Abstract
Long interspersed element-1 elements compose on average one-fifth of mammalian genomes. The expression and retrotransposition of L1 is restricted by a number of cellular mechanisms in order to limit their damage in both germ-line and somatic cells. L1 transcription is largely suppressed in most tissues, but L1 mRNA and/or proteins are still detectable in testes, a number of specific somatic cell types, and malignancies. Down-regulation of L1 expression via premature polyadenylation has been found to be a secondary mechanism of limiting L1 expression. We demonstrate that mammalian L1 elements contain numerous functional splice donor and acceptor sites. Efficient usage of some of these sites results in extensive and complex splicing of L1. Several splice variants of both the human and mouse L1 elements undergo retrotransposition. Some of the spliced L1 mRNAs can potentially contribute to expression ofopen reading frame 2-related products and therefore have implications for the mobility of SINEs even if they are incompetent for L1 retrotransposition. Analysis of the human EST database revealed that L1 elements also participate in splicing events with other genes. Such contribution of functional splice sites by L1 may result in disruption of normal gene expression or formation of alternative mRNA transcripts.
INTRODUCTION
Long interspersed element-1 or LINE-1 (L1) is a non-long terminal repeat (non-LTR), autonomous retroelement currently active in mammalian genomes that composes 17 and 20% of the human and mouse genomes, respectively (1,2). L1 inserts in the forward orientation are depleted in genes, probably due to their deleterious effects on gene expression (3–5). Even though L1 activity has been detected in somatic cells (6–9), L1 is believed to undergo preferential expression and retrotransposition in the germ-line (10,11). Suppression of L1 activity is partly attributed to promoter regulation, either through tissue-specific transcription factors (12,13), or methylation of the L1 promoter that is often released upon malignant transformation (14–16). L1 expression is also attenuated via premature polyadenylation at internal polyadenylation [poly(A)] sites (17). This mechanism is redundant and cannot be easily overcome by removal of a few internal poly(A) signals. A model of hindered polymerase II elongation along the A-rich L1 sequence was put forward as an additional explanation for poor expression through L1 elements (18).
L1 transcription uses an internal RNA pol II promoter to encode a full-length (FL) L1 bicistronic mRNA that produce open reading frame (ORF) 1 and 2 proteins that are essential for retrotransposition (19). This FL transcript is retrotranspositionally competent (20), generating new L1 copies via target-primed reverse transcription (21). The majority of the 500 000 L1 copies found in mammalian genomes are 5′ truncated (1) and/or rearranged (1,22). Thus, only about 100 human elements are capable of expressing full-length RNA that codes for functional ORF1 and ORF2 proteins (23).
The signals necessary for RNA splicing include both cis elements and trans factors, some of which are more conserved and well characterized then others. RNA splicing [reviewed in (24)] involves a splice donor site (SD or 5′ splice), a splice acceptor site (SA or 3′ splice) and a conserved cis element 20–50 bp 5′ to the SA site. Trans-acting factors include five snRNAs (U1, U2, U4, U5 and U6) and at least 150 identified proteins that form a functional spliceosome (25). Additionally, there are exonic and intronic splice enhancers (ESE and ISE) and silencers (ESS and ISS) that can modulate splice site usage. A consensus sequence for the most often occurring 5′and 3′ ESE is G/AAAGAA (26). Deviation from the canonical SD or SA sequences may either lead to exon skipping, or it may result in the usage of cryptic splice sites in the vicinity. Both constitutive and alternative splicing are responsible for the 3-fold increase in protein diversity compared with the number of protein-encoding genes in humans (27,28) with 35–65% of human genes undergoing alternative splicing (27,29). Differential splicing is a tissue-, developmental- and cancer-specific process (30).
L1 elements have generally been considered to produce unspliced mRNA. However, studies on L1 RNA have been confounded by low expression levels and the detection of numerous low-molecular weight, L1-related transcripts that were presumed to be created from the many truncated genomic copies incorporated into other transcripts (31). Here we report that L1 contains multiple predicted SD and SA sites in both sense and antisense strands of its genome. Some of these sites are functional and their usage leads to a widespread, complex splicing pattern for most L1 transcripts. This processing results in weakening of full-length L1 expression and, like Alu, exonization (32), leads to aberrant splicing of genes (5,33,34).
MATERIALS AND METHODS
Cell culture and transfections
NIH 3T3 (ATCC CRL-1658), Ntera2 (ATCC #CRL-1973) and HeLa (ATCC CCL2) cells were maintained as described elsewhere (17). MCF7 cells (ATCC #HTB-22) were maintained in MEM (Gibco) supplemented with 10% bovine serum (Gibco), sodium pyruvate, essential and nonessential amino acids and l-glutamine. Sk-Br-3 cells (ATCC HTB-30) were maintained in RPMI medium1640 supplemented with 15% fetal bovine serum (Gibco). Human mammary epithelial (HME) cells (CRL-4010) were maintained in MEBM (Clonetics) supplemented with MEGM SingleQuots (Clonetics). Transfections of all cell lines were performed byLipofectamine with Plus reagent (Invitrogen) as reported previously (17). Briefly, two T75 flasks with 4–5 × 106 cells were seeded and transfected with 6 µg of CsCl purified DNA 18–20 h later. Total RNA was isolated by TRiZol reagent 24 h post transfection (Invitrogen) followed by chloroform extraction and isopropanol precipitation. Total RNA was poly(A) selected with poly(A) selection kit (Promega) according to the manufacturer's protocol. Poly(A)-selected RNAs were precipitated overnight in isopropanol. Northern blot analysis was performed as described elsewhere (17). The results of the northern blot assays were quantified on a Fuji Phosphorimager. DNA template for the probe was produced by PCR with the primers that amplified either LINE-1.3 5′-untranslated region (5′-UTR), the second exon of the neoR cassette, the intron of the neoR cassette [as described in (17)], the first 100 bp (5′UTR100 probe) (5′-GGAGCCAAGATGGCCGAATAGGAACAGCT-3′ and 5′-ACCTCAGATGGAAATGCAG-3′) or 583–698 bp region (5′UTR600 probe) (5′-GCAGTAACCTCTGCAGAC-3′ and 5′-CCACTTGAGGAGGCAG-3′) of the 5′-UTR. The T7 promoter sequence was included in the reverse primer of each pair.
Site-directed mutagenesis
The QuikChange Site-Directed Mutagenesis kit (STRATAGENE) was used to change the position 97 splice site sequence from T to C at position 99 of L1.3 as described elsewhere (17). The 1M mutation in the L1neo and L1notag vectors was the same as published previously (17).
RT–PCR
Total RNA from HeLa or NIH 3T3 cells transfected with L1notag vector was extracted and poly(A) selected as described elsewhere (17). First-strand synthesis was performed with 3′-UTR(−) (5′-GGTTAGTTACATATGTATAC-3′ and ORF2(−) (5′-CTGTGTCTTTTAATTGCAGAATTTAGTCC-3′) primers with an RT–PCR kit (Promega) according to the manufacturer's protocol followed by PCR with 48(+) primer 5′-GGAGCCAAGATGGCCGAATAGGAACAGCT-3′. The 3′ end of the ORF2(−) primer is complementary to the position 2038 and 1359 of L1.3. PCR products were fractionated on a 1% low-melting agarose gel. The isolated DNA fragments were sequenced (TGEN, AZ).
Human EST database search
To identify examples of endogenous L1 expressed sequence tags (ESTs) that participated in splicing events, NCBI dbEST was searched via BLAST (blastn, E = 1) (35) with the first 210 bp of L1.3 consensus sequence, which encompassed the position 97 SD site. Matches where the similarity with the L1 consensus discontinued within 3 bp of the 97 SD position were retained for additional analysis. Candidate splices were subsequently located in the genome using BLAT (36) and examined for the position and orientation of L1 relative to the gene or other sequences participating in the splice event. In addition, sequences were manually examined for the usage of the 97 bp L1 SD and associated SA site. Finally, in order to exclude the possibility that the putative L1 splice event was the result of transcription from a genomic sequence that mirrored the splice form (either due to spurious deletions or previously retrotransposed spliced RNA), all candidate splices were checked via BLAST and BLAT for identical contiguous matches to genomic DNA.
RESULTS
LINE-1 elements contain functional splice sites
The BDGP program (http://www.fruitfly.org/seq_tools/splice.html) predicted numerous 5′ and 3′ splice sites distributed throughout the sense strand of both the human L1.3 (L19088) and mouse L1spa (AF016099) elements (Figure 1A). The same program also predicted multiple SD and SA sites in the antisense sequence of both elements (data not shown).
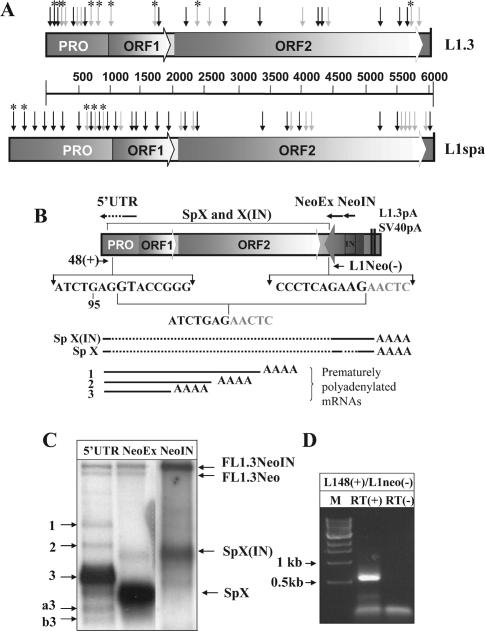
Figure 1.
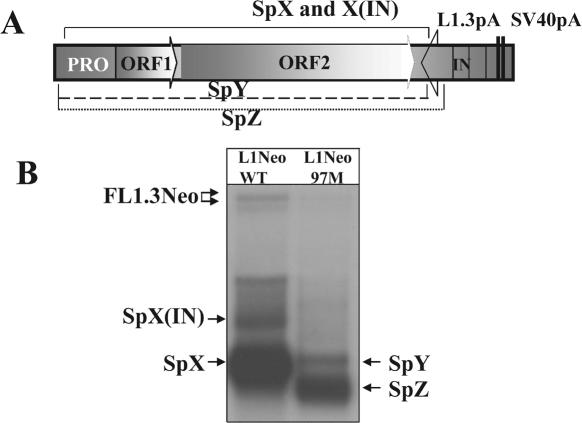
LINE-1 elements contain multiple splice sites. (A) A schematic representation of putative splice sites identified by the BDGP program in the sense strand of human L1.3 and mouse L1spa. Black (SD) and gray (SA) arrows mark splice sites using a default cutoff value of 0.4. Asterisks mark SD and SA sites that have been identified as functional based upon sequence analysis of spliced transcripts recovered by RT–PCR or as determined by sequence analysis of spliced products found re-integrated into the human and mouse genomes. PRO corresponds to the L1 promoter region, ORF1 and ORF2 identify ORFs 1 and 2. (B) A schematic representation of the L1.3 neomycin-resistance (L1.3Neo) expression cassette. The position and orientation of the NeoR gene are shown by an arrow. It is interrupted by an intron (IN) in the same orientation as the L1 ORFs. The cassette ends in L1.3 and SV40 polyadenylation signals (L1.3pA and SV40pA, respectively). The L1.3 portion that is missing in transcripts SpX and X(IN) is marked above the cassette. Sequences below the schematic of the vector demonstrate the 5′ and 3′ splice sites with invariant GT and AG dinucleotides shown in larger font. The resulting sequence of the splicing event is listed under the junctions. Arrows above the L1.3 expression cassette (marked 5′-UTR, NeoEx and NeoIN) represent positions of the strand-specific RNA probes used for L1 RNA detection. The arrows indicate the sense of the probes. Because of the length of the 5′-UTR probe (900 bp) in vitro transcription products are a mix of truncated transcripts, which are enriched for the 3′ end of the L1.3 5′-UTR (solid portion of the arrow). Below the sequences is a schematic representation of the splice products generated between the L1.3 and the Neo cassette [SpX(IN) and SpX] and the premature polyadenylated, unspliced L1.3 mRNAs (1–3). Solid black lines represent portions of the L1.3 and Neo cassette sequences included in the transcripts. Dotted lines denote parts that were removed by splicing. (C) Northern blot analysis of poly(A)-selected mRNAs from NIH 3T3 cells transfected with the L1.3Neo expression vector. Strand-specific probes corresponding to the L1.3 5′-UTR (5′UTR), second exon of the NeoR gene (NeoEx) and intron of the NeoR gene (NeoIN) were designed to detect plus-strand mRNAs. Full-length L1.3Neo mRNAs with spliced and unspliced NeoR introns are marked as FL1.3Neo and FL1.3NeoIN, respectively. Numbers 1 through 3 indicate products of premature polyadenylation (17). Potential splice species are labeled as SpX, SpX(IN), ‘a3’ and ‘b3’ [‘a’ and ‘b’ denotes splicing events (Figure 2) and a number corresponds to the poly(A) sites used to generate the 3′ end of the transcripts]. Note that the most abundant band in the NeoEx lane is barely detected with the 5′-UTR probe (5′UTR lane) indicating that it contains a small portion of L1.3. In vitro-generated 5′-UTR probe is not all full-length and therefore the detection is skewed toward the 3′ end of the 5′-UTR. Results of blotting with the strand-specific probe to the NeoR intron (NeoIN) indicate that SpX(IN) contains the intron while SpX does not. (D) RT–PCR analysis of the L1/NeoR splice junction. Positions of 48(+) and L1Neo(−) primers are shown in (B). RT(+) and (−) indicate the presence or absence of RT in the respective RT–PCRs.
To characterize some of the mRNAs produced by the L1.3 element tagged with the neomycin-resistance (NeoR) cassette (L1.3Neo) (20,37) (Figure 1B), we used a strand-specific probe to the second exon and the intron of the NeoR gene (Figure 1B and C, lane NeoEx and NeoIN) to detect the L1 sense strand transcripts. Full-length mRNAs were detected with, and without, the intron interrupting the NeoR cassette (Figure 1C, bands FL1.3NeoIN and FL1.3Neo). Highly abundant, faster-migrating bands were also detected with both probes. These bands contained NeoR gene sequences, but were too small to include much L1.3 sequence. One transcript did not contain the intron of the NeoR cassette as detected by the intron-specific probe for the Neo resistance gene (Figure 1B and C, SpX) while the slower band contained the intron [Figure 1B and C, SpX(IN)]. The estimated size of the SpX and X(IN) products approximately corresponded to the sizes of the spliced and unspliced NeoR gene, respectively. The Sp(X) band is only weakly detected by a 5′-UTR probe that is biased towards the 3′ end of the 5′-UTR (Figure 1B and C), suggesting that much of the 5′-UTR sequence is not present in this transcript, possibly due to splicing. To confirm the identity of these products, we used an upstream primer corresponding to the beginning of the L1.3 5′-UTR and the downstream primer complementary to the beginning of the second exon of the NeoR gene to perform RT–PCR on poly(A)-selected RNAs from transfected NIH 3T3 cells (Figure 1D). A single band of about 650 bp was detected. Sequence analyses of five independent clones demonstrated that the L1.3 sequence is joined to the sequence of the NeoR gene in the manner consistent with conserved cis elements of mammalian splicing (Figure 1B). Thus, L1.3 contains at least one functional SD site that can be utilized with SA sites downstream of its genome. Both SpX and SpX(IN) bands (Figure 1B) require full-length transcription of the L1.3 mRNA prior to splicing. This may represent the primary difference between the levels of full-length transcripts from the L1.3Neo and L1-notag (which mimics endogenous L1 elements) constructs [(17) and Figure 2].
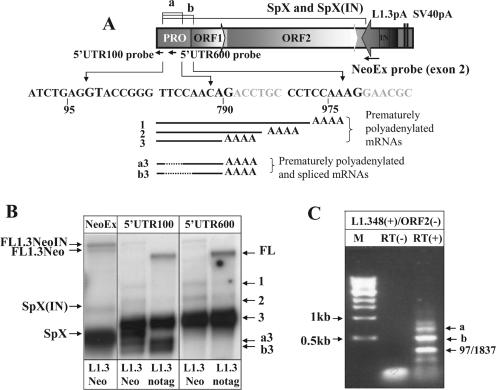
Figure 2.
L1.3 mRNA undergoes splicing at multiple sites. (A) L1.3Neo splicing. The portions of the L1.3 that are removed in splice products ‘a’ and ‘b’ are annotated above the cartoon of the expression cassette with splice site sequences listed underneath. Strand-specific 100 bp probes corresponding to positions 1–100 (5′UTR100) and 583–698 (5′UTR600) of the L1.3 sequence are shown under the promoter (Pro) portion of the L1.3 with the arrow denoting the sense of the probes. Strand-specific NeoEx probe is the same as in Figure 1. Underneath the sequences is a schematic representation of the prematurely polyadenylated L1 mRNAs [1–3, that are detected with both 5′UTR100 and 5′UTR600 probes in (B)] and prematurely polyadenylated and spliced transcripts [a3 and b3, that are detectable only with the 5′UTR100 probe in (B)]. Solid black lines represent parts of L1.3 sequence included in the transcripts. Dotted black lines correspond to the L1.3 sequences removed by splicing. SpX(IN) and SpX products are the same as in Figure 1. (B) Northern blot analysis of poly(A)-selected mRNAs from NIH 3T3 cells transfected with vectors expressing either L1.3Neo (L1Neo) or only L1.3 sequences (L1.3-notag). The 5′UTR100 strand-specific probe detected premature poly(A) products (bands 1 through 3), as well as the splice products SpX in the L1.3Neo RNA and the ‘a3’ and ‘b3’ products for the L1.3Neo and L1.3-notag constructs [where ‘a’ refers to the splice and 3 to the poly(A) site used to create the transcript]. The SpX, ‘a3’ and ‘b3’ bands were not detected by a strand-specific 5′UTR600 probe complementary to the portion of L1.3 expected to be spliced from these transcripts. FL annotates the full-length L1.3 mRNA. (C) RT–PCR analysis of poly(A)-selected mRNAs from NIH 3T3 cells transiently transfected with L1.3 expression cassette. The 48(+) forward primer described in Figure 1B and ORF2(−) reverse primer located in L1 ORF2 were used. Note that ORF2 primer can also anneal at position 1359 of L1.3 sequence therefore products of splices ‘a’ and ‘b’ are smaller than expected when the primer anneals at the position 2038 in ORF2 of L1.3.
RNA splicing limits production of the full-length L1 mRNA
To determine whether there are other functional SD and SA sites in the L1.3 sequence, we probed L1 RNAs with a strand-specific RNA probe complementary to the first 100 bp of the L1.3 5′-UTR (5′UTR100 probe) (Figure 2A and B). If the SD site in the beginning of the L1.3 5′-UTR was utilized for L1 splicing, the 5′UTR100 probe would allow quantitative comparison of the amounts of prematurely terminated transcripts versus spliced products. Northern blot analyses with the 5′UTR100 probe detected the SpX band for the L1Neo construct and two additional faster-migrating bands (a3 and b3,‘a’ and ‘b’ denote splicing events and the number corresponds to the poly(A) sites used to generate the 3′ end of the transcripts) for both L1Neo and L1-notag constructs (Figure 2A and B and Supplementary Figures 1 and 2 that help clarify the nomenclature of the complex group of RNA species formed by the concurrent use of both variable splicing and polyadenylation). These two smaller bands were consistent with splicing within L1.3 mRNA and were as abundant as the previously reported major, prematurely polyadenylated species (17). A strand-specific RNA probe complementary to the 600–700 bp region of the L1.3 5′-UTR (5′UTR600 probe) did not detect bands ‘a3’ and ‘b3’, confirming the loss of this sequence in these bands (Figure 2B). To determine which of the predicted splice sites are used, we performed an RT–PCR analysis of RNA species produced by the L1.3-notag construct in NIH 3T3 cells with primers located at the beginning of the L1.3 sequence and at the 5′ end of ORF2. Sequence analysis of the bands produced in this experiment confirmed usage of splices ‘a’ and ‘b’ (Figure 2C and Supplementary Figures 1 and 2) and detected an additional functional SD site at position 54 of the L1.3 element and five SA sites (Figures 2C and 1, splice sites are marked by an asterisk). One of the functional SA sites is located at position 1837 of the L1.3 sequence. Any mRNA resulting from the usage of this splice site would completely lack ORF1 sequence but would have the potential to produce ORF2 protein.
To determine whether L1.3 splicing detected in NIH 3T3 cells is supported by human cells, the L1.3 expression cassette was transiently transfected in transformed (HeLa and MCF7) and normal (HME) human cells. Northern blot analysis of poly(A)-selected RNAs with the 5′UTR100 strand-specific RNA probe detected mRNA profiles identical to those characterized in the mouse cells (Figure 3A).
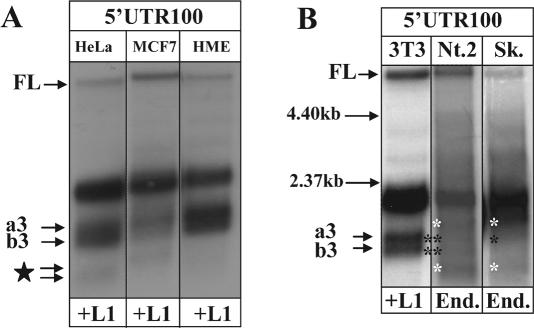
Figure 3.
Transiently transfected and endogenously expressed L1s undergo splicing in human cells. (A) Northern blot analyses of RNA species produced by the L1.3-notag expression cassette transiently transfected in HeLa, MCF7 and HME cells. Poly(A)-selected total mRNAs from the cell lines were detected with the strand-specific RNA probe complementary to the first 100 bp of the L1.3 5′UTR (5′UTR100). FL corresponds to the full-length L1.3 element, a3 and b3 and c4 and b4 indicate spliced and prematurely polyadenylated products. Note that spliced mRNAs are detected in both normal (HME) and cancer cells transfected with L1.3 expression cassette. The star denotes bands with an uncharacterized combination of splicing and polyadenylation. (B) Northern blot analyses of RNA species produced by the L1.3-notag expression cassette transiently transfected in NIH 3T3 cells (Lane 3T3/+L1) or endogenously expressed human L1s from Ntera2 (Nt.2/End.) and Sk-Br-3 (Sk/End.) cells. Poly(A)-selected total mRNAs from the cell lines were detected with the strand-specific RNA probe complementary to the first 100 bp of the L1.3 5′UTR (5′UTR100). FL corresponds to the full-length L1.3 element detected in NIH 3T3 cells or to the endogenous L1s detected in human cell lines. a3 and b3 indicate spliced and prematurely polyadenylated products. Black asterisks mark truncated mRNA products in human cells exhibiting similar migration patterns to the spliced mRNAs identified in NIH 3T3 cells. White asterisks point out truncated mRNA products in Ntera2 and Sk-Br-3 cells that are not detected in transiently transfected mouse cells. Black horizontal arrows correspond to the position of the size markers.
To evaluate RNA profiles of the endogenous human L1 elements, we performed northern blot analysis of RNAs extracted from human Ntera2 (38) and Sk-Br-3 cancer cells that express naturally high levels of L1 elements. The 5′UTR100 probe detected RNA species consistent with ‘a’ and ‘b’ splice products detected in transient transfection of mouse and human cells in both cell types (Figure 3B). Additional faster-migrating bands that were not detected in transient transfections were observed in Ntera2 and Sk-Br-3 cells. These bands are consistent with the expected heterogeneity of the endogenous L1 elements; they could also be tissue- or cancer-specific splice and/or polyadenylation products.
To identify additional functional splice sites in the human L1, and to confirm that endogenous L1 elements undergo splicing, we used a pair of primers located in the beginning and the end of the L1.3 sequence for RT–PCR analysis of poly(A)-selected RNAs from NIH 3T3 cells transfected with the L1.3-notag construct, and endogenous RNAs from HeLa cells (Figure 4). Although there were some variations consistent with the expected heterogeneity of endogenous L1 elements, sequence analysis of some of the bands detected a common functional SA site at the end of the L1 element (position 5721) that was used with SD sites in the beginning of the 5′-UTR by both transfected and endogenous L1 elements (Figure 1A). RT–PCR targeting of other regions of the L1 sequence produced bands consistent with splicing, suggesting that there are almost certainly many other functional L1 SD and SA sites (data not shown).
Figure 4.
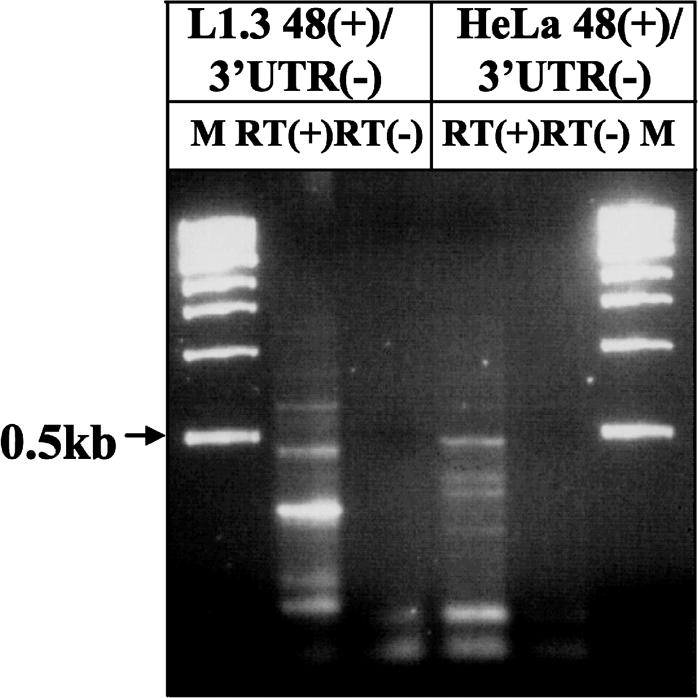
Endogenous L1 elements expressed in HeLa cells undergo splicing. RT–PCR analysis of poly(A)-selected RNAs from HeLa cells and L1-notag transfected NIH 3T3 cells was carried out with 48(+) upstream primer and 3′-UTR(−) downstream primer located in the L1.3 3′-UTR. RT(+) and (−) indicate reactions with and without reverse transcriptase.
The relationship between splicing and premature polyadenylation within LINE-1
It has been reported previously that there is competition among, and between (39–41), different splice sites (42,43) and poly(A) signals (44). It appears that the L1 sequence is riddled with both splice and poly(A) sites. To determine the relationship between these signals, we compared RNA species produced by the wild type (WT) and mutant of the strongest functional internal poly(A) site (1M) for both L1.3Neo and L1-notag (17). This mutant is biologically relevant because one of the ‘hot’ L1 elements, AL137845, (23) is lacking this poly(A) site. We performed a northern blot analysis with the strand-specific 5′UTR100 probe of RNAs from NIH 3T3 cells transfected with WT and 1M L1.3-notag elements. In the WT background, splice variants ‘a3’ and ‘b3’ are prematurely terminated at the strongest poly(A) site at the end of ORF1 (Figure 5A and B). When the strongest poly(A) site is not present in the L1.3 sequence, the 5′UTR100 probe detects a slower-migrating doublet (Figure 5A and B, a4 and b4). This doublet is consistent with the ‘a’ and ‘b’ splice products utilizing poly(A) sites (4) located further downstream in the L1.3 sequence (Figure 5A). Additionally, two new products occur in the 1M mutant for both the WT L1.3 (Figure 5B) and the L1.3Neo constructs (Figure 5C). The small size of these new L1-related RNA species and the fact that they are not detected with the 5′UTR600 strand-specific probe (data not shown) is consistent with the usage of alternative SA/poly(A) sites and/or an increase in production of the splice variants that are made by the WT L1.3 in much lower quantities. It appears that mutations of functional poly(A) sites result in not only increased utilization of the poly(A) signals nearby (17) but also in quantitative alterations in the use of specific splice sites.
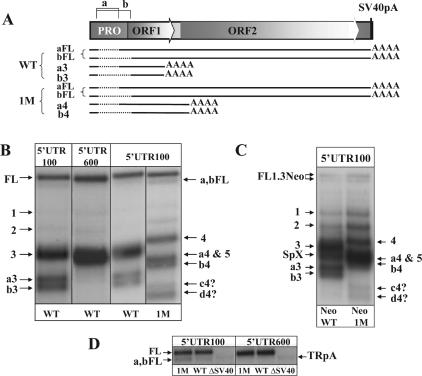
Figure 5.
The relationship between polyadenylation and splicing of L1 transcripts. (A) A diagram of the L1.3-notag construct. Diagrams of the major splice variants detected by northern blot analysis of the wild type (WT) and mutant of the strongest poly(A) site (1M) L1.3 elements are shown underneath the construct. Solid black lines represent L1.3 sequences included into RNA transcripts. Dotted lines represent regions of L1.3 sequence removed by splicing. Some ‘a’ and ‘b’ splicing events may use poly(A) sites at the end of the L1 genome (a,bFL in WT and 1M elements), while others may prematurely terminate at the internal L1 poly(A) signals (‘a3’ and ‘b3’ in the WT and ‘a4’ and ‘b4’ in the 1M L1.3). (B) Northern blot analysis of the WT and 1M L1.3-notag constructs transiently transfected in NIH 3T3 cells with 5′UTR100 and 5′UTR600 strand-specific probes. Full-length L1.3 mRNA and spliced L1.3 mRNA terminated at the end of the L1 genome are marked as FL and a,bFL, respectively. Spliced and prematurely truncated mRNAs are marked as ‘a3’, ‘b3’, a4, b4. The star denotes bands with an as yet uncharacterized combination of splicing and polyadenylation that arise in the 1M mutant. (C) Northern blot analysis of the WT and 1M L1.3Neo constructs transiently transfected in NIH 3T3 cells with 5′UTR100 strand-specific probe. Full-length L1.3Neo mRNA is labeled FL1.3Neo. The splice variant specific to this vector is shown as SpX. Spliced and prematurely truncated mRNAs are identified as described in (B). (D) Northern blot analysis of the full-length and faster-migrating L1.3 mRNAs produced by the WT, 1M and ΔSV40 (L1.3 with no pA site at the end of the element) L1.3 constructs with 5′UTR100 and 5′UTR600 strand-specific probes. TRpA stands for the truncated polyadenylated L1.3 mRNA. Other products are marked as described above.
Some human and mouse L1 splice products are retrotranspositionally active
The 5′UTR100 probe also detected a slightly faster-migrating product than the full-length L1.3 mRNA (Figure 5B and D, a,bFL). The relative amount of this band increased in the 1M mutant of L1.3-notag. The 5′UTR600 strand-specific probe failed to identify the a,bFL band, but the truncated prematurely polyadenylated product (TRpA) produced by the L1.3ΔSV40 construct was detected (Figure 3D). The size of the a,bFL RNA is consistent with either splice ‘a’ and/or ‘b’ that terminated at the poly(A) site at the end of the L1.3 element (Figure 5A). Splice ‘a’ would result in L1 mRNA containing both ORFs and could potentially be retrotranspositionally active. Using a BLAST search with the splice junctions corresponding to the splice ‘a’ (Figure 2A), we identified four sequences on chromosome #1 (AL031985), #3 (AC093006), #9 (AL137022) and #11 (AP00560) that were flanked by target-site duplications, a hallmark of endonuclease-dependant L1 retrotransposition. Alignment of these sequences demonstrated that AC093006 belongs to the Ta family while the others were from older subfamilies (Supplementary Figure 3). Splice ‘b’ would produce a L1 mRNA that could make a truncated ORF1 protein, by utilizing an in-frame AUG downstream of the wt translation initiation codon (Figure 2A). A BLAST search of the human genome with the sequence corresponding to the splice junction ‘b’ identified at least 10 matching hits (Supplementary Table 1). Alignment of these sequences demonstrated that one, AL807813, belongs to the Ta family (Supplementary Figure 4). Additionally, we detected at least one sequence that matches L1 splice 97–303 on chromosome #20 (HSJ581I13). Because L1 constructs in which the 5′-UTR has been almost completely deleted are found to retrotranspose highly efficiently (20), RNAs that splice out portions of the 5′-UTR would also be expected to be capable of autonomous retrotransposition. We also searched the mouse genome with sequences corresponding to some of the splicing events at the predicted splice sites in the L1spa element (Figure 1). We found 22 matches to several of the splicing events predicted to produce retrotranspositionally competent L1spa mRNAs (SD sites at positions 27 and 239 and SA sites at 1514, 1597 and 1702 of the L1spa, Supplementary Table 1 and Supplementary Figures 5–7).
L1 splicing is redundant
The SD site at position 97 of the L1.3 genome appears to be the most commonly used 5′ splice site. We introduced a point mutation that destroyed the conserved GU element of the splice site (97M construct). Northern blot analysis with the strand-specific NeoEx probe detected the SpY band of the size similar to the size of the SpX band, but much lower intensity, and almost complete disappearance of the SpX(IN) band (Figure 6). Detection of the SpY band is consistent with either the usage of a cryptic splice site near the mutated SD site or utilization of the SD site at position 54 of the L1 genome. Use of this SD site would result in production of a transcript of almost the same size as SpX. Additionally, another major, smaller band, SpZ, was identified (Figure 6) consistent with the usage of one of the cryptic SA sites in the exon 2 of the NeoR gene (45). Quantitative analysis detected no increase in the amount of the full-length L1.3 RNA in proportion to the truncated RNA species between the WT and the 97M splice mutant elements. The 97M splice mutant retrotransposed at ∼60% of the efficiency of the wild-type element as determined by a retrotransposition assay in HeLa cells. This result was consistent with a reproducible decrease in the amount of RNA generated by the 97M splice mutant (Figure 6). The 97 splice site overlaps with a Runx3-binding site that regulates L1 promoter activity and the mutations we used to silence the splice site have been shown previously to silence this Runx3 site as well (13). L1.3Neo contains a CMV promoter, but the L1 promoter is also present and may explain changes in RNA levels in this mutant. Alteration in the splicing pattern of the 97M splice mutant, however, demonstrates that the removal of one splice site from the L1.3 sequence results in the more efficient usage of another splice signal. This compensation of the L1 splicing process is similar to the previously reported redundancy of the premature polyadenylation (17).
Figure 6.
Mutation of one of the splice sites in the L1.3 sequence results in the more efficient utilization of another splice signal. (A) An illustration of the major splicing events between either wt L1.3 [marked SpX and X(IN)] or a mutant of the 97 SD site, 97M, (marked SpY and Z) and the NeoR gene produced by the L1Neo construct. The dotted lines labeled SpY and SpZ represent predicted splices corresponding to those bands in the northern blot. (B) A northern blot analysis of the above depicted constructs with the NeoEx strand-specific probe.
L1 splice sites are utilized for hybrid splicing with human genes
L1 insertions into human genes can interfere with normal gene expression in numerous ways, often leading to a disease [reviewed in (46)]. Therefore, they are poorly tolerated, particularly when L1s are inserted in the forward orientation. We wished to determine whether functional splice sites in the L1 sequence can be utilized in combination with the splice sites of the human genes in which they insert. We performed a BLAST search (35) of the human EST database with the 210 bp fragment of the beginning of the L1.3 5′-UTR. Out of the total 1700 hits, 200 ESTs contained L1 sequence terminating precisely at the splice site at the position 97 of the L1.3. Of these ESTs 39 involved clear splicing events between L1 SD site at position 97 and SA sites of 21 different human genes (Table 1). Most of the other ESTs identified had sequence characteristics of authentic splices, but into sequences other than known exonic SAs. Identified splicing events between L1 elements and human genes came from libraries generated from different human tissues (bladder, brain, stomach and others) indicating that the process is not limited to any particular tissue type. We hypothesize that the number of identified ESTs of L1/gene splicing events is underrepresented due to (i) normalization of the majority of the libraries prior to cDNA synthesis, (ii) potential instability of the hybrid mRNAs, and (iii) most likely rapid elimination of the L1 insertion events that significantly interfere with the normal gene expression (disease or potential lethality in utero).
Table 1.
dbEST examples of L1 SD (97 bp) participating in splicing events with adjacent genic sequence
| Gene ID | Number of ESTs | Subfamilya | L1 insert location | Gene SA | Number of exonsb |
|---|---|---|---|---|---|
| BNI3PL | 2 | L1PA2 | intron 3 | exon 4 | 6 |
| C14ORF161 | 1 | L1PA2 | intron 15 | exon 15 | 26 |
| CYSLTR2 | 3 | L1PA2 | 5′ of gene | exon 1c | 1 |
| DOCK3 | 1 | L1PA3 | intron 5 | exon 6 | 53 |
| DST | 1 | L1PA3 | intron 1 | exon 2 | 22 |
| FLJ10986 | 1 | L1PA3 | intron 3 | exon 4 | 16 |
| FLJ22028 | 1 | L1PA2 | 5′ of gene | exon 2 | 12 |
| FLJ30851 | 1 | L1PA6 | 5′ of gene | exon 2 | 13 |
| FLJ39873 | 2 | L1PA3 | 5′ of gene | exon 2 | 4 |
| GABRR1 | 3 | L1PA2 | 5′ of gene | exon 3 | 10 |
| GFM1 | 1 | L1PA2 | 5′ of gene | 5′ UTRc | 17 |
| GUCY1B2 | 2 | L1PA6 | 5′ of gene | exon 14 | 16 |
| IGSF11 | 6 | L1PA3 | introns 2,3d | exon 3 | (7,9)e |
| KRTAP4-10 | 1 | L1PA2 | 5′ of gene | 5′-UTRc | 2 |
| LOC196394 | 6 | L1P-AN | intron 1 | intron 1c | 7 |
| MS4A5 | 1 | L1PA2 | intron 4 | 3′-UTRc | 5 |
| Myosin 1D | 2 | L1PA2 | intron 17 | 17 | 21 |
| PDGFRA | 1 | L1PA3 | intron 1 | 2 | 20 |
| RGL1 | 1 | L1PA3 | intron 1 | intron 1c | 18 |
| STIM1 | 1 | L1PA3 | intron 2 | 3 | 12 |
| TAF9L | 1 | L1PA4 | 5′ of gene | 1 | 7 |
Abbreviations: ID—GenBank gene identifier; SA—splice acceptor.
aL1 subfamily assignment was performed with RepeatMasker (www.repeatmasker.org).
bThe numbers in the last column of the table correspond to the total number of the exons identified in the gene.
cIndicates that L1 SD spliced to a cryptic SA within the gene region indicated.
dDenotes the presence of an L1 that spans two different introns due to a portion of its sequence being incorporated into an exon.
eIndicates alternative exons numbering based on different splice products.
DISCUSSION
Because only full-length L1 elements had been seen as capable of retrotransposition (20), it had been widely assumed that L1 makes only a single RNA species (31). This was called into question with the demonstration that the majority of L1 RNAs are truncated by premature polyadenylation (17). Our current data demonstrate that L1 RNAs are also involved in extensive RNA splicing that would radically alter the diversity of expressed RNA forms from these elements, as well as influence their impact on gene expression upon genomic insertion.
Relevance to the L1 life cycle
The presence of extensive and complex splicing of the L1 mRNA has many potential impacts on the life cycle of L1. Because of the observed cis preference of L1 RNA for its translation products (47), RNAs that do not encode both ORFs would not retrotranspose well and therefore almost all of the L1 splicing events will result in reduction of retrotransposition. The potential exceptions are the splices that primarily remove the 5′-UTR sequences (e.g. splices ‘a’ and ‘b’ in Figure 2A). These splice variants could express both ORFs and therefore be retrotranspositionally competent. Finding a number of full-length L1 elements that have inserted in the genome precisely missing those ‘intronic’ sequences demonstrates that these spliced mRNAs have undergone retrotransposition. Because the splicing events remove most of the promoter (19), any copies inserted by this mechanism would be less capable of further retrotransposition.
The products of splicing appear to be similar in quantity to the abundant premature polyadenylation transcripts. However, we cannot be sure that all spliced RNAs would have similar stabilities to the full-length RNAs. In particular, some would have very poor translational potential and, therefore, they might be subject to degradation by pathways such as nonsense-mediated decay (48,49). Thus, our observations represent a minimum estimate of L1 silencing by splicing.
Whether splicing has any major influence on L1 retrotransposition other than lessening expression is not clear. Between premature polyadenylation and splicing, we would expect production of mRNAs that could translate either ORF1 or ORF2 alone, as well as various truncated versions of these proteins. Production of the ORF2 protein via splicing is most likely not required for L1 retrotransposition because of the cis preference of L1 for its translation products (50). However, it would be expected to be sufficient to drive Alu retrotransposition (51). It is also possible that some of the other translation products may serve to either assist, or hinder, the L1 retrotransposition process.
Although we commonly think of splicing in terms of mRNA maturation, it is worth considering that L1 must return to the nucleus in order to be inserted and may be re-exposed to parts of the splicing apparatus. One observation that supports this association is that L1 elements commonly fuse during integration to spliceosome-associated U6 snRNA (52). Such chimeras can arise by a template switching mechanism, possibly facilitated by U6 snRNA being bound to the L1 mRNA molecule undergoing retrotransposition (52,53).
The genomic impact of L1 splicing
A number of studies have demonstrated that extant Alu elements contribute to extensive alternative splicing of genes through a process termed Alu exonization (54). Splice sites donated by Alu arise from mutations in the sequence of these elements that create consensus splice sites. In contrast, L1 elements already contain functional splice sites in their sequences prior to integration. Our finding of multiple examples of splicing events between L1 elements and human genes in the human EST database is consistent with several previous reports of genetic defect-causing hybrid splicing between L1 elements in either orientation and nearby genes in both human and mouse (5,33,34). We believe that our study is biased against the hybrid splicing events that severely compromise normal gene expression and splicing events that result in unstable transcripts. Plausible scenarios for L1 interference with gene expression include exon skipping via splicing between intronic L1s or an L1 and a SA site of a gene. These events would result in frame shift/nonsense mutations or in production of a protein with potential dominant mutant function. For example, previously reported splicing between L1 sequence and estrogen receptor (ER) gene produces a tumor-specific transcript encoding a protein that lacks hormone-binding domain of the normal ER (55). At least one of the genes in Table 1, GFM1, was reported as utilizing an alternative promoter to generate an alternative exon 1. This alternative exon is derived from the L1 promoter region.
Because L1 elements contain splice sites in both the sense and antisense strands, we would speculate that altered splicing of genes due to L1 elements inserted in introns could be one of their major negative impacts. The most commonly occurring 5′ and 3′ ESE is G/AAAG/AAA (26), suggesting that the A-rich sense strand of L1 elements may have a potential to support more efficient splicing. An ESE analysis program that predicts ESE hexamers (http://genes.mit.edu/burgelab/rescue-ese/) (26,56) identified four times as many ESEs in the sense strand of L1.3 as in the antisense. This suggests that there might be a difference in the strength of the splice sites of L1 strands which is consistent with the general finding that the limited L1 sequences found in introns are preferentially located in the antisense orientation (3,4). Predicted ESEs in the A-rich L1 sequence have a potential to influence the strength of the SA and SD sites of genes they have inserted. The presence of functional splice sites in the L1 genome may also contribute to the previously demonstrated decrease of transcripts containing L1 fragments (18).
The heterogeneity associated with L1 splicing, and its potential to negatively impact both the L1 life cycle and host genes, makes it seem unlikely that most of the splicing observed evolved for a specific purpose. We favor the hypothesis that the A-richness of the L1 coding regions may contribute to the ability of L1 RNAs to splice. Thus, the A-richness may be the cause of multiple forms of silencing of, and by, L1 sequences (17,18,57).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
We would like to thank Dr A. Engel and the members of the Deininger laboratory for helpful discussions. This work was supported by grants from Department of Defense Breast Cancer Research Program, DAMD17-02-1-0597 (V.P.B.), the National Institutes of Health, R01GM45668 (P.L.D), National Science Foundation, EPS-0346411 (P.L.D), and the State of Louisiana Board of Regents Support Fund (P.L.D). The authors gratefully acknowledge the help of Mark Batzer, Harold Silverman and other colleagues at Louisianna State University during the Katrina evacuation. Funding to pay the Open Access publication charges for this article was provided by NIH, R01 GM45668.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Medstrand P., van de Lagemaat L.N., Mager D.L. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 5.Murphy L.C., Dotzlaw H., Hamerton J., Schwarz J. Investigation of the origin of variant, truncated estrogen receptor-like mRNAs identified in some human breast cancer biopsy samples. Breast Cancer Res. Treat. 1993;26:149–161. doi: 10.1007/BF00689688. [DOI] [PubMed] [Google Scholar]
- 6.Benihoud K., Bonardelle D., Soual-Hoebeke E., Durand-Gasselin I., Emilie D., Kiger N., Bobe P. Unusual expression of LINE-1 transposable element in the MRL autoimmune lymphoproliferative syndrome-prone strain. Oncogene. 2002;21:5593–5600. doi: 10.1038/sj.onc.1205730. [DOI] [PubMed] [Google Scholar]
- 7.Bratthauer G.L., Cardiff R.D., Fanning T.G. Expression of LINE-1 retrotransposons in human breast cancer. Cancer. 1994;73:2333–2336. doi: 10.1002/1097-0142(19940501)73:9<2333::aid-cncr2820730915>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Ergun S., Buschmann C., Heukeshoven J., Dammann K., Schnieders F., Lauke H., Chalajour F., Kilic N., Stratling W.H., Schumann G.G. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 9.Muotri A.R., Chu V.T., Marchetto M.C., Deng W., Moran J.V., Gage F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 10.Branciforte D., Martin S.L. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell. Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trelogan S.A., Martin S.L. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl Acad. Sci. USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tchenio T., Casella J.F., Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang N., Zhang L., Zhang Y., Kazazian H.H. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asch H.L., Eliacin E., Fanning T.G., Connolly J.L., Bratthauer G., Asch B.B. Comparative expression of the LINE-1 p40 protein in human breast carcinomas and normal breast tissues. Oncol. Res. 1996;8:239–247. [PubMed] [Google Scholar]
- 15.Takai D., Yagi Y., Habib N., Sugimura T., Ushijima T. Hypomethylation of LINE1 retrotransposon in human hepatocellular carcinomas, but not in surrounding liver cirrhosis. Jpn. J. Clin. Oncol. 2000;30:306–309. doi: 10.1093/jjco/hyd079. [DOI] [PubMed] [Google Scholar]
- 16.Thayer R.E., Singer M.F., Fanning T. Undermethylation of specific LINE-1 sequences in human cells producing a LINE-1-encoded protein. Gene. 1993;133:273–277. doi: 10.1016/0378-1119(93)90651-i. [DOI] [PubMed] [Google Scholar]
- 17.Perepelitsa-Belancio V., Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nature Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 18.Han J.S., Szak S.T., Boeke J.D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 19.Swergold G.D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 21.Cost G.J., Feng Q., Jacquier A., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skowronski J., Singer M.F. The abundant LINE-1 family of repeated DNA sequences in mammals: genes and pseudogenes. Cold Spring Harb. Symp. Quant. Biol. 1986;51:457–464. doi: 10.1101/sqb.1986.051.01.055. [DOI] [PubMed] [Google Scholar]
- 23.Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 25.Jurica M.S., Moore M.J. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 26.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 27.Modrek B., Lee C. A genomic view of alternative splicing. Nature Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 28.Woodley L., Valcarcel J. Regulation of alternative pre-mRNA splicing. Brief. Funct. Genomic. Proteomic. 2002;1:266–277. doi: 10.1093/bfgp/1.3.266. [DOI] [PubMed] [Google Scholar]
- 29.Mironov A.A., Fickett J.W., Gelfand M.S. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo G., Holste D., Kreiman G., Burge C.B. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skowronski J., Fanning T.G., Singer M.F. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell. Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meischl C., Boer M., Ahlin A., Roos D. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur. J. Hum. Genet. 2000;8:697–703. doi: 10.1038/sj.ejhg.5200523. [DOI] [PubMed] [Google Scholar]
- 34.Mulhardt C., Fischer M., Gass P., Simonchazottes D., Guenet J.L., Kuhse J., Betz H., Becker C.M. The spastic mouse-aberrant splicing of glycine receptor-beta subunit messenger-RNA caused by intronic insertion of L1 element. Neuron. 1994;13:1003–1015. doi: 10.1016/0896-6273(94)90265-8. [DOI] [PubMed] [Google Scholar]
- 35.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sassaman D.M., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr Many human L1 elements are capable of retrotransposition [see comments] Nature Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 38.Skowronski J., Singer M.F. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc. Natl Acad. Sci. USA. 1985;82:6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson M.L., Bryman M.B., Peiter M., Cowan C. Exon size affects competition between splicing and cleavage- polyadenylation in the immunoglobulin mu gene. Mol. Cell. Biol. 1994;14:77–86. doi: 10.1128/mcb.14.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batt D.B., Rapp L.M., Carmichael G.G. Splice site selection in polyomavirus late pre-mRNA processing. J. Virol. 1994;68:1797–1804. doi: 10.1128/jvi.68.3.1797-1804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Y., Carmichael G.G. Splice site choice in a complex transcription unit containing multiple inefficient polyadenylation signals. Mol. Cell. Biol. 1991;11:5291–5300. doi: 10.1128/mcb.11.10.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roca X., Sachidanandam R., Krainer A.R. Determinants of the inherent strength of human 5′ splice sites. RNA. 2005;11:683–698. doi: 10.1261/rna.2040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C.D., Helfman D.M. Donor site competition is involved in the regulation of alternative splicing of the rat beta-tropomyosin pre-mRNA. RNA. 1999;5:290–301. doi: 10.1017/s1355838299980743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batt D.B., Luo Y., Carmichael G.G. Polyadenylation and transcription termination in gene constructs containing multiple tandem polyadenylation signals. Nucleic Acids Res. 1994;22:2811–2816. doi: 10.1093/nar/22.14.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulpa D.A., Moran J.V. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 46.Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 47.Moran J.V., Gilbert N., Boeke J., Kazazian H., Ostertag E., Loon S., Wei W. Human L1s retrotransposition: cis-preference vs trans-complementation. Am. J. Hum. Genet. 2000;67:199. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 50.Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 52.Buzdin A., Ustyugova S., Gogvadze E., Vinogradova T., Lebedev Y., Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics. 2002;80:402–406. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- 53.Ostertag E.M., Kazazian H.H. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dotzlaw H., Alkhalaf M., Murphy L.C. Characterization of estrogen receptor variant mRNAs from human breast cancers. Mol. Endocrinol. 1992;6:773–785. doi: 10.1210/mend.6.5.1603086. [DOI] [PubMed] [Google Scholar]
- 56.Fairbrother W.G., Yeo G.W., Yeh R., Goldstein P., Mawson M., Sharp P.A., Burge C.B. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han J.S., Boeke J.D. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.