Abstract
Kinesin superfamily proteins are ubiquitous to all eukaryotes and essential for several key cellular processes. With the establishment of genome sequence data for a substantial number of eukaryotes, it is now possible for the first time to analyze the complete kinesin repertoires of a diversity of organisms from most eukaryotic kingdoms. Such a “holistic” approach using 486 kinesin-like sequences from 19 eukaryotes and analyzed by Bayesian techniques, identifies three new kinesin families, two new phylum-specific groups, and unites two previously identified families. The paralogue distribution suggests that the eukaryotic cenancestor possessed nearly all kinesin families. However, multiple losses in individual lineages mean that no family is ubiquitous to all organisms and that the present day distribution reflects common biology more than it does common ancestry. In particular, the distribution of four families—Kinesin-2, -9, and the proposed new families Kinesin-16 and -17—correlates with the possession of cilia/flagella, and this can be used to predict a flagellar function for two new kinesin families. Finally, we present a set of hidden Markov models that can reliably place most new kinesin sequences into families, even when from an organism at a great evolutionary distance from those in the analysis.
INTRODUCTION
Eukaryotic cells contain three core classes of molecular motors: kinesins, dyneins, and myosins. Each class constitutes a superfamily of proteins of which an individual organism might encode numerous members. Kinesins and dyneins interact with microtubules, whereas myosins move along actin microfilaments, but all three classes use energy derived from ATP hydrolysis to generate a force that can be used by the cell for various ends, including transport of cargoes, segregation of organelles, destabilizing microtubules, alteration of morphology, or movement of the entire cell. Of the three classes, the kinesins tend to be the largest group within any organism. Moreover, although dynein (Lawrence et al., 2001) and myosin (Matsuzaki et al., 2004; Richards and Cavalier-Smith, 2005) superfamilies have both been entirely lost from particular eukaryotic lineages, members of the kinesin superfamily are encoded by all eukaryotes thus far analyzed.
Kinesin superfamily proteins (kinesins; previously also named KRPs, KLPs, or KIFs) are related by a conserved globular motor domain that defines the superfamily as a whole. The motor or “head” domain is often found at the N terminus of the protein, although it may be positioned at any position along the primary sequence, and in certain kinesin types it is more commonly associated with the C terminus, or middle of the sequence (Miki et al., 2005). Outside of this motor domain, most kinesins possess a “tail” domain (so called because in some kinesins, it has been shown to form an extended coiled-coil structure). This domain is important for interactions with kinesin light chain proteins, cargoes, or other macromolecules (e.g., chromatin). Unlike the motor domain, the tail domains of most kinesins are highly divergent. For some kinesins, a “neck” region has been defined between the motor and tail domains. Although less conserved than the motor itself, this domain regulates the activity of the motor region and in certain proteins can determine the motor polarity (Case et al., 1997; Endow and Waligora, 1998).
The kinesin superfamily can be further divided into paralogous protein families that share not only a common ancestor but also often a conserved cellular role (Dagenbach and Endow, 2004; Hirokawa and Takemura, 2004; Miki et al., 2005). Several kinesin types coexist in a single organism presumably to fulfill specific biological functions. Thus, classification of the kinesin repertoire of organisms can be used to infer the presence of particular cellular pathways as well as enabling the prediction of function for newly identified kinesin superfamily members. Moreover, recent analysis of the myosin superfamily has demonstrated how an understanding of paralogue distribution can be used to infer deep-level phylogenetic information (Richards and Cavalier-Smith, 2005). For these reasons, an understanding of the kinesin repertoire across eukaryotes could provide information regarding both eukaryote evolution and individual organism biology.
Several kinesin phylogenies have been described in an attempt to identify kinesin (sub)families (Goodson et al., 1994; Moore and Endow, 1996; Hirokawa et al., 1998; Kim and Endow, 2000; Lawrence et al., 2002; Siddiqui, 2002; Kollmar and Glockner, 2003; Dagenbach and Endow, 2004), culminating recently in the publication of a uniting standardized nomenclature encompassing 14 kinesin families (Lawrence et al., 2004). However to date, kinesin phylogenies have been limited by three factors: First, few of the analyses have sampled the full kinesin repertoires from individual eukaryotes so that it has not been possible to examine the distribution of kinesin families in the context of eukaryotic evolution. Second, there is an understandable bias in most analyses toward sequences from animals (in particular, mammals) and to a lesser extent higher plants. Sufficient taxon sampling is critical when inferring phylogenies from paralogous sequences if the true relationships are to be inferred (Baldauf, 2003) and a biased taxon set may create false associations as well as missing groups. Third, because of the potentially enormous size of the data set, most analyses have confined themselves to relatively unsophisticated models of sequence evolution and tree searching techniques. Such approaches tend to perform poorly in situations where sequences have diverged for long times (Holder and Lewis, 2003), as is predicted to be the case for kinesin families.
Here, we present the first kinesin phylogeny built entirely from complete or near-complete kinesin repertoires. These repertoires are from 19 diverse organisms spanning five of the six proposed eukaryotic supergroups (Cavalier-Smith, 2004; Simpson and Roger, 2004). By using a supercomputing cluster, we have incorporated relatively sophisticated phylogenetic models into the tree inference as well as some bootstrap support. From this analysis, we expand the membership of 11 of the previously defined kinesin families, and we define three new kinesin families, two new phylum-specific groups, and one superfamily. We find that the distribution of kinesin families among the organisms is heavily influenced by the occurrence of flagella/cilia and propose a role in this organelle for two of the new kinesin families. We also present a set of hidden Markov models for prediction of kinesin family membership without the need for full phylogenetic reconstruction.
MATERIALS AND METHODS
Data Set Construction and Tree Inference
All sequences were analyzed at the amino-acid level only. From the predicted proteomes of each of the 19 model organisms, we used the following search criteria to identify putative kinesin superfamily proteins: 1) For annotated genomes, any predicted protein annotated as possessing a Pfam kinesin motor domain (Pfam family PF00225) and/or annotated as “kinesin,” “kinesin-like,” or “kinesin-related”. 2) For genomes unannotated at the time of analysis (Thalassiosira pseudonana and Tetrahymena thermophila), any predicted protein with a BLASTp hit of expectation value <10–10 to either human uKHC (an N-terminal kinesin) or Saccharomyces cerevisiae Kar3p (a C-terminal kinesin). This identifies 486 kinesin-like sequences, excluding splice variants or multiple gene models, which are listed in Supplemental File 1. From this data set, we reduced the redundancy by eliminating 18 sequences from duplicated genes that encode proteins predicted to be identical or nearly identical (>95% identity at the amino acid level) to other sequences from the same organism. We also removed nine fragmentary sequences (8 from the incomplete genome of Chlamydomonas reinhardtii and HsKif26A), which are unlikely to represent full kinesin sequences.
The position and quality of the kinesin motor domain were predicted using profile hidden Markov models (HMMs; see below). On the basis of the distribution of scores among known kinesin repertoires, sequences with a score <238 were excluded as representing unlikely kinesin motors. The 400 remaining sequences were trimmed to 100 aa either side of the kinesin motor domain (as defined by the HMM) or the protein terminus if <100 aa away, allowing the inclusion in phylogenetic inference of any signal from alignable regions adjacent to the core motor. Protein alignments were performed using ClustalX software (Thompson et al., 1997) and then extensively manually refined (Supplemental File 2). Two matrices were used for phylogenetic inference: one matrix of 403 characters (×400 taxa) representing all the regions of the motor domain and surrounding regions that could be aligned with confidence; and one matrix of 293 characters made up of the most conserved regions of the first matrix.
Bayesian trees were inferred from alignments of proteins using Metropolis-coupled Markov chain Monte Carlo method as implemented by the program MrBayes3b4 (Ronquist and Huelsenbeck, 2003). The WAG substitution matrix was used (Whelan and Goldman, 2001) with a gamma-distributed variation in substitution rate approximated to four discrete categories. A covarion model was also implemented (Galtier, 2001) to allow characters invariant in one protein family to be evolving elsewhere in the tree. A “full” phylogeny was inferred using the 403 × 400 matrix. Eight Markov chains were run for 1,000,000 generations from a random starting tree sampling every 1000 generations and with a “temperature” of 0.2. Tree likelihoods seemed to reach stationary phase at around generation 700,000, and the last 300,000 generations were used to construct the consensus tree shown in Figure 2. To provide topology support, a further 10 partial replicates were run using the smaller 293 × 400 matrix, and 800,000 generations (burnin = 700,000; all other parameters as for the full analysis). Distance-based neighbor-joining trees and maximum parsimony trees were inferred from the full matrix using the software PAUP4b10 (Swofford, 1998).
Figure 2.
A Bayesian kinesin motor domain phylogeny encompassing 400 nonredundant sequences from 19 diverse organisms with complete or near-complete genome sequence (see Materials and Methods for details). The tree is arbitrarily rooted using ScSmy1. Prefixes are as follows: At, A. thaliana; Ce, C. elegans; Cm, C. merolae; Cp, C. parvum; Cr, C. reinhardtii; Dd, D. discoideum; Dm, D. melanogaster; Ec, E. cuniculi; Eh, E. histolytica; Gl, G. lamblia; Hs, H. sapiens; Lm, L. major; Pf, P. falciparum; Sc, S. cerevisiae; Sp, S. pombe; Ta, T. annulata; Tb, T. brucei; Tp, T. pseudonana; and Tt, T. thermophila. Nodes that were also found in a majority of 10 Bayesian partial replicates are indicated (thick lines). Support for the inferred topology from 500 neighbor-joining bootstrap replicates is indicated: >50% (empty circles); >75% (filled circles). For clarity, bootstrap values for some higher nodes are omitted, but in all cases the deepest node in any family with >50% support is indicated. Family annotation: family described by standard kinesin nomenclature (dark blue bracket), subfamily (light blue bracket), new kinesin family (solid orange bracket), and new phylum-specific group (dotted orange bracket). Sequences within these groups are emboldened. We find no consensus for monophyly of the proposed families Kinesin-15 and -16 (previously described Kinesin-12 family; asterisks). Predictive HMMs: sequences from Tetrahymena (Tt) are highlighted green or red on the basis of passing or failing the HMM threshold for their respective families (see text for details).
Profile Hidden Markov Models
All HMM construction and searching was done using the software package HMMERv2.3.2 (http://hmmer.wustl.edu/). The general kinesin motor domain HMM was constructed using the PF00225.11 Pfam-A alignment of kinesin motor domains (http://www.sanger.ac.uk/Software/Pfam/) as a seed, but it was built with local versus local alignments with only one domain “hit” allowed per sequence (hmmbuild -s), because this most closely reflects the observed distribution of the motor domain in proteins. The HMM was calibrated and used to search the nonredundant data set of 459 kinesin-like sequences (468 211 aa).
Individual protein family and subfamily HMMs were constructed from manually edited alignments of full kinesin sequences (i.e., not just the motor domain) either with or without the inclusion of sequences from Tetrahymena thermophila. These were calibrated and used to search the nonredundant kinesin data set—again, either with or without the inclusion of T. thermophila sequences. The distribution of scores from these searches was used to define trusted cut-off (TC) and noise cut-off (NC) scores. The gathering cut-off (GA) was defined as (TC + 3*NC)/4. HMMs from the 15 herein defined kinesin families, two subfamilies, and two phylum-specific groups were appended into two HMM databases—one database of which had been constructed using all the available sequences (Supplemental File 3) and one database of which had no experience of T. thermophila sequence. The latter of these was used to search a database of T. thermophila kinesins using either GA or NC thresholds.
RESULTS
Defining a Kinesin Data Set
To assess the kinesin repertoire across as broad a range of eukaryotes as possible, we selected 19 disparate organisms for which complete or very near-complete genome sequences are publicly available. These organisms were the Metazoa Homo sapiens (Lander et al., 2001), Drosophila melanogaster (Adams et al., 2000), and Caenorhabditis elegans (The C. elegans Sequencing Consortium, 1998); the yeasts S. cerevisiae (Goffeau et al., 1996) and Schizosaccharomyces pombe (Wood et al., 2002); the microsporidian Encephalitozoon cuniculi (Katinka et al., 2001); the Amoebozoa Entamoeba histolytica (Loftus et al., 2005) and Dictyostelium discoideum (Eichinger et al., 2005); the kinetoplastids Trypanosoma brucei (Berriman et al., 2005) and Leishmania major (Ivens et al., 2005); the diplomonad Giardia lamblia (www.mbl.edu/Giardia); the ciliate T. thermophila (www.tigr.org/tdb/e2k1/ttg/); the diatom Thalassiosira pseudonana (Armbrust et al., 2004); the Apicomplexa Plasmodium falciparum (Gardner et al., 2002), Cryptosporidium parvum (Abrahamsen et al., 2004), and Theileria annulata (Pain et al., 2005); the red alga Cyanidioschyzon merolae (Matsuzaki et al., 2004); the green alga Chlamydomonas reinhardtii (genome.jgi-psf.org/chlre2); and the higher plant Arabidopsis thaliana (Arabidopsis Genome Initiative, 2000). These genome sequences were selected to provide, as much as is currently possible, a good breadth of sequence from across the whole of the Eukarya without including many sequences from closely related organisms or from any single kingdom. The 19 included organisms span five of the six proposed eukaryotic supergroups (Cavalier-Smith, 2004; Simpson and Roger, 2004) with the most closely related thought to be the two kinetoplastids, T. brucei and L. major, which diverged ∼250 Mya (Douzery et al., 2004). At the time of analysis, all but three of these organisms—C. reinhardtii, G. lamblia, and T. thermophila—had completed genome sequences. In total, these 19 eukaryotes possess 486 genes encoding kinesin-like proteins (predicted splice variants typically encode very similar proteins and were not considered). Excluding fragmentary sequences and near identical sequences (>95% identity) arising from duplicated genes, these sequences encompass a nonredundant data set of 459 sequences (see Materials and Methods). A full list of the 486 sequences along with synonymous names, accession numbers, and annotations is provided as Supplemental File 1.
Only the kinesin motor domain is common to all the proteins in the kinesin superfamily. Divergent tail domains are generally not alignable, even sometimes within an individual kinesin family. Thus, to attain meaningful alignments across the kinesin superfamily, sequences must be trimmed to a sequence around the alignable motor region. Large-scale kinesin phylogenies are further hindered by the presence of a small number of kinesins with highly divergent “motor” domains (most of which have probably lost their function as molecular motors). Such divergent sequences frequently cause artifacts in phylogenetic inference, such as the well-documented “long-branch attraction” (Bergsten, 2005), and several attempts by us to build trees including divergent sequences resulted in inconsistent phylogenies (our unpublished data). For these reasons, we chose to exclude the most divergent (and hence also the most unlikely) kinesin sequences (see below).
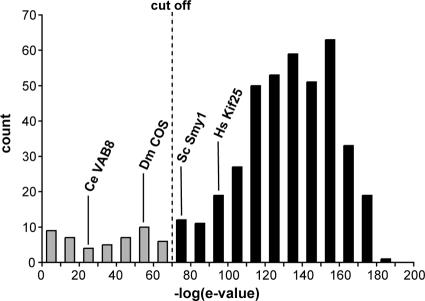
Putative kinesin sequences were screened using the position-specific scoring matrices encompassed by profile HMMs (Krogh et al., 1994). Such scoring techniques provide a robust, relatively fast method for both delineating and assessing the quality of protein domains, as amply demonstrated by the Pfam (Bateman et al., 2004) and SMART (Letunic et al., 2004) databases. We used a kinesin motor domain HMM to parse our set of putative kinesins (see Materials and Methods). Figure 1 shows the distribution of expectation values for motor domains in each of the 459 nonredundant sequences. The majority contain motor domains with very low expectation values, as expected for a well-conserved domain (e = 10–100–10–180; score 340–605). There is then a tail of increasingly weak hits, some of which have exceptionally high expectation values (e > 1; score less than –1). We imposed a condition of e < 10–70 (score >238) for sequences to be included in further analysis (Figure 1). This threshold is sufficiently liberal to include all the previously identified kinesins from well-studied organisms such as H. sapiens, S. pombe, and S. cerevisiae (including the divergent kinesin ScSmy1). It also includes all kinesins except the atypical Cos2 kinesin from D. melanogaster—a protein that may have no motor activity, binding to microtubules in an apparently ATP-independent manner (Sisson et al., 1997)—and all but the very highly divergent VAB8 (KLP5) from C. elegans. From the nonredundant set of 459 kinesins, 400 pass the e < 10–70 cut-off. This refined data set still represents considerable sequence diversity—sequences share as little as 8% identity (14% similarity) across the motor domain (average: 23% identity, 38% similarity). A manually edited alignment of the motor domains is provided in Supplemental File 2.
Figure 1.
The “quality” of the kinesin motor domains in a data set of 459 nonredundant kinesin-like proteins (468 211 aa) from 19 diverse organisms as assessed by the hit to Pfam motor domain profile PF00225 (see Materials and Methods). Sequences passing the threshold, e < 10–70, are in black. The positions of four divergent kinesins from well-studied models are shown for information.
A “Holistic” Kinesin Phylogeny
We used our refined data set of 400 kinesin sequences for phylogenetic inference. Our primary analysis used a likelihood-based Bayesian approach. Such approaches to tree estimation have gained favor in recent years because, among other advantages, they can incorporate more realistic (parameter-rich) models of sequence evolution while taking account of the unreliability of parameter estimation (Huelsenbeck et al., 2002; Holder and Lewis, 2003). They can also, if allowed to run for long enough, provide measures of the clade support without the need to run multiple analyses. However, the large size of the kinesin data set presents specific problems for phylogenetic inference. First, large data sets are obviously more demanding computationally. Second, with such a large number of sequences, the treespace (i.e., the number of possible trees) is so phenomenally large that the potential for getting stuck in local optima is high. Unfortunately, the exceptionally long run times that would demonstrate that the analysis had overcome the latter problem are prohibited at present by the former. To achieve a level of confidence in the inferred phylogeny on a realistic time scale, we instead used a partial replicate strategy, in which support for the topology in one tree-building estimate was provided by 10 smaller Bayesian analyses (see Materials and Methods). We also analyzed the data set with nonparametric bootstrapping under simpler models of sequence evolution. Even on this scale, the analysis requires the equivalent to ∼560 d of continuous calculations on a high-end (3.6-GHz) single processor computer and was only feasible by parallelizing the process on a supercomputing cluster. The estimated Bayesian consensus tree along with topology support is shown in Figure 2.
The standardized nomenclature for the kinesin superfamily comprises 14 families (Lawrence et al., 2004) named Kinesin-1 (conventional KHC) to Kinesin-14 (C-terminal motor kinesins). The phylogeny presented here confirms the monophyly of the families Kinesin-1 to -9 and Kinesin-13 and -14. Our inclusion of newly available sequences from a more diverse range of organisms expands the membership of several families, for example, the Kinesin-8 (Kip3) and Kinesin-9 (Kif9) families, bringing in previously unassigned sequences. More importantly, greater taxon sampling identifies new kinesin groups, including three new kinesin families. In the following, by extension to the standard kinesin nomenclature, we refer to these new families as Kinesin-15, -16, and -17 (also see note in Discussion). However, to avoid unnecessary additions to the standard nomenclature, this new nomenclature should be viewed as provisional until the families identified have been corroborated by independent analyses.
First, greater sampling of sequences resolves the Kinesin-12 family into two new monophyletic groups—the proposed new Kinesin-15 (containing HsKif15 and AtPAKRP1) and Kinesin-16 (containing HsKif12) families. At the time of naming of the Kinesin-12 family (Lawrence et al., 2004), it was equivocal as to whether all the members were monophyletic. With the addition of more data, we find strong evidence for two groups, but no evidence that these groups form a single clade to the exclusion of other kinesin families (i.e., for the Kinesin-15 and -16 families to be subfamilies of a Kinesin-12 clade). Moreover, the proposed Kinesin-15 and -16 families have a very different distribution among the organisms in this analysis and different suggested biological roles (see below).
Second, we identify an entirely new cross-kingdom kinesin clade—the proposed Kinesin-17 family (Figure 2). This group is very strongly supported under all the phylogenetic models tested (monophyletic in all Bayesian trees; 98% bootstrap support under neighbor-joining model), and we find no evidence for this group being part of a previously identified family. It is likely that this clade has not been previously identified simply as a result of the lack of sequence data available for the organisms that contribute to it: kinetoplastids, an alga, a ciliate, and a diplomonad. Although all unicellular, these organisms are very diverse; the family spans at least three of the six eukaryotic supergroups (Cavalier-Smith, 2004; Simpson and Roger, 2004). Alongside this kinesin family, we identify two new clades of kinesins that are specific to the two kinetoplastids (T. brucei and L. major) included in the analysis (Figure 2). At present, these new clades do not constitute kinesin families because they are only found in one phylum (Lawrence et al., 2004).
Third, our phylogeny unites the Kinesin-4 and -10 families. This association has been observed elsewhere. For example, in the maximum likelihood analysis of Lawrence et al. (2002), these two families formed a single, well-supported clade, within which a subclade containing sequences from the Kinesin-4 family was also well-supported. Monophyly for the three sequences from the Kinesin-10 family was less well supported in their analysis. Significantly, although greater sampling across a family might be expected to increase support for a true clade, our analyses do not support monophyly for the Kinesin-10 family. Instead, we find a monophyletic Kinesin-4/10 clade, within which falls the Kinesin-4 group (Figure 2). Rebuilding this region of the tree with all the Kinesin-4/10 kinesins and a small number of sequences from outside of the group (i.e., in a smaller treespace), also gave a consensus for Kinesin-10 paraphyly in all analyses (Bayesian, neighbor-joining, and maximum parsimony; data not shown) and did not support the inclusion of HsKif22 in the Kinesin-4/10 clade as has previously been suggested (Lawrence et al., 2004).
As well as the families mentioned above, our analysis also identifies two clear subfamilies within the large Kinesin-14 family. The Kar3 clade is similar to the previously described Kinesin-14A subfamily, whereas the KatD clade sequences fall within the Kinesin-14B subfamily (Lawrence et al., 2002). From the distribution of the subfamilies, Kinesin-14A seems to be primary, occurring in all organisms that possess a Kinesin-14 family member, whereas the Kinesin-14B subfamily has a much more limited distribution (see below). However, we cannot unambiguously group all of the identified Kinesin-14 members into these two subfamilies on the basis of our analyses. For example, the human sequences KifC2, KifC3, and Kif25 (Kinesin-14B subfamily members) do not consistently fall within the KatD clade described here (Figure 2), suggesting that the family may be more complex that two simple subclades. Again, rebuilding this region of the phylogeny in a smaller treespace did not greatly alter the topology (our unpublished data). This, and the small-scale analysis for the Kinesin-4/10 group mentioned above, indicates that the topology shown in Figure 2 is likely to be a good approximation to the optimal tree for this alignment.
In all, we were able to place with reasonable confidence 78% (312 of 400) of the kinesins in our data set into either previously identified kinesin families or the new families identified by this study. Including the two kinetoplastid-specific groups, this rises to 84% (335 of 400). Our analysis encompasses 11 known families, three proposed new kinesin families, a Kinesin-4/10 “superfamily,” and two new phylum-specific groups. The only family unaccounted for in our analysis is Kinesin-11 (divergent kinesin-I). We find no evidence for a monophyletic Kinesin-11 clade. In part, this is entirely expected, because our kinesin data set excludes two founder members (the extremely divergent CeVAB8, and the fragmentary HsKif26A). However, it should be noted that an association between highly divergent sequences under simple models of sequence evolution is suggestive of artifact.
Distribution of Kinesin Families
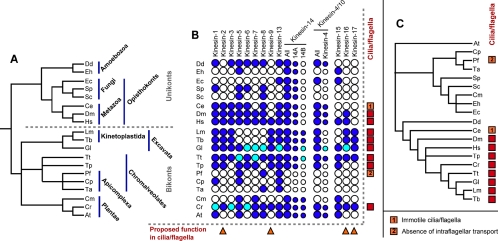
Using data from diverse organisms with complete or near-complete genome sequences allows the analysis not only of kinesin family groups but also the distribution of these groups between organisms (Figure 3B and Table 1). The first, rather surprising, result from such an analysis is that, no single kinesin family is ubiquitous to all organisms. The most common kinesin families are associated with nuclear division—Kinesin-5 (BimC), Kinesin-13 (MCAK), and the Kinesin-14A (Kar3) subfamily—but each of these groups is absent from particular lineages. It is possible for some of the organisms in this analysis that divergent orphan (ungrouped) kinesins are compensating for the absence of a more conserved family member. For example, it is credible that the ScSmy1p protein is a divergent Kinesin-1 orthologue as suggested previously (Lawrence et al., 2002). However, for two organisms—C. merolae and T. annulata—we are able to classify their entire kinesin repertoire into two nonoverlapping sets: Kinesin-5 (BimC), -7 (CENP-E), -14 (C-terminal), and the proposed Kinesin-15 (PAKRP1) family for C. merolae; and Kinesin-8 (Kip3) and -13 (MCAK) for the tiny two kinesin repertoire of T. annulata. This implies that either other motors associated with other processes are able to substitute for the missing family members or that the biological function of each individual kinesin family can be dispensed.
Figure 3.
Distribution of kinesin families among eukaryotes. (A) Cladogram showing the probable evolutionary relationships of the 19 organisms analyzed. (B) Taxonomic distribution of kinesin families: presence of paralogue (dark blue dot), absence of a paralogue from an incomplete genome (light blue dot), and absence from a complete genome (open dot). Subfamilies are represented by smaller dots. Kinesin families with a proposed role in cilia/flagella are indicated (triangle), as are organisms that build cilia/flagella (square). (C) Consensus of the 10 most parsimonious trees accounting for the observed kinesin paralogue distribution using family presence/absence as a binary character. See legend to Figure 2 for organism abbreviations.
Table 1.
Number of kinesin genes possessed by 19 diverse eukaryotes and their classification into families
| Organisma | At | Ce | Cm | Cp | Cr | Dd | Dm | Ec | Eh | Gl | Hs | Lm | Pf | Sc | Sp | Ta | Tb | Tp | Tt | All |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Previously identified families | ||||||||||||||||||||
| Kinesin-1 | 1 | 1 | 0 | 1 | 0 | 4 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 9 | 23 |
| Kinesin-2 | 0 | 3 | 0 | 0 | 2 | 0 | 3 | 0 | 0 | 2 | 4 | 2 | 0 | 0 | 0 | 0 | 2 | 1 | 6 | 25 |
| Kinesin-3 | 0 | 3 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 2 | 6 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 21 |
| Kinesin-5 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 2 | 1 | 0 | 0 | 1 | 0 | 18 |
| Kinesin-6 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 10 |
| Kinesin-7 | 15 | 0 | 1 | 0 | 3 | 2 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 28 |
| Kinesin-8 | 2 | 1 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 1 | 2 | 1 | 0 | 0 | 10 | 25 |
| Kinesin-9 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 4 | 13 |
| Kinesin-13 | 2 | 1 | 0 | 1 | 1 | 1 | 3 | 1 | 0 | 1 | 2 | 5 | 1 | 0 | 0 | 1 | 5 | 2 | 3 | 30 |
| Kinesin-14 | 18 | 3 | 2 | 0 | 4 | 1 | 1 | 1 | 1 | 2 | 4 | 2 | 0 | 1 | 2 | 0 | 2 | 5 | 5 | 54 |
| Subfamily: 14A | 2 | 2b | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 0 | 1 | 3 | 2 | (23) |
| Subfamily: 14B | 12 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | (16) |
| Proposed new families | ||||||||||||||||||||
| Kinesin-4/10c | 3 | 2 | 0 | 0 | 4 | 1 | 3 | 1 | 0 | 1 | 3 | 2 | 0 | 0 | 0 | 0 | 2 | 4 | 1 | 27 |
| Subfamily: Kinesin-4 | 3 | 1 | 0 | 0 | 2 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | (13) |
| Kinesin-15d | 6 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 17 |
| Kinesin-16d | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 5 | 13 |
| Kinesin-17 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 8 |
| New phylum-specific groups | ||||||||||||||||||||
| Kinetoplastid group1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 6 |
| Kinetoplastid group2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 17 |
| Total grouped | 51 | 16 | 5 | 3 | 23 | 13 | 23 | 4 | 3 | 13 | 30 | 31 | 4 | 4 | 7 | 2 | 28 | 21 | 54 | 335 |
| Total orphan | 5 | 1 | 0 | 2 | 2 | 0 | 0 | 2 | 1 | 6 | 1 | 10 | 4 | 2 | 2 | 0 | 9 | 2 | 16 | 65 |
| Total othere | 2 | 1 | 0 | 4 | 7 | 0 | 1 | 0 | 1 | 6 | 0 | 11 | 1 | 0 | 0 | 0 | 14 | 4 | 7 | 59 |
Prefixes are as described in legend for Figure 2. Near identical (>95% aa identity) and fragmentary sequences are excluded.
The motor domains of CeKLP15 and KLP17 are sufficiently divergent to place the sequences outside of the Kinesin-14A (Kar3) subfamily in phylogenetic reconstructions, but HMMs built using the full sequences place them in the Kinesin-14A group.
We find no consensus for a monophyletic Kinesin-10 family.
We find no consensus for the previously identified Kinesin-12 family (containing HsKif12, Kif15 and AtPAKRP1), these sequences instead being divided between the proposed Kinesin-15 (Kif15/PAKRP1) and Kinesin-16 (Kif12) families.
Highly divergent proteins failing the imposed HMM threshold but possessing homology to kinesins.
At the other extreme from the very small kinesin repertoires in Apicomplexa, some organisms possess a huge diversity of kinesins—the ciliate T. thermophila seems to have the largest repertoire thus far, contributing 70 nonredundant kinesin sequences to the refined data set. However, no organism contains members of all of the kinesin families. The organisms with the most kinesin families are humans (13 of 14 families as defined here) and the ciliate T. thermophila (12 of 14 families). Also, the slime mold D. discoideum encodes nine of the 10 families that are not associated with flagella/cilia (see below), organelles that it lacks in all life cycle stages.
The distribution of the kinesin families among eukaryotes contrasts starkly with that for myosin families. There may be as many as 37 myosin families, the majority of which are specific to individual eukaryotic lineages (Richards and Cavalier-Smith, 2005). By definition, any clade of kinesins constituting a family occurs in at least two kingdoms (Lawrence et al., 2004). Moreover, outside of the two kinetoplastid-specific kinesin groups, there are very few well-supported clades within the orphan kinesins, even from organisms within a kingdom (Figure 2). If we assume all of the families in Figure 2 arise from a single ancestral sequence (i.e., that none of the families is an artifact caused by sequence convergence), then the last common ancestor of all eukaryotes would have had to possess all families except the new Kinesin-17 family, which is only found in bikonts (Figure 3). This would suggest that the eukaryotic cenancestor was placing even more emphasis onto its tubulin cytoskeleton than do most present day eukaryotes. Coincidentally, this would give it the cenancestor the same kinesin family repertoire as humans.
Putative Function for Two New Kinesin Families
In a recent analysis, Richards and Cavalier-Smith (2005) showed that the distribution of myosin paralogues among diverse eukaryotes provides support for monophyly of the unikonts and hence for a primary unikont/bikont split very early in eukaryote evolution (Figure 3A). For the kinesin superfamily, although there are a small number of lineage-specific losses (e.g., the loss of Kinesin-14 from the Apicomplexa), in our analysis we find no clear patterns in the distribution of paralogues that could be used for inference of early eukaryote evolution. Indeed, if the presence/absence of a paralogue is treated as a binary character state, the most parsimonious tree is one that bears little connection to the known or most probable evolutionary relationships (compare Figure 3, A and C). This tree instead reflects common biology. In particular, organisms that build flagella/cilia are brought together across taxonomic supergroups. Hence, although each kinesin family has been lost in specific eukaryotic lineages, individual kinesin families do still seem to be linked to specific biological functions.
A close alliance of specific biology with particular homologous proteins is, of course, the expected situation. Comparative genomic analysis can be used to infer the presence/absence of particular biological pathways from the presence/absence of particular proteins (for example, see Briggs et al., 2004). In this context, the distribution of two of the newly identified kinesin families may be used to infer a putative function for the family members. Both the proposed Kinesin-16 and -17 families are found in organisms that build flagella/cilia but not in any organisms that lack this ability (Figure 3B). Based on the number of kinesins encoded by different organisms (Table 1), the probabilities of building these clades by chance from kinesins solely from organisms that possess flagella/cilia are 0.01 and 0.07 for the Kinesin-16 (n = 13) and Kinesin-17 (n = 8) families, respectively. This suggests (especially for Kinesin-16) that the correlation is more than coincidence. Moreover, the only other kinesin families consisting solely of sequences from organisms that possess flagella/cilia are the Kinesin-2 (KRP85/95) and Kinesin-9 (Kif9) families. Although some Kinesin-2 family members have been recruited into roles as diverse as melanosome movement (Rogers et al., 1997; Tuma et al., 1998), membrane transport (Le Bot et al., 1998), cytokinesis (Fan and Beck, 2004), and mRNA transport (Betley et al., 2004), the Kinesin-2 family is primarily a motor of intraflagellar transport (Marszalek and Goldstein, 2000) and has been found in every organism thus far analyzed that builds cilia/flagella—with the exception of Plasmodium that build flagella by an independent, cytoplasmic mechanism—but is absent from those that lack axonemes (Figure 3; Briggs et al., 2004). The function of the Kinesin-9 family is more equivocal, but the association of one of its founder members, CrKLP1, with the axonemal central pair in Chlamydomonas (Bernstein et al., 1994; Yokoyama et al., 2004) is strongly suggestive of a flagellar function. At present, there are very few functional data regarding members of the new Kinesin-16 and -17 families that could substantiate or contest our prediction of a flagellar function for these new families. However, recent data on the Kinesin-16 family member HsKif12 demonstrate that both its expression pattern (Katoh, 2005) and its genetic association with polycystic kidney disease (Mrug et al., 2005) are compatible with our suggestion of an axonemal role.
Predictive HMMs
Phylogenetic inference using large data sets requires a large amount of computing power and a high degree of user intervention. It is also unable to use sequence information that cannot be reliably aligned across all sequences. Pairwise alignment tools such as BLAST, in contrast, are fast, automatible, and can take advantage of all available sequence information. Unfortunately, they are also very poor at correctly assigning sequences to individual groups within multigene families. The problem is particularly acute when no reliable annotation for a closely related organism exists.
We considered that it might be possible to use phylogenetic information to create position-specific scoring profiles that would accurately place newly identified proteins into kinesin families without the need for full tree estimation. Such profiles could use information from outside of the kinesin motor domain, and most importantly, could be calibrated to provide a family-specific threshold for inclusion. We tested the validity of this approach using the T. thermophila kinesin repertoire. This ciliate was selected because 1) it contains the largest number of kinesins of any organism in this study (70 passing the imposed e < 10–70 threshold; Table 1); 2) it encodes members of a wide range, but not all, of the kinesin families identified; and 3) it is one of the most taxonomically divergent organisms in our analysis and hence provides one of the greatest challenges for family prediction.
Using the families defined in Figure 2, we created seed alignments for each family, excluding any T. thermophila sequences. These alignments were used to create HMMs for each family (see Materials and Methods), which were scanned against all identified kinesins except those from T. thermophila. The lowest scoring family member and the highest scoring nonfamily member were then used to define TCs and NCs for the profile (Figure 4). A gathering cut-off was set at a quarter of the distance between the two. These HMMs were then used to predict the placement of the 70 T. thermophila sequences within the tree. At the gathering cut-off, the HMMs correctly place 50% of sequences (27 of 54) into their respective kinesin families. Dropping the threshold to the noise cut-off increases this to 75% of sequences (40 of 54). Importantly, none of the sequences, including the 16 orphan (ungrouped) kinesins, was incorrectly assigned to any family at either cut-off, meaning that the HMMs underpredict but do not produce false positives. In comparison, a cautious examination of the best BLASTp hits for the same proteins incorrectly assigns 42% (23 of 54) of the sequences and also erroneously groups the 16 orphan sequences. Examining the position of the kinesins that were placed into kinesin families by phylogenetic analysis but that were below the family HMM cut-off (Figure 2, red highlight) shows that most are either near the root of the given family or have accrued a lot of substitutions (indicated by longer branches). It is unsurprising that sequences such as these would fail the threshold until more divergent sequences are included in the groups.
Figure 4.
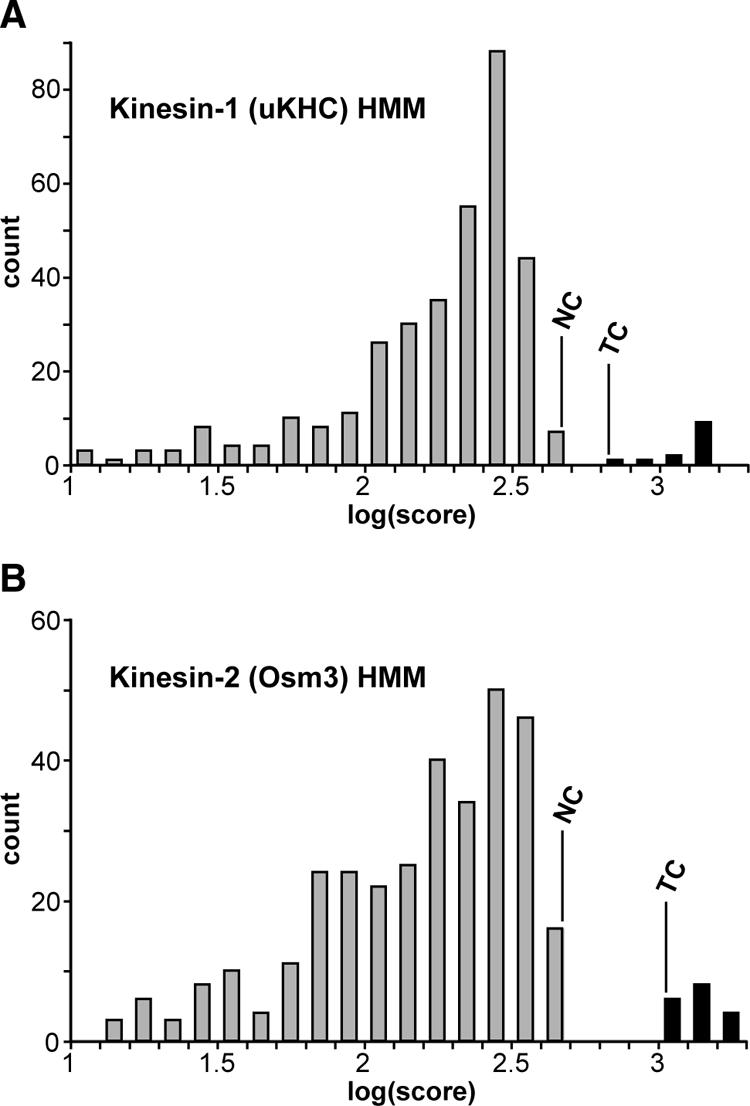
Performance of Kinesin-1 and -2 profiles against the data set of 459 nonredundant kinesin-like proteins. The performance of the other HMMs is similar. Sequences used in the HMM seed alignment are in black. TCs and NCs are illustrated.
On the basis of the relative success of the test HMMs (excluding T. thermophila), we have created alignments and analytical HMMs including the T. thermophila sequences and defined a set of cut-offs for prediction of family membership (Supplemental File 3). All of these HMMs are self-consistent, i.e., they produce a higher score hit to all of the seed sequences than they do to any other sequences (Figure 4), with the exception of the following: 1) CeKLP15 and CeKLP17 give very good hits to the Kinesin-14A subfamily HMM. It seems that although the motor domains of these proteins have diverged sufficiently to place the sequences outside of the Kinesin-14A clade in trees, conservation in the N-terminal “tail” is high enough for the HMMs to recognize them as members. This is consistent with other analyses that have put CeKLP15/17 into the Kinesin-14A group (Lawrence et al., 2002; Dagenbach and Endow, 2004) and also with the distribution of the Kinesin-14A subfamily (Figure 3B). It also demonstrates how information in regions that are not readily used for tree building can be used by HMMs. 2) Both HsKif25 and At5g65460 fail the Kinesin-14 family HMM threshold. Interestingly, both of these proteins have very short N-terminal “tails.” It is unclear whether these sequences possess C-terminal–like motors in an N-terminal context or represent errors in the gene models for these organisms. 3) CeKLP13 and Tp133437 fail their respective family thresholds, probably because of erroneous “tail” truncations in the predicted protein sequence. Inclusion/exclusion of the above-mentioned sequences makes very little difference to the performance of the respective HMMs (our unpublished data). Significantly, none of the orphan kinesins, or any of the kinesin-like sequences that failed the e < 10–70 threshold for inclusion in the refined data set, scores above the noise cut-off of any of the HMMs.
In total, we have created analytical HMMs for 19 kinesin groups: 14 families, two Kinesin-14 subfamilies, the Kinesin-4/10 “superfamily,” and two phylum-specific groups. These HMMs, with defined cut-offs, are available for download in Supplemental File 3.
DISCUSSION
With the growing availability of complete or near complete eukaryotic genome sequences, it is now becoming possible to analyze the complete repertoires of multigene families across diverse eukaryotes. The greater taxon sampling can reveal previously undiscovered relationships between sequences. Also, the use of complete organismal data sets allows a comparison of the distribution of (sub)families. Here, we have analyzed a large data set of kinesin superfamily sequences from a set of 19 eukaryotes using current state-of-the-art phylogenetic methods. This has allowed us to confirm and expand upon several previously identified kinesin families, adding significantly to the diversity of sequences contained within each family. It also suggests some important additions to the recently published standard kinesin relationships (Lawrence et al., 2004), most significant among which are the identification of three new kinesin families, and a combined family. The identification of new kinesin families obviously necessitates some modification to the standard kinesin nomenclature—a provision explicitly allowed for in its construction (Lawrence et al., 2004). However, it is very important that the standard nomenclature represents a consensus view of kinesin phylogeny and that unnecessary changes are avoided. For this reason, the nomenclature used here should be viewed as provisional. If the three new kinesin families presented here are verified in the analyses of others, then we propose they be added to the standard nomenclature as Kinesin-15, -16, and -17 families, along the following lines: The equivocal Kinesin-12 family is divided between the proposed Kinesin-15 (for the presence of HsKif15) and -16 families. We have purposely avoided “overwriting” the Kinesin-12 family, in spite of the presence of the mnemonic HsKif12 in the proposed Kinesin-16 family. This is to prevent inevitable confusion between the old Kinesin-12 and any potential new family of the same name. It would also allow the reuniting of the Kinesin-15 and -16 families at a later date, if the need arose. By simple extension, the kinesin family based around GlKLP5 and KLP10, which contains no sequences previously assigned to other families, is named Kinesin-17.
The distribution of kinesin families among the organisms chosen (but not the phylogenetic relationship within each family) reflects shared biology more closely than likely evolutionary relationships. The paralogue distribution pattern is particularly influenced by the ability of organisms to build flagella/cilia. This pattern suggests a possible flagellar function for two of the new kinesin families—Kinesin-16 and -17—in spite of a paucity of functional data for the members of either. It also strengthens the assertion that the Kinesin-9 family is also principally a motor of flagella/cilia (Yokoyama et al., 2004; Miki et al., 2005), an important finding for a family whose function is still uncertain. The assignment of such putative functions is only possible from a meta-analysis encompassing complete kinesin repertoires from several diverse organisms, and it will be interesting to see whether the addition of more data from a wider variety of organisms will produce any other instances. In particular, inclusion of sequence when it becomes available from the little-studied Rhizaria (the only eukaryotic supergroup to have no representative in this analysis) may provide further surprises in kinesin phylogeny.
Given the close association between particular kinesin paralogues and specific biological functions, the correct assignment of kinesin sequences to families is paramount if meaningful inferences are to be made from annotations. The phylogenetic analysis presented here (summarized in Figure 2) was computationally demanding; equivalent to ∼560 d of continuous calculations on a high-end (3.6-GHz) single processor computer. Such analyses are only really feasible on supercomputer clusters. These analyses will undoubtedly become easier as computing performance and phylogenetic techniques improve, but they will struggle to keep up with the expected rise in the number of available kinesin sequences. In contrast to phylogenetic analyses, scanning the 459 sequence (468,211 aa) nonredundant data set with the 19 analytical HMMs (Supplemental File 3), requires only 9 min on the same 3.6-GHz processor (a 90,000-fold improvement in speed). The performance of test HMMs (excluding T. thermophila) against the real, complex data set of the 70 T. thermophila kinesins was very encouraging given the evolutionary distance between this organism and the others in the analysis. Moreover, the full analytical HMMs (including T. thermophila) should significantly outperform the test set because of the inclusion of more divergent sequence.
The underprediction of family membership (i.e., overassignment of “orphan” status) by the test HMMs demonstrates that such profiles do not replace the need for large-scale phylogenetic analyses. However, underprediction, although a problem, is undoubtedly preferable in most situations to misannotation, because it neither implies a specific biological function nor excludes the possibility of being a more distant relative of a specific family. We feel that the analytical HMMs provided for download in Supplemental File 3 offer a good alternative to top-BLAST-hit type identification because of their ability to fail sequences on the basis of probabilistic score derived from multiple sequences. This is particularly apposite when the sequences concerned are from an organism at some evolutionary remove from comparison sequences in the database. They also require less user intervention than either phylogenetic analysis or manual screening of BLAST hits, making them particularly amenable to analysis of large data sets, such as annotation of newly sequenced genomes. Such models can also be readily modified to incorporate new sequences as they emerge.
Supplementary Material
Acknowledgments
We thank Joe Pitt-Francis (University of Oxford) for help with parallelizing MrBayes. Predicted protein sequence data were obtained from the following: At, The Arabidopsis Information Resource (www.arabidopsis.org); Ce, WormBase (www.wormbase.org); Cm, The National Institute of Genetics Japan (merolae.biol.s.u-tokyo.ac.jp/); Cp, Ec, and Gl, GenBank database (www.ncbi.nlm.nih.gov/GenBank/); Cr and Tp: The Joint Genome Institute (genome.jgi-psf.org); Dd, Eh, Lm, Pf, Sc, Sp, Ta, and Tb, The Sanger Institute (www.genedb.org); Dm and Hs, Ensembl (www.ensembl.org); and Tt, The Institute for Genomic Research (www.tigr.org/tdb/e2k1/ttg/). We thank each of these organizations for making sequence and annotation data publicly available. We also acknowledge the use of facilities at the Oxford Supercomputing Centre (www.osc.ox.ac.uk) in tree estimation. This work was supported by a program grant from the Wellcome Trust.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1090) on February 15, 2006.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Abrahamsen, M. S., et al. (2004). Complete genome sequence of the apicomplexan Cryptosporidium parvum. Science 304, 441–445. [DOI] [PubMed] [Google Scholar]
- Adams, M. D., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Armbrust, E. V., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. [DOI] [PubMed] [Google Scholar]
- Baldauf, S. L. (2003). Phylogeny for the faint of heart: a tutorial. Trends Genet. 19, 345–351. [DOI] [PubMed] [Google Scholar]
- Bateman, A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten, J. (2005). A review of long-branch attraction. Cladistics 21, 163–193. [DOI] [PubMed] [Google Scholar]
- Bernstein, M., Beech, P. L., Katz, S. G., and Rosenbaum, J. L. (1994). A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 125, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman, M., et al. (2005). The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422. [DOI] [PubMed] [Google Scholar]
- Betley, J. N., Heinrich, B., Vernos, I., Sardet, C., Prodon, F., and Deshler, J. O. (2004). Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 14, 219–224. [DOI] [PubMed] [Google Scholar]
- Briggs, L. J., Davidge, J. A., Wickstead, B., Ginger, M. L., and Gull, K. (2004). More than one way to build a flagellum: comparative genomics of parasitic protozoa. Curr. Biol. 14, R611–R612. [DOI] [PubMed] [Google Scholar]
- Case, R. B., Pierce, D. W., HomBooher, N., Hart, C. L., and Vale, R. D. (1997). The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell 90, 959–966. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. (2004). Only six kingdoms of life. Proc. R. Soc. Lond., Ser. B Biol. Sci. 271, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenbach, E. M., and Endow, S. A. (2004). A new kinesin tree. J. Cell Sci. 117, 3–7. [DOI] [PubMed] [Google Scholar]
- Douzery, E. J. P., Snell, E. A., Bapteste, E., Delsuc, F., and Philippe, H. (2004). The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Nat. Acad. Sci. USA 101, 15386–15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger, L., et al. (2005). The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow, S. A., and Waligora, K. W. (1998). Determinants of kinesin motor polarity. Science 281, 1200–1202. [DOI] [PubMed] [Google Scholar]
- Fan, J., and Beck, K. A. (2004). A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J. Cell Sci. 117, 619–629. [DOI] [PubMed] [Google Scholar]
- Galtier, N. (2001). Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol. Biol. Evol. 18, 866–873. [DOI] [PubMed] [Google Scholar]
- Gardner, M. J., et al. (2002). Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau, A., et al. (1996). Life with 6000 genes. Science 274, 546–&. [DOI] [PubMed] [Google Scholar]
- Goodson, H. V., Kang, S. J., and Endow, S. A. (1994). Molecular phylogeny of the kinesin family of microtubule motor proteins. J. Cell Sci. 107, 1875–1884. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N., Noda, Y., and Okada, Y. (1998). Kinesin and dynein superfamily proteins in organelle transport and cell division. Curr. Opin. Cell Biol. 10, 60–73. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N., and Takemura, R. (2004). Kinesin superfamily proteins and their various functions and dynamics. Exp. Cell Res. 301, 50–59. [DOI] [PubMed] [Google Scholar]
- Holder, M., and Lewis, P. O. (2003). Phylogeny estimation: traditional and Bayesian approaches. Nat. Rev. Genet. 4, 275–284. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P., Larget, B., Miller, R. E., and Ronquist, F. (2002). Potential applications and pitfalls of Bayesian inference of phylogeny. Syst. Biol. 51, 673–688. [DOI] [PubMed] [Google Scholar]
- Ivens, A. C., et al. (2005). The genome of the kinetoplastid parasite Leishmania major. Science 309, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka, M. D., et al. (2001). Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450–453. [DOI] [PubMed] [Google Scholar]
- Katoh, M. (2005). Characterization of KIF12 gene in silico. Oncol. Rep. 13, 367–370. [PubMed] [Google Scholar]
- Kim, A. J., and Endow, S. A. (2000). A kinesin family tree. J. Cell Sci. 113, 3681–3682. [DOI] [PubMed] [Google Scholar]
- Kollmar, M., and Glockner, G. (2003). Identification and phylogenetic analysis of Dictyostelium discoideum kinesin proteins. BMC Genomics 4, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., Brown, M., Mian, I. S., Sjolander, K., and Haussler, D. (1994). Hidden Markov models in computational biology: applications to protein modeling. J. Mol. Biol. 235, 1501–1531. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. J., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J., Malmberg, R. L., Muszynski, M. G., and Dawe, R. K. (2002). Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 54, 42–53. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. J., Morris, N. R., Meagher, R. B., and Dawe, R. K. (2001). Dyneins have run their course in plant lineage. Traffic 2, 362–363. [DOI] [PubMed] [Google Scholar]
- Le Bot, N., Antony, C., White, J., Karsenti, E., and Vernos, I. (1998). Role of Xklp3, a subunit of the Xenopus kinesin II heterotrimeric complex, in membrane transport between the endoplasmic reticulum and the Golgi apparatus. J. Cell Biol. 143, 1559–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., Ponting, C. P., and Bork, P. (2004). SMART 4.0, towards genomic data integration. Nucleic Acids Res. 32, D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus, B., et al. (2005). The genome of the protist parasite Entamoeba histolytica. Nature 433, 865–868. [DOI] [PubMed] [Google Scholar]
- Marszalek, J. R., and Goldstein, L. S. B. (2000). Understanding the functions of kinesin-II. Biochim. Biophys. Acta Mol. Cell Res. 1496, 142–150. [DOI] [PubMed] [Google Scholar]
- Matsuzaki, M., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657. [DOI] [PubMed] [Google Scholar]
- Miki, H., Okada, Y., and Hirokawa, N. (2005). Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 15, 465–476. [DOI] [PubMed] [Google Scholar]
- Moore, J. D., and Endow, S. A. (1996). Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays 18, 207–219. [DOI] [PubMed] [Google Scholar]
- Mrug, M., Li, R. H., Cui, X. Q., Schoeb, T. R., Churchill, G. A., and Guay-Woodford, L. M. (2005). Kinesin family member 12 is a candidate polycystic kidney disease modifier in the cpk mouse. J. Am. Soc. Nephrol. 16, 905–916. [DOI] [PubMed] [Google Scholar]
- Pain, A., et al. (2005). Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science 309, 131–133. [DOI] [PubMed] [Google Scholar]
- Richards, T. A., and Cavalier-Smith, T. (2005). Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113–1118. [DOI] [PubMed] [Google Scholar]
- Rogers, S. L., Tint, I. S., Fanapour, P. C., and Gelfand, V. I. (1997). Regulated bidirectional motility of melanophore pigment granules along microtubules in vitro. Proc. Natl. Acad. Sci. USA 94, 3720–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3, Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. [DOI] [PubMed] [Google Scholar]
- Siddiqui, S. S. (2002). Metazoan motor models: kinesin superfamily in C. elegans. Traffic 3, 20–28. [DOI] [PubMed] [Google Scholar]
- Simpson, A. G. B., and Roger, A. J. (2004). The real `kingdoms' of eukaryotes. Curr. Biol. 14, R693–R696. [DOI] [PubMed] [Google Scholar]
- Sisson, J. C., Ho, K. S., Suyama, K., and Scott, M. P. (1997). Costal2, a novel kinesin related protein in the hedgehog signaling pathway. Cell 90, 235–245. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. (1998). Phylogenetic analysis using parsimony (*and other methods), Sunderland, MA: Sinauer.
- The C. elegans Sequencing Consortium. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Gibson, T .J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma, M. C., Zill, A., Le Bot, N., Vernos, I., and Gelfand, V. (1998). Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J. Cell Biol. 143, 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum likelihood approach. Mol. Biol. Evol. 18, 691–699. [DOI] [PubMed] [Google Scholar]
- Wood, V., et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415, 871–880. [DOI] [PubMed] [Google Scholar]
- Yokoyama, R., O'Toole, E., Ghosh, S., and Mitchell, D. R. (2004). Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA 101, 17398–17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.