Abstract
Introns are shorter in housekeeping genes than in tissue- or development-specific genes. Differing explanations have been offered for this phenomenon: selection for economy (in housekeeping genes), mutation bias or “genomic design.” The large-scale implementation in this present paper of a rigorous local sequence alignment algorithm revealed an unprecedented fraction of evolutionarily conserved DNA in human–mouse introns (∼60% of human and ∼70% of mouse intron length remained after masking for lineage-specific repeats). The length distributions of both conserved and nonconserved regions are very broad but show peaks close to nucleosomal and dinucleosomal DNA. Both the fraction of conserved sequence and its absolute length were higher in introns of tissue-specific genes than housekeeping genes. This difference remained after control for between-species identity of the conserved fraction, mutation rate, and GC content. In a more direct control, the product of the conserved sequence fraction and the between-species identity of this fraction (which can be considered to be the fraction of conserved nucleotides) was greater in introns of tissue-specific genes than housekeeping genes. Neither the fraction of intron length covered by repeats nor the balance of small insertions and deletions (indels) can explain the greater length of introns in tissue-specific genes. The length of the conserved intronic DNA in a gene is correlated with the number of functional domains in the protein encoded by that gene. These results suggest that the greater length of introns in tissue-specific genes is not due to selection for economy or mutation bias but instead is related to functional complexity (probably mediated by chromatin condensation), and that the evolution of the bulk of noncoding DNA is not completely neutral.
An important problem of modern genomics is the relevance of a poorly understood 99% of the human genome (noncoding part; Venter et al. 2001) to the functioning of a much better understood 1% (coding part). Variation in the amount of noncoding DNA in regard to gene expression can shed light on this problem. It is known that in rather diverse multicellular organisms (human, fruitfly, nematode), introns are longer in tissue- and development-specific genes than in housekeeping genes (Castillo-Davis et al. 2002; Eisenberg and Levanon 2003; Urrutia and Hurst 2003; Vinogradov 2004). Different explanations are proposed for this phenomenon: selection for economy (in housekeeping genes), mutation bias and “genome design.” The “selection for economy” hypothesis implicitly assumes a neutralist (permissive) interpretation of the accumulation of DNA in eukaryotic genomes. The widely expressed genes are supposed to “slim down” (selection condition), whereas those that work less intensively “get fat” (permissive condition) (Castillo-Davis et al. 2002; Eisenberg and Levanon 2003; Urrutia and Hurst 2003). In contrast, the “genome design” hypothesis suggests that the greater amount of intra- and intergenic noncoding DNA, in which the tissue-specific genes are embedded, is involved in the more complex regulation and chromatin-mediated suppression of these genes (Vinogradov 2004, 2005). In other words, the adaptationist “genome design” model postulates that the length of genomic elements is determined by their function. In contrast, the semineutralist “selection for economy” model assumes that the variation of the length of genomic elements is determined by the overall mutation pressure for greater length counteracted by economy selection in actively transcribed genes, whereas the neutralist “mutation bias” model suggests that it is determined only by the within-genome variation of mutation pressure.
In the present paper, the large-scale implementation of a rigorous local sequence alignment algorithm was used for revealing a maximum possible fraction of evolutionarily conserved DNA in the human–mouse comparison of introns in genes with different among-tissues breadth of expression. These results should help to sort out different explanations proposed for the within-genome variation in amount of noncoding DNA.
Results
The length of conserved regions
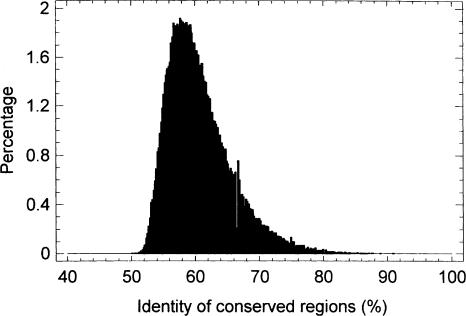
The total length of conserved regions (i.e., nonoverlapped consecutive local alignments) found in the present work constitutes 57.3% of intron length remaining after masking for lineage-specific repeats (44.4% of total intron length) in the human and 69.8% (52.0% of total intron length) in the mouse. Previously, in a much more limited comparison of human–mouse introns (77 genes), only 23% of intron length was found to be conserved (Jareborg et al. 1999). Similar figures (20%–30%) were obtained for conservation of intergenic noncoding DNA (Shabalina et al. 2001; Kondrashov and Shabalina 2002). The difference for the introns arises because in the previous work (Jareborg et al. 1999), an identity threshold (60%) was used, in contrast to a statistical significance threshold (P <10–6) applied in the present work (see Methods section). The data set used in the previous work (Jareborg et al. 1999) was analyzed here for comparison, and a similar fraction of conserved sequence (∼25% of total intronic length) was found only if matches with identity above 60% were taken. The fraction of conserved sequence rose to about 47% if all the statistically significant matches were taken, with the lowest identity level being about 53% (see Supplemental data: Tables 1 and 2, and the representative alignments for the most complex cases, i.e., for introns containing the largest numbers of local alignments). It should be noted that the identity threshold seems to be more arbitrary than the statistical significance threshold. In the work on the intergenic noncoding DNA, a threshold of 50% identity was used (Shabalina et al. 2001; Kondrashov and Shabalina 2002). For the total data set studied in the present work, the identity of matched regions (found on the basis of a statistical significance threshold) was generally in the range of 51%–85% (Fig. 1).
Figure 1.
Histogram of identities of conserved regions. (The conserved regions are the nonoverlapped local alignments.)
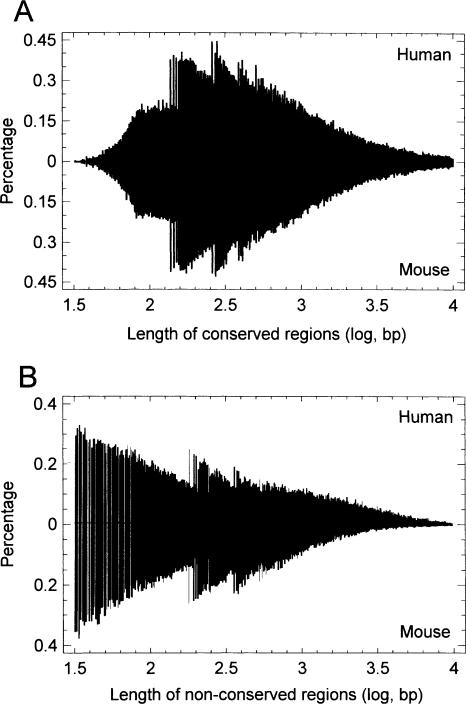
The distribution of the lengths of conserved regions shows peaks close to those of nucleosomal and dinucleosomal DNA, or their intermediates (Fig. 2A). It is known that the nucleosome core involves a DNA sequence of ∼150 nt length, whereas the nucleosome linker varies, so that the total nucleosomal DNA can be in the range of 170–220 nt (Mohd-Sarip and Verrijzer 2004; Nemeth and Langst 2004). Therefore, the dinucleosomal DNA can in general be in the range from 320 nt (two nucleosome cores plus one minimal linker) to 510 nt (two cores plus three maximal linkers). The right-skewed form of the observed peaks suggests that their left edges serve as boundaries of sequence identity decay. Surprisingly, the distribution of nonconserved regions shows similar peaks, albeit slightly shifted to the longer length (Fig. 2B). The whole picture indicates that the nucleosomal and dinucleosomal DNA sequences might be structural elements of identity conservation or decay (i.e., they are predominantly conserved or decayed as a whole). The slightly higher length corresponding to peaks of nonconserved regions (Fig. 2B) suggests that nucleosome linker regions (external linkers in the case of dinucleosomes) might be included in a predominantly decayed element but not in a predominantly conserved element. The nucleosome formation potential was on average higher in conserved regions compared to nonconserved (1.32 ± 0.01 vs. 1.21 ± 0.01, respectively).
Figure 2.
Histograms of lengths of conserved (A) and nonconserved (B) regions. (The conserved regions are the nonoverlapped local alignments; the nonconserved regions are sequence segments between the two nearest consecutive local alignments or between the extreme alignment and the corresponding intron end.)
The fraction of conserved sequence
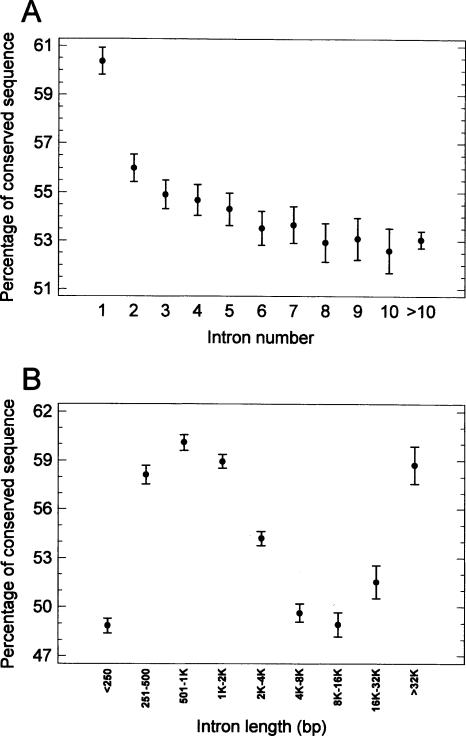
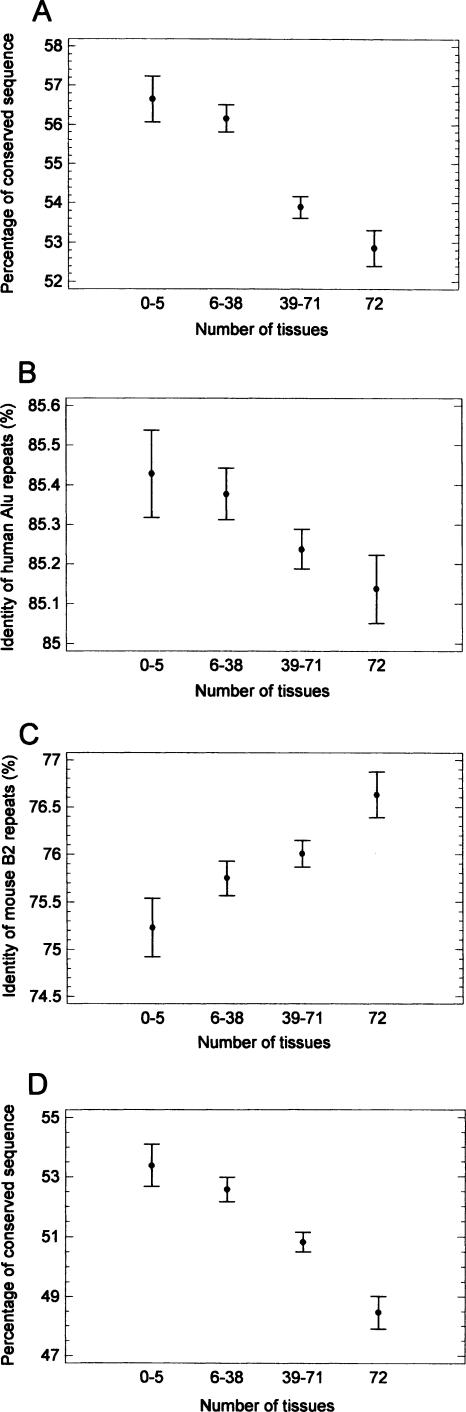
The fraction of conserved sequence was higher in the first intron (Fig. 3A). There is a complex (sigmoid) relationship between the intron length and the fraction of conserved sequence (Fig. 3B). In general, these parameters correlated weakly negatively (legend to Fig. 3B). However, introns of tissue-specific genes, which are longer than introns of housekeeping genes (Castillo-Davis et al. 2002; Eisenberg and Levanon 2003; Urrutia and Hurst 2003; Vinogradov 2004), show not only a higher absolute length of conserved sequence (not shown) but also a higher fraction of it (Fig. 4A). (The difference of conserved fraction between the gene groups was similar if it was calculated in regard to the total intron length. i.e., not only to the length remaining after masking for lineage-specific repeats, as in Fig. 4A.) The difference was even greater if the fraction of conserved sequence was calculated not on a by-intron basis (as in Fig. 4A) but as the ratio of the total conserved length to the total intron length (remaining after masking for lineage-specific repeats) in a given gene group. In the latter case, the percentage of conserved sequence varied from 60.1% in the genes expressed in zero to five tissues to 52.1% in the genes expressed in 72 tissues.
Figure 3.
The fraction of the conserved sequence (ratio of the conserved sequence length to intron length remaining after masking for lineage-specific repeats) in human introns (means with LSD intervals). (A) in first (and other) introns. (B) in different intron length classes. (For the correlation of conserved fraction with intron length, if all the introns were taken, Spearman r = –0.04, P <10–6, n = 65,432; if only introns containing the conserved fraction, Spearman r = –0.33, P < 10–6, n = 56,539. It should be noted that introns longer than 16 kb that show the rightmost upward trend in part B represent only <6% of the total intron number.)
Figure 4.
The fraction of conserved sequence and the identity of lineage-specific repeats (compared to their consensus sequence) in introns of genes expressed in different numbers of human tissues (means with LSD intervals). (A) fraction of conserved sequence in human introns. (B) identity of human Alu repeats. (C) identity of mouse B2 repeats. (D) fraction of conserved sequence in human introns, corrected simultaneously for identity of human Alu repeats, average between-species identity of conserved fraction and intron GC content, using the general linear model (GLM). (Only introns containing Alu repeats were taken in the latter case; the nonconserved regions were assumed to have zero identity.) For the effect of the number of tissues in the GLM, P <10–8. The picture was similar if the correction parameters were included in the model separately (one-by-one).
Can the higher fraction of conserved sequence in introns of tissue-specific genes be because of the lower mutation rate in them? The mutation rate (small insertions, deletions and nucleotide substitutions) was estimated by divergence of (intron-located) lineage-specific repeats from their ancestor copies. The picture was complicated. The human Alu repeats showed a higher divergence in the introns of housekeeping genes compared to tissue-specific genes (Fig. 4B). The mouse B4 repeats showed no dependence on tissue-specificity (not shown), whereas the mouse B2 repeats showed a lower divergence in the introns of housekeeping genes (Fig. 4C). It is known from the human–chimp comparison that primate lineage-specific repeats (Alu) located within introns have higher mutation rates than the rest of the intron (Fig. 2 in Kondrashov et al. 2006). This fact indicates that these relatively “fresh” insertions are not functionally constrained (or constrained to a lesser extent compared to the rest of intron), and thus they probably show a rate of divergence closer to the real mutation rate. The other intronic parts (including old, nonlineage specific repeats) can already be co-opted for some function and therefore evolutionarily constrained. Hence, the lineage-specific repeats seem to be the most sensitive indicators of mutation rate.
To make a conservative test of the effect shown in Figure 4A, the divergence of human Alu repeats (which showed a higher divergence in introns of housekeeping genes compared to tissue-specific, i.e., in the direction opposite to the effect shown in Fig. 4A) was used for correcting the mutation rate. After control for this parameter (together with control for between-species identity of conserved fraction and intronic GC content, taken as covariates in the general linear model, GLM), the fraction of conserved sequence still remained higher in introns of tissue-specific genes (Fig. 4D).
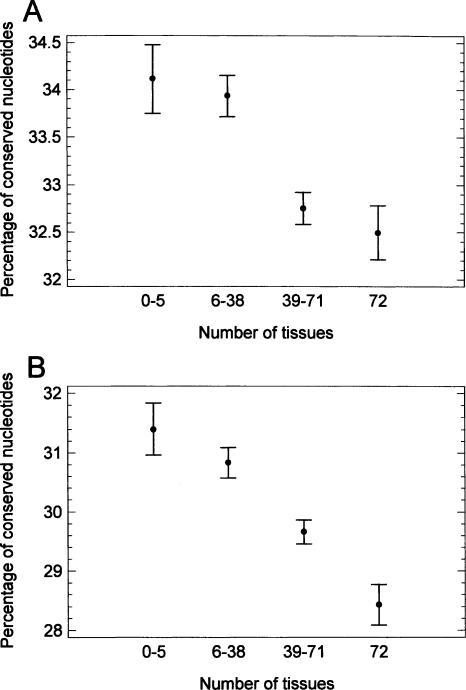
In a more direct variant of control for between-species identity of conserved fraction, the products of the conserved sequence fraction and the between-species identity of this fraction were compared. (This product can be considered as the fraction of sequence with 100% identity or the fraction of conserved nucleotides.) This parameter (with and without correction for mutation bias and intron GC content) was also higher in introns of tissue-specific compared to housekeeping genes (Fig. 5A,B). Also, the picture remained qualitatively similar after using the Jukes and Cantor (1969) method to correct the between-species identity of the conserved sequence for multiple and reversed mutations (not shown).
Figure 5.
The product of the conserved sequence fraction and the between-species identity of this fraction (which can be considered as the fraction of conserved nucleotides) in introns of genes expressed in different numbers of human tissues (means with LSD intervals). (A) without correction. (B) corrected for identity of human Alu repeats and intron GC content, using the general linear model (GLM). (Only introns containing Alu repeats were taken in the latter case.) For the effect of the number of tissues in the GLM, P <10–8.
Mutation bias
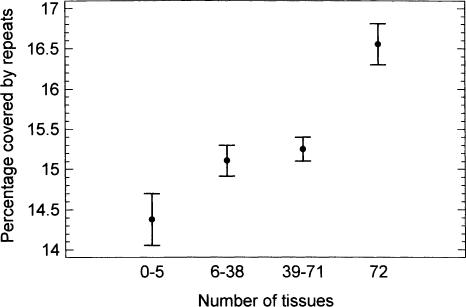
The fraction of intron length covered by lineage-specific repeats was higher in housekeeping genes (Fig. 6). Although the balance of small insertions and deletions (indels) was biased in favor of deletions, and the corresponding decrease of intron length was slightly higher in housekeeping genes (estimated using human Alu repeats), the difference between the extreme gene groups (i.e., expressed in 0–5 tissues and in 72 tissues) was minimal (<0.2%). Therefore, this difference cannot compensate for the difference in intron length caused by insertion of lineage-specific repeats (Fig. 6). For the fraction of intron length covered by all detectable repeats (i.e., not only those inserted after the human–mouse split), no dependence on tissue-specificity was found (not shown). As there is an increase of genome size in human compared to mouse (∼15%), it was interesting to compare the human–mouse balance of indels in the conserved regions of introns in genes with different expression breadth. The overall increase of conserved sequence length in human compared to mouse was ∼4.5%, with a minimal difference between the gene groups (4.3% in genes expressed in 0–5 tissues vs. 4.6% in genes expressed in 72 tissues). All these facts contradict the “mutation bias” explanation for longer introns in tissue-specific genes compared to housekeeping genes.
Figure 6.
The fraction covered by lineage-specific repeats in introns of human genes expressed in different numbers of tissues (means with LSD intervals).
Relation to protein complexity
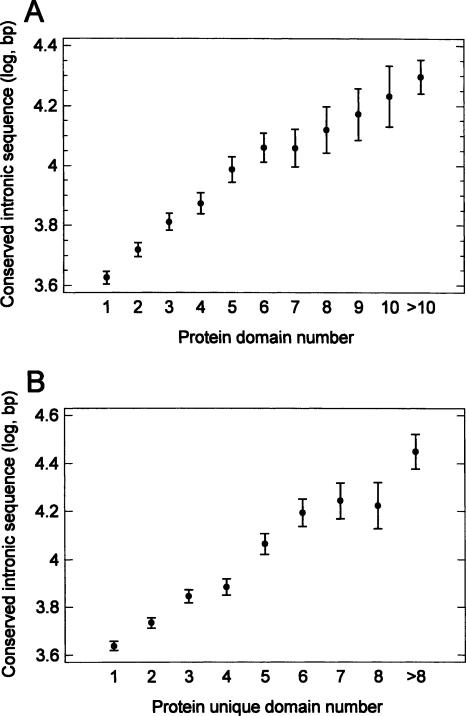
The length of the conserved intronic sequence was higher in the genes encoding for proteins with a higher number of functional domains, including unique (different) domains (Fig. 7A,B). This fact suggests that the higher protein functional complexity correlates with the higher amount of conserved noncoding DNA (as it was assumed in the “genome design” model).
Figure 7.
The length of conserved intronic sequence in human genes encoding for proteins with different numbers of functional domains (means with LSD intervals). (A) total number of domains. (B) number of unique (different) domains.
The problem of hidden exons
There is the problem of unknown alternatively spliced variants or antisense genes, the presence of which could be more likely in the longer introns. However, there is a general (albeit weak) decrease of conserved fraction with the increase of intron length (see Fig. 3B legend). Also, as human intronic sequences are on average 24-fold longer than exonic (Venter et al. 2001), only about 4% of intron length could be covered by alternatively spliced or antisense exons. At the same time, the difference in conserved fractions between the extreme gene groups (expressed in 0–5 tissues and in 72 tissues) is also about 4% (Fig. 4 A,D). To explain this difference by unknown exons hidden in introns, all introns of the first (the most tissue-specific) gene group should be completely covered by alternatively spliced variants or antisense genes while no coverage should occur in the housekeeping genes, which is very unlikely. In addition, the alternative splicing is poorly conserved between human and mouse (Nurtdinov et al. 2003). The exons of antisense genes usually overlap with exons of the sense genes, not with the introns (Chen et al. 2005a). When they overlap with introns (in the “antisense-like” genes), the introns of the sense gene are unusually long (Chen et al. 2005a), and exons of the antisense gene could cover only a negligible part of them. Moreover, if the conserved fraction was taken as the ratio of the total conserved length to total intron length, the difference between the extreme gene groups was ∼8%. This is twice as high as the putative hidden exonic length in the case of complete coverage of introns by alternatively spliced variants or antisense genes in the first (the most tissue-specific) gene group and the complete absence of this coverage in the housekeeping genes.
Discussion
The results presented here suggest that the greater length of introns in tissue-specific genes is not due to mutation bias or economy selection (in housekeeping genes). In the latter case, the fraction of conserved sequence should be higher in introns of housekeeping genes due to shrinkage of dispensable, nonconservative parts of introns. In contrast, the presented results show that the fraction of conserved intronic sequence is greater in the tissue-specific genes. This difference remains if the fraction of conserved nucleotides (i.e., only sequence with 100% identity) is considered. Furthermore, even if there was no difference, it would already indicate in favor of the “genome design” model (because it would mean that introns of both housekeeping and tissue-specific genes are loaded with function in proportion to their length). It should be noted that in the yeast Saccromyces cerevisiae (and probably some other unicellulars, judging on the correlation between intron length and frequency of optimal codons), the longer introns are in the highly expressed genes (Vinogradov 2001), which also contradicts the “economy selection” model. It was recently shown in a comparison of Drosophila melanogaster with D. simulans that the intron length correlated negatively with between-species sequence divergence (Haddrill et al. 2005). The longer introns are found in fruitfly development- and condition-specific genes compared to housekeeping (Vinogradov 2004); therefore, this result also supports the “genome design” model.
Recently, an examination of exon–intron proportions in human sense and antisense genes allowed rejection of the “mutation bias” and “economy selection” models for the case of bidirectional genes (Chen et al. 2005a,b). The sense (major) genes were found to have unusually long introns and unusually high intronic to exonic length ratios compared to nonbidirectional genes (Chen et al. 2005a,b). This is probably necessary for accommodation of the antisense genes on the opposite DNA strand (in agreement with the “genome design” model). These facts suggest that intronic sequence is not mere “junk” (as the “mutation bias” and “economy selection” models implicitly assume), because the presence of exons on the opposite strand results in the extension of intronic length. The antisense (minor) genes have a lower intronic to exonic length ratio compared to sense genes, which was interpreted as evidence for selection for rapid transcription (“efficiency selection”) required by presumably regulatory function of antisense genes (Chen et al. 2005a,b). Another possible explanation—the antisense genes are shorter because they should be accommodated within the loci of the sense (major) genes—is consistent with the “genome design” model.
Generally, the fraction of conserved sequence in human–mouse introns revealed in the present work constitutes roughly one half of (total) intronic length, which poses a question of its function. The conserved sequence seems to be related to chromatin condensation and functional complexity (Figs. 2, 7). It has long been argued that a bulk of eukaryotic noncoding DNA might be necessary for correct chromatin structure because exons are under selection for encoded information (e.g., Trifonov 1993; Zuckerkandl 1997; Levitsky et al. 2001; Vinogradov 2005). It was even shown in several cases that after experimentally removing the introns, genes lose the ability to form nucleosomes (Lauderdale and Stein 1992; Liu et al. 1995). Furthermore, it is known that transcription factor binding sites (TFBS) are of low informational content by themselves because of their short length and that they spuriously occur in many places in the genome (Wray et al. 2003; Frith et al. 2004). Therefore, it is possible that a synergistic interplay between TFBS and orderly chromatin structure is necessary for correct transcriptional regulation, and that the noncoding DNA can be selected (albeit weakly) in regard to chromatin structure.
There is a large literature on the sequence-dependence of nucleosome positioning (e.g., Ioshikhes et al. 1996; Kiyama and Trifonov 2002; Kato et al. 2003; Cioffi et al. 2004; Thastrom et al. 2004; Levitsky et al. 2005). It was recently shown for the yeast genome that the majority (∼70%) of nucleosomes are well positioned, i.e., occupy the same location in every cell (Yuan et al. 2005; see also Marx 2005). In turn, a high local concentration of nucleosomes is necessary for higher-order chromatin condensation, which is a distinct level of transcriptional regulation (Jenuwein and Allis 2001; Horn and Peterson 2002; Gilbert et al. 2004; Nemeth and Langst 2004). Dinucleosome formation is the first step in the organization of higher-order chromatin structure (Kato et al. 2003; Cioffi et al. 2004). The higher nucleosome formation potential was found in vertebrate noncoding DNA (including introns) compared to exons, and in human tissue-specific genes compared to housekeeping genes (Levitsky et al. 2001; Ganapathi et al. 2005; Vinogradov 2005; Vinogradov and Anatskaya 2006), which support the notion that noncoding DNA is loaded with function in regard to chromatin structure and involved in chromatin-mediated gene regulation. In the present study, the higher nucleosome formation potential was found in the conserved fraction compared to nonconserved. The higher-order nuclear organization can also involve matrix attachment regions, which are presented among regions of conserved noncoding DNA (Glazko et al. 2003).
There is now a boom in regard to the functional significance of transcription of noncoding DNA, including even intergenic regions (e.g., Mattick 2001, 2004; Vinogradov 2003; Frith et al. 2005; Johnson et al. 2005). It was even suggested that these transcripts are involved in parallel digital regulatory networks, which determine eukaryotic complexity (Mattick 2001, 2004; Mattick and Gagen 2005). However, the known noncoding RNAs with regulatory function are very short (Kawasaki et al. 2004; Mattick and Makunin 2005; Storz et al. 2005). Therefore, they can probably explain only a small part of conserved noncoding sequence. Furthermore, there is a notion that transcription of a bulk of noncoding DNA may just serve for chromatin remodeling because the RNA polymerase II complex might “piggyback” enzymatic remodeling complexes that set epigenetic marks by histone modifications (Drewell et al. 2002; Rank et al. 2002).
The greater fraction of conserved sequence found in the first intron (Fig. 3A) suggests also a more direct regulatory involvement of conserved regions. It is known that TFBS are more frequently found in the first intron compared to others (Majewski and Ott 2002; Keightley and Gaffney 2003). Gene knockout experiments revealed that a high percentage (∼80%) of yeast genes seemed to be “nonessential” under laboratory conditions but can be necessary in nature (Papp et al. 2004). The percentage may be even higher for TFBS (which should be more condition-dependent), especially in the more complex organisms. Therefore, it is possible that a much higher number of functionally significant TFBS exist in the human genome than are thought now (on the ground of experiments with laboratory cell cultures).
All this suggests that a considerable portion of noncoding DNA can be under selection. There is a problem of genetic load that should be high in this case. The multi-site (truncation) selection could alleviate this problem. Thus, it is known that nearly one in 300 nt is polymorphic in humans with a minor allele frequency >1% (Hinds et al. 2005).
Methods
Gene sequences and overall statistics
Gene sequences were extracted from the RefSeq database (Pruitt et al. 2005). The homology between human and mouse genes was established using the HomoloGene database (Wheeler et al. 2005). Only genes that are present in the Gene Expression Atlas (Su et al. 2004) were used. It is known that for the human–mouse pair, only a few intron losses occurred in the mouse lineage but no intron losses in the human or intron gains in either lineage were found (Roy et al. 2003). Therefore, introns of homologous genes were treated as homologous if both genes had the same number of introns. Only the internal introns (that reside within the CDS) were taken for consistency (because the complete mRNAs may not be known for all genes). Before the search for conserved regions, introns were masked for lineage-specific repeats (that were inserted after the human–mouse split) using the standalone RepeatMasker and DateRepeats programs (A.F.A. Smit, R. Hubley, and P. Green, unpubl., http://repeatmasker.org.). The total was 65,432 introns in 7258 genes, with the total length of 317.9 Mb in the human and 258.3 Mb in the mouse. After masking for lineage-specific repeats, there remains 246.2 Mb in the human and 192.3 Mb in the mouse.
Sequence alignment and analyses
The matching of homologous introns was done using the very rigorous (but very slow) Huang-Miller algorithm for local sequence alignment (Huang and Miller 1991) (which is a variant of the Waterman-Eggert algorithm; Waterman and Eggert 1987), implemented in the LALIGN program from the FASTA package (Pearson 1999). After obtaining all possible local alignments, the longest chain of nonoverlapped sequential (consecutive) local alignments was taken for each pair of homologous introns. The total length of local alignments in this chain was used as a length of conserved sequence in the intron. Also, the average identity of conserved sequence (weighted for the length of local alignments) was calculated for each intron. The LALIGN program was used with the significance level for spurious match being set to a conservative threshold P <10–6 (the other parameters were used as default; the maximum possible number of displayed alignments was set to 1,000,000; for details see the Supplemental data). Under these conditions, the alignment of randomized homologous introns (including reversed sequences, i.e., randomized with preservation of possible low complexity regions and local variation in GC content) showed the total length of “conserved” sequence <0.5% (regions masked for lineage-specific repeats were not involved in randomization). If an intron length was >140 kb (the limit of LALIGN program), it was split into 140 kb-long frames with a step of 10 kb, and all the between-species frame combinations were matched. (The total computing time was about five months on Pentium-4 2.8 GHz with 2 Gb RAM.) Some representative chains of nonoverlapped local alignments are shown in the Supplemental data.
The lineage-specific repeats were used for estimation of local mutation rate. For this purpose, the percentage of nucleotide substitutions, small insertions and deletions was estimated in regard to consensus sequence of a given repeat family that approximated an active ancestral copy (using the RepeatMasker program), as was done previously (Vinogradov 2002).
The nucleosome formation potential was determined for conserved and nonconserved regions in randomly chosen 10,000 human introns using the RECON program (Levitsky 2004).
Gene expression breadth
The data on gene expression were taken from the Gene Expression Atlas (Su et al. 2004). They present the results of oligonucleotide microarray experiments performed uniformly with 72 normal human tissues. The signals from probes on the chip corresponding to the same gene were averaged; the replicates representing the same tissue were also averaged. As recommended (Su et al. 2004), a gene was regarded as expressed if its signal level exceeded a conservative threshold of the data set median. For those genes that have references to the SWISS-PROT (UniProt) database (6442 proteins were found), the number of distinct functional domains in the encoded proteins was estimated using the SwissPfam database (Bateman et al. 2004).
Statistical analyses
The comparison of the average fraction of conserved sequence in introns of genes with different among-tissues breadth of expression was made using the Statgraphics software package (Statistical Graphics Corp.). Correction for the weight-averaged identity of conserved sequence, mutation bias, and intron GC content was done using the general linear model (GLM, which can be considered as a generalization of multifactor analysis of variance, ANOVA) implemented in Statgraphics, with correction variables being added as covariates in the model. In addition, in the special variant of analysis, the product of the conserved sequence fraction and the between-species identity of this fraction (which can be considered as the fraction of conserved nucleotides or the fraction of sequence with 100% identity) was used as a factor variable in GLM (with and without the other correction variables—mutation bias and GC content). This variant of analysis tested directly the combination of the length and the identity of conserved fraction.
Acknowledgments
I thank Fyodor A. Kondrashov, Herbert R. Halvorson, Vladimir M. Tchernajenko, and two anonymous reviewers for helpful comments. This work was supported by the Russian Foundation for Basic Research (RFBR) and by the Programme of the Presidiumof the Russian Academy of Sciences “Molecular and Cellular Biology” (MCB RAS).
[Supplemental material is available online at www.genome.org.]
E-mail aevin@mail.cytspb.rssi.ru; fax 07 812 247 0341.
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4318206.
References
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32 D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis, C.I., Mekhedov, S.L., Hartl, D.L., Koonin, E.V., and Kondrashov, F.A. 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31 415–418. [DOI] [PubMed] [Google Scholar]
- Chen, J., Sun, M., Hurst, L.D., Carmichael, G.G., and Rowley J.D. 2005a. Human antisense genes have unusually short introns: Evidence for selection for rapid transcription. Trends Genet. 21 203–207. [DOI] [PubMed] [Google Scholar]
- Chen, J., Sun, M., Rowley, J.D., and Hurst, L.D. 2005b. The small introns of antisense genes are better explained by selection for rapid transcription than by “genomic design.” Genetics 171 2151–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi, A., Dalal, Y., and Stein, A. 2004. DNA sequence alterations affect nucleosome array formation of the chicken ovalbumin gene. Biochemistry 43 6709–6722. [DOI] [PubMed] [Google Scholar]
- Drewell, R.A., Bae, E., Burr, J., and Lewis, E.B. 2002. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. 99 16853–16858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, E. and Levanon, E.Y. 2003. Human housekeeping genes are compact. Trends Genet. 19 362–365. [DOI] [PubMed] [Google Scholar]
- Frith, M.C., Fu, Y., Yu, L., Chen, J.F., Hansen, U., and Weng, Z. 2004. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 32 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith, M.C., Pheasant, M., and Mattick, J.S. 2005. The amazing complexity of the human transcriptome. Eur. J. Hum. Genet. 13 894–897. [DOI] [PubMed] [Google Scholar]
- Ganapathi, M., Srivastava, P., Das Sutar, S.K., Kumar, K., Dasgupta, D., Pal Singh, G., Brahmachari, V., and Brahmachari, S.K. 2005. Comparative analysis of chromatin landscape in regulatory regions of human housekeeping and tissue specific genes. BMC Bioinformatics 6 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., Boyle, S., Fiegler, H., Woodfine, K., Carter, N.P., and Bickmore, W.A. 2004. Chromatin architecture of the human genome: Gene-rich domains are enriched in open chromatin fibers. Cell 118 555–566. [DOI] [PubMed] [Google Scholar]
- Glazko, G.V., Koonin, E.V., Rogozin, I.B., and Shabalina, S.A. 2003. A significant fraction of conserved noncoding DNA in human and mouse consists of predicted matrix attachment regions. Trends Genet. 19 119–124. [DOI] [PubMed] [Google Scholar]
- Haddrill, P.R., Charlesworth, B., Halligan, D.L., and Andolfatto, P. 2005. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 6 R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds, D.A., Stuve, L.L., Nilsen, G.B., Halperin, E., Eskin, E., Ballinger, D.G., Frazer, K.A., and Cox, D.R. 2005. Whole-genome patterns of common DNA variation in three human populations. Science 307 1072–1079. [DOI] [PubMed] [Google Scholar]
- Horn, P.J. and Peterson, C.L. 2002. Molecular biology. Chromatin higher order folding–wrapping up transcription. Science 297 1824–1827. [DOI] [PubMed] [Google Scholar]
- Huang, X. and Miller, W. 1991. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 12 337–357. [Google Scholar]
- Ioshikhes, I., Bolshoy, A., Derenshteyn, K., Borodovsky, M., and Trifonov, E.N. 1996. Nucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequences. J. Mol. Biol. 262 129–139. [DOI] [PubMed] [Google Scholar]
- Jareborg, N., Birney, E., and Durbin, R. 1999. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 9 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein, T. and Allis, C.D. 2001. Translating the histone code. Science 293 1074–1080. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Edwards, S., Shoemaker, D., and Schadt, E.E. 2005. Dark matter in the genome: Evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 21 93–102. [DOI] [PubMed] [Google Scholar]
- Jukes, T.H. and Cantor, C.R. 1969. Evolution of protein molecules. In Mammalian protein metabolism (ed. Munro H.N.), pp. 21–123. Academic Press, New York.
- Kato, M., Onishi, Y., Wada-Kiyama, Y., Abe, T., Ikemura, T., Kogan, S., Bolshoy, A., Trifonov, E.N., and Kiyama, R. 2003. Dinucleosome DNA of human K562 cells: Experimental and computational characterizations. J. Mol. Biol. 332 111–125. [DOI] [PubMed] [Google Scholar]
- Kawasaki, H., Wadhwa, R., and Taira, K. 2004. World of small RNAs: From ribozymes to siRNA and miRNA. Differentiation 72 58–64. [DOI] [PubMed] [Google Scholar]
- Keightley, P.D. and Gaffney, D.J. 2003. Functional constraints and frequency of deleterious mutations in noncoding DNA of rodents. Proc. Natl. Acad. Sci. 100 13402–13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama, R. and Trifonov, E.N. 2002. What positions nucleosomes? A model. FEBS Lett. 523 7–11. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A.S. and Shabalina, S.A. 2002. Classification of common conserved sequences in mammalian intergenic regions. Hum. Mol. Genet. 11 669–674. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F.A., Ogurtsov, A.Y., and Kondrashov, A.S. 2006. Selection in favor of nucleotides G and C diversifies evolution rates and levels of polymorphism at mammalian synonymous sites. J. Theor. Biol. (in press). [DOI] [PubMed]
- Lauderdale, J.D. and Stein, A. 1992. Introns of the chicken ovalbumin gene promote nucleosome alignment in vitro. Nucleic Acids Res. 20 6589–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitsky, V.G. 2004. RECON: A program for prediction of nucleosome formation potential. Nucleic Acids Res. 32 W346–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitsky, V.G., Podkolodnaya, O.A., Kolchanov, N.A., and Podkolodny, N.L. 2001. Nucleosome formation potential of exons, introns, and Alu repeats. Bioinformatics 17 1062–1064. [DOI] [PubMed] [Google Scholar]
- Levitsky, V.G., Katokhin, A.V., Podkolodnaya, O.A., Furman, D.P., and Kolchanov, N.A. 2005. NPRD: Nucleosome positioning region database. Nucleic Acids Res. 33 D67–D70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K., Sandgren, E.P., Palmiter, R.D., and Stein, A. 1995. Rat growth hormone gene introns stimulate nucleosome alignment in vitro and in transgenic mice. Proc. Natl. Acad. Sci. 92 7724–7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski, J. and Ott, J. 2002. Distribution and characterization of regulatory elements in the human genome. Genome Res. 12 1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx, J. 2005. Nucleosomes help guide yeast gene activity. Science 308 1724. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. 2001. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick, J.S. 2004. RNA regulation: A new genetics? Nat. Rev. Genet. 5 316–323. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. and Gagen, M.J. 2005. Accelerating networks. Science 307 856–858. [DOI] [PubMed] [Google Scholar]
- Mattick, J.S. and Makunin, I.V. 2005. Small regulatory RNAs in mammals. Hum. Mol. Genet. 14 R121–R132. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip, A. and Verrijzer, C.P. 2004. A higher order of silence. Science 306 1484–1485. [DOI] [PubMed] [Google Scholar]
- Nemeth, A. and Langst, G. 2004. Chromatin higher order structure: Opening up chromatin for transcription. Brief. Funct. Genomic Proteomic 2 334–343. [DOI] [PubMed] [Google Scholar]
- Nurtdinov, R.N., Artamonova, I.I., Mironov, A.A., and Gelfand, M.S. 2003. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 12 1313–1320. [DOI] [PubMed] [Google Scholar]
- Papp, B., Pal, C., and Hurst, L.D. 2004. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature 429 661–664. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. 1999. Flexible similarity searching with the FASTA3 program package. In Bioinformatics methods and protocols (eds. S. Misener and S.A. Krawetz), pp. 185–219. Humana Press, Totowa, NJ.
- Pruitt, K.D., Tatusova, T., and Maglott, D.R. 2005. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33 D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank, G., Prestel, M., and Paro, R. 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell Biol. 22 8026–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S.W., Fedorov, A., and Gilbert, W. 2003. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc. Natl. Acad. Sci. 100 7158–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina, S.A., Ogurtsov, A.Y., Kondrashov, V.A., and Kondrashov, A.S. 2001. Selective constraint in intergenic regions of human and mouse genomes. Trends Genet. 17 373–376. [DOI] [PubMed] [Google Scholar]
- Storz, G., Altuvia, S., and Wassarman, K.M. 2005. An abundance of RNA regulators. Annu. Rev. Biochem. 74 199–217. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K.A., Block, D., Zhang, J., Soden, R., Hayakawa, M., Kreiman, G., et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 101 6062–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrom, A., Bingham, L.M., and Widom, J. 2004. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J. Mol. Biol. 338 695–709. [DOI] [PubMed] [Google Scholar]
- Trifonov, E.N. 1993. Spatial separation of overlapping messages. Comput. Chem. 117 27–31. [Google Scholar]
- Urrutia, A.O. and Hurst, L.D. 2003. The signature of selection mediated by expression on human genes. Genome Res. 13 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291 1304–1351. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A.E. 2001. Intron length and codon usage. J. Mol. Evol. 52 2–5. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A.E. 2002. Growth and decline of introns. Trends Genet. 18 232–236. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A.E. 2003. Silent DNA: speaking RNA language? Bioinformatics 9 2167–2170. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A.E. 2004. Compactness of human housekeeping genes: Selection for economy or genomic design? Trends Genet. 20 248–253. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A.E. 2005. Noncoding DNA, isochores and gene expression: Nucleosome formation potential. Nucleic Acids Res. 33 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov, A.E. and Anatskaya, O.V. 2006. Genome size and metabolic intensity in tetrapods: A tale of two lines. Proc. Roy. Soc. Lond. B 273 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman, M.S. and Eggert, M. 1987. A new algorithm for best subsequences alignment with application to tRNA-rRNA comparisons. J. Mol. Biol. 197 723–728. [DOI] [PubMed] [Google Scholar]
- Wheeler, D.L., Barrett, T., Benson, D.A., Bryant, S.H., Canese, K., Church, D.M., DiCuccio, M., Edgar, R., Federhen, S., Helmberg, W., et al. 2005. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 33 D39–D45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G.A., Hahn, M.W., Abouheif, E., Balhoff, J.P., Pizer, M., Rockman, M.V., and Romano, L.A. 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20 1377–1419. [DOI] [PubMed] [Google Scholar]
- Yuan, G.C., Liu, Y.J., Dion, M.F., Slack, M.D., Wu, L.F., Altschuler, S.J., and Rando, O.J. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309 626–630. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl, E. 1997. Junk DNA and sectorial gene repression. Gene 205 323–343. [DOI] [PubMed] [Google Scholar]