Abstract
Post-translational modification of mitochondrial proteins by phosphorylation or dephosphorylation plays an essential role in numerous cell signaling pathways involved in regulating energy metabolism and in mitochondria-induced apoptosis. Here we present a phosphoproteomic screen of the mitochondria matrix proteins and begin to establish the protein phosphorylations acutely associated with calcium ions (Ca2+) signaling in porcine heart mitochondria. Forty-five phosphorylated proteins were detected by gel electrophoresis/mass spectrometry of Pro-Q Diamond staining while many more Pro-Q Diamond stained proteins were below mass spectrometry detection. Time dependent 32P incorporation in intact mitochondria confirmed the extensive matrix protein phosphoryation and revealed the dynamic nature of this process. Classes of proteins detected included all of the mitochondrial respiratory chain complexes, as well as enzymes involved in intermediary metabolism, such as pyruvate dehydrogenase (PDH), citrate synthase and acyl-CoA dehydrogenases. These data demonstrate that the phosphoproteome of the mitochondria matrix is extensive and dynamic. Ca2+ has previously been shown to activate various dehydrogenases, promote reactive oxygen species (ROS) generation, and initiate apoptosis via cytochrome c release. To evaluate the Ca2+ signaling network, the effects of a Ca2+ challenge sufficient to release cytochrome c were evaluated on the mitochondrial phosphoproteome. Novel Ca2+-induced dephosphorylation was observed in manganese superoxide dismutase (MnSOD) as well as the previously characterized PDH. A Ca2+ dose dependent dephosphorylation of MnSOD was associated with a ∼2-fold maximum increase in activity; neither the dephosphorylation nor activity changes were induced by ROS production in the absence of Ca2+. These data demonstrate the use of a phosphoproteome screen in determining mitochondrial signaling pathways and reveal new pathways for Ca2+ modification of mitochondrial function at the level of MnSOD.
Mitochondria are thought to be the result of an early interaction of two lines of cellular life, the bacterium and eukaryotic cell (1;2). At this point in time, mitochondria play a critical role in energy metabolism, apoptosis and cell signaling pathways in the cell. However, the acute and chronic regulatory mechanisms of this organelle remain poorly defined. One approach to assessing the function and regulation of the mitochondrion is an evaluation of the mitochondrial proteome. Estimates predict up to 3000 proteins (3;4) in mitochondria, however, recent large-scale screening studies by Taylor (5) and Mootha (6) identified only about 600 distinct mitochondrial proteins. Many have used proteomic approaches to evaluate differential protein expression in mitochondria to provide insight into chronic responses to perturbations and disease (for examples see (7;8)). The rapid response by mitochondria to changes in energy demand and other environmental factors suggests that acute regulatory pathways are also important in mitochondrial function. Phosphorylation events regulated by networks of kinases and phosphatases are currently believed to be among the most prevalent acute regulatory modifications within the cell (9-11). Many mitochondrial proteins have been demonstrated or proposed to be regulated by protein phosphorylation, including pyruvate dehydrogenase (PDH) (12) and components of the respiratory chain complexes (13-18). A phosphoproteome screen of potato mitochondria membranes using radiolabeled ATP found a wide range of dynamically phosphorylated proteins suggesting that the phosphorylation mechanism is extensively used in the mitochondria matrix(19). Information on the distribution of kinases and phosphatases within mitochondria is limited. Until recently, mitochondrial enzymes PDH kinase and branched-chain alpha-ketoacid dehydrogenase kinase were thought to be the main kinases functioning in mitochondria (20). Recent studies indicate that several cytosolic kinases translocate into mitochondria, including protein kinase A, protein kinase C δ and ε isoforms, stress-activated protein kinase, and A-Raf kinase (21;22). Several of these kinases are activated by calcium (Ca2+), a signaling molecule involved in activation of dehydrogenases (23), generation of reactive oxygen species (ROS)(24), and initiation of apoptosis (25;26).
The purpose of this study was to characterize the phosphoproteome of porcine heart mitochondria, as detected by Pro-Q Diamond stain using two-dimensional (2D) gel electrophoresis and 32P radioisotopic analysis as well as perform an initial screen for mitochondrial kinases and phosphatases associated with these protein phosphorylations. Following establishment of steady-state conditions, the effects of acute alterations in extramitochondrial Ca2+ sufficient to initiate mitochondria-induced apoptosis were evaluated on the mitochondrial phosphoproteome to provide insight into the signaling pathways associated with the complex action of Ca2+ on mitochondrial function.
EXPERIMENTAL PROCEDURES
Mitochondrial isolation:
Mitochondria were isolated from pig hearts, cold-perfused in situ to remove blood and extracellular Ca2+ (27). Briefly, were harvested from anesthetized and heparinized (10,000 units iv) animals. The was perfused via the aorta in a retrograde fashion in situ with ∼400 ml of ice-cold isolation buffer [0.28 M sucrose, 10 mM N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid (HEPES), and 0.2 mM EDTA, pH 7.21 to remove blood and reduce free calcium for mitochondrial isolation. The perfused heart was weighed, and the left ventricle was dissected free of fat, large vessels, and the right ventricular free wall. Sections of the left ventricle (4-5 g) were minced in 20 ml isolation buffer. Trypsin (2.5 mg) was then added, and the tissue was incubated for 15 min at 4°C. The digestion was stopped by adding 20 ml of isolation buffer with 1 mg/ml bovine serum albumin (BSA) and 13 mg trypsin inhibitor. The suspension was decanted, and the remaining tissue was resuspended in 20 ml of ice-cold isolation buffer with 1 mg/ml BSA. The tissue was homogenized with a loose-fitting Teflon homogenizer (2 times) followed by a tight-fitting Teflon pestle (5 times). The homogenate was centrifuged at 600 g for 10 min at 4°C and the supernatant was decanted and centrifuged at 8,000 g for 15 min. The buffy coat was removed, and the pellet was resuspended in 20 ml of ice-cold isolation buffer with 1 mg/ml BSA. The wash-and-centrifugation step was repeated twice, once in the presence of 1 mg/ml BSA and the final time in the absence of BSA. The final pellet was resuspended in 137 mM KCl, 10 mM HEPES, and 2.5 mM MgCls at pH 7.2 (experimental buffer) and stored on ice. It should be noted that this preparation created with a trypsin digestion represents a mixed population of mitochondria from the heart (28). Experiments on mitochondria isolated with the same procedure without trypsin resulted in a much lower yield and a very different protein profiles (not shown) suggesting that different pools of mitochondrial proteomes are present in the heart consistent with previous studies(29;30). No evidence of protein fractionation by trypsin was evident in comparing the tyrpsin and non-trypsin preparations suggesting that the trypsin treatment was not significantly influencing these results. All procedures performed were in accordance with the guidelines described in the Animal Care and Welfare Act (7 U.S.C. 2142 § 13).
Mitochondrial function and cytochrome c release:
The rate of mitochondrial oxygen consumption was determined at 37°C using a closed water-jacketed reaction chamber containing a Clark oxygen electrode as previously described (27). Most experiments were conducted in an oxygen-saturated buffer containing 125 mM KCl, 15 mM NaCl, 20 mM HEPES, 1 mM EGTA, 1 mM EDTA, 5 mM MgCl2 at pH 7.1 (buffer A). Mitochondria were allowed to equilibrate in the reaction chamber with buffer A for 6 minutes to permit Ca2+ depletion before adding carbon substrates (glutamate (5 mM) and malate (5 mM))(27).
Ca2+-dependent cytochrome c (cyt c) release was used as a marker of mitochondrial induction of apoptosis. Cyt c release was determined spectrophotometrically by quantifying the removal of cyt c from mitochondria pellets. After each experimental perturbation mitochondria were pelleted at 15,800 g and stored at -80°C for later analysis. Mitochondria pellets were resuspended (1 nmole cytochrome a (cyt a)/ml) in buffer A containing 5 μM Antimycin A, 5mM glutamate/malate, and 1% Triton X-100. Antimycin A was added to prevent electron flow to cyt c, resulting in highly oxidized states. Glutamate/malate was used to maximally reduce cytochrome b and FAD. Triton X-100 was used to minimized light scattering (31). The mitochondrial cyt c (550 nm) and cyt a (605 nm) content was determined from difference absorption spectra of the suspension in the presence and absence of sodium hydrosulfite to maximally reduce cyt c and cyt a. Mitochondria cyt c content is reported as the relative 550 nm peak area versus the 605 nm peak area of cyt a. It is important to note that since cytochrome b was held fully reduced in both conditions that it did not interfere with this determination.
The dependence of Ca2+-induced cyt c release on ATP, ADP and Pi is highly variable in the literature. Thus, we determined these interdependencies for this preparation. Combinational dose response curves for cyt c release at 5 minutes after addition of Ca2+ were conducted using Pi, ADP, ATP, and Ca2+. These studies revealed that ADP had little or no effect on this process, while 5 mM Pi and 10 mM ATP were found to generate an optimal release of cyt c in the presence of 100 μM free Ca2+ (see Results). Free Ca2+ levels were determined using the MaxChelator software for the elements in Buffer A. In separate experiments the time course of cyt c release was evaluated under these optimal conditions (buffer A with 5 mM Pi, 10 mM ATP and 100 μM Ca2+) and found to plateau approximately 5 minutes after Ca2+ addition. Thus, the conditions used for evaluating Ca2+-induced cyt c release were 100 μM free Ca2+, 5 mM Pi and 10 mM ATP added after the 6 minute depletion conditions outlined above. The controls were identical with the omission of Ca2+. Inhibition of Complex I was achieved by adding 6 μM rotenone and 3 mM succinate in lieu of Ca2+ and incubating for 5 minutes.
32P Labeling experiments:
To investigate the dynamics of 32P labeling of mitochondrial matrix proteins, experiments were performed to expose matrix proteins to physiological levels of ATP labeled in the gamma position with 32P (32PγATP). The experimental rationale was to add 32P as inorganic phosphate (Pi) to fully energized mitochondria. The Pi is transported into the matrix and used to synthesize 32PγATP by oxidative phosphorylation. It was assumed that this would provide a very high specific activity of the millimolar matrix ATP. This was accomplished by adding 0.75 m Currie of 32Pi to 15 mg of mitochondrial protein in 3 mls of buffer A in the absence of ATP or cold Pi in the presence of 5 mM G/M. The mitochondria were allowed to incubate for 5 to 20 minutes at which time an aliquot was removed and reaction was quenched with 5% TCA at 0° C with 5 mM KF. In some samples 0.1 mM dinitrophenol was added after the 20 minute labeling period with 32Pi and the incubation extended for additional 5 minutes to uncouple mitochondria, the sample was then quenched as described above. Samples were pelleted at 10,000 g. Mitochondrial pellets (3 mg protein) were solubilized with 100 μl of 1% SDS (w/v) in 100 mM Tris-HCl, pH 7.0 at 95°C. Pellets were incubated at 95°C for 5 min followed by cooling on ice for 5 min. A chloroform/methanol precipitation was performed to remove salts, lipids and free 32Pi or 32PγATP (32) by adding 6 ml methanol, 150 μl chloroform, and 450 μl dH20 to each pellet, vortexing between each addition. Samples were centrifuged for 5 min at 12,000 g and the supernatant was discarded. Precipitated protein was washed again by centrifuging in 450 μl methanol.
2D gel electrophoresis and gel staining:
Samples were run differently for the radioisotopic and Pro-Q Diamond and Sypro Ruby straining procedures. For Pro Q staining mitochondrial pellets (1 nmol cyt a) were solubilized with 100 μl of 1% SDS (w/v) in 100 mM Tris-HCl, pH 7.0 at 95°C. Pellets were sonicated 5 times for 3 sec each or until dissolved. Pellets were incubated at 95°C for 5 min followed by cooling on ice for 5 min. A chloroform/methanol precipitation was performed to remove salts and lipids (32) by adding 600 μl methanol, 150 μl chloroform, and 450 μl dH20 to each pellet, vortexing between each addition. Samples were centrifuged for 5 min and the supernatant was discarded. Precipitated protein was washed again by centrifuging in 450 μl methanol. The supernatant was discarded and pellets were re-suspended in 100 μl of buffer containing 30 mM Tris-HCl, 7 M urea, 2 M thiourea, and 4% CHAPS (w/v). Samples were pooled at this stage to obtain adequate protein (500 μg/gel) for paired 2D gel analysis. Because protein is lost during this precipitation procedure, the correlation between cyt a content and total protein may no longer be valid. Therefore, total protein of each sample was determined using the Amersham Quant kit (Amersham Biosciences, Piscataway, NJ). For each sample, 500 μg total protein in 440 μl of rehydration solution [7 M urea, 2 M thiourea, and 4% CHAPS (w/v), 1% De-streak reagent (v/v), and 2% (pH 3-10NL) Pharmalyte (v/v)] were loaded onto 24-cm Immobiline DryStrip gels (pH 3-10 NL). Isoelectric focusing was achieved by active rehydration for 12 h at 30V followed by stepwise application of 500 V, 1000 V, and 8,000 V for a total of ∼70,000 Vh (Ettan IPG Phor, Amersham). Immobiline DryStrip gels were equilibrated in 10 ml SDS equilibration solution (50 mM Tris·HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS) for 10 minutes, first containing 0.5% DTT then with 4.5% iodoacetemide. Gel strips were applied to 12.5% SDS-PAGE gels and sealed with 0.5% agarose containing bromophenol blue. Electrophoresis was performed in an Ettan DALT-12 tank (Amersham) in electrophoresis buffer consisting of 25mM Tris, pH 8.3, 192 mM glycine, and 0.2% SDS until the dye front advanced completely (∼1750 Vhrs). Gels were fixed overnight in 500 ml in a solution of 10% TCA and 30% methanol. Fix solution was changed once. Following 4 15-minute washes in 1 L warm water each, gels were stained with 500 ml Pro-Q Diamond (Molecular Probes, Eugene, OR) for 3 hours and de-stained using 4 1-hour washes with 500 mL of de-stain containing 50 mM sodium acetate and 10% acetonitrile. Following image acquisition, gels were stained with Sypro Ruby protein gel stain (Bio-Rad Laboratories, Hercules, CA). For the radioisotope studies mitochondrial protein was suspended to a concentration of 500 μg in 500 μl of a solution containing rehydration buffer (8M urea, 2% CHAPS, 15 mM DTT, 0.2% ampholytes pH 3-10, and 0.001% orange G). The 500 μl protein dilutions were loaded onto IPG strips (24 cm, linear pH 3-10) by overnight, passive rehydration at room temperature. Isoelectric focusing was performed simultaneously on all IPG strips using the Protean IEF Cell (BioRad), by a program of progressively increasing voltage (150 V for 2 h, 300 V for 4 h, 1500 V for 1 h, 5000 V for 5 h, 7000 V for 6 h, and 10,000V for 3 h) for a total of 100,000 Vh. A computer-controlled gradient casting system was used to prepare second-dimension SDS gradient slab gels (20 × 25 × 0.15 cm) in which the acrylamide concentration varied linearly from 8% to 15% T. First-dimension IPG strips were loaded directly onto the slab gels following equilibration for 10 minutes in Equilibration Buffer I and 10 minutes in Equilibration Buffer II (Equilibration Buffer I: 6M urea, 2% SDS, 0.375M Tris-HCl pH 8.8, 20% Glycerol, 130mM DTT; Equilibration Buffer II: 6M urea, 2% SDS, 0.375M Tris-HCl pH 8.8, 20% Glycerol, 135mM iodoacetamide). Second-dimension slab gels were run in parallel at 4°C for 18 h at 160V. Slab gels were stained using a colloidal Coomassie Blue G-250 procedure. Gels were fixed in 1.5 L of 50% ethanol/2% phosphoric acid overnight followed by three 30 min washes in 2 L of deionized water. Gels were transferred to 1.5 L of 30% methanol/17% ammonium sulfate/3% phosphoric acid for 1 h followed by addition of 1 g of powdered Coomassie Blue G-250 stain(33). After 96 h, gels were washed several times with water. Gels were allowed to equilibrate overnight in a 5% glycerol solution and then dried in a large format gel dryer for 6 hours at 65° C under a vacuum. Dried gels were placed in a film development cassette (Kodak) for 5 days with 3 sheets of 8 × 10 maximum sensitivity film (Kodak).
Image acquisition and analysis:
For Pro-Q Diamond and Supro Ruby analysis gels were scanned on a Typhoon 9400 variable mode imager (Amersham) at a resolution of 100 μm. Excitation was at 532 nm with emission filters of 610BP30 for Sypro Ruby and 580BP30 for Pro-Q Diamond. Image analysis was performed using single stain analysis with intelligent noise correction algorithm (INCA) processing by Progenesis Discovery software (Nonlinear Dynamics, Newcastle upon Tyne, UK). Radiograms and dried gels were scanned on a Epson CX5400 high resolution scanner The Ettan Spot Handling Workstation performed automated extraction and in gel trypsin digestion of selected protein spots according to Amersham instructions. Peptides were analyzed using a mass spectrometer (4700 Proteomics Discovery System, Applied Biosystems, Foster City, CA) using MALDI-TOF and tandem MS/MS. At least two peptides were obtained for each protein using MS/MS. Proteins were identified from the acquired spectra using the MASCOT database search function.
Enzyme activity assays:
The activity of manganese superoxide dismutase (MnSOD) was measured spectroscopically using a commercially available assay kit (Trevigen, Gaithersburg, MD). Superoxide anions generated by the conversion of xanthine to uric acid and hydrogen peroxide by xanthine oxidase in turn convert NBT to NBT-diformazan, which absorbs light at 550 nm. MnSOD activity was measured in both control and high Ca2+-treated mitochondria pellets by the reduction of NBT-diformazan, as indicated by a decrease in A550.
PDH activity was determined by following NADH production in the presence of pyruvate, coenzyme A and NAD (34). Mitochondria pellets from control and high Ca2+ experiments were resuspended in small volume and pulverized to disrupt membranes. Mitochondria matrix elements were exposed by freezing the mitochondria suspension in liquid nitrogen and pulverizing the frozen pellet using a tissue Bessman pulverizer (BioSpec Products Inc., Bartlesville, OK). The thawing and freeze-pulverizing cycle was repeated 2 times. PDH activity was assayed in a reaction mixture (pH 8.0) containing 50 mM Tris, 10 mM pyruvate, 0.2 mM Coenzyme A, 2 mM NAD, 2 mM cocarboxylase, 1 mM MgCl2, and pulverized mitochondria at a concentration of 0.2-0.4nmol cyt a/mL. The reaction was carried out at 37°C and was initiated with coenzyme A. Production of NADH was measured spectrophotometrically by monitoring A350.
H2O2 Production:
H2O2 was measured fluorometrically using the Amplex Red Hydrogen Peroxide Assay Kit (Molecular Probes). The production of H2O2, as indicated by the conversion of Amplex Red to resorufin, was monitored under control and high Ca2+ conditions for 10 minutes. Fluorescence intensity was measured with a fluorometer (FL3-22, Jobin-Yvon Horiba, Edison, NJ) using excitation and emission wavelengths of 545 nm and 590 nm, respectively.
Screen for kinases and phosphatases:
Mitochondria pellets were suspended in lysis buffer (20 mM Tris, 40 mM glycerophosphate, 30 mM sodium fluoride, 20 mM sodium pyrophosphate, 5 mM EDTA, 2 mM EGTA, 1 mM sodium orthovanadate, 0.5% Triton X-100 and 1 mM DTT) supplemented with 1 mM phenylmethanesulfonylfluoride, 2 mg/ml leupeptin, 4 mg/ml aprotinin and 1 mg/ml pepstatin A, and sonicated for 15 sec. Debris was removed by centrifugation at 100,000 rpm for 30 min at 4°C. Protein concentration of the resulting supernatant was determined using the Amersham Quant kit. Kinetworks analyses (Kinexus Bioinformatics Corp., Vancouver, Canada) were performed on 300-600 μg protein/sample by SDS-PAGE and subsequent immunoblotting with panels of up to three primary antibodies per channel in a 20-lane multiblotter. The Kinetworks analyses screened for 75 kinases (KPKS 1.2) and 25 phosphatases (KPPS 1.2).
RESULTS
Initial studies were conducted to determine what proteins of the mitochondrial proteome are resolved and detected using our 2D gel electrophoresis system. From gels stained for total protein with Sypro Ruby we identified mitochondrial proteins of various functions, including intermediary metabolism, β-oxidation, amino acid biosynthesis, complexes of oxidative phosphorylation, transport proteins including chaperones, etc., consistent with what has previously been reported in mouse brain, heart, kidney and liver (6) and human heart (5) mitochondria. Because the pig genome has not been fully sequenced, we were unable to identify some proteins based on existing porcine sequence data and therefore used other mammalian database information because many mitochondrial proteins are highly conserved across species. Some proteins were unable to be identified using these methods, despite a relative protein abundance, suggesting extensive gene-splicing or post-translational modifications complicating the identifications. Similar problems have been noted in prior studies (6;7).
Two strategies were used for detecting phosphorylated proteins. Pro-Q Diamond was used to stain for phosphorylated proteins independent of protein turnover. 32P labeling was used to examine the dynamics of matrix protein phosphorylation and provide confirmation of protein phosphorylations found in the more indirect Pro-Q Diamond staining approach for those proteins with adequate phosphate turnover. The sensitivity of Pro-Q Diamond for serine, threonine and tyrosine phosphorylation has been validated in several systems (35;36). Most recently, it has been used to characterize the global effects on protein phosphorylation in response to alterations of cellular kinases (37). A representative gel of mitochondrial proteins stained with Pro-Q Diamond is shown in Figure 1. Automatic spot detection showed about 200 phosphorylated spots per gel with Pro-Q Diamond staining. However, the total number of proteins was less than 200 since many proteins had a distribution of spots generated by the isoelectric focusing caused by multiple phosphorylations, as observed with aconitase or pyruvate dehydrogenase, or other phenomena such as differential oxidation. We considered a positive identification to be indicated by >95% confidence in the MASCOT identification. Using these criteria, 45 separate proteins were identified by mass spectrometry analysis, accounting for a majority of the observed proteins. Proteins identified included all of the complexes of oxidative phosphorylation, numerous enzymes of intermediary metabolism as well as enzymes involved in reactive oxygen species metabolism (Table 1). While many of these phosphorylations have been previously described, several of these phosphorylated enzymes, to our knowledge, have not been previously reported in the literature and represent unique observations. These include several complex I subunits, enzymes involved in fatty acid metabolism, and the gamma subunit of the F1-ATPase (γF1). MnSOD has been shown to be phosphorylated in potato mitochondria (19) using radiolabeled ATP, but we have not found evidence of this phosphorylation described in mammalian systems. The Pro-Q Diamond-stained gel shows 4 distinct spots that were each identified as MnSOD using MS/MS, which have similar molecular weight but different isoelectric points, consistent with multiple phosphorylation states.
Fig. 1.

Two-dimensional gel electrophoresis and staining of the phosphoproteome of porcine heart mitochondria with Pro-Q Diamond phosphoprotein gel stain. Proteins are separated by isoelectric point (pI), from pH ∼4-9 along the horizontal axis, and by molecular weight, from ∼100 to 10 kD, vertically. Numbers refer to the protein identifications presented in Table 1. Not all Pro-Q Diamond-stained proteins were identified due to not reaching statistical significance in the mass spectrometry analysis.
Table 1.
Porcine heart mitochondrial proteins stained with Pro-Q Diamond
| Functional Category | Spot Number | Protein Name | NCBI Accession Number |
|---|---|---|---|
| Oxidative Phosphorylation | |||
| Complex I | 11, 12 | NADH dehydrogenase (ubiquinone) Fe-S protein 1, (75kDa) | 51858651 |
| 22 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | 23574759 | |
| 60 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 (39kDa) | 13097156 | |
| 65 | NADH dehydrogenase 24 kDa subunit | 1364245 | |
| 73 | NADH-ubiquinone oxidoreductase 23 kDa subunit, mitochondrial precursor | 2499325 | |
| 78 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa | 28461255 | |
| 84 | NADH-ubiquinone oxidoreductase 15 kDa subunit (Complex I-15 kDa) | 400587 | |
| Complex II | 13-17 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor | 1352262 |
| 72 | Succinate dehydrogenase Ip subunit | 27716317 | |
| Complex III | 26,27 | Ubiquinol-cytochrome-c reductase complex core protein I, mitochondrial precursor | 10720406 |
| 40, 43 | Ubiquinol-cytochrome c reductase core protein II | 27807143 | |
| 67 | Ubiquinol-cytochrome-c reductase, Rieske iron-sulfur protein precursor | 111883 | |
| 83 | Ubiquinol-cytochrome c reductase binding protein | 34866011 | |
| Complex IV | 81, 82 | Cytochrome c oxidase polypeptide Va | 117097 |
| F0F1-ATPase | 23 | ATP synthase, mitochondrial F1 complex, alpha subunit | 15030240 |
| 24, 35 | ATP synthase, mitochondrial F1 complex, beta subunit precursor | 32189394 | |
| 62 | ATP synthase gamma subunit precursor | 162717 | |
| 19, 20 | Chain C, bovine mitochondrial F0F1-Atpase | 33357743 | |
| 79, 80 | H+-ATPase subunit, oligomysin sensitivity conferring protein | 913531 | |
| Intermediary Metabolism | |||
| Krebs Cycle | 51-53 | Aspartate aminotransferase, mitochondrial precursor | 112985 |
| 2-7 | Aconitate hydratase, mitochondrial precursor | 113159 | |
| 39 | Citrate Synthase, chain B | 230994 | |
| 42, 49, 50 | Isocitrate dehydrogenase (NADP-dependent) | 284570 | |
| 56-59 | Malate dehydrogenase | 65932 | |
| 1 | 2-Oxoglutarate dehydrogenase | 2160381 | |
| 69, 70 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex | 266684 | |
| 28-34 | Pyruvate dehydrogenase, E1 alpha subunit | 448580 | |
| Fatty Acid Oxidation | 47 | Acyl-coA dehydrogenase, long-chain specific, mitochondrial precursor (LCAD) | 2829676 |
| 21 | Dihydrolipoyl dehydrogenase, mitochondrial precursor | 1706444 | |
| 71 | Electron transfer flavoprotein, beta subunit precursor | 35384838 | |
| 68 | Enoyl coenzyme A hydratase, short chain | 49257190 | |
| 8-10 | Gastrin-binding protein | 47522754 | |
| 61 | Chain C, pig heart short chain L-3-Hydroxyacyl coA dehydrogenase | 6435806 | |
| 77 | Hydroxyacyl-coenzyme A dehydrogenase, type II | 27805907 | |
| 44, 45 | Long-chain 3-ketoacyl-coA thiolase | 47522760 | |
| 46 | Chain A, medium chain acyl-Coa Dehydrogenase (MCAD) | 640350 | |
| 48 | Short-chain acyl-CoA dehydrogenase (SCAD) | 47522686 | |
| Transport | |||
| 18 | Chaperonin; mitochondrial protein P1 precursor | 90207 | |
| 55 | Voltage-dependent anion channel 1 | 47522750 | |
| 54 | Voltage-dependent anion channel 2 | 55664661 | |
| Antioxidant | |||
| 74-76 | Mn superoxide dismutase | 15082142 | |
| 66 | Thioredoxin-dependent peroxide reductase, mitochondrial precursor | 2507170 | |
| Other | |||
| 37 | Sarcomeric mitochondrial creatine kinase precursor | 4502855 | |
| 35, 36, 41 | Creatine kinase, mitochondrial 2 | 38174368 | |
| 63, 64 | Prohibitin | 4505773 | |
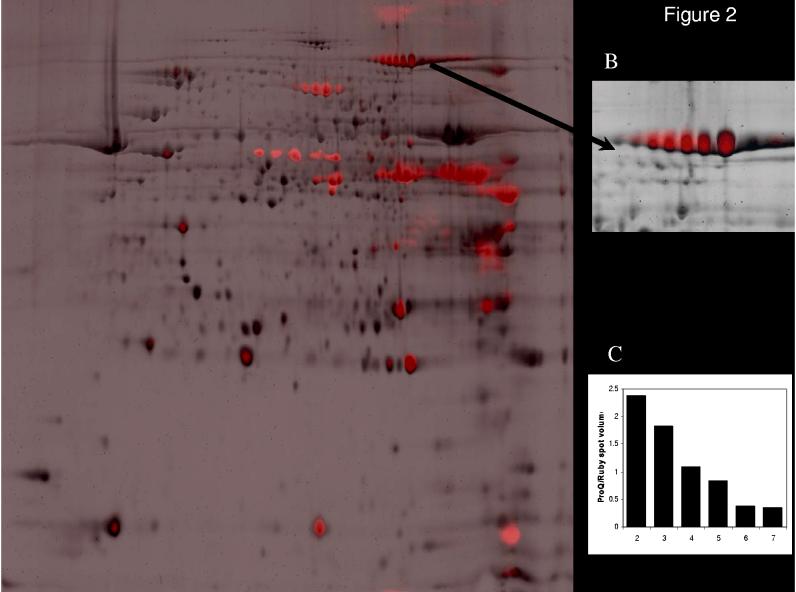
To relate the level of phosphorylation to protein content, Figure 2A shows an overlay of the Pro-Q (red) and Sypro Ruby (black) images, indicating the relative intensity of phosphorylation compared to the total amount of protein present for each spot. Intensely red spots are highly phosphorylated low abundance proteins. Multiple aconitase spots (Figure 2B) reveal the relative degree of phosphorylation changing with the isoelectric focusing pH, revealed by a ratiometric approach (Fig. 2C). The low abundance of some phosphorylated proteins hampered mass spectrometry identification and suggested that some proteins were better detected with Pro-Q Diamond than Sypro Ruby. A similar observation was made between Comassie stain and 32P labeling below.
Fig. 2.
Overlay of Sypro Ruby total protein (black) and Pro-Q Diamond phosphoprotein (red) staining of mitochondrial proteome in the absence of Ca2+ (A). The relative amplitude of the two channels was arbitrarily set. The majority of proteins detected by Sypro Ruby were not detected with Pro-Q Diamond resulting in a predominance of pure black spots. Proteins heavily labeled with Pro-Q Diamond appear red with essentially no Sypro Ruby signal (for example PDH, spots 27-33). We used the ratio of the Sypro Ruby and Pro-Q Diamond signals as a quantitative method for determining the degree of protein phosphorylation. As a control for this approach, the multiple phosphorylation states of aconitase (spots 2-7) were evaluated in panels B and C. The enhanced phosphorylation of aconitase is associated with an acid shift in its isoelectric focusing pH, taking the ratio of the Sypro Ruby stain and Pro-Q Diamond revealed a quantitative relationship between this ratio and isoelectic focus for this single protein.
A representative phosphor image of 32P labeled mitochondrial proteins with the corresponding Coomassie stained gel are shown in Figure 3A,B. Due to the wide range of 32P labeling any one exposure is not adequate to reveal all of the sites without over or under exposure of the film or contrast/brightness setting in software. We have selected an intermediate exposure for this example. The proteins labeled using this approach included: SOD-2, PDH E1α, citrate synthase, inhibin, MCAD, LCAD and Rieske iron sulfur protein (RISP) with details in the figure legend. Many of the 32P labeled protein corresponded to observation in ProQ-Diamond. However, there were many notable differences between 32P and Pro-Q Diamond staining. Many more proteins were labeled with 32P where there was no corresponding Pro-Q Diamond, Coomassie/Sypro Ruby staining, leaving a much different overall pattern in all three staining approaches. The direct comparison of the 32P labeling (red) with Coomassie (green) is seen in the overlay presented in Figure 3C. At this exposure the 32P labeling was overexposed in the PDH E1α region. The region around MnSOD and RISP has been expanded in all of the panels. The correlation of the Coomassie with the 32P labeling is generally poor suggesting many low abundance proteins with significant number of phosphorylation sites with high turnover. These observations suggest that the overall sensitivity of the 32P method is significantly higher than Pro-Q Diamond especially for proteins with high phosphate turnover rates while Pro-Q Diamond is more sensitive to more abundant proteins with slow turnover rates. In addition, the absolute sensitivity for Pro-Q Diamond for all phosphorylation sites should not be considered constant, as it surely is for 32P labeling, since the confirmation of the relative sensitivity of Pro-Q Diamond has been limited to a handful of proteins. The dependence on phosphorylation turnover can limit 32P detection of phosphorylation as illustrated by the effects of incubation times of 5 and 20 minutes on 32P labeling in Figure 4. Clearly, a longer incubation time results in more detectable phosphorylation sites. The labeling of a significant fraction of proteins in 5 to 20 minutes suggested that the turnover of the phosphorylation events was quite rapid for many proteins. To confirm the off-rate, we treated the mitochondria with uncoupler that would stimulate breakdown and inhibit synthesis of matrix 32PγATP. After only 5 minutes, the overall 32P labeling was significantly reduced supporting the notion of a rapidly turning over pool of phosphorylated proteins (4C). The complete time dependence of this process is outside the scope of the current report, but this approach can clearly be applied to obtain 32P turnover rates for many of these proteins. One interesting omission from the 32P data was any detectable turnover of phosphorylation in aconitase or succinate dehydrogenase. Both Pro-Q Diamond and the isoelectric shift pattern of these proteins are consistent with the phosphorylation. The lack of 32P labeling of these proteins suggests a very slow turnover of much more than 20 minutes in this preparation.
Fig. 3.

Two-dimensional gel electrophoresis and staining of the phosphoproteome of porcine heart mitochondria with P32-PO4. Proteins are separated by isoelectric point (pI), from pH ∼4-9 along the horizontal axis, and by molecular weight, from ∼100 to 10 kD, vertically. A) Autoradiogram of gel. The MnSOD-Rieske iron sulfur region is expanded in this and all other panels at optimal contrast/brightness. B) Coomassie stain of same gel. C) Color overlay of autoradiogram (red) and Coomassie stain (green). Amplitude of both gels was arbitrarily set.
Fig. 4.
Two-dimensional gel electrophoresis and staining of the phosphoproteome of porcine heart mitochondria with 32P PO4. Proteins are separated by isoelectric point (pI), from pH ∼4-9 along the horizontal axis, and by molecular weight, from ∼100 to 10 kD, vertically. The mitochondria were harvested after incubation with 32P-PO4 for either 5 minutes (A), 20 minutes (B) or 20 minutes then 5 minutes with dinitrophenol, a mitochondrial uncoupler (C). The incubation conditions are outlined in the Methods section.
Ca2+ is a well-recognized second messenger in the control of mitochondrial function both under normal and pathophysiological conditions (23;38). Ca2+ action has often been linked to protein phosphorylation events via Ca2+-sensitive kinases and phosphatases. Thus, we applied this phosphoprotein screen to evaluate the acute effects of extramitochondrial Ca2+ on mitochondrial protein phosphorylation. The concentration of Ca2+ used was selected to be sufficient to induce cyt c release from mitochondria, the initial step of mitochondria-induced apoptosis. Because it was difficult to predict the optimal extramitochondrial conditions to cause cyt c release with Ca2+ from the literature, we determined the extramitochondrial conditions of maximal Ca2+-induced cyt c release for our system by exposing mitochondria to various concentrations of Ca2+ in the presence of glutamate and malate, Pi, and adenine nucleotides (ATP or ADP) while respiration and cyt c release were monitored. Maximal cyt c release occurred in the presence of 5 mM Pi, 10 mM ATP, and excess of 100 μM free Ca2+. Mitochondria released 65.7 ± 5.0% of total cyt c under these conditions, compared to 6.9 ± 2.7% (P<0.001) release in the absence of Ca2+ (n=4). Ca2+ concentrations above 100 μM did not result in a significant increase in the percent of cyt c released, suggesting that the amount released during this acute perturbation plateaus around 70% of the total cyt c content. Mitochondria released only 14.1 ± 2.2% of the total cyt c (P <0.01, n=4) in 100 μM Ca2+ without Pi and ATP, demonstrating that cyt c release depends on Pi and ATP in addition to high free Ca2+. Pre-incubation of mitochondria in Cyclosporin A, an agent that inhibits mitochondria-induced apoptosis by blocking the mitochondria permeability transition pore, before exposure to high Ca2+, Pi, and ATP, resulted in release of only 36.6 ± 3.8% (P <0.01, n=4) of total cyt c, a blockage of nearly 68% of the cyt c release occurring under the high Ca2+ conditions in the absence of this agent (data not shown). It is important to note that in these studies with 10 mM ATP in the extramitochondrial space that significant ATP depletion would likely not occur even under uncoupled conditions. This is in contrast to the 32P studies were the total concentration of ATP was limited by the very low concentration of 32P added to the sample.
The effects of Ca2+ on mitochondria protein phosphorylation were determined by incubating mitochondria under the control conditions in the absence and presence of 100 uM free Ca2+. Intensity of staining was given by the spot volume corrected for noise using the Progenesis Intelligent Noise Correction Algorithm (INCA), normalized for total protein content using the total spot volume of the corresponding Sypro Ruby image. Due to the signal to noise ratio of the Pro-Q Diamond data, we only considered a significant change in phosphorylation when the normalized area changed more than 30% in these initial studies. While most of the proteins did not exhibit a significant change in phosphorylation state with this perturbation, several proteins showed a dramatic change between control and high Ca2+ conditions. Again, we focused on the proteins undergoing the largest change in phosphorylation state in this perturbation. These highly significant changes include PDH, MnSOD and the γ F1. It is important to note that these are the proteins exhibiting large alterations with Ca2+, and that more subtle changes in phosphorylation or effects on proteins at low concentrations were ignored in this initial screen. We did not detect a dephosphorylation of RISP in response to Ca2+ as suggested by isoelectric focusing shifts previously observed in liver mitochondria(39).
We focused on PDH and MnSOD for the remaining portion of this study since one, PDH, served as a control as a well known protein phosphorylation affected by matrix Ca2+ while MnSOD is an important enzyme in ROS metabolism that was previously unknown as a phosphorylated protein in mammalian mitochondria.
Previous selective non-screening studies demonstrated Ca2+-induced dephosphorylation of the E1α subunit of PDH and activation of PDH (12;40). Consistent with these results, Pro-Q Diamond staining showed a dramatic dephosphorylation of the E1α subunit of PDH in the presence of Ca2+ (Fig. 5A) that had the appropriate sensitivity to the extramitochondrial Ca2+ level. Four distinct PDH E1α subunit spots present in all Pro-Q Diamond stained gels showed an average 63.1% decrease in phosphorylation with Ca2+ (n=9, P <0.05; Fig. 5B). Activity of PDH was increased by 203.6% (n=5, P <0.05) under the high Ca2+conditions that induced the dephosphorylation of the E1α subunit (Fig. 5C). This confirmation of the well known effects of Ca2+ on PDH phosphorylation provides a useful confirmation of this phosphoprotein screen.
Fig. 5.

Effect of Ca2+ on PDH phosphorylation and activity. Representative images of gels stained with Pro-Q Diamond indicate the degree of PDH E1α phosphorylation of individual proteins under conditions of zero (top panel) and high free Ca2+ (bottom panel) (A). Multiple protein spots of pyruvate dehydrogenase E1 alpha subunit stain with Pro-Q Diamond more intensely under control conditions than under high Ca2+ conditions. The degree of phosphorylation under each condition was calculated as the ratio of intensity of Pro-Q staining for each spot to the total Sypro Ruby spot volume for that gel to normalize for any difference in total protein loaded in the gel and is given as the mean ± S.E.M. (B). Because these proteins are highly phosphorylated but not abundant, matching spots from Pro-Q Diamond to Sypro Ruby images was difficult and therefore total spot volume of the gel was used to normalize to amount of protein. PDH enzyme activity increased in the presence of high Ca2+ relative to control conditions (C).
Ca2+ exposure yielded an average 50.8 % dephosphorylation of MnSOD (n=8, P <0.05; Fig. 6A, B). To our knowledge, the phosphorylation of MnSOD has not previously been described in mammalian mitochondria. This enzyme is believed to play a critical role in the scavenging of ROS in the mitochondrial matrix. Thus, we further investigated the functional consequences of this dephosphorylation. The Ca2+ challenge resulted in a ∼10 fold increase in mitochondrial H2O2 generation. Associated with this Ca2+-induced increase in H2O2 production was a 59.1% increase in MnSOD activity (n=3, P <0.05; Fig. 6C). The extramitochondrial Ca2+ K50 of MnSOD activity was ∼10 μM (Fig. 6D). Since ROS production was increased by Ca2+, we hypothesized that ROS could be directly responsible for the activation of MnSOD, independent of Ca2+. This hypothesis was tested by generating similar levels of H2O2 production independent of Ca2+ and examining the effects on MnSOD phosphorylation and activity. Rotenone and succinate were titrated to increase H2O2 generation rate (41) to levels similar to those achieved under high Ca2+ conditions (Fig. 7A). Unlike the high Ca2+, the rotenone/succinate addition did not induce significant release of cyt c (Fig. 7B), indicating that high ROS production alone is not sufficient to induce mitochondrial apoptosis, nor did it change MnSOD activity or phosphorylation state (Fig. 7C, D). These data are consistent with a Ca2+-dependent activation of MnSOD via dephosphorylation. In addition, the screen failed to detect any large changes in protein phosphorylation associated with the rotenone/succinate condition with the large increase in H2O2 production, suggesting that acute ROS generation alone was not effective in modulating the phosphorylation state of PDH, γF1 or other mitochondrial phosphoproteins detected (data not shown).
Fig. 6.

The effect of Ca2+ on MnSOD phosphorylation and activity. A) MnSOD also showed less intense staining with Pro-Q Diamond under high Ca2+ conditions (bottom panel) compared to control (top panel). Quantification of the degree of phosphorylation under each condition was determined by the intensity of Pro-Q Diamond staining normalized to the corresponding Sypro Ruby intensity for that MnSOD spot (B). The activity of MnSOD normalized to cyt a content under control and high Ca2+ conditions shows increased enzyme activity with the addition of Ca2+ (C). This increase is dependent on Ca2+ concentration. The dose-response curve of MnSOD activity over Ca2+ conditions ranging from 0 to 100 μM free Ca2+, expressed as the percent activity under control conditions, show that the K50 is ∼10μM (D).
Fig. 7.

The effects of matrix ROS production on MnSOD phosphorylation and activity. Rate of H2O2 production per minute, normalized to cyt a content, shows that treatment of mitochondria with rotenone and succinate (R/S) increased rate of H2O2 production significantly over control levels, similar to the increase induced by the high Ca2+ conditions (A). Cyt c is released from mitochondria in the presence of high Ca2+, but not with R/S, indicating that the increased H2O2 production does not induce apoptosis (B). MnSOD spots in gels of mitochondria exposed to R/S show no change in Pro-Q Diamond staining intensity relative to control (C). MnSOD activity shows no significant difference under control and R/S conditions (D).
The dose dependence of Ca2+ on PDH and MnSOD phosphorylation was evaluated at four concentrations 0, 0.6, 40 and 100 uM free Ca2+ in paired experiments to determine whether these effects were due to the gross metabolic insult associated with Ca2+ induced cyt c release. In addition, we could roughly compare the MnSOD phosphorylation level with activity to further establish cause and effect. The 0.6 uM concentration was selected as the maximum Ca concentration for activating dehydrogenase and F1-ATPase activity in this preparation (27) without evidence of uncoupling or cyt c release. The 40 uM was selected as an intermediate value. The results of this dosing study are presented in Figure 8. Both Mn-SOD and PDH had very similar responses to the addition of Ca2+ with the largest effect occurring with the addition of 0.6 μM, or the level activating ATP production under these Ca2+ depleted conditions. This high Ca2+ affinity for dephosphorylation of MnSOD is consistent with the dose dependence of activity noted above (Figure 6D) with a ∼10 μM K50. Clearly, a low Ca2+ dose that does not induce uncoupling or cyt c release resulted in a significant decrease in phosphorylation. This result suggests that the dephosphorylation in these two proteins was not necessarily limited to the high levels of Ca used to mimic the apoptotic effects and potentially generate gross metabolic consequences.
Fig. 8.
The [Ca2+] dose dependence of PDH and MnSOD phosphorylation Experiments were conducted under identical conditions as in Figures 5 and 6 with the free [Ca2+] of 0, 0.6, 40 and 100 μM.
The large number of phosphorylated enzymes detected in the mitochondrial matrix implies a very diverse and active system of kinases and phosphatases that might play a key role in the regulation of mitochondrial function, much like has been extensively described for PDH. To begin to unravel the mitochondria kinase/phosphatase interactions, we conducted an initial screen for kinases, phosphatases, and sites of phosphorylation using a commercial antibody-based screening procedure. This screening procedure positively identified 11 kinases and 3 phosphatases (Table 2). Due to numerous confounding factors, such as dependence on antibody specificity, cross-reactivity between mouse-specific antibodies and porcine proteins, and individual protein concentrations, this list cannot be considered comprehensive at this time. These results confirm previous studies localizing several of these kinases to mitochondria including the MAP kinase system, Raf kinases, and lyn kinase (21;22). Most kinases localized to the mitochondria in this study have been associated with apoptosis signaling events including p38 MAP kinase, stress-activated protein kinase (42), DNA-activated protein kinase (43), casein kinase II α̣(44), IκBα̣(45), ribosomal S6 protein kinase 1 (46), and protein kinase C β (47), however, how these might be linked to the phosphoproteome and effects of Ca2+ has yet to be resolved. The acute effects of extramitochondrial Ca2+ on the mitochondrial phosphoproteome resulted almost exclusively in dephosphorylation events, thus phosphatases sensitive to Ca2+, or conditions generated by Ca2+addition, will be likely candidates for further investigation in this signaling process. Finally, though the mitochondria preparation appears to be quite pure based on the proteomic profiles obtained, we can also not be certain that some of these postive results for kinases and phosphatases could be due to adhesion to the outside of the mitochondria and not present in the matrix, or caused by small contaminating structures from the cytosol. Confirmation of the localization of these enzymes within the matrix will be required.
Table 2.
Antibody-based screen for kinases and phosphatases in isolated mitochondria
| Protein Name | |
|---|---|
| Kinases | |
| p38 Hog CT | p38 alpha MAP kinase (Hog) |
| PKC-β1 | Protein kinase C beta 1 |
| Mek1 | MAP kinase kinase 1 |
| DNAPK | DNA-activated protein kinase |
| Mek6 | MAP kinase kinase 6 |
| Rsk1 (C21) | Ribosomal S6 kinase 1 |
| CK2α-III | Casein kinase 2 alpha |
| IKKα (H744) | Inhibitor NF kB kinase alpha |
| Lyn (H-6) | Oncogene Lyn |
| Raf1 (C20) | Oncogene Raf 1 |
| JAK1 (HR-785) | Janus kinase 1 |
| Phosphatases | |
| PP2A/A | Protein phosphatase 2A - A regulatory subunit |
| PP2A/C | Protein phosphatase 2A - catalytic subunit |
| MKP-1 (V-15) | MAP kinase phosphatase 1 |
| VHR | Dual specificity phosphatase 3 |
DISCUSSION
The current study extends the knowledge of the mitochondrial phosphoproteome in the porcine heart using the Pro-Q Diamond staining procedure in conjunction with 2D gel electrophoresis/mass spectrometry approaches. Forty-five phosphorylated proteins were identified in total extracts of mitochondria, covering a wide range of functional attributes from membrane transport events, to energy and ROS metabolism. Many of these phosophorylations are previously unknown, suggesting phosphorylation may be a more prominent regulatory mechanism in mitochondria than previously thought. The 32P protein labeling in intact mitochondria confirmed the extensive nature of matrix protein phosphorylation as well as its dynamic nature required for a acute signaling network. In this study, we examined the interaction of Ca2+ and ROS, which are believed to activate complex metabolic and functional networks and are thought to play an important role in energy metabolism regulation as well as mitochondria-initiated apoptosis.
The sensitivity of Pro-Q Diamond has been validated in several systems (35;36). The major advantage of Pro-Q Diamond that determines the total protein phosphorylation is that it functions in the steady state not requiring phosphate turnover, as required for 32P labeling experiments, and can work in the presence of high concentrations of ATP without concern of competition. However, many limitations exist for this type of screen. Gel-based techniques are inherently biased to detect the most abundant proteins and the most dramatic changes. Screening total protein phosphorylation limits detection if only few of many phosphorylation sites on one protein are affected by a signaling pathway. In addition, some have reported weak non-specific protein staining with Pro-Q Diamond (48). 32P labeling confirmed many of the Pro-Q phosphorylation sites including: PDH, MnSOD, inhibin, MCAD, LCAD and Rieske iron protein but the patterns of 32P labeling and Pro-Q Diamond were, not surprisingly, very different as will be discussed further below. Finally, the detection limits of Pro-Q Diamond for protein phosphorylation have not been extensively determined with regard to the number of phosphorylation sites/protein required. However, in some cases we found that the Pro-Q Diamond stain was more sensitive for protein detection than Sypro Ruby staining (for example PDH and cytochrome oxidase in Figure 2C) while several Pro-Q Diamond stained proteins are yet to be determined due to sensitivity limits of the mass spectroscopy suggesting. The latter results suggest that the sensitivity of Pro-Q Diamond for some protein phosphorylations is very good.
As discussed above the 32P labeling resulted in the detection of an extensive dynamic pool of phosphorylated proteins in the mitochondrial matrix. To our knowledge this is the first demonstration of such extensive phosphorylation likely due to the nature of the protocol that generates predictably near a 100% 32P specific activity for the γ phosphate of ATP in the mitochondria matrix. In these initial studies, we confirmed many of the proteins detected using Pro-Q Diamond but also detected many others that were in regions where Coomassie staining revealed no proteins, implying a high phosphate turnover of a very small protein pool, ideal for acute signaling purposes. The initial time courses of labeling confirm a rapid phosphorylation turnover in the matrix (Figure 4). Adding uncoupler, that rapidly depletes the tiny matrix ATP pool, resulted in a dramatic decrease in overall 32P labeling also confirmed that the protein phosphorylations were turning over at least on the minute time scale (Compare Figures 4B and 4C). This result also implies that the protein phosphorylation state could be sensitive to the matrix ATP levels providing yet another potential feedback signal for energy metabolism.
The advantage of the 32P labeling approach is that it is the gold standard with regard to the proof of association of a phosphate with a protein but it also provides unique information on the turnover of the protein phosphorylation that is key in understanding signaling networks. One of the disadvantages is that care must be taken to avoid competition with cold phosphate limiting the concentration of physiological substrates such as phosphate and ATP. This forces the 32P labeling to be conducted under non-physiological conditions and be very sensitive to the metabolic state of the mitochondria since exogenous ATP cannot be provided. The identity of many of the proteins seen in the 32P screen will be difficult to unravel due to low abundance, however, these studies reveal a large network of protein phosphoryaltions that may play a key role in the acute and chronic regulation of mitochondrial function.
A dose of extramitochondrial Ca2+ was selected to induce cyt c release and simulate the initial stages of mitochondria-induced apoptosis. This high dose of Ca2+ was used with the expectations that both the more sensitive energy metabolism activation processes as well as processes related to apoptosis and cyt c release could be captured in a single screen. We determined the optimal conditions to generate cyt c release for this preparation due to the wide variation in conditions found in the literature. We found a dependence of Ca2+-induced cyt release on millimolar concentrations of both Pi and ATP. The mechanisms associated with the Pi and ATP requirements of Ca2+ induced cyt c release remain poorly defined.
Extramitochondrial Ca2+ was found to dephosphorylate PDH, MnSOD and γF1. The dephosphorylation and activation of PDH serves as a useful control since the Ca2+ activation of pyruvate dehydrogenase phosphatase I, resulting in PDH dephosphorylation and activation, has been well established (12). The extensive phosphorylation of PDH observed with Pro-Q was confirmed in the 32P labeling experiments as the most extensive phosphorylation site. In addition the turnover of PDH phosphorylation was very fast based on the limited time course and rapid dephosphorylation with uncoupler. The dose dependence of PDH dephosphorylation reaching a maximum at 600 nM was consistent with the metabolic actions of Ca, stimulation previously established in this preparation, and not with the release of cyt c and general metabolic failure.
MnSOD, a matrix protein, converts superoxide to hydrogen peroxide and represents the primary mitochondrial defense against damage induced by superoxide radicals (49). Several lines of evidence support the notion that MnSOD is phosphorylated: 1) MnSOD was labeled with 32P confirming earlier work in potato mitochondria (50), 2) MnSOD stained with Pro-Q Diamond, 3) Four protein spots with similar molecular weights but different isoelectric points were identified as MnSOD consistent with protein phosphorylation. 4) The activity of MnSOD was inversely correlated with protein phosphorylation in the Ca2+ dose response experiments (Figure 6D and Figure 8). It is also interesting to note that the 32P labeling was concentrated in the most acid shifted form of MnSOD consistent with the highest level of phosphorylation (see Figure 3). The 32P labeling of MnSOD was not as extensive as other proteins suggesting a relatively slow turnover of the phosphorylation site under steady state conditions. The Ca2+-sensitive dephosphorylation and activation of MnSOD is a novel finding and suggests that MnSOD activity may be controlled to regulate matrix levels of superoxide or hydrogen peroxide for other signaling processes. Potentially, the ROS generation stimulated by Ca2+ might be “buffered” by a parallel activation of the ROS scavenging MnSOD. Although the sites of phosphorylation of MnSOD remain to be determined, we speculate that the phosphorylation of Tyr 34 may be a mechanism of inhibition of MnSOD activity. It was previously shown that reactive nitrogen species attack Tyr 34 in the active site of MnSOD, causing nitration of the amino acid and subsequent inhibition of the enzyme(51;52). Phosphorylation of this tyrosine could protect the residue from nitration but be made rapidly available by dephosphorylation when needed for enzyme activation. Attempts to dephosphorylate MnSOD with alkaline phosphatase, protein phosphatase 1 were unsuccessful; suggesting that a specific phosphatase is likely responsible for the Ca2+ actions on the enzyme while the kinase also remains unknown at this time.
With the discovery of the Ca2+ effects on MnSOD activity, we tested the hypothesis that matrix ROS generation alone could alter MnSOD phosphorylation and activity. Under the conditions of our study we found little effects of ROS generation on overall matrix protein phosphorylation. Specifically, we found no large effect of ROS generation on PDH and MnSOD phosphorylation or any other phosphorylated protein detected in the Pro-Q Diamond staining. We have not attempted these experiments on the turnover experiments with 32P labeling. These data suggest that the secondary formation of ROS alone with Ca2+ is not responsible for the dephosphorylation of PDH or MnSOD under these experimental conditions.
In summary, we have shown that the phosphoproteome of the intact mitochondria matrix is extensive and dynamic. Most of the major metabolic pathways within the matrix possess dynamic protein phosphorylation sites, while many of the sites observed have not yet been identified. These results are consistent with protein phosphorylation in the matrix playing a major role in acute cellular signaling for energy metabolism, as well as the numerous other functions of the mitochondrion.
Acknowledgments
Acknowledgements: Intramural Funding of the Division of Intramural Research, NHLBI, NIH, DHHS and NIH Grant DK47844 to RH.
References
- 1.Gray MW. The endosymbiont hypothesis revisited. Int. Rev. Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann. N. Y. Acad. Sci. 1987;503:55–71. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- 3.Westermann B, Neupert W. ‘Omics’ of the mitochondrion. Nat. Biotechnol. 2003;21:239–240. doi: 10.1038/nbt0303-239. [DOI] [PubMed] [Google Scholar]
- 4.Richly E, Chinnery PF, Leister D. Evolutionary diversification of mitochondrial proteomes: implications for human disease. Trends Genet. 2003;19:356–362. doi: 10.1016/S0168-9525(03)00137-9. [DOI] [PubMed] [Google Scholar]
- 5.Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, Ghosh SS. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 6.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 7.Kernec F, Unlu M, Labeikovsky W, Minden JS, Koretsky AP. Changes in the mitochondrial proteome from mouse hearts deficient in creatine kinase. Physiol Genomics. 2001;6:117–128. doi: 10.1152/physiolgenomics.2001.6.2.117. [DOI] [PubMed] [Google Scholar]
- 8.Liu XH, Qian LJ, Gong JB, Shen J, Zhang XM, Qian XH. Proteomic analysis of mitochondrial proteins in cardiomyocytes from chronic stressed rat. Proteomics. 2004;4:3167–3176. doi: 10.1002/pmic.200300845. [DOI] [PubMed] [Google Scholar]
- 9.Hunter T. Signaling--2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 10.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 11.Cohen P. The origins of protein phosphorylation. Nat. Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 12.Linn TC, Pettit FH, Reed LJ. Alpha-keto acid dehydrogenase complexes. X. Regulation of the activity of the pyruvate dehydrogenase complex from beef kidney mitochondria by phosphorylation and dephosphorylation. Proc. Natl. Acad. Sci. U. S. A. 1969;62:234–241. doi: 10.1073/pnas.62.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azarashvili TS, Tyynela J, Odinokova IV, Grigorjev PA, Baumann M, Evtodienko YV, Saris NE. Phosphorylation of a peptide related to subunit c of the F0F1-ATPase/ATP synthase and relationship to permeability transition pore opening in mitochondria. J. Bioenerg. Biomembr. 2002;34:279–284. doi: 10.1023/a:1020204518513. [DOI] [PubMed] [Google Scholar]
- 14.Papa S, Sardanelli AM, Cocco T, Speranza F, Scacco SC, Technikova-Dobrova Z. The nuclear-encoded 18 kDa (IP) AQDQ subunit of bovine heart complex I is phosphorylated by the mitochondrial cAMP-dependent protein kinase. FEBS Lett. 1996;379:299–301. doi: 10.1016/0014-5793(95)01532-9. [DOI] [PubMed] [Google Scholar]
- 15.Bender E, Kadenbach B. The allosteric ATP-inhibition of cytochrome c oxidase activity is reversibly switched on by cAMP-dependent phosphorylation. FEBS Lett. 2000;466:130–134. doi: 10.1016/s0014-5793(99)01773-1. [DOI] [PubMed] [Google Scholar]
- 16.Chen R, Fearnley IM, Peak-Chew SY, Walker JE. The phosphorylation of subunits of complex I from bovine heart mitochondria. J. Biol. Chem. 2004;279:26036–26045. doi: 10.1074/jbc.M402710200. [DOI] [PubMed] [Google Scholar]
- 17.Hojlund K, Wrzesinski K, Larsen PM, Fey SJ, Roepstorff P, Handberg A, Dela F, Vinten J, McCormack JG, Reynet C, Beck-Nielsen H. Proteome analysis reveals phosphorylation of ATP synthase beta-subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J. Biol. Chem. 2003;278:10436–10442. doi: 10.1074/jbc.M212881200. [DOI] [PubMed] [Google Scholar]
- 18.Schulenberg B, Aggeler R, Beechem JM, Capaldi RA, Patton WF. Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J. Biol. Chem. 2003;278:27251–27255. doi: 10.1074/jbc.C300189200. [DOI] [PubMed] [Google Scholar]
- 19.Bykova NV, Egsgaard H, Moller IM. Identification of 14 new phosphoproteins involved in important plant mitochondrial processes. FEBS Lett. 2003;540:141–146. doi: 10.1016/s0014-5793(03)00250-3. [DOI] [PubMed] [Google Scholar]
- 20.Harris RA, Popov KM, Zhao Y, Kedishvili NY, Shimomura Y, Crabb DW. A new family of protein kinases--the mitochondrial protein kinases. Adv. Enzyme Regul. 1995;35:147–162. doi: 10.1016/0065-2571(94)00020-4. [DOI] [PubMed] [Google Scholar]
- 21.Goldenthal MJ, Marin-Garcia J. Mitochondrial signaling pathways: a receiver/integrator organelle. Mol. Cell Biochem. 2004;262:1–16. doi: 10.1023/b:mcbi.0000038228.85494.3b. [DOI] [PubMed] [Google Scholar]
- 22.Thomson M. Evidence of undiscovered cell regulatory mechanisms: phosphoproteins and protein kinases in mitochondria. Cell Mol. Life Sci. 2002;59:213–219. doi: 10.1007/s00018-002-8417-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balaban RS. Cardiac energy metabolism homeostasis: role of cytosolic calcium. J. Mol. Cell Cardiol. 2002;34:1259–1271. doi: 10.1006/jmcc.2002.2082. [DOI] [PubMed] [Google Scholar]
- 24.Dykens JA. Isolated cerebral and cerebellar mitochondria produce free radicals when exposed to elevated CA2+ and Na+: implications for neurodegeneration. J. Neurochem. 1994;63:584–591. doi: 10.1046/j.1471-4159.1994.63020584.x. [DOI] [PubMed] [Google Scholar]
- 25.Crompton M. The mitochondrial permeability transition pore and its role in cell death. Biochem. J. 1999;341(Pt 2):233–249. [PMC free article] [PubMed] [Google Scholar]
- 26.Mattson MP, Chan SL. Calcium orchestrates apoptosis. Nat. Cell Biol. 2003;5:1041–1043. doi: 10.1038/ncb1203-1041. [DOI] [PubMed] [Google Scholar]
- 27.Territo PR, Mootha VK, French SA, Balaban RS. Ca(2+) activation of heart mitochondrial oxidative phosphorylation: role of the F(0)/F(1)-ATPase. Am. J. Physiol Cell Physiol. 2000;278:C423–C435. doi: 10.1152/ajpcell.2000.278.2.C423. [DOI] [PubMed] [Google Scholar]
- 28.Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL. Mitochondrial dysfunction in cardiac disease: ischemia--reperfusion, aging, and heart failure. J. Mol. Cell Cardiol. 2001;33:1065–1089. doi: 10.1006/jmcc.2001.1378. [DOI] [PubMed] [Google Scholar]
- 29.Palmer JW, Tandler B, Hoppel CL. Biochemical differences between subsarcolemmal and interfibrillar mitochondria from rat cardiac muscle: effects of procedural manipulations. Arch. Biochem. Biophys. 1985;236:691–702. doi: 10.1016/0003-9861(85)90675-7. [DOI] [PubMed] [Google Scholar]
- 30.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J. Biol. Chem. 1977;252:8731–8739. [PubMed] [Google Scholar]
- 31.Balaban RS, Mootha VK, Arai A. Spectroscopic determination of cytochrome c oxidase content in tissues containing myoglobin or hemoglobin. Anal. Biochem. 1996;237:274–278. doi: 10.1006/abio.1996.0239. [DOI] [PubMed] [Google Scholar]
- 32.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 33.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 34.Robertson JG, Barron LL, Olson MS. Effects of alpha-ketoisovalerate on bovine heart pyruvate dehydrogenase complex and pyruvate dehydrogenase kinase. J. Biol. Chem. 1986;261:76–81. [PubMed] [Google Scholar]
- 35.Schulenberg B, Goodman TN, Aggeler R, Capaldi RA, Patton WF. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis. 2004;25:2526–2532. doi: 10.1002/elps.200406007. [DOI] [PubMed] [Google Scholar]
- 36.Chou CL, Christensen BM, Frische S, Vorum H, Desai RA, Hoffert JD, de Lanerolle P, Nielsen S, Knepper MA. Non-muscle myosin II and myosin light chain kinase are downstream targets for vasopressin signaling in the renal collecting duct. J. Biol. Chem. 2004;279:49026–49035. doi: 10.1074/jbc.M408565200. [DOI] [PubMed] [Google Scholar]
- 37.Unwin RD, Sternberg DW, Lu Y, Pierce A, Gilliland DG, Whetton AD. Global effects of BCR/ABL and TEL/PDGFRbeta expression on the proteome and phosphoproteome: identification of the Rho pathway as a target of BCR/ABL. J. Biol. Chem. 2005;280:6316–6326. doi: 10.1074/jbc.M410598200. [DOI] [PubMed] [Google Scholar]
- 38.Gunter TE, Yule DI, Gunter KK, Eliseev RA, Salter JD. Calcium and mitochondria. FEBS Lett. 2004;567:96–102. doi: 10.1016/j.febslet.2004.03.071. [DOI] [PubMed] [Google Scholar]
- 39.He L, Lemasters JJ. Dephosphorylation of the Rieske iron-sulfur protein after induction of the mitochondrial permeability transition. Biochem. Biophys. Res. Commun. 2005;334:829–837. doi: 10.1016/j.bbrc.2005.06.170. [DOI] [PubMed] [Google Scholar]
- 40.Pettit FH, Roche TE, Reed LJ. Function of calcium ions in pyruvate dehydrogenase phosphatase activity. Biochem. Biophys. Res. Commun. 1972;49:563–571. doi: 10.1016/0006-291x(72)90448-2. [DOI] [PubMed] [Google Scholar]
- 41.Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem. J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 43.Wang S, Guo M, Ouyang H, Li X, Cordon-Cardo C, Kurimasa A, Chen DJ, Fuks Z, Ling CC, Li GC. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1584–1588. doi: 10.1073/pnas.97.4.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo C, Yu S, Davis AT, Wang H, Green JE, Ahmed K. A potential role of nuclear matrix-associated protein kinase CK2 in protection against drug-induced apoptosis in cancer cells. J. Biol. Chem. 2001;276:5992–5999. doi: 10.1074/jbc.M004862200. [DOI] [PubMed] [Google Scholar]
- 45.Fujihara S, Jaffray E, Farrow SN, Rossi AG, Haslett C, Hay RT. Inhibition of NF-kappa B by a cell permeable form of I kappa B alpha induces apoptosis in eosinophils. Biochem. Biophys. Res. Commun. 2005;326:632–637. doi: 10.1016/j.bbrc.2004.11.090. [DOI] [PubMed] [Google Scholar]
- 46.Harada H, Andersen JS, Mann M, Terada N, Korsmeyer SJ. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9666–9670. doi: 10.1073/pnas.171301998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao MY, Shinjo F, Heinrichs S, Soh JW, Jongstra-Bilen J, Jongstra J. Inhibition of anti-IgM-induced translocation of protein kinase C beta I inhibits ERK2 activation and increases apoptosis. J. Biol. Chem. 2001;276:24506–24510. doi: 10.1074/jbc.M103883200. [DOI] [PubMed] [Google Scholar]
- 48.Murray J, Marusich MF, Capaldi RA, Aggeler R. Focused proteomics: monoclonal antibody-based isolation of the oxidative phosphorylation machinery and detection of phosphoproteins using a fluorescent phosphoprotein gel stain. Electrophoresis. 2004;25:2520–2525. doi: 10.1002/elps.200406006. [DOI] [PubMed] [Google Scholar]
- 49.Fridovich I. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 50.Bykova NV, Stensballe A, Egsgaard H, Jensen ON, Moller IM. Phosphorylation of formate dehydrogenase in potato tuber mitochondria. J. Biol. Chem. 2003;278:26021–26030. doi: 10.1074/jbc.M300245200. [DOI] [PubMed] [Google Scholar]
- 51.MacMillan-Crow LA, Crow JP, Thompson JA. Peroxynitrite-mediated inactivation of manganese superoxide dismutase involves nitration and oxidation of critical tyrosine residues. Biochemistry. 1998;37:1613–1622. doi: 10.1021/bi971894b. [DOI] [PubMed] [Google Scholar]
- 52.Yamakura F, Taka H, Fujimura T, Murayama K. Inactivation of human manganese-superoxide dismutase by peroxynitrite is caused by exclusive nitration of tyrosine 34 to 3-nitrotyrosine. J. Biol. Chem. 1998;273:14085–14089. doi: 10.1074/jbc.273.23.14085. [DOI] [PubMed] [Google Scholar]