Abstract
The plant toxin ricin is transported retrogradely from the cell surface to the endoplasmic reticulum (ER) from where the enzymatically active part is retrotranslocated to the cytosol, presumably by the same mechanism as used by misfolded proteins. The ER degradation enhancing α-mannosidase I-like protein, EDEM, is responsible for directing aberrant proteins for ER-associated protein degradation. In this study, we have investigated whether EDEM is involved in ricin retrotranslocation. Overexpression of EDEM strongly protects against ricin. However, when the interaction between EDEM and misfolded proteins is inhibited by kifunensin, EDEM promotes retrotranslocation of ricin from the ER to the cytosol. Furthermore, puromycin, which inhibits synthesis and thereby transport of proteins into the ER, counteracted the protection seen in EDEM-transfected cells. Coimmunoprecipitation studies revealed that ricin can interact with EDEM and with Sec61α, and both kifunensin and puromycin increase these interactions. Importantly, vector-based RNA interference against EDEM, which leads to reduction of the cellular level of EDEM, decreased retrotranslocation of ricin A-chain to the cytosol. In conclusion, our results indicate that EDEM is involved in retrotranslocation of ricin from the ER to the cytosol.
INTRODUCTION
The toxin ricin is a heterodimeric lectin produced in the seeds of the castor oil plant, Ricinus communis. It is very potent to mammalian cells, being able to fatally disrupt protein synthesis by attacking the 28S ribosomal subunit (reviewed in Sandvig et al., 2005). Entry of ricin into cells involves a series of steps. The toxin first binds via its ricin B-chain (RTB) to cell surface glycolipids or glycoproteins having β-1.4–linked galactose residues. Then, ricin is endocytosed and enters early endosomes where a small fraction is sorted by retrograde transport to the trans-Golgi network (TGN) and further to the endoplasmic reticulum (ER). The disulfide bond connecting the ricin A-chain (RTA) and the RTB can be reduced in ER, followed by a partial unfolding of RTA, rendering it competent to cross the ER membrane, presumably via the Sec61p translocon in a similar manner as misfolded ER proteins. In the cytosol, RTA must refold into its biologically active conformation to inactivate the ribosomes. Although ricin has been shown to interact with Sec61p (Wesche et al., 1999), and it has been reported that protein disulfide isomerase is involved in reduction of ricin in the ER (Spooner et al., 2004), little is known about the role of other ER proteins in ricin retrotranslocation to the cytosol.
A recently discovered protein, ER degradation enhancing α-mannosidase I-like protein (EDEM), has been shown to be involved in retrotranslocation and degradation of misfolded ER proteins in a process called ER-associated degradation (ERAD) (Hosokawa et al., 2001). Two groups have shown that EDEM is able to extract misfolded glycoproteins, but not glycoproteins undergoing productive folding, from the calnexin cycle (Molinari et al., 2003; Oda et al., 2003). In their study, EDEM overexpression was shown to result in faster release of folding-incompetent proteins from the calnexin cycle and earlier onset of their degradation, whereas EDEM down-regulation prolonged folding attempts and delayed ERAD. It has been established that formation of the mannose-N-acetylglucosamine Man8GlcNAc2 isomer B of N-linked oligosaccharides is the critical luminal event as a signal for ERAD (Hosokawa et al., 2001). Selective inhibition of ER α-mannosidase I with the alkaloid kifunensin prevents hydrolysis of the Man9 structure, retards release of terminally misfolded glycoproteins from the calnexin cycle, and delays their degradation (Molinari et al., 2003). Interestingly, EDEM is a membrane-bound protein in COS cells (Hosokawa et al., 2001), whereas it seems to be a luminal protein in human embryonic kidney (HEK)293 cells (Mast et al., 2004; Olivari et al., 2005).
In this study, we have investigated the role of EDEM for retrotranslocation of ricin to the cytosol. Our data suggest that when EDEM is available for ricin and the ER channels are not occupied with newly synthesized or misfolded proteins, EDEM promotes retrotranslocation of ricin from the ER to the cytosol. We showed that RNA-mediated interference against EDEM reduced transport of ricin A-chain to the cytosol. Furthermore, a high expression level of EDEM protected against ricin retrotranslocation, perhaps because of an increased flow of misfolded proteins across the translocon. This hypothesis was supported by the observation that overproduction of model misfolded proteins decreased ricin retrotranslocation from the ER to the cytosol. Importantly, when the interaction of EDEM with misfolded proteins and therefore also their retrotranslocation from the ER was inhibited by kifunensin, high levels of EDEM sensitized the cells to ricin, and more A-chain was transported to the cytosol. Puromycin, which inhibits protein synthesis and keeps the translocation channels open, also increased ricin retrotranslocation to the cytosol in EDEM-transfected cells. Immunoprecipitation experiments confirmed that after kifunensin and puromycin treatment more ricin could interact both with EDEM and with the Sec61p translocon. Together, our data indicate a direct role of EDEM in ricin transport from the ER to the cytosol.
MATERIALS AND METHODS
Materials
Ricin, HEPES, lactose, puromycin were all obtained from Sigma-Aldrich (St. Louis, MO), whereas kifunensin was from Toronto Research Chemicals (Toronto, Ontario, Canada). [3H]Leucine and Na2 35SO4 were purchased from GE Healthcare (Princeton, NJ). Na-125I came from DuPont (Brussels, Belgium). The rabbit anti-ricin was obtained by standard immunization and has been described previously (Sandvig and Olsnes, 1979). The rabbit polyclonal anti-hemagglutinin (HA) antibody was obtained from Upstate Biotechnology (Waltham, MA), whereas the mouse monoclonal anti-HA antibody came from Covance Research Products (Denver, CO). The rabbit anti-Sec61α and mouse anti-myc antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), whereas the mouse anti-α1-anti-trypsin antibody came from US Biological (Swampscott, MA). The mouse anti-calnexin (BD Biosciences, Palo Alto, CA) was used in Western blots and immunofluorescence, anti-calreticulin (BioSite, San Diego, CA) and anti-α-tubulin (Sigma-Aldrich) were used in Western blots, whereas a sheep anti-TGN46 (Serotec, Oxford, United Kingdom) was used only in immunofluorescence. The secondary antibodies anti-rabbit horseradish peroxidase (HRP), anti-mouse HRP, anti-mouse Cy-2, and anti-sheep Cy-2 were all obtained from The Jackson Laboratory (Bar Harbor, MA).
DNA Constructs and Transfections
cDNA encoding the mouse EDEM fused to an HA-tag in the pCMV-SPORT2 vector was a kind gift from Prof. K. Nagata and Dr. N. Hosokawa (Institute of Frontier Medical Science, Kyoto University, Kyoto, Japan). For details of the cloning, refer to Hosokawa et al., 2001. cDNA encoding α1-anti-trypsin wild type (wt) and Hong Kong mutant, BACE 501, BACE 457, and BACE 457Δ were a kind gift of Dr. P. Paganetti (Novartis Pharma, Basel, Switzerland). The small interfering RNAs (siRNAs) against EDEM were constructed using a shuttle vector containing a U6 promoter sequence (Grimmer et al., 2005). The sense sequences selected for human and mouse EDEM were as follows: RNAi1, 5′-GCAGTCAAGTTAGTGATCA-3′; and RNAi2, 5′-GAATCCCTTCTACCTCCAT-3′. For each sequence, a pair of primers (5′→ 3′ and 3′→ 5′ orientated) were designed. They contain these sequences in sense and anti-sense orientations linked by a KpnI sequence (loop of the hairpin) and BamHI and XhoI overhangs, 5′ phosporylated. These pairs were annealed and the resulting duplexes were cloned into our shuttle vector predigested with the same enzymes. As a negative control, a U6 promoter fused to a degenerated hairpin was used. All constructs were sequenced. HEK293 cells were transiently transfected with FuGENE transfection reagent according to manufacturer's procedure (Roche Diagnostics, Indianapolis, IN).
Cell Culture
HEK293 cells were grown in complete medium: DMEM (Cambrex Bio Science Walkersville, Walkersville, MD) supplemented with 10% fetal calf serum (FCS) (Integro, Zaandam, The Netherlands), 2 mM glutamine, 25 U/ml penicillin, and 25 μg/ml streptomycin (all from Cambrex Bio Science Walkersville) under 5% CO2 in a 37°C incubator. HeLa-203, HeLa-204, and HeLa-MSCV stably transfected with Derlin-1GFP, Derlin-2GFP, and empty pMSCV puro vector, respectively (Lilley and Ploegh, 2004) were grown in RPMI 1640 medium (Cambrex Bio Science Walkersville) supplemented with 10% FCS (Integro), 2 mM glutamine, 25 U/ml penicillin, 25 μg/ml streptomycin (all from Cambrex Bio Science Walkersville), and 0.5 μg/ml puromycin (Sigma-Aldrich) under 5% CO2 in a 37°C incubator. HeLa-203 and HeLa-204 were tested by Western blot to demonstrate that the two cell lines have equal amounts of Derlin-green fluorescent protein (GFP). Confocal microscope revealed that all cells in the cultures express the constructs.
Immunofluorescence Microscopy
HEK293 cells were grown on coverslips and transiently transfected with the EDEM gene alone or in combination with GFP. Three days posttransfection, cell were fixed in 3% paraformaldehyde and then permeabilized in 0.1% Triton X-100 and blocked in 5% FCS before the cells were labeled with rabbit anti-HA together with mouse anti-calnexin or sheep anti-TGN46 and detected by the appropriate secondary antibodies. Confocal images were acquired with a Leica TCS-NT digital scanning confocal microscope equipped with a Zeiss LSM 410 (Carl Zeiss, Thornwood, NY) with a 100× Zeiss Plan Apochromat oil immersion objective numerical aperture 1.4.
Ricin Toxicity and Measurements of Protein Synthesis
HEK293 cells (1 × 104 cells in each well) were seeded in 24-well plates and transfected with either the EDEM cDNA or a control empty vector. Three days posttransfection, cells were washed in leucine-free medium and incubated with different concentrations of ricin or ricin sulf-1 for 3 h. Optionally, the cells were preincubated with inhibitors for 30 min before ricin was added. Concentrations of inhibitors are indicated in the figure legends (Figures 6 and 8). The cells were then incubated in leucine free medium supplemented with 1 μCi/ml [3H]leucine for 20 min at 37°C. The medium was then removed, and the cells were incubated twice with a 5% (wt/vol) trichloroacetic acid solution for 10 min at room temperature. Finally, acid precipitable proteins were solubilized in 0.1 M KOH, and the radioactivity was measured. The results are expressed in percentage of [3H]leucine incorporated into cells incubated without toxin. Deviations between duplicates did not vary by >10%.
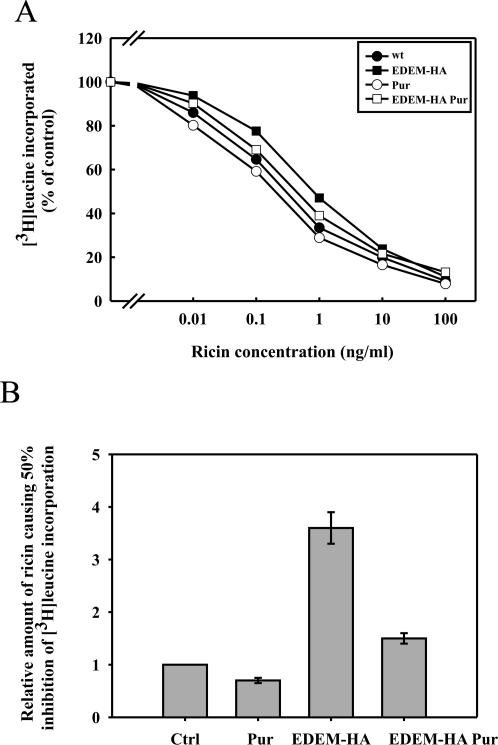
Figure 6.
EDEM-transfected cells are sensitized to ricin in the presence of puromycin. (A) Effect of puromycin on the toxicity of ricin in mock and EDEM-transfected cells. Cells were preincubated with puromycin (1 μg/ml) for 30 min before addition of increasing amounts of ricin. Protein synthesis was measured 3 h later. EDEM-HA, closed squares; mock-transfected, closed circles; EDEM-HA with puromycin, open squares; mock-transfected with puromycin, open circles. One representative example of an experiment is shown. (B) Average data with SD from five independent experiments.
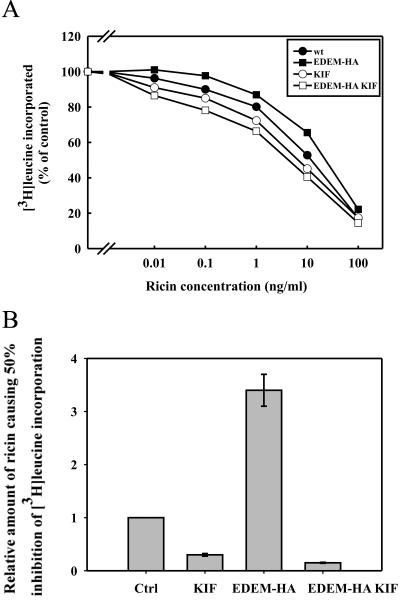
Figure 8.
EDEM-transfected cells are sensitized to ricin in the presence of the ER α-mannosidase I inhibitor kifunensin. (A) Effect of kifunensin on the toxicity of ricin in mock and EDEM-transfected cells. Cells were preincubated with kifunensin (0.2 mM) for 30 min before addition of increasing amounts of ricin. Protein synthesis was measured 3 h later (see Materials and Methods). EDEM-HA, closed squares; mock transfected, closed circles; EDEM-HA with kifunensin, open squares; and mock transfected with kifunensin, open circles. One representative example of an experiment is shown. (B) Average data with SD from five independent experiments.
Sulfation of Ricin Sulf-1 and Permeabilization of Cells
Ricin A-chain sulf-1 containing a sulfation site in the carboxy terminus was produced, purified, and reconstituted with ricin B-chain to form ricin sulf-1 as described previously (Rapak et al., 1997). Cells (1 × 105/plate) were seeded in 5-cm plates and transfected after 24 h. Three days posttransfection, cells were incubated with 0.2 mCi/ml Na2 35SO4 in DMEM without sulfate for 3 h before ∼500 ng/ml ricin sulf-1 was added, and the incubation was continued for 3 h at 37°C. In some cases, chemical inhibitors were added together with ricin sulf-1. Concentrations of inhibitors are indicated in the figure legends (Figures 7 and 9). The cells were then washed with a 0.1 M lactose solution at 37°C. For direct lysis, cells were washed once with ice-cold phosphate-buffered saline (PBS) and lysed (lysis buffer: 0.1 M NaCl, 10 mM Na2HPO4, 1 mM EDTA, and 1% Triton X-100, pH 7.4) in the presence of a protease inhibitor mixture (Roche Diagnostics, Basel, Switzerland). For permeabilization, the cells were first washed once at room tempered PBS and incubated for 5 min at room temperature with KOAc buffer (115 mM CH3COOK, 25 mM HEPES, and 2.5 mM MgCl2, pH 7.4) containing 3 μg/ml digitonin followed by a 30-min incubation on ice to allow cytosolic proteins to diffuse into the medium. The supernatants were centrifuged to remove cell debris and nuclei for 10 min at 5000 rpm in an Eppendorf centrifuge (Eppendorf, Hamburg, Germany). Sulfated ricin sulf-1 was immunoprecipitated from the supernatant with rabbit anti-ricin antibodies immobilized on protein A-Sepharose CL-4B (GE Healthcare). Finally, the beads were washed with ice-cold PBS supplemented with 0.35% Triton X-100, and the adsorbed material was analyzed by SDS-PAGE (12%) under reducing conditions. For the detection of 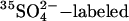 ricin, the proteins were transferred to a polyvinylidene diflouride (PVDF) membrane (Millipore, Billerica, MA) by Mini Trans-Blot cell (Bio-Rad, Hercules, CA). Dried membranes were exposed to Kodak BioMax MR film (Eastman Kodak, Rochester, NY) at room temperature. Signal intensities of the bands were quantified using ImageQuant 5.0 software (GE Healthcare).
ricin, the proteins were transferred to a polyvinylidene diflouride (PVDF) membrane (Millipore, Billerica, MA) by Mini Trans-Blot cell (Bio-Rad, Hercules, CA). Dried membranes were exposed to Kodak BioMax MR film (Eastman Kodak, Rochester, NY) at room temperature. Signal intensities of the bands were quantified using ImageQuant 5.0 software (GE Healthcare).
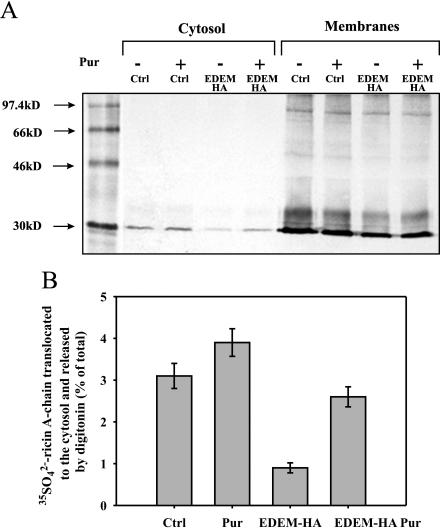
Figure 7.
Puromycin increases retrotranslocation of ricin A-chain to the cytosol in EDEM-transfected cells. Retrotranslocation of ricin A-chain to the cytosol in EDEM and mock-transfected cells. Puromycin (1 μg/ml) and 500 ng/ml ricin sulf-1 were added to cells already incubated with Na 352SO4 for 3 h, and then the incubation was continued for 3 h more. (A) One representative experiment is shown. SDS-PAGE was performed under reducing conditions after incubation of the cells with and without inhibitor. Molecular mass markers are shown on the left. (B) Average data with SD from five independent experiments. For more details about the release of cytosol by digitonin, see Materials and Methods. Pur, puromycin.
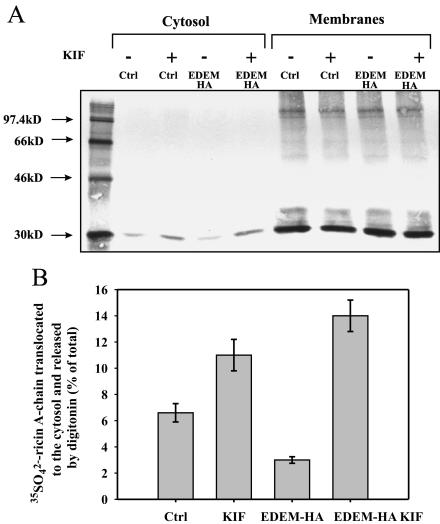
Figure 9.
Kifunensin increases retrotranslocation of ricin A-chain to the cytosol in EDEM-transfected cells. Retrotranslocation of ricin A-chain to the cytosol in EDEM and mock-transfected cells. Kifunensin (0.2 mM) and 500 ng/ml ricin sulf-1 were added to cells already incubated with Na 352SO4 for 3 h and then the incubation was continued for the next 3 h (see Materials and Methods). (A) One representative experiment is shown. SDS-PAGE was performed under reducing conditions after incubation of the cells with and without inhibitor. Molecular mass markers are shown on the left. (B) Average data with SD from five independent experiments. For more details about the release of cytosol by digitonin, see Materials and Methods. KIF, kifunensin.
The radioactivity in the cell lysates was measured to detect possible differences in the total amount of isotope incorporated under the different conditions (our unpublished data). The efficiency of permeabilization was estimated by measuring release of the cytosolic enzyme lactate dehydrogenase (LDH) to the cytosolic fraction as well as in the remaining fraction. Measurements were performed in Technicon SRA 2000 (Technicon Instrument, Tarrytown, NY) equipment following the protocol provided by the manufacturer. The data obtained indicate that 20–25% of the total cellular LDH was present in the extracted cytosolic fraction. The effect of cell permeabilization on the release of resident ER proteins has been studied by Western blotting analysis performed in every experiment. Absence of the ER proteins calnexin and calreticulin in the cytosolic fraction confirmed that digitonin, used at an appropriate low concentration (3 μg/ml), forms pores selectively in the plasma membrane without disturbing the ER membrane.
RNA Isolation and Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, CA) was used for total RNA extraction, following the protocol provided by the manufacturer. One microgram of total RNA was then reverse transcribed into cDNA using iScript cDNA synthesis kit (Bio-Rad). For amplification, 10 ng of cDNA was mixed with iQ SYBR Green Supermix (Bio-Rad) and with 0.3 μM of each primer: forward, 5′-GCAGGTGCTAG-3′; and reverse, 5′-CAGTTATATCTCTC-3′. The PCR reaction was performed in an iCycler thermal cycler with an iCycler IQ real-time detection system (Bio-Rad) under the following conditions: 3 min 95°C to activate the enzyme followed by: 50 cycles at 95°C denaturation for 10 s and 48°C annealing and extension for 35s. A melt-curve analysis was added to exclude nonspecific amplification. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control. Primers for GAPDH amplification were as follows: forward, 5′-GGAAGG TGAAGGTCGGAGGTC-3′; and reverse, 5′-GGAAGATGGTGATGGGATTTC-3′. In all experiments, cells were harvested for real-time RT-PCR 3 d after transfection.
Immunoprecipitation
HEK293 cells (8 × 105/plate) were seeded in 10-cm plates and transfected after 24 h with either the EDEM cDNA or a control empty vector. Three days posttransfection, cells were incubated with 80 ng/ml 125I-labeled ricin for 3 h. Ricin was 125I-labeled according to Fraker and Speck (1978) to a specific activity of 3 × 104–5 × 104 cpm/ng. Optionally, cells were preincubated with inhibitors for 30 min before 125I-ricin was added. Concentrations of inhibitors are indicated in the legend to Figure 10. The cells were washed with 0.1 M lactose solution at 37°C and then with ice-cold HEPES-buffered saline (HBS: 20 mM HEPES and 150 mM NaCl, pH 6.8) and lysed in a buffer containing 2% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS) in HBS, pH 6.8, 20 mM N-ethylmalemide, and protease inhibitor mixture (Roche Diagnostics, Basel). The supernatants were centrifuged to remove cell debris and nuclei for 10 min at 14,000 rpm in an Eppendorf centrifuge (Eppendorf). Lysates were immunoprecipitated for 1 h at 4°C using mouse anti-HA or rabbit anti-Sec61α antibody coupled to protein G Dyna beads (Dynal Biotech, Oslo, Norway). The beads were washed three times with HBS buffer, pH 6.8, containing 0.5% CHAPS and 0.1% Tween 20. Samples were resolved by 12% nonreducing SDS-PAGE. For the detection of 125I-labeled ricin, the immunoprecipitated proteins were transferred to a PVDF membrane (Millipore) by Mini Trans-Blot cell (Bio-Rad). Dried membranes were exposed to Kodak BioMax MS film (Eastman Kodak) at –80°C. Signal intensities of the bands were quantified using ImageQuant 5.0 software (GE Healthcare). The same membranes were reprobed with either anti-HA or anti-Sec61α antibodies.
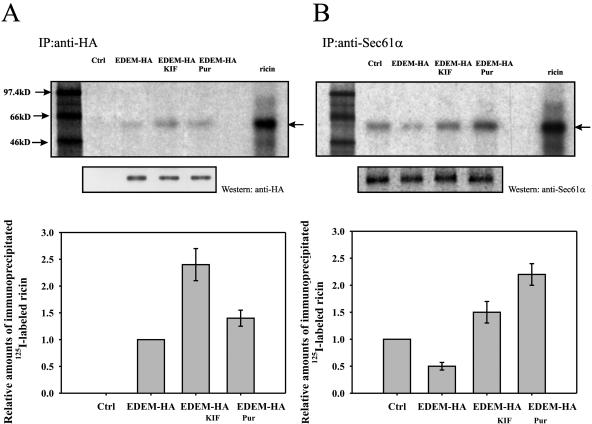
Figure 10.
Ricin coimmunoprecipitates with EDEM and with Sec61α. Immunoprecipitation of 125I-labeled ricin with anti-HA (A) or anti-Sec61α antibody (B). Kifunensin (0.2 mM) or puromycin (1 μg/ml) were added to EDEM-transfected cells 30 min before addition of 80 ng/ml 125I-labeled ricin, and then the incubation was continued for the next 3 h (see Materials and Methods). Top, representative examples of experiments. SDS-PAGE was performed under nonreducing conditions after incubation of the cells with and without inhibitors. Molecular mass markers are shown on the left. Membranes were reprobed with anti-HA or anti-Sec61α antibody to confirm equal immunoprecipitation. Bottom, average data with SD from five independent experiments. The level of immunoprecipitated ricin from EDEM-transfected cells (A) or mock-transfected cells (B) is marked as 1, and the amounts of immunoprecipitated ricin under other conditions are plotted as relative values compared with this number. The arrows indicate holotoxin. KIF, kifunensin; Pur, puromycin.
RESULTS
High Expression of EDEM Protects against Ricin
To investigate whether EDEM, which is involved in ERAD, is also involved in ricin transport to the cytosol, HEK293 cells overexpressing EDEM-HA and control cells were incubated with different concentrations of ricin for 3–4 h, and the protein synthesis was then measured. As shown in Figure 1A, overexpression of EDEM protects 3–4 times (IC50) against ricin toxicity in HEK293 cells.
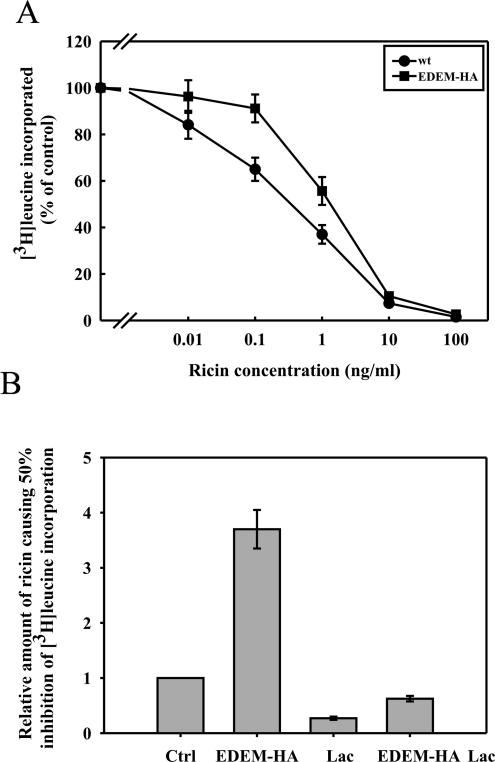
Figure 1.
High expression of EDEM protects against ricin toxicity. (A) The effect of overexpression of EDEM on the toxicity of ricin. EDEM-HA, closed squares; mock-transfected, closed circles. (B) Effect of the proteasomal inhibitor lactacystin on the toxicity of ricin in mock- and EDEM-transfected cells. The level of 50% incorporation of [3H]leucine in mock-transfected cells without lactacystin is marked as 1, and the concentration of ricin required to induce a similar inhibition under other conditions are plotted as relative values compared with this control (ctrl). The other columns represent: EDEM-HA, cells transfected with HA-tagged EDEM; Lac, mock-transfected cells incubated with lactacystin; and EDEM-HA Lac, cells transfected with EDEM-HA and incubated with lactacystin. Columns below control represent sensitization, and columns above the control represent protection against ricin. Data with SD from five independent experiments.
There was a possibility that the observed protection against ricin in EDEM-transfected cells might be because of increased degradation of ricin in the cytosol. It has been show previously that ricin degradation can be inhibited by lactacystin (Wesche et al., 1999), which inhibits degradation by proteosomes and by a newly discovered protease complex (Geier et al., 1999). However, EDEM-expressing cells were 2–3 times less sensitive to ricin than control cells even in the presence of lactacystin (Figure 1B), which sensitized both control and EDEM-expressing cells to the toxin. Thus, these data suggest that the observed protection of EDEM-transfected cells was not because of an increased degradation of ricin after retrotranslocation to the cytosol. As expected, another toxin, diphtheria toxin, which enters from acidified endosomes (Sandvig and Olsnes, 1981), was equally toxic to control and EDEM-transfected cells (our unpublished data).
High Expression of EDEM Does Not Affect the Reductive Release of Ricin A-Chain but Decreases Retrotranslocation of Ricin A-Chain into the Cytosol
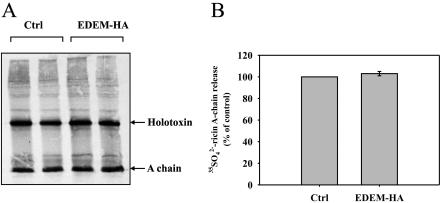
To investigate whether high expression of EDEM affects the reduction of the disulfide bond and the release of A-chain from the holotoxin, EDEM-transfected cells were incubated with ricin sulf-1 (Rapak et al., 1997), a modified ricin molecule containing a sulfation site in the A-chain. By incubating cells in the presence of 35SO2–4, the A-chain becomes radioactively labeled due to the sulfotransferase in the TGN, and one can then study the fate of the 35SO2–4-labeled ricin molecule. As shown in Figure 2, no significant effect of EDEM expression on A-chain release was observed after 3 h. The same was the case after 90 min with the toxin (our unpublished data). Thus, overexpression of EDEM does not have any effect on the reduction of the disulfide bond connecting the A- and B-chain of the toxin.
Figure 2.
High expression of EDEM does not affect release of the ricin A-chain. Tyrosine sulfated ricin sulf-1 in EDEM- and mock-transfected cells after 3-h incubation with the toxin run on SDS-PAGE under nonreducing conditions. (A) One representative gel is shown. (B) Data with SD from five independent experiments. The experiments were done in duplicates.
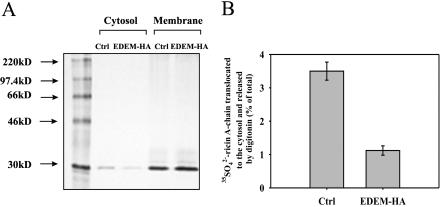
Because high expression of EDEM did not affect the release of the A-chain, we wanted to examine whether the observed protection of EDEM-transfected cells against ricin was because of decreased retrotranslocation of toxin from the ER to the cytosol. Cells treated with 35SO2–4 sulfate for 3 h and then incubated with ricin sulf-1 for 3 h were subjected to permeabilization with digitonin to separate the cytosolic fraction from the ER membranes. As shown in Figure 3, the cytosolic fraction of EDEM transfected cells contains an ∼3 times lower amount of ricin A-chain than mock-transfected cells. This was not because of reduced ricin sulfation, because both EDEM and mock-transfected cells contain equal amounts of ricin in membrane fractions (Figure 3A).
Figure 3.
High expression of EDEM reduces retrotranslocation of ricin A-chain to the cytosol. Sulfated ricin A-chain in the cytosol and membrane fractions (ER and Golgi) of EDEM and mock-transfected cells. (A) One representative experiment is shown. The incubations and SDS-PAGE were performed as described in Materials and Methods. Molecular mass markers are indicated on the left. (B) Average data with SD from five independent experiments. For more details about the release of cytosol by digitonin, see Materials and Methods.
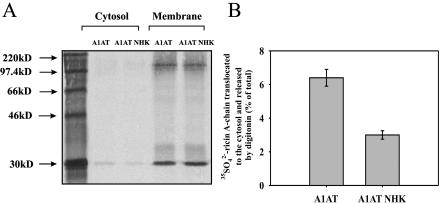
A possible explanation for the reduced retrotranslocation of ricin to the cytosol could be that the high expression of EDEM increases extraction of misfolded proteins from the calnexin cycle (Molinari et al., 2003) and increases ERAD, thereby inhibiting access of ricin to the translocon. It has been shown that overexpression of EDEM increases ERAD by accelerating the degradation of model misfolded proteins (Hosokawa et al., 2001; Molinari et al., 2003). To investigate whether increased ERAD can lead to decrease in retrotranslocation of ricin A-chain from the ER to the cytosol, we have studied transport of 35SO2–4-labeled ricin after overproduction of model misfolded proteins. HEK293 cells were transfected with the β-site amyloid precursor protein cleaving enzyme isoform, membrane BACE 457, luminal BACE 457Δ (Molinari et al., 2003), and with α1-antitrypsin (A1AT) variant null (Hong Kong, NHK; Hosokawa et al., 2001). For the control experiments, cells were transfected with properly folded BACE 501 and A1AT wild type. BACE 457, BACE 457Δ, and A1AT NHK become misfolded in the ER and are subsequently directed to degradation by the cytoplasmic proteasome. Permeabilization-sulfation experiments, which were performed as described previously, revealed that retrotranslocation of ricin A-chain to the cytosol is more than twofold decreased in cells expressing high level of A1AT NHK (Figure 4). This decrease was counteracted by kifunensin (our unpublished data). Similar results were obtained after overproduction of BACE 457 and BACE 457Δ (our unpublished data). Thus, increased ERAD can decrease ricin retrotranslocation to the cytosol. Overexpression of A1AT NHG does not have any effect on the reduction of the disulfide bond connecting the A- and B-chain of the toxin (our unpublished data).
Figure 4.
High expression of α1-anti-trypsin null Hong Kong mutant reduces retrotranslocation of ricin A-chain to the cytosol. Sulfated ricin A-chain in the cytosol and membrane fractions (ER and Golgi) of cells transfected with α1-anti-trypsin and α1-anti-trypsin null Hong Kong. (A) One representative experiment is shown. The incubations and SDS-PAGE were performed as described in Materials and Methods. Molecular mass markers are indicated on the left. (B) Average data with SD from five independent experiments. For more details about the release of cytosol by digitonin, see Materials and Methods. A1AT, α1-anti-trypsin; A1AT NHK, α1-anti-trypsin null Hong Kong mutant.
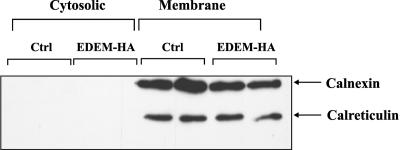
Finally, as a control, we tested whether the ER proteins remained in the ER after permeabilization. Therefore, we studied the distribution of calnexin, an ER membrane protein and calreticulin, a soluble ER protein. As shown in Figure 5, these protein markers were not released from the ER to the cytosolic fraction, indicating that ER membranes remained intact. Calculations based on detection levels of the ER proteins in our assay revealed that a possible leakage from the ER must account for <0.4% of total proteins present in the ER membranes (our unpublished data). In contrast, up to ∼14% of total ricin A-chain present in the ER was translocated to the cytosolic fraction in our experiments cells. It should be noted that only ∼20–25% of the cytosol was released by our mild digitonin-treatment (see Materials and Methods). Thus, presence of ricin in the cytosolic fraction cannot be due to digitonin-induced leakage from the ER.
Figure 5.
Calnexin and calreticulin are not released to the cytosolic fraction. Distribution of the resident ER proteins, calnexin and calreticulin after permeabilization with digitonin. As demonstrated by SDS-PAGE and Western blot with anti-calnexin and anti-calreticulin antibody, there was no visible leakage of calnexin and calreticulin to the cytosolic fraction. The conditions were the same as those described in Figure 3. The experiments were done in duplicates.
Retrotranslocation of Ricin to the Cytosol Is Affected by Inhibition of Protein Synthesis
To further investigate the mechanism for EDEM-induced protection and decreased retrotranslocation of ricin, we performed several experiments in the presence of puromycin and kifunensin. By inhibiting the import of proteins into ER one might expect the Sec61 channels to be more accessible for export of ricin to the cytosol. Puromycin inhibits protein synthesis by blocking peptidyl transferase. It releases nascent chains from ribosomes but leaves the ribosomes bound to the ER membrane with the protein channels open (Simon and Blobel, 1991). In fact, puromycin was found to sensitize both control and EDEM-transfected cells to ricin (Figure 6, A and B). Importantly, in EDEM-transfected cells the observed sensitization was almost fivefold higher than the sensitization seen in mock-transfected cells (Figure 6, A and B). These results suggest that EDEM promotes retrotranslocation of ricin A-chain to the cytosol under puromycin treatment. There might be a competition between ricin, newly synthesized proteins, and misfolded proteins for retrotranslocation across the ER membrane.
To investigate more directly whether puromycin affected the transport of ricin to the cytosol, we studied retrotranslocation of 35SO2–4-ricin A-chain in the presence of this inhibitor. As before, we permeabilized the plasma membrane with digitonin to measure translocation of toxin to the cytosol. Control experiments confirmed that the permeabilization procedure did not generate leakage of the residual ER proteins out of the ER (our unpublished data). Incubation of EDEM and mock-transfected cells with puromycin resulted in increased retrotranslocation of ricin A-chain to the cytosol (Figure 7, A and B). However, in EDEM-transfected cells puromycin increased the retrotranslocation of ricin A-chain much more than in control cells (Figure 7, A and B). The increased retrotranslocation of ricin to the cytosol in the presence of puromycin could be because of lack of competition between EDEM and newly synthesized proteins, but it could also be because puromycin keeps the channels in an open state (Simon and Blobel, 1991). These results are consistent with toxicity experiments and support the idea that after treatment with puromycin, when EDEM is overproduced, more ricin is retrotranslocated to the cytosol, thereby increasing its toxic effect. Thus, these data suggest that EDEM might target ricin to the translocon.
To strengthen this hypothesis, we investigated the effect of specific inhibition of mannose trimming on ricin toxicity and retrotranslocation of 35SO2–4-labeled ricin. We used kifunensin, which specifically inhibits ER α-mannosidase I. Trimming of Man9 to the B isoform of Man8 catalyzed by ER mannosidase I is a critical step in ERAD of misfolded proteins (Tokunaga et al., 2003). Kifunensin inhibits recognition of ERAD candidates by EDEM and thereby delays both the release of misfolded proteins from the calnexin cycle and the degradation of the ERAD candidates (Molinari et al., 2003). Kifunensin treatment might therefore increase the ability of EDEM to recognize and interact with ricin. In agreement with this hypothesis, toxicity experiments revealed that EDEM-transfected cells became highly sensitive to ricin upon addition of kifunensin, and importantly, they became even more sensitive than control cells treated with kifunensin (Figure 8, A and B). The observed effect was not because of the changed viability of the cells, because protein synthesis measured in samples without ricin addition was equal both in the control and EDEM-transfected cells treated with kifunensin (our unpublished data).
Studies of retrotranslocation of 35SO2–4-labeled ricin revealed that kifunensin induced an ∼50% increase in A-chain retrotranslocation to the cytosol in control cells. However, in EDEM-transfected cells we observed an even stronger increase (∼5-fold; Figure 9, A and B). Thus, a considerable fraction of ricin was retrotranslocated to the cytosol in cells expressing EDEM in the presence of kifunensin. These data show that EDEM can promote retrotranslocation of ricin to the cytosol when the interaction between EDEM and misfolded proteins is disrupted.
Ricin Coimmunoprecipitates with EDEM and with Sec61α
To confirm that EDEM is directly involved in ricin retrotranslocation from the ER to the cytosol, we analyzed the interaction between ricin and EDEM and between ricin and the Sec61p translocon. We coimmunoprecipitated 125I-labeled ricin with anti-HA or anti-Sec61α antibodies from lysates of EDEM-HA and mock-transfected HEK293 cells. Also, EDEM-transfected cells were incubated with kifunensin and puromycin (see Materials and Methods) to investigate whether these two inhibitors affected interactions between EDEM or the Sec61p translocon and ricin. As shown in Figure 10, ricin coimmunoprecipitates both with EDEM and with Sec61α (Figure 10, A and B). Importantly, 2.5-fold more ricin interacts with EDEM after kifunensin treatment compared with the nontreated EDEM-transfected cells (Figure 10A). These data confirm that when the interaction between EDEM and misfolded proteins is inhibited, more EDEM is able to interact with ricin. Importantly, after kifunensin treatment, not only is there an increased interaction of ricin with EDEM, but more ricin is coimmunoprecipitated with the Sec61 translocon. This was shown by incubating cells with 125I-labeled ricin (Figure 10B). As shown, also puromycin increased the interactions between EDEM and ricin and between the Sec61p and ricin. Treatment of HA-EDEM-transfected cells with puromycin resulted in a 40% increase in the amount of ricin coimmunoprecipitated with anti-HA antibody whereas puromycin had an even larger effect on the amount of ricin coimmunoprecipitated with the Sec61α antibody (compared with the amount of ricin found in untreated EDEM-transfected cells) (Figure 10, A and B). On the other hand, Sec61p is less accessible for ricin when EDEM is overproduced and the cells are not treated with the inhibitors. As shown in Figure 10B, there was an ∼50% decrease in the amount of ricin coimmunoprecipitated with Sec61α in EDEM-transfected cells in comparison with mock-transfected cells. Thus, overexpression of EDEM decreases the interaction of ricin with the Sec61p translocon. This finding is in agreement with our data showing that after EDEM overexpression less ricin is retrotranslocated to the cytosol (Figure 3). Control experiment with a monoclonal mouse antibody targeted against c-myc confirmed that the observed immunoprecipitation of ricin was not because of unspecific binding of ricin to either antibodies or beads (our unpublished data). Together, these results show that EDEM directly interacts with ricin and this interaction seems critical for ricin retrotranslocation from the ER to the cytosol.
Reduced Amounts of EDEM Decrease Retrotranslocation of Ricin A-Chain to the Cytosol
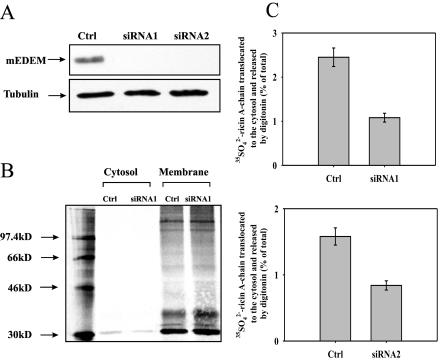
To reduce the intracellular concentration of EDEM and determine the effect of this down-regulation on ricin A-chain retrotranslocation, RNA interference (RNAi) was used. We transfected HEK293 cells with two different vectors expressing short hairpin RNA (see Materials and Methods). The two siRNA constructs were designed against both mouse and human EDEM. We tested the efficiency of our constructs by cotransfection with HA-tagged mouse EDEM. Figure 11A demonstrates a very efficient knockdown of mouse EDEM for both constructs. Reduction in expression of endogenous human EDEM was confirmed by real-time RT-PCR, which revealed that there was from 70 to 95% reduction in the mRNA level of this gene (our unpublished data).
Figure 11.
Retrotranslocation of ricin A-chain into the cytosol is decreased in cells with reduced level of EDEM. (A) The presence of EDEM-HA was determined by Western blot analysis with anti-HA antibody. Cells were cotransfected with HA-tagged mouse EDEM and siRNA constructs against EDEM. Membranes were reprobed with anti-tubulin antibody for equal loading control. (B) Sulfated ricin A-chain in the cytosol and membrane fractions of siRNA and mock-transfected cells. A representative example of the gel with siRNA construct number 1 (siRNA1). SDS-PAGE was performed under reducing conditions. Molecular mass markers are shown on the left. (C) Average data with SD from five independent experiments with siRNA1 and siRNA2. For more details about the release of cytosol by digitonin, see Materials and Methods.
Experiments with 35SO2–4-labeled ricin performed with cells transfected with siRNA vectors showed that a reduced amount of EDEM decreases more than twofold the retrotranslocation of ricin A-chain to the cytosol (Figure 11, B and C). This is not because of a reduced level of Sec61 in cells transfected with siRNA against EDEM, because the amount of Sec61 was equal in these cells compared with control cells (our unpublished data). We also observed a twofold protection against ricin toxicity in cells with reduced levels of EDEM (our unpublished data). These results are in agreement with the suggestion that EDEM is directly involved in retrotranslocation of ricin from the ER to the cytosol.
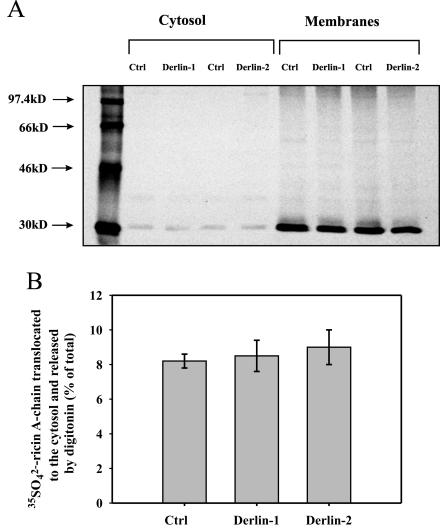
Retrotranslocation of Ricin A-Chain from the ER to the Cytosol Is Derlin-1 and Derlin-2 Independent
It was previously shown that Derlin-1 is involved in the retrotranslocation of major histocompatibility complex (MHC) class I heavy chains from the ER to the cytosol (Ye et al., 2004; Lilley and Ploegh, 2004). It is likely that this protein also plays a more general role in the extraction of certain aberrantly folded proteins from the ER. Derlin-2 is a protein related to Derlin-1 (Ye et al., 2004; Lilley and Ploegh, 2004). Recent studies have provided evidence that Derlin-2 can be associated with retrotranslocation out of the ER, in a similar way as the Derlin-1 protein (Lilley and Ploegh, 2005). To investigate whether Derlin-1 and Derlin-2 are involved in ricin retrotranslocation to the cytosol, we have studied the transport of 35SO2–4–ricin A-chain from the ER to the cytosol in HeLa cells stably transfected with an empty vector and dominant-negative constructs of Derlin-1 (Derlin-1GFP) and Derlin-2 (Derlin-2GFP) (Lilley and Ploegh, 2004). As described in Materials and Methods, the cell lines were checked for the expression of the constructs. As described above, we permeabilized the plasma membrane with digitonin to separate the cytosolic fraction from the ER membranes. Again, control experiments showed that our mild digitonin treatment did not cause leakage of the ER proteins (our unpublished data). Importantly, there were no significant differences in the retrotranslocaton of ricin A-chain to the cytosol between mock and Derlin-1GFP– or Derlin-2GFP–transfected cells (Figure 12). Thus, Derlin-1 and Derlin-2 do not seem to be involved in the retrotranslocation of ricin A-chain from the ER to the cytosol. However, we cannot exclude that other protein channels in addition to Sec61p might be involved in ricin retrotranslocation to the cytosol.
Figure 12.
Retrotranslocation of ricin to the cytosol is not affected by dominant negative versions of Derlin-1 and Derlin-2 proteins. Retrotranslocation of ricin A-chain to the cytosol in Derlin-1GFP and Derlin-2GFP–transfected cells and mock-transfected cells (see Materials and Methods). Ricin sulf-1 (500 ng/ml) was added to cells already incubated with Na 352SO4 for 3 h, and then the incubation was continued for the next 3 h (see Materials and Methods). (A) One representative experiment is shown. SDS-PAGE was performed under reducing conditions. Molecular mass markers are shown on the left. (B) Average data with SD from five independent experiments. For more details about the release of cytosol by digitonin, see Materials and Methods. Derlin-1, Derlin-1GFP; Derlin-2, Derlin-2GFP.
DISCUSSION
The results presented in this study indicate that EDEM is directly involved in retrotranslocation of ricin from the ER to the cytosol. Ricin coimmunoprecipitates both with EDEM and Sec61α (the major component of the translocation complex). Experiments with RNA-mediated interference against EDEM showed that a reduced level of EDEM decreased retrotranslocation of ricin to the cytosol. Moreover, our data indicate that EDEM can promote retrotranslocation of ricin to the cytosol when the interaction between EDEM and misfolded proteins is disrupted.
In our study, we also used HEK293 cells with overexpression of HA-tagged mouse EDEM (Hosokawa et al., 2001). We confirmed that EDEM-HA, as shown previously for COS-7 cells (Hosokawa et al., 2001), was localized in the ER. The protein was found to colocalize with the ER chaperone calnexin but not with the Golgi marker TGN46 in HEK293 cells (our unpublished data).
Our results demonstrate that overexpression of EDEM decreases retrotranslocation of ricin A-chain to the cytosol. This was shown both by quantifying the amount of sulfated ricin that reached the cytosol and by measuring the toxicity of ricin. In the permeabilization-sulfation experiments, the RTA present in the cytosolic fraction released by digitonin does not represent the total amount of A-chain translocated the cytosol, because our mild digitonin treatment released only 20–25% of the cytosol. A low concentration of digitonin was used to avoid leakage from other organelles. Thus, sulfated RTA present in the “membrane fraction” consists of RTA not only in the ER but also of RTA partially or completely translocated to the cytosol. RTA in the cytosol might to some extent be polyubiquitinated, and this could be the reason why in some figures RTA in the membrane fractions occurs as a smear. Because ubiquitination of ricin is not required for retrotranslocation to the cytosol (Deeks et al., 2002), we did not investigate this topic in more detail. The bands showing ricin in the membrane fraction were in some cases saturated. This was because of the long exposure of the film required to show differences in the amount of ricin present in the cytosol. We have, however, quantified data from a large number of experiments (3–10), and we have also studied and quantified bands that were not saturated. Such measurements confirm the data presented.
If ricin interacts with EDEM before retrotranslocation to the cytosol, one might have expected that overexpression of EDEM would increase toxin transport to the cytosol and sensitize the cells to ricin. However, high levels of EDEM are likely to increase binding to Man8 residues of misfolded proteins and thereby also increase retrotranslocation of such proteins through the ER membrane. An increased transport of misfolded proteins could in this manner occupy channels otherwise used for retrotranslocation of ricin. This suggestion was supported by the observation that after overproduction of model misfolded proteins retrotranslocation of ricin to the cytosol was significantly decreased.
The protein synthesis inhibitor puromycin keeps the translocation channels open (Simon and Blobel, 1991) and revealed much larger sensitization of EDEM-expressing cells than control cells in toxicity experiments. These differences were reflected by the amount of ricin found in the cytosol. Nevertheless, puromycin treatment might give rise to unfinished nascent chains of secretory and membrane proteins, and these truncated and folding-incompetent polypeptides in the ER could possibly compete with RTA for EDEM binding. However, immunoprecipitation experiments showed that more ricin could interact with EDEM after puromycin treatment.
Kifunensin, which prevents binding of misfolded proteins to EDEM, increased transport of ricin to the cytosol to a larger extent in EDEM-transfected cells than in control cells, demonstrating that EDEM promotes retrotranslocation of ricin in the presence of this inhibitor. Kifunensin would prevent misfolded proteins from filling the ER channels, and immunoprecipitation experiments revealed that kifunensin significantly increased the interactions between ricin and EDEM and between ricin and the Sec61p.
How can ricin interact with EDEM? Ricin A-chain purified from plants contains two N-linked oligosaccharide chains (Rutenber and Robertus, 1991). However, these are not essential for the protection against ricin provided by overexpressed EDEM because there was a similar protection against ricin sulf-1 (our unpublished data), which does not contain carbohydrates in the A-chain (modified ricin A-chain produced in bacteria lacks oligosaccharides). This result is important because ricin sulf-1 was used in all permeabilization-sulfation experiments. It is possible that ricin could interact with EDEM by its B-chain. An interaction between the galactose-specific binding site of RTB and galactosyl groups present on the ER chaperone calreticulin has been described previously (Day et al., 2001). However, this does not seem to be the case for the ricin–EDEM interaction. Ricin could still be immunoprecipitated with EDEM after blocking the RTB galactose-specific binding site by lactose (our unpublished data). Moreover, the fact that inhibition of interaction between EDEM and misfolded proteins increases the ricin-EDEM interaction suggests that ricin interacts with EDEM in a different way than misfolded proteins. It is possible that ricin interacts with EDEM in a carbohydrate-independent manner. EDEM has in fact been demonstrated to interact directly with proteins via hydrophobic domains (Wang and Hebert, 2003).
Although siRNA against EDEM quite efficiently reduced the level of mouse EDEM expressed in the HEK293 cells and also strongly decreased the mRNA level of endogenous EDEM, retrotranslocation of ricin was reduced no more than twofold. One might have expected a stronger effect. There are several possible explanations for that. It is important to remember that ricin very efficiently inhibits protein synthesis after retrotranslocation to the cytosol. Therefore, few molecules are sufficient to inhibit protein synthesis, and the EDEM still present after siRNA treatment might be sufficient for the retrotranslocation obtained. Furthermore, several recent studies have provided evidence that there is more than one isoform of EDEM. In addition to the EDEM1 variant protein (Hosokawa et al., 2001), EDEM2 and EDEM3 homologues also exist in the ER lumen (Mast et al., 2004; Olivari et al., 2005). When overexpressed, EDEM2 accelerates ERAD of terminally misfolded glycoproteins by facilitating their extraction from the calnexin cycle. Both EDEM2 and EDEM3 could be involved in ricin retrotranslocation. It is possible that ricin as well as other proteins may use several channels/translocons for their transport to the cytosol. Ricin (Wesche et al., 1999) as well as other protein toxins (Koopmann et al., 2000; Schmitz et al., 2000) have been shown to interact with the Sec61p complex, and it has been assumed that it is the main pathway used by these toxins to enter the cytosol. However, it has now been reported that there are several pathways involved in transporting misfolded proteins from the ER to the cytosol. Similarly, ricin may exploit more than one mechanism. Recent studies have provided evidence that Derlin-1 and Derlin-2 are important factors for the extraction of aberrantly folded proteins from the ER (Lilley and Ploegh, 2004, 2005; Ye et al., 2004). Our studies revealed however, that ricin retrotranslocation to the cytosol are not dependent on Derlin-1 and Derlin-2 proteins.
In conclusion, our data demonstrate a novel role for EDEM. The results suggest that EDEM, which seems to be crucial for extraction of terminally misfolded proteins from the calnexin cycle (Molinari et al., 2003; Oda et al., 2003), directing them for ERAD, is also important for ricin retrotranslocation to the cytosol.
Acknowledgments
We thank Jorunn Jakobsen, Anne Engen, Anne-Gro Bergersen, Hilde Raa, Hanne Rønning, and Guro Johansen for excellent technical assistance. We are grateful to Prof. K. Nagata and Dr. N. Hosokawa for cDNA encoding the mouse EDEM. We are grateful to Dr. M. Molinari (Institute for Research in Biomedicine, Bellinzona, Switzerland) and Dr. P. Paganetti for cDNA encoding α1-antitrypsin wt and Hong Kong mutant, BACE 501, BACE 457, and BACE 457Δ. We are grateful to Dr. H. Ploegh (Harvard Medical School, Boston, MA) for cells transfected with Derlin-1GFP and Derlin-2GFP. We thank Dr. Hege Holte Slagsvold and Audrun Utskarpen for help with the real-time RT-PCR. The present study was supported by the Norwegian Cancer Society, the Norwegian Research Council for Science and the Humanities, the Jahre Foundation, Jeanette and Søren Bothners legacy, and by the National Programme for Research in Functional Genomics in Norway. M.S.-W. was supported by European Molecular Biology Organization postdoctoral long-term fellowship ALTF 49-2003. S. W. was supported by Federation of European Biochemical Societies and Fond National Suisse de la Recherche Scientifique from 2003 to 2004 and since 2004 by a postdoctoral fellowship from the Medical Faculty of Oslo.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-10-0961) on February 1, 2006.
References
- Day, P. J., Owens, S. R., Wesche, J., Olsnes, S., Roberts, L. M., and Lord, J. M. (2001). An interaction between ricin and calreticulin that may have implications for toxin trafficking. J. Biol. Chem. 276, 7202–7208. [DOI] [PubMed] [Google Scholar]
- Deeks, E. D., Cook, J. P., Day, P. J., Smith, D. C., Roberts, L. M., and Lord, J. M. (2002). The low lysine content of ricin A chain reduces the risk of proteolytic degradation after translocation from the endoplasmic reticulum to the cytosol. Biochemistry 41, 3405–3413. [DOI] [PubMed] [Google Scholar]
- Fraker, P. J., and Speck, J. C., Jr. (1978). Protein and cell membrane iodinations with a sparingly soluble chloroamide, 1,3,4,6-tetrachloro-3a,6a-diphrenylglycoluril. Biochem. Biophys. Res. Commun. 80, 849–857. [DOI] [PubMed] [Google Scholar]
- Geier, E., Pfeifer, G., Wilm, M., Lucchiari-Hartz, M., Baumeister, W., Eichmann, K., and Niedermann, G. (1999). A giant protease with potential to substitute for some functions of the proteasome. Science 283, 978–981. [DOI] [PubMed] [Google Scholar]
- Grimmer, S., Ying, M., Walchli, S., van, D. B., and Sandvig, K. (2005). Golgi vesiculation induced by cholesterol occurs by a dynamin- and cPLA-dependent mechanism. Traffic 6, 144–156. [DOI] [PubMed] [Google Scholar]
- Hosokawa, N., Wada, I., Hasegawa, K., Yorihuzi, T., Tremblay, L. O., Herscovics, A., and Nagata, K. (2001). A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmann, J. O., Albring, J., Huter, E., Bulbuc, N., Spee, P., Neefjes, J., Hammerling, G. J., and Momburg, F. (2000). Export of antigenic peptides from the endoplasmic reticulum intersects with retrograde protein translocation through the Sec61p channel. Immunity 13, 117–127. [DOI] [PubMed] [Google Scholar]
- Lilley, B. N., and Ploegh, H. L. (2004). A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429, 834–840. [DOI] [PubMed] [Google Scholar]
- Lilley, B. N., and Ploegh, H. L. (2005). Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 102, 14296–14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast, S. W., Diekman, K., Karaveg, K., Davis, A., Sifers, R. N., and Moremen, K. W. (2004). Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 15, 421–436. [DOI] [PubMed] [Google Scholar]
- Molinari, M., Calanca, V., Galli, C., Lucca, P., and Paganetti, P. (2003). Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science 299, 1397–1400. [DOI] [PubMed] [Google Scholar]
- Oda, Y., Hosokawa, N., Wada, I., and Nagata, K. (2003). EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science 299, 1394–1397. [DOI] [PubMed] [Google Scholar]
- Olivari, S., Galli, C., Alanen, H., Ruddock, L., and Molinari, M. (2005). A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 280, 2424–2428. [DOI] [PubMed] [Google Scholar]
- Rapak, A., Falnes, P. O., and Olsnes, S. (1997). Retrograde transport of mutant ricin to the endoplasmic reticulum with subsequent translocation to cytosol. Proc. Natl. Acad. Sci. USA 94, 3783–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutenber, E., and Robertus, J. D. (1991). Structure of ricin B-chain at 2.5 A resolution. Proteins Struct. Funct. Genet. 10, 260–269. [DOI] [PubMed] [Google Scholar]
- Sandvig, K., Lauvrak, S. U., and van Deurs, B. (2005). Host cell penetration and trafficking of protein toxins. In: Microbial Toxins: Molecular and Cellular Biology, ed. Thomas Proft, Norfolk, NJ: Horizon Press, 473–496.
- Sandvig, K., and Olsnes, S. (1981). Rapid entry of nicked diphtheria toxin into cells at low pH. Characterization of the entry process and effects of low pH on the toxin molecule. J. Biol. Chem. 256, 9068–9076. [PubMed] [Google Scholar]
- Sandvig, K. and Olsnes, S. (1979). Effect of temperature on the uptake, excretion and degradation of abrin and ricin by HeLa cells. Exp. Cell Res. 121, 15–25. [DOI] [PubMed] [Google Scholar]
- Schmitz, A., Herrgen, H., Winkeler, A., and Herzog, V. (2000). Cholera toxin is exported from microsomes by the Sec61p complex. J. Cell Biol. 148, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, S. M., and Blobel, G. (1991). A protein-conducting channel in the endoplasmic reticulum. Cell 65, 371–380. [DOI] [PubMed] [Google Scholar]
- Spooner, R. A., Watson, P. D., Marsden, C. J., Smith, D. C., Moore, K. A., Cook, J. P., Lord, J. M., and Roberts, L. M. (2004). Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem. J. 383, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga, F., Hara, K., and Koide, T. (2003). N-linked oligosaccharide processing, but not association with calnexin/calreticulin is highly correlated with endoplasmic reticulum-associated degradation of antithrombin Glu313-deleted mutant. Arch. Biochem. Biophys. 411, 235–242. [DOI] [PubMed] [Google Scholar]
- Wang, T., and Hebert, D. N. (2003). EDEM an ER quality control receptor. Nat. Struct. Biol. 10, 319–321. [DOI] [PubMed] [Google Scholar]
- Wesche, J., Rapak, A., and Olsnes, S. (1999). Dependence of ricin toxicity on translocation of the toxin A-chain from the endoplasmic reticulum to the cytosol. J. Biol. Chem. 274, 34443–34449. [DOI] [PubMed] [Google Scholar]
- Ye, Y., Shibata, Y., Yun, C., Ron, D., and Rapoport, T. A. (2004). A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429, 841–847. [DOI] [PubMed] [Google Scholar]