Abstract
Little is known about what dictates the round shape of the yeast Saccharomyces cerevisiae nucleus. In spo7Δ mutants, the nucleus is misshapen, exhibiting a single protrusion. The Spo7 protein is part of a phosphatase complex that represses phospholipid biosynthesis. Here, we report that the nuclear protrusion of spo7Δ mutants colocalizes with the nucleolus, whereas the nuclear compartment containing the bulk of the DNA is unaffected. Using strains in which the nucleolus is not intimately associated with the nuclear envelope, we show that the single nuclear protrusion of spo7Δ mutants is not a result of nucleolar expansion, but rather a property of the nuclear membrane. We found that in spo7Δ mutants the peripheral endoplasmic reticulum (ER) membrane was also expanded. Because the nuclear membrane and the ER are contiguous, this finding indicates that in spo7Δ mutants all ER membranes, with the exception of the membrane surrounding the bulk of the DNA, undergo expansion. Our results suggest that the nuclear envelope has distinct domains that differ in their ability to resist membrane expansion in response to increased phospholipid biosynthesis. We further propose that in budding yeast there is a mechanism, or structure, that restricts nuclear membrane expansion around the bulk of the DNA.
INTRODUCTION
The nucleus has a distinct organization, characterized by the presence of internal subcompartments. Moreover, in most, but not all, cell types the nucleus adopts a round shape. Understanding how this organization and shape are achieved is of major biological and medical interest. Certain cell types, such as those found in blood lineages (e.g., neutrophils, monocytes, and eosinophils), undergo dramatic nuclear shape changes as they differentiate (Gartner et al., 1990). Cancerous states are often associated with changes in nuclear morphology, most frequently nucleolar enlargement, changes in nuclear shape, or a combination of the two (Zink et al., 2004). Although these changes provide useful diagnostic markers of cancer progression, their mechanistic basis is still poorly understood. Other diseases are also associated with changes in nuclear shape and organization. For example, Hutchinson-Gilford progeria syndrome, which leads to premature aging, is caused by a specific mutation in the gene encoding A-type lamins and is associated with nuclear shape changes (Gruenbaum et al., 2005). Interestingly, progeria-like changes in nuclear shape are part of the normal aging process of nonneuronal cells in Caenorhabditis elegans (Haithcock et al., 2005). Genetic defects in the nuclear lamina are also associated with several types of muscular dystrophy (Gruenbaum et al., 2005). Nuclear lamins and their associated proteins provide both a rigid structure that helps shape the nuclear membrane and a platform onto which protein complexes and chromatin can bind (Holaska et al., 2002). Although much has been learned in recent years about the function of nuclear lamins, many significant questions remain. For example, it is not known how disease-associated changes in nuclear shape affect cellular and nuclear processes, or how the nuclear lamina, and other factors that may regulate nuclear shape, respond to signals associated with cellular growth.
The study of many questions in cell biology has been aided by the use of genetic tools available in the yeast Saccharomyces cerevisiae. In spite of the absence of nuclear lamins in yeast, there are reasons to believe that mechanisms common to yeast and higher eukaryotes are likely to be involved in the regulation of nuclear shape. For example, when nuclear lamins are expressed in yeast they properly localize to the nuclear periphery, suggesting that certain lamin-interacting factors may be conserved (Smith and Blobel, 1994). In addition, the interphase yeast nucleus, like its counterpart in higher eukaryotes, has a round shape, with the nuclear membrane juxtaposed to the DNA mass. Similar to higher eukaryotes, yeast must change their nuclear shape in response to various cues. For example, the shape of the yeast nucleus is restructured in response to mating pheromone (Stone et al., 2000). In addition, because the yeast nuclear envelope does not break down during mitosis, the nucleus adopts an hourglass shape during this stage of the cell cycle to accommodate the movement of the segregating chromosomes. The absence of a lamin equivalent raises the question of how the round shape of the nucleus is achieved: is it a property of the nuclear envelope, is it dependent on the underlying chromatin, or does it require a yet unidentified structural element?
In fact, the budding yeast nucleus has a specific internal organization that is suggestive of structural elements associated with the nuclear envelope. This is highlighted by the enrichment of certain DNA sequences, such as telomeres or late firing origins of replication, at the nuclear periphery (Heun et al., 2001; Hediger and Gasser, 2002). In addition, the nucleolus is limited to a crescent-shaped region, covering about one-third of the nuclear surface (Molenaar et al., 1970; Smitt et al., 1972). The nucleolus is a self-organizing structure that assembles by virtue of its specialized function in ribosome biogenesis (Dundr and Misteli, 2001). There is also evidence to suggest that both RNA polymerase I, which is involved in rRNA synthesis, and chromosomal elements flanking the rDNA region are important for the peripheral localization of the nucleolus (Oakes et al., 1998). Still, the molecular basis for this peripheral localization and how the crescent shape is achieved are poorly understood.
The yeast nucleus undergoes specific structural changes when the expression of certain proteins or growth conditions is altered. For example, in mutants with reduced levels of acetyl-CoA carboxylase activity, or mutants in certain early secretory pathway genes, the outer nuclear membrane becomes extended (Schneiter et al., 1996; Kimata et al., 1999; Matynia et al., 2002). Because mutants that block the secretory pathway at later steps do not show nuclear alterations, the latter phenotype is probably due to the accumulation of extra endoplasmic reticulum (ER) membranes in early secretory mutants. In addition, certain nuclear pore mutants, such as nup84Δ, exhibit multiple small outgrowths that include both the inner and outer nuclear membranes (Fabre and Hurt, 1997). Overexpression of particular ER membrane proteins, the best studied of which is Hmg1p, leads to the formation of stacked ER membrane pairs called “karmellae” (Wright et al., 1988). These stacks usually surround the nucleus and are largely devoid of nuclear pores. Finally, in ∼5–10% of log-phase cells, the nuclear envelope at the nucleus–vacuole junction folds into teardroplike blebs that protrude into the vacuole (Kohlwein et al., 2001; Roberts et al., 2003). These blebs, which are eventually converted into intravacuolar vesicles, are involved in microautophagy of nuclear constituents, including nucleolar material, and their prevalence increases in starved cells (Roberts et al., 2003).
Of interest to us was the altered nuclear shape first described by Siniossoglou et al. (1998) that was caused by the inactivation of either of two proteins, Spo7p or Nem1p. These proteins form an ER-associated phosphatase complex that negatively regulates phospholipid biosynthesis (Siniossoglou et al., 1998; Santos-Rosa et al., 2005). Deletion of either the SPO7 or NEM1 gene results in yeast cells containing nuclei with a single region of extra growth extending into the cytoplasm (Siniossoglou et al., 1998). These extensions, which can be quite dramatic, contain nuclear pore complexes but lack the bulk of DNA, as assayed by 4,6-diamidino-2-phenylindole (DAPI) staining (Siniossoglou et al., 1998). The role of Spo7p and Nem1p in inhibiting phospholipid biosynthesis suggests that the formation of nuclear extensions is due to an expansion of the nuclear membrane (Santos-Rosa et al., 2005).
Given that the nuclear extension in spo7Δ mutants is limited to a single outgrowth, we reasoned that this phenotype would afford us with an opportunity to explore functional differences between the various nuclear membrane domains. Here, we report that the extra membrane growth in spo7Δ mutants occurs in the nuclear membrane that is associated with the nucleolus but not in the nuclear membrane that is associated with the bulk of the DNA. The nuclear membrane is contiguous with the ER, and we find that the peripheral ER of spo7Δ mutants is also expanded. Our findings have implications for how nucleolar shape is determined and for the mechanisms that restrain nuclear membrane expansion.
MATERIALS AND METHODS
Strains and Plasmids
All strains were in the W303 background. Gene disruptions and epitope tagging were done according to Longtine et al. (1998) and Goldstein and McCusker (1999). OCF1533-1A: MATa ura3 leu2 his3; JCY565: same as OCF1533-1A but also spo7Δ::nat; JCY617 and JCY619 are the same as JCY565 and OCF1533-1A, respectively, except that they contain the Pus1p-GFP plasmid (pPUS1-URA). JCY620 and JCY622 are the same as JCY565 and OCF1533-1A, respectively, except that they contain the Sec63p-GFP plasmid (pJK59). JCY663: same as OCF1533-1A but spo7Δ::URA3; JCY680: same as OCF1533-1A but spo7-11; JCY681: same as OCF1533-1A but spo7-12; JCY720: MATa/MATα ura3/ura3 leu2/LEU2 his3/HIS3 trp1/TRP1; JCY721: same as JCY720 but also spo7Δ::nat/spo7Δ::nat; OCF2189: MATa bar1 ura3 leu2 ade2 his3 trp1 MLP1-13Myc::kan MLP2-GFP(S65T)::his5+; JCY545: same as OCF2189 but also spo7Δ::nat. OCF2276-1-9B: MATα ura3 trp1 NSR1-GFP::his5+; OCF2276-1-9D; MATa ura3 leu2 trp1 NSR1-GFP::his5+; OCF2276-2–3D same as OCF2276-1-9D but also spo7Δ::nat; OCF2276-1-12C MATα ura3 ade2 NSR1-GFP::his5+ spo7Δ::nat; OCF2284 MATa/MATα ura3/ura3 NSR1-GFP::his5+/NSR1-GFP::his5+ spo7Δ::nat/spo7Δ::nat ADE2/ade2 LEU2/leu2 TRP1/trp1; OCF2285 MATa/MATα ura3/ura3 NSR1-GFP::his5+/NSR1-GFP::his5+ LEU2/leu2 TRP1/trp1. Strain NOY891 MATa ade2 ura3 trp1 his3 leu2 can1 rdnΔΔ::HIS3, carrying plasmid NOY353, was a generous gift from Masayasu Nomura (UC Irvine, Irvine, CA) and Alex Strunnikov (NICHD, NIH, Bethesda, MD) and was described in Wai et al. (2001). Strains JCY831 and JCY832 are two independent isolates of NOY891 also containing spo7Δ::nat. JCY991 and JCY992 are independent isolates of NOY891 in which MLP1 was tagged with 13Myc::kan and transformed with pNUP49-URA. JCY994 and JCY995 are independent isolates of JCY831 in which MLP1 was tagged with 13Myc::kan and transformed with pNUP49-URA.
Construction of spo7 Temperature-sensitive (ts) Strains. To generate ts alleles of SPO7, the SPO7 gene was cloned into the polylinker of a URA3 CEN plasmid (pRS316; Sikorski and Hieter, 1989). It was then subjected to PCR mutagenesis using a GeneMorph II Random Mutagenesis kit from Stratagene (La Jolla, CA) and primers flanking the polylinker of pRS316. The PCR fragment was then transformed along with pRS315 (Sikorski and Hieter, 1989) that had been linearized by digestion in its polylinker into a leu2 spo7Δ::NAT strain (JCY565). Recombinants between the PCR fragment and the linearized plasmid were selected on leucine drop-out plates. Because spo7Δ mutants are sensitive to 0.25 M calcium (our unpublished observations), we were able to screen for ts mutants by selecting for colonies that were calcium resistant at 23°C and calcium sensitive at 37°C. The alleles used in this study, spo7-11 and spo7-12, contain multiple mutations, and we currently do not know which mutations are responsible for the ts phenotype. The wild-type SPO7 gene in the chromosome was then replaced with the ts alleles. This was accomplished by constructing a strain (JCY663 same as OCF1533-1A but spo7Δ::URA3), transforming it with a PCR fragment containing the ts allele, and selecting for 5-fluoroorotic acid-resistant colonies. These colonies were then screened to identify those that were calcium resistant at 23°C and calcium sensitive at 37°C. The appropriate gene replacement was also verified by PCR. The resulting two strains, JCY680 (same as OCF1533-1A but spo7-11) and JCY681 (same as OCF1533-1A but spo7-12), were then transformed with pPUS1-URA.
Plasmids. pASZ11-ADE2-PUS1-GFP, encoding PUS1-GFP (Hellmuth et al., 1998), and pASZ11-ADE2-NUP49-GFP, encoding NUP49-GFP (Belgareh and Doye, 1997), were gifts from Ed Hurt (University of Heidelberg, Heidelberg, Germany). These plasmids were modified by PCR-mediated replacement of the ADE2 gene with the URA3 gene to yield plasmids pPUS1-URA and pNUP49-URA, respectively. pJK59 is a centromeric plasmid that codes for SEC63-GFP and URA3. It was a gift from William Prinz (NIDDK, NIH, Bethesda, MD) and was described in Prinz et al. (2000). NOY353 (Wai et al., 2001) is a 2μ-based plasmid carrying GAL7-35S rDNA 5S rDNA TRP1 ampr.
Media and Growth Conditions
Cells containing plasmids bearing green fluorescent protein (GFP) fusions to PUS1, NUP49, or SEC63 were grown in casamino acid-URA media (2% casamino acid, 2% glucose, 0.17% yeast nitrogen base, and amino acid mix lacking uracil [obtained from Bio101, Carlsbad, CA]). Cells without plasmid were gown at 30°C in YPD (1% yeast extract, 2% peptone, and 2% glucose). rdnΔΔ::HIS3 strains containing the NOY353 plasmid were grown on 2% galactose + 2% raffinose as a carbon source. For the hydroxyurea (HU) arrest/release experiment, cells were grown at 23°C in casamino acid-URA to early log phase, spun down, resuspended in YPD containing 0.2 M HU (Sigma-Aldrich, St. Louis, MO), and incubated at 23°C. A complete cell cycle arrest (as judged by cell morphology) was obtained after ∼5 h. Once the cells were fully arrested, the culture was shifted to 37°C for 30 min, washed four times in prewarmed YPD, and released into YPD at 37°C. Samples were taken at the indicated time points.
Cytology
With the exception of the Sec63p-GFP, all images were of cells that were fixed in 1× phosphate-buffered saline (PBS) containing 4% paraformaldehyde (Electron Microscopy Services, Fort Washington, PA) for 1 h at 23°C, washed with 1× PBS, and stored at 4°C. Immediately before observation, the fixed cells were incubated briefly in 0.1% Triton X-100 and mixed with an equal volume of Vectashield with DAPI (Vector Laboratories, Burlingame, CA). Images were taken using an Olympus BX61 microscope, UPlanApo 100×/1.35 lens, Qimaging Retiga EX camera, and IPLab version 3.6.3 software (Scanalytics, Fairfax, VA). Image overlays were done with the pseudocolored images using the IPLab software. Sec63p-GFP images were done with live cells, using a Nikon E800 microscope equipped with a PerkinElmer Ultraview LCI CSU10 scanning unit, an argon/krypton ion laser (Meller Griot, Carlsbad, CA), and an ORCA ER cooled charge-coupled device camera (Hamamatsu Photonics, Hamamatsu, Japan), operated by OpenLab 3.0 software (Improvision, Lexington, MA). For immunofluorescence, cells were fixed for 1 h in 4% paraformaldehyde and treated as described in Kilmartin and Adams (1984), except that cells were spheroplasted in 50 μg/ml Zymolyase 100T (ICN, Aurora, OH) for 20 min at 23°C. Mlp1p-myc was detected using the 9E10 anti-Myc antibodies (Covance, Berkeley, CA) at a 1:500 dilution and Alexa Fluor 568 goat anti-mouse antibodies (Invitrogen, Carlsbad, CA) at a 1:200 dilution. For cell counting, fixed cells were sonicated gently, treated with Triton X-100 and DAPI as described above, and examined with a Nikon E-800 microscope using a Plan Fluor 100× differential interference contrast objective. Fluorescence in situ hybridization (FISH) and FISH + immunofluorescence experiments were done as described in Lorenz et al. (2002) and Fuchs and Loidl (2004)). Images were taken using a Zeiss Axioskop microscope with a 100×/1.30 Plan-Neofluar objective and a Quantix camera (Photometrics, Tucson, AZ) controlled by IPLab software. Image overlays were done with the Adobe Photoshop software (Adobe Systems, Mountain View, CA).
RESULTS
Characterization of the spo7Δ Nuclear “Flares”
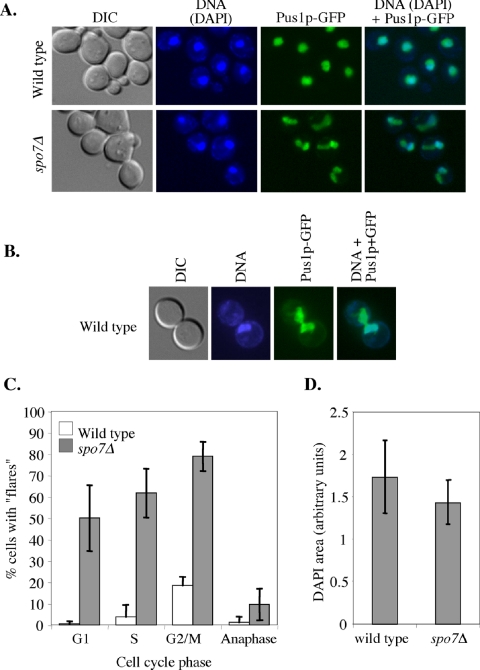
The inactivation of Spo7p, an inhibitor of phospholipid biosynthesis, leads to the appearance of a single “flare”like nuclear protrusion that can be seen using a soluble nuclear protein, such as Pus1p, fused to GFP (Siniossoglou et al., 1998; Santos-Rosa et al., 2005; Figure 1A). The most parsimonious explanation for this phenotype is that the increase in phospholipid biosynthesis leads to nuclear membrane expansion (Santos-Rosa et al., 2005). We were intrigued by the fact that this nuclear expansion is limited to a single outgrowth. In a budded spo7Δ cell, the flare is in the same cell body as the DNA mass, but the flare itself is largely devoid of DAPI staining material (Siniossoglou et al., 1998; Figure 1A), suggesting that the bulk of the DNA does not enter the flare. We compared the appearance of flares in wild-type and spo7Δ cells. Under standard growth conditions (rich media; 30°C), nuclear regions devoid of DAPI staining material could be detected, at low frequency, in wild-type cells (flares were observed in 7.2 ± 1.3% of wild-type cells compared with 56.3 ± 6.0% of spo7Δ mutant cells; n = 500 and 1000 for wild-type and spo7Δ strains, respectively). However, flares of wild-type cells were almost always in large budded preanaphase cells, where the DNA mass and the DAPI-free nuclear region are on different sides of the mother-bud neck (Figure 1, B and C). This phenotype, which is likely to be caused by preanaphase nuclear transits (Palmer et al., 1989), is distinct from the spo7Δ flares where the DNA mass and the DAPI-free nuclear compartment are in the same cell and can be readily detected in unbudded and small budded cells (compare Figure 1, A and B).
Figure 1.
The presence of flares in the spo7Δ mutant is cell cycle regulated. (A) Strains JCY619 (SPO7 pPUS1-GFP) and JCY617 (spo7Δ pPUS1-GFP) were grown to early log phase and imaged for GFP (nucleus, green) and DAPI (chromosomes, blue). “Flares” were defined as nuclear protrusions that do not contain DAPI staining material. (B) A typical flare of wild-type cells (JCY619). (C) Cell cycle distribution of flares. The data are shown as the percentage of cells with flares out of the total number of cells in a particular cell cycle phase. G1, unbudded cells with a single nucleus; S, small budded cells with a single nucleus; G2/M, large budded cells with a single nucleus; anaphase, large budded cells with two DNA masses. White columns, wild type; gray columns, spo7Δ. Strains are as described in A. (D) Measurements of the area occupied by DAPI staining material in wild-type and spo7Δ strains, described in A. Stacked images were taken at 2-μm intervals, and the DAPI area was measured at the focal plane with largest circumference. The number of nuclei counted was 24 and 20 for wild-type and spo7Δ cells, respectively.
Siniossoglou et al. (1998) concluded that the spo7Δ mutation does not lead to a change in the shape of the DAPI staining mass, which corresponds to the bulk of the chromosomal DNA. However, they did not rule out the possibility that the membrane surrounding the DNA expanded in a uniform manner, thus maintaining the overall round shape of the DAPI-stained area. To examine this issue in a more quantitative manner, we determined the area occupied by the DNA by using fixed cells and measuring the circumference of the DAPI-stained region at the focal plane where the circumference was at its largest. We found that the area occupied by the DNA is similar in wild-type and spo7Δ mutant cells (Figure 1D), and this was also the case if this area was calculated as a fraction of the cell size (our unpublished data). Because, apart from the flare, the nuclear membrane was always in tight association with the DNA (see below), we conclude that in the absence of Spo7p the organization of the bulk of the DNA, along with its associated nuclear membrane, is not perturbed. Thus, as previously suggested by Siniossoglou et al. (1998), nuclear membrane expansion in the spo7Δ mutant is limited to a single region of the nuclear envelope.
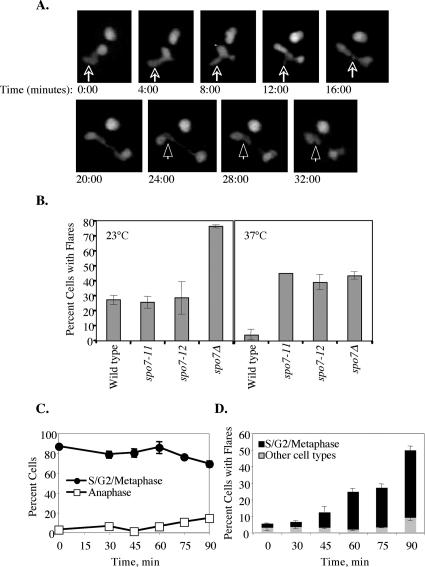
When examining the correlation between cell cycle stage and the existence of a flare (Figure 1C), we noticed a paucity of flares in spo7Δ cells that were in anaphase. Because the majority of G2/M cells have flares, we carried out time-lapse microscopy of spo7Δ cells expressing PUS1-GFP to determine what happens to flares as cells undergo anaphase. A typical example is shown in Figure 2A. At time 0, the nucleus shown in the lower left part of the image had a flare (indicated by the arrow) and was about to enter anaphase. Nuclear elongation was detected at the 8:00-min time point. The flare, which persisted up to the 12:00-min time point, became visibly shorter, and it seemed to be absorbed by the elongating nucleus. By the 16:00-min time point, when the nucleus was still in anaphase, the flare was completely absorbed. We did not observe flares occurring during anaphase in any of our time courses. We conclude that if deregulated phospholipid synthesis because of Spo7p inactivation takes place during anaphase, it does not result in the appearance of flares, and the majority of flares that existed before anaphase get incorporated into the elongating nucleus.
Figure 2.
Flare formation before and after anaphase. (A) Strain JCY617 (spo7Δ pPUS1-GFP) were embedded on 25% gelatin slabs prepared in YPD and visualized by confocal microscopy, taking a series of stacks every 4 min. A 32-min segment is shown. Note that at time 0, there are two nuclei in the field: the nucleus in the upper right corner remained in interphase for the duration of the experiment, whereas the nucleus in the lower left corner underwent nuclear division. The arrow points to the flare in the latter nucleus, and the arrowhead points to a remnant of the nuclear membrane that failed to be removed after nuclear division. The nucleus in the lower right corner at the 32:00-min time point also contains a flare, but because of the geometry of the plane of division it was not possible to determine whether this flare formed “de novo” or whether it was also a result of the failure to remove excess nuclear membrane after nuclear division. (B) Wild-type (JCY619), spo7-11 (JCY680), spo7-12 (JCY681), and spo7Δ (JCY617) strains, all containing the pPUS1-GFP plasmid, were grown at either 23 or 37°C for 16 h to mid-log phase and scored for the appearance of flares as described in Figure 1. (C) Cell cycle distribution of spo7-12 (JCY681) cells that were released from an S-phase arrest at 37°C at time 0; samples were taken at the indicated time points. The S/G2/metaphase cells represent large budded cells with a single nucleus, whereas anaphase cells are large budded with an elongated nucleus or two separate nuclei. A significant fraction of cells were in anaphase at 120 min after the release (our unpublished data). (D) For each time point shown in C, the percentage of cells with flares was scored as a function of cell cycle stage (for each time point, n > 50 cells were scored in each of three separate experiments). Shown is the overall percentage of cells that had flares as well as the types of cells with flares (S/G2/metaphase versus other cell types), at each time point. Similar results were obtained with the spo7-11 allele (our unpublished data).
Our time-lapse experiments also yielded clues as to how flares may form. In Figure 2A, the dividing nucleus reached full extension at the 24:00-min time point. In a wild-type cell, at this point, the nucleus would divide, resulting in two round daughter nuclei. This process entails the reorganization of the nuclear membrane that connected the two daughter nuclei before their separation. We occasionally observed that spo7Δ mutant cells were defective in removing this excess nuclear membrane. This can be seen in the nucleus shown in Figure 2A, where the extra membrane (arrowhead) remains as a flare in one of the two daughter nuclei. Thus, in spo7Δ mutant cells, flares can form due the inability to properly absorb the extranuclear membrane after nuclear division. Might this be the only mechanism for flare formation? To address this question, we created conditional spo7 mutant alleles (spo7-11 and spo7-12; see Materials and Methods). For unknown reasons, the appearance of flares, in both wild-type and spo7Δ cells, was temperature dependent: as mentioned, at 30°C wild-type and spo7Δ cells showed a 7.2 and 56.3% occurrence of flares, respectively, and both these percentages increased when cells were grown at 23°C and decreased when cells were grown at 37°C (Figure 2B). Nonetheless, cells carrying either the spo7-11 or spo7-12 alleles showed wild-type levels of flares at 23°C but spo7Δ levels of flares at 37°C (Figure 2B). To determine whether flares can form independently of nuclear division, spo7-12 cells were grown at 23°C, arrested in S phase with HU, shifted to 37°C, and released from the S phase arrest. Cell cycle progression and flare formation were monitored by DAPI staining and Pus1p-GFP, respectively. If flares were solely a consequence of a membrane remnant formed during nuclear division, they would occur only after the completion of anaphase. However, at 60–90 min after the release from S phase arrest, although most of the cells in the population had yet to enter anaphase (Figure 2C), a significant number of preanaphase nuclei exhibited flares (Figure 2D). Flares formed equally well when spo2-12 cells were released from the S-phase arrest into 37°C media containing the microtubule-depolymerizing drug nocodazole, indicating that flare formation is independent of microtubules (our unpublished data). Importantly, the spo7-12 flares that formed before anaphase had the same characteristics as flares that form in spo7Δ mutants (see below). Thus, flares can form independently of nuclear division.
The spo7Δ Flare Colocalizes with the Nucleolus
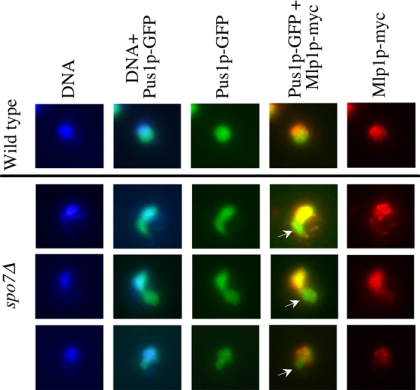
To further understand the effect of the spo7Δ mutation on nuclear organization, we examined the localization of the Mlp1 protein in wild-type and spo7Δ mutant cells. In wild-type cells, Mlp1p localizes to the nuclear periphery, but it is excluded from the nucleolus (Galy et al., 2004; Figure 3, top row). We found that in spo7Δ cells, Mlp1p is also localized at the nuclear periphery and surrounds the bulk of the DNA, but almost no Mlp1p was detected in flares (Figure 3, bottom rows, arrows). This observation, when combined with the aforementioned lack of change in the DAPI staining mass, is in further support of the idea that apart from the flare, the nucleus in spo7Δ mutant cells maintains its general organization despite nuclear membrane expansion. This result also suggested that the flare of the spo7Δ mutant is occupied by the nucleolus.
Figure 3.
The Mlp1 protein is excluded from the flare. Strains OCF2189 (SPO7 MLP1-MYC pPUS1-GFP) and JCY545 (spo7Δ MLP1-MYC pPUS1-GFP) were grown to early log phase and processed for immunofluorescence using antibodies against the Myc tag, while maintaining the fluorescence of Pus1-GFP. Mlp1p-Myc, red; DNA, blue; and Pus1-GFP, green. Three typical images of spo7Δ cells are shown.
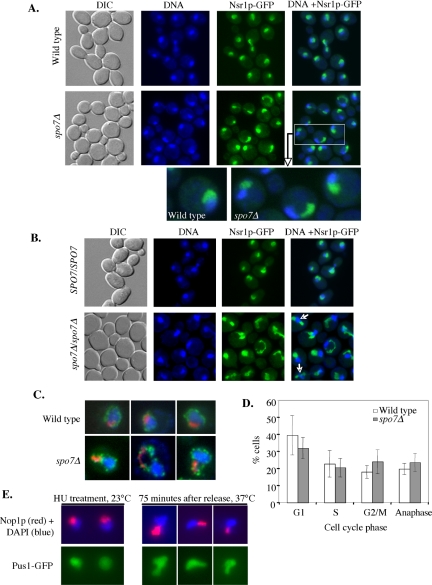
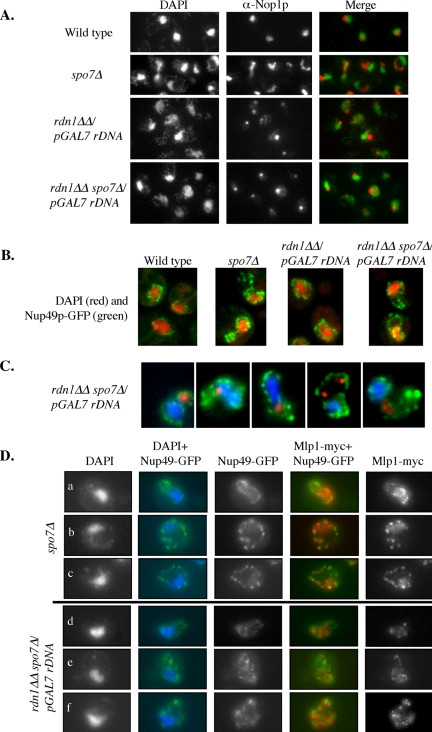
If indeed the flare was occupied by the nucleolus, one would expect that the shape of a spo7Δ mutant nucleolus would differ considerably from that of a wild-type cell. To examine nucleolar morphology directly, various nucleolar proteins, including Nsr1p, Utp10p, Nop6p, and Srp40p, were tagged with GFP and expressed from their native promoter in either wild-type or spo7Δ cells. While in wild-type cells these proteins localized in a typical crescent shape, we observed that in spo7Δ cells these nucleolar proteins were abnormally localized with respect to the DNA mass (Figure 4A; our unpublished data). The abnormal nucleolar shape was manifested both in a narrower region of contact between the nucleolus and DAPI-stained DNA mass, and in the shape of the nucleolus, which often exhibited the flare-like morphology seen with Pus1p-GFP (Figure 4A, enlarged images). Interestingly, in diploid cells, the complete absence of Spo7p resulted in the frequent appearance of two separate nucleoli, as opposed to the single nucleolus typically seen in wild-type cells (Figure 4B, the percentage of nuclei with two nucleoli was 22.5 ± 2.6% and <1.5% for spo7Δ/spo7Δ and SPO7/SPO7 cells, respectively).
Figure 4.
The flare colocalizes with the nucleolus. (A) Strains OCF2276-1–9B (SPO7 NSR1-GFP) and OCF2276-1-12C (spo7Δ NSR1-GFP) were grown to early log phase, fixed, and imaged for DNA (blue) and Nsr1p-GFP (green). Enlarged images of a wild-type and several spo7Δ cells are also shown. Identical results were obtained with Utp10p, Nop6p, and Srp1p tagged with GFP (our unpublished data). (B) Diploid strains OCF2285 (SPO7/SPO7 NSR1-GFP/NSR1-GFP) and OCF2284 (spo7Δ/spo7Δ NSR1-GFP/NSR1-GFP) were grown to early log phase, fixed, and examined for DNA (blue) and Nsr1p-GFP (green) as described in text. Note the appearance of two nucleoli in some of the spo7Δ/spo7Δ cells (arrows). (C) Strains OCF1533-1A (SPO7) and JCY565 (spo7Δ), both containing a plasmid coding for the nuclear pore protein Nup49p fused to GFP (pNUP49-URA), were grown to early log phase and processed for immunofluorescence using antibodies against the nucleolar protein Nop1p, while maintaining the GFP fluorescence of Nup49-GFP. Images taken for DAPI (blue), Nup49-GFP (green), and Nop1p (red) were overlaid. Three typical examples are shown. (D) Cell cycle distribution of wild-type and spo7Δ cultures. Cells were grown to early log phase, fixed, treated with DAPI, and counted for cell cycle stage. Data are shown as the average percentage of cells in a particular cell cycle stage out of the total number of cells counted. (E) spo7-12 pPUS1-GFP (JCY 681) cells, used in the HU experiment described in Figure 2, C and D, were collected at the HU arrest point (23°C) and at 75 min after release from the HU arrest (37°C) and analyzed by indirect immunofluorescence using antibodies against Nop1p while maintaining the Pus1-GFP fluorescence. Typical examples are shown.
To determine what fraction of flares were associated with the nucleolus, haploid cells expressing a nuclear pore protein, Nup49p, fused to GFP, were subjected to indirect immunofluorescence using antibodies against the nucleolar protein Nop1p. The Nup49-GFP patterns revealed only one flare per haploid nucleus (Figure 4C), and in 94% of cases the flare was associated with the nucleolus. Interestingly, spo7Δ mutant cells did not exhibit a significant cell cycle delay in mitosis (Figure 4D), suggesting that despite their abnormal shape, spo7Δ nucleoli divided with roughly wild-type kinetics. We also examined whether in spo7-12 mutants grown at 37°C after an S-phase arrest (Figure 2, C and D) the flares were associated with an abnormal nucleolar shape. Indeed, in cells that were harvested 75 min after the release from the S-phase arrest, the flares were associated with misshapen nucleoli (Figure 4E). This is in contrast the normal nucleolar morphology of spo7-12 cells grown at 23°C (Figure 4E). Finally, we observed a similar relationship between the presence of flares and alteration in nucleolar morphology in smp2Δ mutants (our unpublished data). Smp2p is a downstream target of Spo7p and its absence leads to increased expression of phospholipid biosynthesis genes (Santos-Rosa et al., 2005). Thus, our results suggest that under conditions of increased phospholipid biosynthesis, only one region of the nuclear membrane, namely, that which is associated with the nucleolus, exhibits expansion. These findings, combined with the observation that the absence of Spo7p function does not affect the nuclear compartment associated with the bulk of the DNA mass, indicates that the membrane associated with the nucleolus and the membrane surrounding the bulk of the DNA differ in their susceptibility to membrane expansion.
The rDNA Region of Chromosome XII Spreads into the spo7Δ Flare
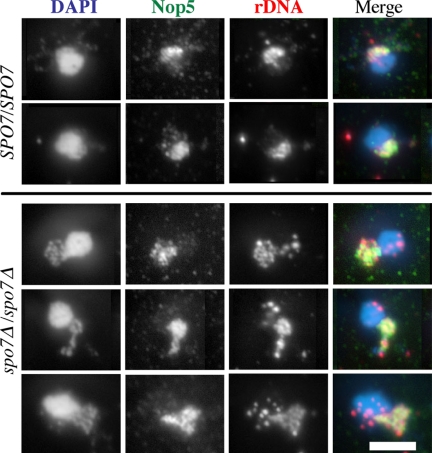
Having established that nucleolar proteins are located throughout the spo7Δ flare, we next examined the localization of the rDNA region of chromosome XII. In wild-type cells, this region loops into the nucleolus, whereas the non-rDNA arms of chromosome XII extend into the nucleoplasm (Fuchs and Loidl, 2004). It was therefore of interest to determine what fraction of the spo7Δ flare is occupied by the rDNA: does the rDNA, like the nucleolar proteins, occupy the entire flare, or does it maintain a wild-type–like organization, occupying only a region that is proximal to the bulk of the DNA? To examine the relationship between the rDNA and the nucleolus, FISH analysis using rDNA probes was combined with immunofluorescence against a nucleolar protein (Nop5p). As reported previously, in wild-type cells the rDNA localizes within the typical crescent-shaped nucleolus (Fuchs and Loidl, 2004; Figure 5, top). In spo7Δ cells, we found that the rDNA extended throughout the whole expanded nucleolar domain (Figure 5, bottom). Thus, not only nucleolar proteins but also the rDNA segment extend throughout the flare.
Figure 5.
The rDNA of spo7Δ mutants extends throughout the flare. Diploid wild type (JCY720) and spo7Δ/spo7Δ (JCY721) cells were grown to early log phase, fixed, and treated as described by Fuchs and Loidl (2004). The DNA was detected by DAPI staining (blue in the overlay), the nucleolus was detected with anti-Nop5p antibodies (green in the overlay), and the rDNA was detected with an rDNA probe (red in the overlay). Note that in the spo7Δ/spo7Δ cells the rDNA extends throughout the abnormally shaped nucleolus. Bar, 5 μm.
Flare Formation in spo7Δ Mutant Cells Is Independent of Nucleolar Expansion
Our results show that the spo7Δ nuclear expansion is tightly associated with the nucleolus. Thus, spo7Δ flares could have formed by one of two mechanisms: 1) the absence of Spo7p could have caused the nucleolus to expand, thereby “pushing” the nuclear membrane away from the main nuclear volume; or 2) the nuclear membrane associated with the nucleolus could be susceptible to expansion in the presence of elevated phospholipid levels, resulting, after nuclear membrane expansion, in the nucleolus “spilling” into the expanded domain. To distinguish between these two possibilities, we examined the effect of Spo7p depletion in a strain that forms small, spherical nucleoli that are largely detached from the nuclear membrane. Such a strain was developed by Oakes et al. (1998), who deleted the rDNA repeats from chromosome XII and placed an rDNA gene driven by a polymerase II (Pol II) promoter on a multicopy plasmid. In this strain, the nucleolus adopts a round shape (with sometimes more than one nucleolar body per cell) that is considerably smaller than a wild-type nucleolus and has limited contact with the nuclear envelope (Oakes et al., 1998; Figure 6A, strain rdn1ΔΔ/pGAL7 rDNA). When we deleted the SPO7 gene in the rdn1ΔΔ/pGAL7 rDNA background, nucleoli remained round and were indistinguishable from rdn1ΔΔ/pGAL7 rDNA nucleoli that formed in the presence of wild-type Spo7p (Figure 6A). However, we found that the rdn1ΔΔ/pGAL7 rDNA spo7Δ cells formed flares that were the same as flares of spo7Δ cells (Figure 6B), in both frequency of occurrence and shape. Because the rdn1ΔΔ/pGAL7 rDNA spo7Δ flares formed in the presence of a small spherical nucleolus, these findings show that nucleolar expansion is not necessary for flare formation. Interestingly, in 87% of flares in the rdn1ΔΔ/pGAL7 rDNA spo7Δ strain, the round nucleolus (or nucleoli) was either near or inside the flare (Figure 6C), a colocalization rate similar to what was found in spo7Δ cells (Figure 4C). Moreover, the flares of rdn1ΔΔ/pGAL7 rDNA spo7Δ cells contained no visible DAPI staining, indicating that the bulk of the chromosomal DNA does not move into the flare, even though it is no longer occupied by the nucleolus. This suggests that the organization of the DNA mass is independent, at least in part, of the nuclear membrane. Whether the DNA mass will maintain its shape if the entire nuclear membrane is somehow removed is an interesting question that remains to be answered.
Figure 6.
The formation of the spo7Δ flare is independent of nucleolar expansion. (A) Strains OCF1533-1A (SPO7), JCY565 (spo7Δ), NOY891 (rdn1ΔΔ pGAL7 rDNA), and JCY831 (rdn1ΔΔ pGAL7 rDNA spo7Δ) were grown to early log phase and processed for immunofluorescence using antibodies against Nop1p. (B) The same strains as in A, also carrying pNup49-URA, were fixed and stained for DAPI. Images are overlays of DAPI (red) and Nup49-GFP (green) images. (C) Strain JCY831 (rdn1ΔΔ pGAL7 rDNA spo7Δ) carrying pNup49-GFP was grown to early log phase and processed for immunofluorescence using antibodies against Nop1p. Images are overlays of DAPI (blue) Nup49-GFP (green), and Nop1p (red) images. For comparison with a spo7Δstrain, see Figure 3B. Images are representative of the different types of flare/nucleolus localizations observed in this strain background. (D) Strains spo7Δ MLP1-myc pNUP49-GFP (JCY831 and JCY832) and rdn1ΔΔ pGAL7 rDNA spo7Δ MLP1-myc pNUP49-GFP (JCY994 and JCY995) were grown to early log phase, fixed, and processed for indirect immunofluorescence as described above. In the overlays, DAPI is in blue, Nup49-GFP is in green, and Mlp1-Myc is in red.
Although our results show that flare formation is not caused by nucleolar enlargement, it is still possible that the nucleolus influences properties of the nuclear membrane with which it is associated, somehow making it more susceptible to expansion. Alternatively, it is possible that the colocalization of the nucleolus and flares is a coincidence generated by the mode of nuclear division (see Discussion).
The Intranuclear Localization Pattern of Mlp1p Is Not Due to Nucleolar Exclusion
A handful of proteins, including Mlp1p and the telomere tethering protein Esc1, localize to the nuclear periphery but are excluded from the nucleolus (Andrulis et al., 2002; Galy et al., 2004; Taddei et al., 2004). The reason for this exclusion is not clear, but one possibility is that the nucleolus itself serves as a barrier, preventing these proteins from interacting with nuclear membrane that is associated with the nucleolus. Alternatively, these proteins may have a higher affinity to the nuclear compartment containing the bulk of the DNA. One way to distinguish between these possibilities is by using an rdn1ΔΔ/pGAL7 rDNA strain, in which the nucleolus no longer occupies the crescent shape region where, in wild-type cells, Mlp1p is typically absent. If, in wild-type cells, Mlp1p is confined to the DNA-containing compartment of the nucleus because of exclusion by the nucleolus, then in an rdn1ΔΔ/pGAL7 rDNA strain Mlp1p should fill the whole nuclear volume. However, we found that because of the narrow width of the crescent-shaped Mlp1p-less region, even in a wild-type strain it is often difficult to determine unambiguously where Mlp1p is not present. We therefore decided to test the localization of Mlp1p in spo7Δ and rdn1ΔΔ/pGAL7 rDNA spo7Δ strains. In spo7Δ strains, the region devoid of Mlp1p colocalized not only with the nucleolus but also with the flare (Figure 3). In the rdn1ΔΔ/pGAL7 rDNA spo7Δ strain, the flares still largely occur at their normal nuclear location (i.e., opposite the spindle pole), but they are no longer filled with the nucleolus, allowing us to examine whether it is the nucleolus that excludes Mlp1p from this nuclear region.
To address this question, we used Nup49p-GFP as a marker for nuclear membrane; flares were identified as nuclear regions devoid of the bulk of the chromosomal DNA (as detected by DAPI staining). In the spo7Δ strain, only a small amount of Mlp1p entered the flare, exhibiting a punctate pattern often associated with site of Nup49p-GFP localization along the flare membrane (Figure 6D, rows a–c). Importantly, even when Mlp1p entered the flare, its distribution differed considerably from the dense Mlp1p pattern associated with the bulk of the DNA. The distribution of Mlp1p in the rdn1ΔΔ/pGAL7 rDNA spo7Δ strain was indistinguishable from that of the spo7Δ strain (Figure 6D, rows d–f). Thus, in the rdn1ΔΔ/pGAL7 rDNA spo7Δ strain, Mlp1p failed to enter the flare in a significant way, even though the flare was not occupied by a nucleolus. This suggests that the localization pattern of Mlp1p is not caused by nucleolar exclusion. Instead, Mlp1p's localization is likely to be determined by factors that are present in the nuclear compartment that contains the bulk of the DNA. Because Mlp1p is known to associate with nuclear periphery proteins (Galy et al., 2004), the finding that Mlp1p remains out of the flare in the rdn1ΔΔ/pGAL7 rDNA spo7Δ strain provides further support for the idea that the nuclear membrane contains domains that are inherently different.
spo7Δ Mutant Cells Exhibit Proliferation of the Peripheral ER
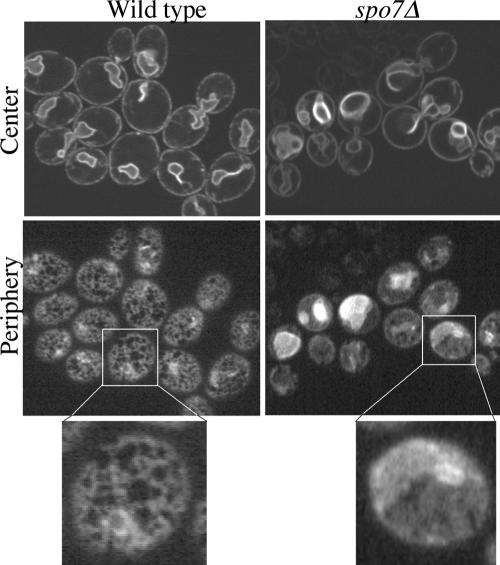
Our data suggest that the nuclear membrane has distinct domains that differ in their susceptibility to membrane expansion: in spo7Δ cells, the nuclear membrane in the nucleolar region expands, whereas the membrane associated with the bulk of the DNA does not. This raises the question of whether the nucleolus-associated membrane is particularly sensitive to expansion or whether the rest of the nuclear membrane is somehow able to resist expansion. Given that the nuclear membrane is contiguous with the ER, we examined whether the membrane of another ER domain, namely, the peripheral ER, was expanded in spo7Δ mutants. The ER membrane was visualized by expressing Sec63p-GFP. As reported previously (Santos-Rosa et al., 2005), the nuclear ER membrane of spo7Δ mutants was more convoluted than that of wild-type cells (Figure 7, top). The peripheral ER membrane of wild-type cells looked like an organized lattice network (Prinz et al., 2000; Figure 7, bottom left). In contrast, we observed that in spo7Δ mutant cells the peripheral ER membranes had a more sheetlike appearance (Figure 7, bottom right), similar to what was observed in strains carrying mutations in components of the COP I complex (Prinz et al., 2000). This phenotype is consistent with the loss of Spo7p function leading to an expansion of the peripheral ER, along with the membrane associated with the nucleolus. Thus, when considering the various ER membranes, the nuclear ER membrane that is adjacent to the DNA mass is unique in its ability to resist membrane expansion.
Figure 7.
spo7Δ mutants exhibit proliferation of the peripheral ER. Wild-type (JCY622) and spo7Δ (JCY620) cells expressing the ER-associated protein Sec63p-GFP from a centromeric plasmid (pJK59) were grown to early log phase and examined for Sec63p-GFP distribution. Confocal sections corresponding to the cell center (top row) and cell periphery (bottom row) are shown.
DISCUSSION
An important question in cell biology is what controls organelle shape. Throughout most of the cell cycle, the yeast nucleus is round, but the underlying structure responsible for this shape is unknown. Here, we analyzed the phenotype of a budding yeast mutant that has a nonround nucleus, displaying a single nuclear flarelike extension. Our results show that the spo7Δ nuclear flare colocalizes with the nucleolus (Figure 4). In a wild-type cell, the membrane adjacent to the nucleolus seems indistinguishable from the membrane that surrounds the rest of the nucleus, although it was previously noted that some nuclear periphery proteins, such as Mlp1p and Esc1p, are excluded from the nucleolus (Andrulis et al., 2002; Galy et al., 2004; Taddei et al., 2004). Our analysis of the spo7Δ mutant phenotype suggests that the nuclear membrane associated with the nucleolus has certain properties that distinguish it from the membrane surrounding the rest of the nucleus, because in the absence of Spo7p function only the nucleolus-associated membrane becomes extended (Figure 4). The involvement of Spo7p in inhibiting phospholipid biosynthesis (Santos-Rosa et al., 2005) strongly suggests that the flare is a result of membrane proliferation due to elevated levels of phospholipids, rather than an expansion of the nucleolus itself. Indeed, we found that when the nucleolus was no longer intimately associated with the nuclear envelope, such as in a strain in which the only rDNA copies were on plasmids and expressed by Pol II (Oakes et al., 1998), Spo7p inactivation did not alter nucleolar shape, but it still led to the appearance of flares (Figure 6). Thus, the nuclear membrane associated with the nucleolus differs from the nuclear membrane associated with the bulk of the DNA in its susceptibility to expansion when phospholipid levels are elevated.
Our findings raise an interesting question as to how the crescent-shaped nucleolus is normally formed. This structure could, in principle, be an inherent property of the nucleolus. However, we observed that when the nuclear membrane expands, such as in a spo7Δ mutant, the nucleolus loses its crescent shape. Moreover, under these conditions, the rDNA, which resides within the nucleolus, spreads throughout the entire flare (Figure 5). Because, as mentioned above, flare formation is independent of nucleolar expansion, we favor the idea that as the flare forms, nucleolar components spread into this extended space. This suggests that the nucleolus does not have an inherent crescent-shaped structure but that it is confined by the nuclear membrane. In wild-type cells, at the end of anaphase, the nuclear membrane must be remodeled by a yet unknown mechanism to absorb or remove the extra membrane that connects the two DNA masses (Figure 8A, wild type). Because the nucleolus divides only at the end of anaphase (Fuchs and Loidl, 2004; Pereira and Schiebel, 2004), trailing behind the rest of the DNA, it is possible that as the nucleus regains its round shape, the nucleolus is compacted by the nuclear membrane into a crescent shape (Figure 8A). In the absence of Spo7p function, this compaction may fail (Figure 8A, spo7Δ), resulting in an extended nucleolus. Thus, the nuclear membrane itself may contribute to the shape of the budding yeast nucleolus, and by limiting the amount of nuclear membrane synthesized, Spo7p may play a role maintaining proper nucleolar shape. The question of what determined nucleolar shape could be further extended to diploid cells, where under wild-type conditions there is only one nucleolus, despite having two copies of chromosome XII, each carrying an rDNA array. How the single nucleolus forms in diploid cells is unknown, but our results show that the nuclear membrane plays an important role in maintaining nucleolar structure, because nuclei of spo7Δ/spo7Δ mutant cells exhibit a high frequency of two nucleoli (Figure 4). Hence, the yeast nucleus and its nucleolus provide an interesting paradigm for elucidating the role of intracellular membranes in ensuring organelle shape.
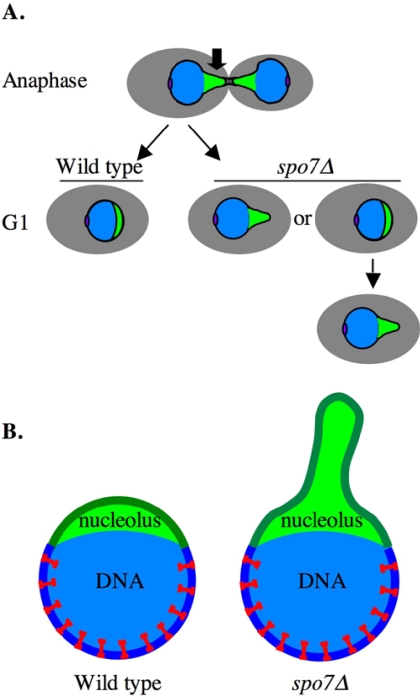
Figure 8.
Schematic model to explain the mechanism of flare formation. (A) The structural changes experienced by the nucleus and nucleolus of dividing cells. Cell body, gray; the bulk of the DNA mass, blue; nucleolus, green; and spindle pole body, purple. Note that in anaphase, the nucleolus trails behind the bulk of the DNA. The arrow indicates the region of the nuclear membrane that has to be removed after the nucleus divides. In spo7Δ mutants, a flare may form due to incomplete removal of this membrane region at the end of anaphase, or it can form some time after nuclear division. (B) Model. The expansion of the nuclear membrane around the bulk of the DNA is restricted by a tethering mechanism, indicated as red staples, that is excluded from the nucleolus.
Our finding that the spo7Δ flares are associated with the nucleolus, whereas the bulk of the DNA remains tightly associated with the nuclear membrane, led us to examine how another ER-associated membrane, the peripheral ER, responds to the absence of Spo7p. We found that in the absence of Spo7p, the peripheral ER is also extended, no longer exhibiting the typical tubular structure (Figure 7). Because the nuclear membrane is contiguous with the ER, our data suggest that all ER membranes, with the exception of the nuclear membrane that is associated with the bulk of the DNA, exhibit membrane expansion in the absence of Spo7p. How does the nuclear membrane associated with the bulk of the DNA resist membrane expansion? It is tempting to speculate that this membrane domain is tethered to the DNA, either directly or indirectly, by a chromosome associated factor(s) that is absent from the region occupied by the nucleolus (Figure 8B). This kind of tethering would be analogous to the role played by lamins in shaping nuclei of higher eukaryotes, although a yeast laminlike protein has yet to be identified. When phospholipid levels increase, this mechanism would restrict membrane expansion around the bulk of the DNA. In budding yeast, several proteins are known to tether DNA, and specifically telomeric regions, to the nuclear periphery. These include Sir4p, the Ku70p/Ku80p complex, and Esc1p (reviewed in Taddei et al., 2005). In an attempt to determine whether these proteins also act to prevent membrane expansion in the spo7Δ mutant, we deleted these genes individually and in combination (i.e., sir4Δ ku70Δ spo7Δ or esc1Δ ku70Δ spo7Δ) and examined what effect these combined mutations have on nuclear structure. In none of the mutant combinations could we detect an exacerbation of the spo7Δ phenotypes, and there was no noticeable membrane detachment from the bulk of the DNA (our unpublished data). This suggests that these proteins are not necessary to resist membrane expansion, although we cannot rule out the possibility that they play a nonessential role in this process. In this regard, a study from the Tartakoff laboratory showed that overexpression of Esc1p leads to dramatic elaborations of the nuclear envelope that include nuclear pores but contain a limited amount of nucleoplasm (Hattier, Andrulis, and Tartakoff, personal communication). The mechanism by which this occurs remains to be determined. We also examined whether the simultaneous deletion of the Mlp1p and Mlp2p proteins, whose localization pattern made them tempting candidates for playing a role in membrane tethering, might affect the spo7Δ phenotype, but found no effect (our unpublished data). It was previously found that the spo7Δ and nup84Δ mutations are synthetically lethal, namely, that cells can survive with either, but not both, mutations (Siniossoglou et al., 1998). The Nup84 protein is part of the nuclear pore, and in its absence nuclear pores cluster into discrete domains rather than distribute throughout the nuclear envelope. The synthetic lethal interaction between spo7Δ and nup84Δ is suggestive of a possible role for nuclear pores in tethering the nuclear envelope of the bulk of the DNA, although additional components must be involved because nuclear pores are present in the membrane associated with the nucleolus. It will be possible to examine the involvement of Nup84p in membrane tethering once an appropriate system for effectively activating and inactivating Spo7p and/or Nup84p is developed.
If there is membrane tethering mechanism, why and how it is excluded from the nucleolus are not known. It is possible that it is advantageous to the cell to keep the nucleolus-associated membrane amenable to expansion. This raises interesting questions as to how the yeast nucleus grows in size as cells undergo division: can membrane components be incorporated at any place on the nuclear membrane, or is there a specific “entry site”? Because the flare can form at various points of the cell cycle, does this mean that the nucleolus remains associated with the same “susceptible” membrane domain? An insight into how proteins partition between the nucleolus and the rest of the nucleus was gained from our experiment examining the localization of Mlp1p in the rdn1ΔΔ/pGAL7 rDNA spo7Δ strain. Because Mlp1p remained associated with the bulk of the DNA and did not spread into the flare despite the existence of a much smaller nucleolus (Figure 6), it seems likely that the intranuclear distribution of Mlp1p is determined not by nucleolar exclusion but by an affinity to one or more components that reside in the nonnucleolar compartment of the nucleolus. Further experiments are needed to determine what these components are.
Why do flares always form in the vicinity of the nucleolus? One possibility is that the expansion in the vicinity of the nucleolus is influenced by the nucleolus itself. Alternatively, the colocalization of the nucleolus and the flare could be a coincidence created by the mechanics of nuclear division. The nucleolus is the last region of DNA to segregate, placing it at the same nuclear membrane region that must be remodeled during nuclear division (Figure 8A). A nuclear landmark that is in a fixed relationship to the nucleolus is the spindle pole body (SPB), which is embedded in the nuclear membrane and serves to nucleate both cytoplasmic and spindle microtubules. The SPB, which is in the leading edge of the dividing nucleus, and the nucleolus, which is at the trailing end of the nucleus, are located on opposite sides of the nucleus and this organization is maintained throughout most of the cell cycle (Yang et al., 1989; Bystricky et al., 2005). Interestingly, we found that in the rdn1ΔΔ/pGAL7 rDNA strain, as in the wild-type and spo7Δ strains, the nucleolus was typically found opposite the spindle pole, thus precluding the determination of whether flare formation is dependent on a particular nuclear membrane domain (i.e., opposite the spindle pole) or whether it depends on the proximity to the nucleolus. However, because the nucleoli in this strain are significantly smaller than nucleoli of wild-type cells, and consequently occupy only a very small fraction of the spo7Δ flare (Figure 6), we favor the possibility that the site of flare formation is independent of the nucleolus. We speculate that the membrane opposite the spindle pole, which has to remodel after the completion of anaphase, has properties that make it distinct from the nuclear membrane that surrounds the DNA. Our results show that the spo7Δ flare may form at the end of anaphase due to incomplete remodeling of the nuclear membrane located between the segregated DNA masses (Figures 2A and 8B). We also find that a flare may form later in the cell cycle, independent of nuclear division, but again coincident with the nucleolus (Figures 2D and 8B). If indeed nuclear division results in a distinct membrane domain that is sensitive to expansion, one possibility is that its colocalization with the nucleolus contributes to its maintenance in this distinct state. This raises an interesting question: does the nucleolus remain associated with a particular membrane region throughout the cell cycle, and if so, what is the functional importance of this association?
Together, our results suggest that the nuclear membrane has discrete domains that respond differently to changes in phospholipid levels. We propose that this is due to an asymmetric distribution of membrane tethering factors that are present throughout the nuclear envelope except in the newly reorganized region. What these factors are and whether the nucleolus plays a role in preventing membrane tethering awaits further analysis.
Acknowledgments
We thank Masayasu Nomura, Ed Hurt, Will Prinz, Alex Strunnikov, and Rohinton Kamakaka for strains and plasmids and John P. Aris for the Nop5 antibody (37C12). We are grateful to Alan Tartakoff and members of his laboratory for sharing unpublished results. We also thank Will Prinz and Kevin O'Connell for invaluable assistance with microscopy. We thank Brenda Peculis, Will Prinz, Paula Fearon, and Armand de Gramont for helpful discussions and comments on the manuscript. J. L. and A. L. were supported by Grant P16282 by the Austrian Science Fund. J.L.C., K.L.W., T. H., and O.C.F. were supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-09-0839) on February 8, 2006.
Abbreviations used: DAPI, 4,6-diamidino-2-phenylindole; ER, endoplasmic reticulum; FISH, fluorescence in situ hybridization; HU, hydroxyurea.
References
- Andrulis, E. D., Zappulla, D. C., Ansari, A., Perrod, S., Laiosa, C. V., Gartenberg, M. R., and Sternglanz, R. (2002). Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 22, 8292–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh, N., and Doye, V. (1997). Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 136, 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky, K., Laroche, T., van Houwe, G., Blaszczyk, M., and Gasser, S. M. (2005). Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., and Misteli, T. (2001). Functional architecture in the cell nucleus. Biochem. J. 356, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, E., and Hurt, E. (1997). Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet. 31, 277–313. [DOI] [PubMed] [Google Scholar]
- Fuchs, J., and Loidl, J. (2004). Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 12, 427–438. [DOI] [PubMed] [Google Scholar]
- Galy, V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63–73. [DOI] [PubMed] [Google Scholar]
- Gartner, L. P., Hiatt, J. L., and Gartner, L. P. (1990). Color Atlas of Histology, Baltimore: Williams & Wilkins.
- Goldstein, A. L., and McCusker, J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Margalit, A., Goldman, R. D., Shumaker, D. K., and Wilson, K. L. (2005). The nuclear lamina comes of age. Nat. Rev. Mol. Cell. Biol. 6, 21–31. [DOI] [PubMed] [Google Scholar]
- Haithcock, E., Dayani, Y., Neufeld, E., Zahand, A. J., Feinstein, N., Mattout, A., Gruenbaum, Y., and Liu, J. (2005). Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102, 16690–16695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger, F., and Gasser, S. M. (2002). Nuclear organization and silencing: putting things in their place. Nat. Cell Biol. 4, E53–E55. [DOI] [PubMed] [Google Scholar]
- Hellmuth, K., Lau, D. M., Bischoff, F. R., Kunzler, M., Hurt, E., and Simos, G. (1998). Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18, 6374–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun, P., Laroche, T., Raghuraman, M. K., and Gasser, S. M. (2001). The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 152, 385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska, J. M., Wilson, K. L., and Mansharamani, M. (2002). The nuclear envelope, lamins and nuclear assembly. Curr. Opin. Cell Biol. 14, 357–364. [DOI] [PubMed] [Google Scholar]
- Kilmartin, J. V., and Adams, A. E. (1984). Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98, 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata, Y., Lim, C. R., Kiriyama, T., Nara, A., Hirata, A., and Kohno, K. (1999). Mutation of the yeast epsilon-COP gene ANU2 causes abnormal nuclear morphology and defects in intracellular vesicular transport. Cell Struct. Funct. 24, 197–208. [DOI] [PubMed] [Google Scholar]
- Kohlwein, S. D., Eder, S., Oh, C. S., Martin, C. E., Gable, K., Bacikova, D., and Dunn, T. (2001). Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., Demarini, D. J., Shah, N. G., Wach, A., Arndt, B., Philippsen, P., and Pringle, J. R. (1998). Additional modules for varsatine and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lorenz, A., Fuchs, J., Trelles-Sticken, E., Scherthan, H., and Loidl, J. (2002). Spatial organisation and behaviour of the parental chromosome sets in the nuclei of Saccharomyces cerevisiae x S. paradoxus hybrids. J. Cell Sci. 115, 3829–3835. [DOI] [PubMed] [Google Scholar]
- Matynia, A., Salus, S. S., and Sazer, S. (2002). Three proteins required for early steps in the protein secretory pathway also affect nuclear envelope structure and cell cycle progression in fission yeast. J. Cell Sci. 115, 421–431. [DOI] [PubMed] [Google Scholar]
- Molenaar, I., Sillevis Smitt, W. W., Rozijn, T. H., and Tonino, G. J. (1970). Biochemical and electron microscopic study of isolated yeast nuclei. Exp. Cell Res. 60, 148–156. [DOI] [PubMed] [Google Scholar]
- Oakes, M., Aris, J. P., Brockenbrough, J. S., Wai, H., Vu, L., and Nomura, M. (1998). Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 143, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, R. E., Koval, M., and Koshland, D. (1989). The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J. Cell Biol. 109, 3355–3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., and Schiebel, E. (2004). Cdc14 phosphatase resolves the rDNA segregation delay. Nat. Cell Biol. 6, 473–475. [DOI] [PubMed] [Google Scholar]
- Prinz, W. A., Grzyb, L., Veenhuis, M., Kahana, J. A., Silver, P. A., and Rapoport, T. A. (2000). Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, P., Moshitch-Moshkovitz, S., Kvam, E., O'Toole, E., Winey, M., and Goldfarb, D. S. (2003). Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa, H., Leung, J., Grimsey, N., Peak-Chew, S., and Siniossoglou, S. (2005). The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter, R., Hitomi, M., Ivessa, A. S., Fasch, E. V., Kohlwein, S. D., and Tartakoff, A. M. (1996). A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol. Cell. Biol. 16, 7161–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou, S., Santos-Rosa, H., Rappsilber, J., Mann, M., and Hurt, E. (1998). A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17, 6449–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S., and Blobel, G. (1994). Colocalization of vertebrate lamin B and lamin B receptor (LBR) in nuclear envelopes and in LBR-induced membrane stacks of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 91, 10124–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smitt, W. W., Vermeulen, C. A., Vlak, J. M., Rozijn, T. H., and Molenaar, I. (1972). Electron microscopic autoradiographic study of RNA synthesis in yeast nucleus. Exp. Cell Res. 70, 140–144. [DOI] [PubMed] [Google Scholar]
- Stone, E. M., Heun, P., Laroche, T., Pillus, L., and Gasser, S. M. (2000). MAP kinase signaling induces nuclear reorganization in budding yeast. Curr. Biol. 10, 373–382. [DOI] [PubMed] [Google Scholar]
- Taddei, A., Gartenberg, M. R., Neumann, F. R., Hediger, F., and Gasser, S. M. (2005). Multiple pathways tether telomeres and silent chromatin at the nuclear periphery: functional implications for sir-mediated repression. Novartis Found Symp. 264, 140–156; discussion 156–165, 227–230. [PubMed] [Google Scholar]
- Taddei, A., Hediger, F., Neumann, F. R., Bauer, C., and Gasser, S. M. (2004). Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai, H., Johzuka, K., Vu, L., Eliason, K., Kobayashi, T., Horiuchi, T., and Nomura, M. (2001). Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol. Cell. Biol. 21, 5541–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R., Basson, M., D'Ari, L., and Rine, J. (1988). Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 107, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. H., Lambie, E. J., Hardin, J., Craft, J., and Snyder, M. (1989). Higher order structure is present in the yeast nucleus: autoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma 98, 123–128. [DOI] [PubMed] [Google Scholar]
- Zink, D., Fischer, A. H., and Nickerson, J. A. (2004). Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687. [DOI] [PubMed] [Google Scholar]