information on the distribution can be found in Supplemental Table 1. The 95th percentile values are shown as central bars from each mean value.
Oceania and the Americas show similar average trends, with most values >0.8, and with the three reference populations providing portable tagSNPs, but the Asian reference has the highest efficiency (fewer markers to achieve a similar power). Populations from Oceania and the Americas have accrued genetic differentiation by drifting from their parental sources; therefore, it may be somewhat surprising that SNPs defined as tagSNPs elsewhere in the world do indeed capture LD patterns in America and Oceania, as well. It should be noticed that the SNPs used in the analyses were never ascertained in the Americas or Oceania, and they had non-extreme frequencies; they can thus tag the common haplotypes, which are the same ones found in other places of the world, especially Asia.
We have conducted two different analyses, and it is worth comparing them. The “blind” is much easier than the “ideal,” which intends to optimize the tagging of the compared population through the information of the reference one. In the majority of within- and across-continent portability analysis, no difference in terms of tagSNP performance is observed between the “ideal” and “blind” test; even when there is difference, it is generally very small. This means that even in situations where a tagSNP was found to be monomorphic or failed in genotyping in the compared population, the set of tagSNPs as a whole can still maintain good power (Figs. 2, 3; Supplemental Table 1).
Discussion
We have studied the portability of tagSNPs across worldwide populations and have found that tagSNPs are often highly portable across human populations, with the partial exception of some populations, mainly African. The tagSNPs defined in the current reference populations used in the HapMap project may be useful not only for other populations of the same geographic regions, but also for populations in the rest of the world. The present results go beyond the expected portability shown in Europe (Mueller et al. 2005), or the similar contour of the LD pattern across the standard three or four populations used in most studies (a single one or two from Europe, Asia, and Africa; Ke et al. 2004; de la Vega et al. 2005) or the Hap-Map project (The International HapMap Consortium 2005) and expands the analysis of portability of tagSNPs to a very wide variety of human groups from all continents (likely to represent most of human variation as seen by the analysis of neutral markers; Rosenberg et al. 2002) with a clear result: the extensive portability of tagSNPs across populations. There is a high portability as a mean, which does not imply portability for each tagSNP as seen by the distribution of maximum r2 values (Supplemental Table 1).
The best portability of tagSNPs is obtained using SNPs that are known to be polymorphic in both the reference populations and all the populations being compared in the same or different continental groups (data not shown). Nonetheless, this is not a real case, and values are artificially inflated. In a realistic situation, SNPs polymorphic in a reference population are not necessarily also polymorphic in a test population, and this is the scenario upon which the present study is based (“blind” and “ideal” tests). In a “blind” test, tagSNPs selected in a reference population are applied to a compared population without regard to whether any of the tags is monomorphic or fails the genotyping, whereas in an “ideal” test, efforts would be made to replace such monomorphic or failed tagSNPs. The results of the two tests are very similar and demonstrate a generally high portability of tags across populations. What is more, compared with the “blind” tests, there is hardly any increase in portability in the “ideal” tests. This further indicates that in a real-world situation, tagSNPs are generally very effective and portable across populations.
The observation that tagSNPs are very effective for distant and differentiated populations is an important one and suggests that new haplotype maps in other populations than those included in the current HapMap initiative are not urgently needed. We note, however, that the present data cover a small fraction of the genome at a density that is slightly less than that of the HapMap, and some sample sizes are small. Studies in other genomic regions, mainly in specific gene regions, and with higher marker density and also in other specific populations with large sample size would therefore be required, but the results here suggest promise for those panels in providing robust coverage in the genetic search for complex traits. In the present work, there are three populations with a sample size <30 chromosomes; this problem is acute in the San with only 14 chromosomes, but affects also Cambodians and Colombians. It is known that r2 is inflated when estimated from a very small number of chromosomes, and, as a result, the portability of tagSNPs will possibly be overestimated. Results about these populations in the present study, therefore, should be interpreted very carefully. It is interesting to note, however, that their behavior is very similar to other populations of the same geographic area with larger sample size.
Beyond the case of Eurasia and Africa, some other population groups deserve particular discussion. In the populations where drift (mainly through founder effect) has been an important factor in producing genetic differences among humans, portability does not seem to diminish. The main source of variation in those populations is the frequency of common haplotypes rather than their haplotype composition, and thus most of those common haplotypes will be captured by the same tagSNPs as in their source population (see references in Bertranpetit et al. 2003). Some of the common haplotypes may be lost by drift, but this seems not to be a major problem in portability. Most of the genetic differences between Native Americans or Oceanians and their founder populations are due to a lower amount of genetic diversity, as exemplified by the dearth of mtDNA, Y chromosome (Jobling et al. 2004), or ABO lineages. In this study we have observed that the sharing of SNPs and their general LD pattern between these populations and the rest of the world led to the portability of tagSNPs identified in other continents to them. Obviously, this is compatible with the existence of local polymorphisms generated after the initial colonization, with their own local patterns of LD and possible biomedical relevance. The situation in Africa may be different, with a longer time for recombination to shape LD patterns and impinge on SNP portability. Africa indeed deserves a more detailed study, and this could also be the case for some other specific populations.
The present results corresponding to a geneless region of chromosome 22 are relevant and are likely to be applied for the genome in general, and for gene regions in particular. It is known that LD patterns are unpredictable in a given region, and the most detailed studies in specific chromosomes (Patil et al. 2001; Dawson et al. 2002; Ke et al. 2004; de la Vega et al. 2005; Myers et al. 2005) have shown that, with marginal exceptions, LD patterns do not correlate with the feature contents of the genome, including genes. Thus, in the present state of knowledge, it seems likely that the present findings of a very high portability of tagSNPs among human populations can be taken as general for the human genome. Further studies that expand to other genome regions, other populations, or other SNP properties (allele frequency, density, distribution) are needed, but the general scenario of high portability seems to open the possibility of applying the tools defined by the HapMap project to any human population.
Methods
Data set
SNPs were selected at 5-kb spacing across a 987,872-bp region of human chromosome 22 (NCBI Build 34; 32600114 bp to 33587986 bp) using dbSNP build 115. To improve experimental success, we applied a hierarchical approach, preferentially selecting SNPs verified in Dawson et al. (2002), those reported by dbSNP to be verified in other studies, followed by those in which both alleles were observed in sequences from at least two different DNA samples. SNPs in which either allele was observed in only one DNA sample were used to fill remaining gaps. The sources of the sequences used to identify these SNPs included: SNP Consortium reads (where the libraries were created from the pooled DNA of 24 individuals from the polymorphism discovery panel); clone overlaps from the human genome sequence; end sequences from the Whitehead Institute/MIT Center for Genome Research fosmid library created from cell line NA15510; reads from four chromosome 22-specific sequencing libraries created at the Sanger Institute by flow-sorting the cell lines NA10470 (African-Pygmy), NA11321 (Chinese), NA17119 (African-American), and NA07340 (European); and the recent versions of the reference genome sequence.
Although it is still unclear whether the effects of natural selection can be wholly avoided, a gene-free region was selected in order to minimize the possible confounding effects of selection and hitchhiking. The 1-Mb region begins at the 3′ end of the Glycosyltransferase-like protein LARGE, which belongs to the Glycosyltransferase family 8; no other known gene maps to this interval. Different classes of repeats have been found in the region, including SINEs, LINEs, LTRs, STRs, and others (Dunham et al. 1999). For SNP assay design we used Spectrotyper (Sequenom) at a multiplex level of four SNPs. In total, we designed 211 assays. Genotyping was carried out with the Homogeneous MassExtend assay and MALDI-TOF mass spectrometry (Sequenom platform). The 211 SNPs were typed against the Human Genome Diversity Cell Line Panel (CEPH-HGDP; see below), including 10% of samples in duplicate, giving a total of 244,760 attempted genotypes. We removed all SNPs with a call rate <70% (34 in total), two with experimental problems, and 14 out of Hardy-Weinberg (HW) equilibrium. We set the threshold for HW failure as follows: The total number of tests with P < 0.05 gave an average of 4.8 populations with P < 0.05 per locus. Assuming that the number of populations that would fail HW for a given SNP follows a Poisson distribution, we removed the SNPs that failed in a number of populations over the 95% tail of the Poisson distribution (in this case, eight populations). In addition, 17 SNPs were monomorphic in all samples. In total, 144 SNPs were selected for further analysis, giving an average density of one SNP per 6.860 kb. For these markers, only 27 genotype discrepancies for specific SNPs in specific individuals were found in 16,704 duplicated genotypes (0.16%), and these results were discarded before the analysis.
The CEPH-HGDP diversity panel contains 1064 individuals representing 51 populations (Cann et al. 2002). Samples were regrouped into 38 populations based on geographic and ethnic criteria to avoid small sample size; we grouped Tuscans and North Italians as Continental Italians (CIT); Dai, Lahu, Miaozu, Naxi, She, Tujia, and Yiku as South Chinese (SCH); and Daur, Hezhen, Mongolian, Orogen, Tu, Uygur, and Xibo as North Chinese (NCH). Populations were clustered in regional groups according to the results obtained by Rosenberg et al. (2002) (Table 1). Nonetheless, other samples with a small number of chromosomes (San, 14; Cambodians, 22; Colombians, 28) were not grouped to others due to the lack of known similarity with other groups (Rosenberg et al. 2002). The importance of population size in estimating the common haplotype frequencies and defining tagSNPs seems to be acceptable for most cases. Two samples were dropped because of their uncertain population origin (HGDP00770 and HGDP00980). Also, HGDP00641, HGDP00652 (Bedouin); HGDP00564, HGDP00576, HGDP00588, HGDP00602 (Druze); and HGDP00682 (Palestine) failed systematically in the analysis and were rejected. Final sample size consists of 38 populations with a mean number of chromosomes of 55.5 and a median of 50.
Probability of being tags and definition of best tagging SNPs
ldSelect (Carlson et al. 2004b) was used for tagSNP selection. According to its algorithm, there were usually multiple alternative tagSNPs within a bin, from which only one was needed for genotyping. In a given population, for each of the tagSNPs in a bin, a probability value was calculated by dividing 1 with the total number of tagSNPs in the bin. Probability values were from 0 (e.g., for a SNP which is not selected as tagSNP) to 1 (unique tagSNP selected in a bin), with intermediate values determined by how many alternative tagSNPs were present. This probability value reflected the underlying LD information of a particular SNP in relation to other SNPs in the same region, and gave us an estimate on how such information was conserved across populations.
To apply tagSNPs from one population to another, best tagSNPs were used. They were defined based on the ldSelect algorithm with the following modification: If there were multiple tagSNPs in a bin, the most common tagSNPs (highest value of MAF) were always selected first because the more common SNP in a population, the higher the chance of it being polymorphic in another. If there were multiple tagSNPs in a bin (having the same highest MAF value), the average pairwise r2 between each of them and all the rest of SNPs in the same bin was calculated. TagSNPs with the highest average r2 values were selected from each bin to create the best tagSNP set. For a given population, tagSNPs were selected with a threshold of r2 > 0.5, 0.64 (default value of ldSelect and results given in the main text), and 0.8.
It may be stressed that r2 is inversely related to the sample sizes required for a given power in association studies (Weiss and Clark 2002), so that, for example, an r2 of 0.50 means that twice as many samples would be needed to achieve the same power as a directly measured causal SNP, given the same modeling assumptions for the indirect and direct SNPs.
Applying tagSNPs across populations
Two types of tests were carried out: “blind” test and “ideal” test. In a blind test, the best tagSNP set was selected from all the common markers (MAF >5%) in one reference population and applied to a test population to examine the tagging performance, disregarding whether any of the tagSNPs were monomorphic or had failed in genotyping. An ideal test was similar to a blind test except that if a tagSNP was found to be monomorphic or failed in genotyping in the test population, a tagSNP re-selection process was followed. In the tagSNP re-selection, all the tagSNPs that were genotyped successfully and were polymorphic in the test population would always be kept as tagSNPs, whereas those tagSNPs that failed in genotyping or were found to be monomorphic (i.e., nonfunctional) in the test population were excluded. This process was repeated until all tagSNPs were polymorphic and therefore functional in the test population.
For each of the two main types of tests, the following statistics were calculated to evaluate the effectiveness in a test population of tagSNPs selected from a reference population for three values of the r2 threshold (0.5, 0.64, and 0.8). For each of the non-tagSNPs in the test population, the pairwise r2 value between it and each of the tagSNPs was calculated. The maximum of such r2 values was regarded as the measure of how effective the tagSNPs as a whole were to that particular non-tagSNP in the test population. Average values of such overall non-tagSNPs (and the corresponding 95th percentile) were then computed as a measure of the overall effectiveness of a tagSNP set in another population. With each testing threshold of r2 (0.50, 0.64, and 0.80), and to have a better description of the distribution of maximum r2 values, we also computed the percentage of non-tagSNPs in a test population that had a maximum r2 value over a given cut point, using the same three r2 values.
Acknowledgments
This study was supported by the European Project QLG2-CT-2002-00916 and by the Ministerio de Ciencia y Tecnología from the Spanish Government (BMC2001-0772 and BFU2004-02002/BMC) and DURSI, Generalitat de Catalunya (Grup de Recerca Consolidat 2001SGR00285 and Distinció per a la Recerca Universitària to J.B.). Additional support was received from the Wellcome Trust and from the European Science Foundation (ESF) Integrated Approaches for Functional Genomics Program. We thank Mònica Vallés (UPF), and Sobia Raza and Benedict Cross (Sanger) for technical support, and Anthony Boyce for providing the unique environment of St. John's College, Oxford.
[Supplemental material is available online at www.genome.org.]
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4138406.
References
- Ardlie, K.G., Kruglyak, L., and Seielstad, M. 2002. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3 299–309. [DOI] [PubMed] [Google Scholar]
- Bertranpetit, J., Calafell, F., Comas, D., González-Neira, A., and Navarro, A. 2003. Structure of linkage disequilibrium in humans: Genome factors and population stratification. In Cold Spring Harb. Symp. Quant. Biol., pp. 79–88. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Cann, H.M., de Toma, C., Cazes, L., Legrand, M.F., Morel, V., Piouffre, L., Bodmer, J., Bonne-Tamir, B., Cambon-Thomsen, A., Chen, Z., et al. 2002. A human genome diversity cell line panel. Science 296 261–262. [DOI] [PubMed] [Google Scholar]
- Carlson, C.S., Eberle, M.A., Rieder, M.J., Smith, J.D., Kruglyak, L., and Nickerson, D.A. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33 518–521. [DOI] [PubMed] [Google Scholar]
- Carlson, C.S., Eberle, M.A., Kruglyak, L., and Nickerson, D.A. 2004a. Mapping complex disease loci in whole-genome association studies. Nature 429 446–452. [DOI] [PubMed] [Google Scholar]
- Carlson, C.S., Eberle, M.A., Rieder, M.J., Yi, Q., Kruglyak, L., and Nickerson, D.A. 2004b. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas, D., Plaza, S., Wells, R.S., Yuldaseva, N., Lao, O., Calafell, F., and Bertranpetit, J. 2004. Admixture, migrations, and dispersals in Central Asia: Evidence from maternal DNA lineages. Eur. J. Hum. Genet. 12 495–504. [DOI] [PubMed] [Google Scholar]
- Crawford, D.C., Carlson, C.S., Rieder, M.J., Carrington, D.P., Yi, Q., Smith, J.D., Eberle, M.A., Kruglyak, L., and Nickerson, D.A. 2004. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am. J. Hum. Genet. 74 610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, E., Abecasis, G.R., Bumpstead, S., Chen, Y., Hunt, S., Beare, D.M., Pabial, J., Dibling, T., Tinsley, E., Kirby, S., et al. 2002. A first-generation linkage disequilibrium map of human chromosome 22. Nature 418 544–548. [DOI] [PubMed] [Google Scholar]
- de la Vega, F.M., Isaac, H., Collins, A., Scafe, C.R., Halldorsson, B.V., Su, X., Lippert, R.A., Wang, Y., Laig-Webster, M., Koehler, R.T., et al. 2005. The linkage disequilibrium maps of three human chromosomes across four populations reflect their demographic history and a common underlying recombination pattern. Genome Res. 15 454–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, I., Hunt, R., Collins, J.E., Bruskiewich, R., Beare, D.M., Clamp, M., Smink, L.J., Ainscough, R., Almeida, J.P., Babbage, A., et al. 1999. The DNA sequence of human chromosome 22. Nature 402 489–495. [DOI] [PubMed] [Google Scholar]
- Gabriel, S.B., Schaffner, S.F., Nguyen, H., Moore, J.M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. 2002. The structure of haplotype blocks in the human genome. Science 296 2225–2229. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. 2003. The International HapMap Project. Nature 426 789–796. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. 2005. A haplotype map of the human genome. Nature 437 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling, M.A., Hurles, M.E., and Tyler-Smith, C. 2004. Human evolutionary genetics: Origins, peoples, and disease. Garland Science, Taylor & Francis, New York.
- Johnson, G.C., Esposito, L., Barratt, B.J., Smith, A.N., Heward, J., Di Genova, G., Ueda, H., Cordell, H.J., Eaves, I.A., Dudbridge, F., et al. 2001. Haplotype tagging for the identification of common disease genes. Nat. Genet. 29 233–237. [DOI] [PubMed] [Google Scholar]
- Ke, X., Hunt, S., Tapper, W., Lawrence, R., Stavrides, G., Ghori, J., Whittaker, P., Collins, A., Morris, A.P., Bentley, D., et al. 2004. The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum. Mol. Genet. 13 577–588. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L. 1999. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 22 139–144. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L. and Nickerson, D.A. 2001. Variation is the spice of life. Nat. Genet. 27 234–236. [DOI] [PubMed] [Google Scholar]
- Mateu, E., Pérez-Lezaún, A., Martínez-Arias, R., Andrés, A.M., Vallés, M., Bertranpetit, J., and Calafell, F. 2002. PKLR-GBA region shows almost complete linkage disequilibrium over 70 kb in a set of worldwide populations. Hum. Genet. 110 532–544. [DOI] [PubMed] [Google Scholar]
- Mueller, J.C., Lohmussaar, E., Magi, R., Remm, M., Bettecken, T., Lichtner, P., Biskup, S., Illig, T., Pfeufer, A., Luedemann, J., et al. 2005. Linkage disequilibrium patterns and tagSNP transferability among European populations. Am. J. Hum. Genet. 76 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, S., Bottolo, L., Freeman, C., McVean, G., and Donnelly, P. 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310 321–324. [DOI] [PubMed] [Google Scholar]
- Patil, N., Berno, A.J., Hinds, D.A., Barrett, W.A., Doshi, J.M., Hacker, C.R., Kautzer, C.R., Lee, D.H., Marjoribanks, C., McDonough, D.P., et al. 2001. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294 1719–1723. [DOI] [PubMed] [Google Scholar]
- Pritchard, J.K. and Przeworski, M. 2001. Linkage disequilibrium in humans: Models and data. Am. J. Hum. Genet. 69 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Soriano, A., Lao, O., Soldevila, M., Calafell, F., Bertranpetit, J., and Comas, D. 2005. Haplotype tagging efficiency in worldwide populations in CTLA4 gene. Genes Immun. 6 646–657. [DOI] [PubMed] [Google Scholar]
- Risch, N. and Merikangas, K. 1996. The future of genetic studies of complex human diseases. Science 273 1516–1517. [DOI] [PubMed] [Google Scholar]
- Rosenberg, N.A., Pritchard, J.K., Weber, J.L., Cann, H.M., Kidd, K.K., Zhivotovsky, L.A., and Feldman, M.W. 2002. Genetic structure of human populations. Science 298 2381–2385. [DOI] [PubMed] [Google Scholar]
- Simoni, L., Calafell, F., Pettener, D., Bertranpetit, J., and Barbujani, G. 2000. Geographic patterns of mtDNA diversity in Europe. Am. J. Hum. Genet. 66 262–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldevila, M., Calafell, F., Helgason, A., Stefansson, K., and Bertranpetit, J. 2005. Assessing the signatures of selection in PRNP from polymorphism data: Results support Kreitman and Di Rienzo's opinion. Trends Genet. 21 389–391. [DOI] [PubMed] [Google Scholar]
- Weiss, K.M. and Clark, A.G. 2002. Linkage disequilibrium and the mapping of complex human traits. Trends Genet. 18 19–24 [DOI] [PubMed] [Google Scholar]