Abstract
We identified a Rho guanine exchange factor (GEF) expressed as two splice variants, which differ only in either having or lacking a Postsynaptic density 95, Disk large, Zona occludens-1 (PDZ) motif. The PDZ adaptor protein synectin bound the longer splice variant, Syx1, which was targeted to the plasma membrane in a synectin-dependent manner. The shorter variant, Syx2, was diffusely distributed in the cytoplasm. Fluorescence resonance energy transfer (FRET) imaging revealed similar differences between the spatial patterns of active RhoA in Syx1 versus Syx2-expressing cells. Expression of Syx1 augmented endothelial cell (EC) migration and tube formation, whereas Syx2 expression did not. It appears, therefore, that synectin-dependent targeting of Syx is critical to its contribution to these EC functions. Although agonist-stimulated global RhoA activity was similar in Syx1- and Syx2-expressing cells, basal RhoA activity was surprisingly higher in the latter. Out of 23 cell types, we found a significant level of endogenous Syx2 expression only in brain tumor cells, which also exhibited high basal RhoA activity. We found that the activity level of JNK, which mediates transcriptional regulation downstream of RhoA, is elevated in a Syx2-dependent manner in these cells, possibly contributing to their tumorigenicity.
INTRODUCTION
Rho guanine exchange factors (GEFs) activate GTPases by facilitating the replacement of GDP by GTP (Erickson and Cerione, 2004). Because GEFs interact directly with their effector GTPases (Snyder et al., 2002), Rho GEF targeting determines the spatial pattern of GTPase activity. This targeting does not appear to be regulated via a common mechanism shared by most Rho GEFs, however, as different domains interacting with other proteins and phospholipids determine the cellular positioning of various members of the Rho GEF family (Rossman et al., 2005).
To date, postsynaptic density 95, disk large, zona occludens-1 (PDZ) binding motifs have been identified in four Rho GEFs, two of which, the Rac1-specific Kalirin-7 (Penzes et al., 2001) and the Rac1/Cdc42 GEF βPIX (Park et al., 2003), have been studied in neurons. Both Kalirin-7 and βPIX interact with multiple postsynaptic density PDZ scaffold proteins. The third, neuroepithelioma transforming gene 1 (Net1), is a RhoA-specific GEF that has not yet been associated with any PDZ adaptor protein (Qin et al., 2005).
The fourth is a GEF with a PDZ-binding motif (GEF720) that was recently identified in a human cDNA database (Kikuno et al., 2002) and classified as RhoA-specific (De Toledo et al., 2001). A rat orthologue of the same GEF (Tech, a transcript highly enriched in cortex and hippocampus) was independently identified and its catalytic properties were characterized (Marx et al., 2005).
In parallel to the above studies, a DNA sequence corresponding to the last 40 C-terminus residues of the mouse orthologue of GEF720/Tech was cloned by yeast two-hybrid screening of a 3T3 fibroblast cDNA library (M. Simons, personal communication) using synectin (Gao et al., 2000) as bait. Synectin is an adaptor protein that contains a single PDZ domain and is expressed early and ubiquitously (Zhang et al., 2004). It is one of the most versatile PDZ proteins known to date, with upwards of 20 binding partners, most of which are transmembrane receptors (e.g., Gao et al., 2000; Blobe et al., 2001; Lou et al., 2001) with type I C-terminus PDZ-binding motifs conforming to the consensus sequence (S/T)-X-(V/A) (Songyang et al., 1997).
We found that the recently identified synectin-binding Rho GEF is expressed as two splice variants that differ only in either having or lacking a PDZ motif, and took advantage of this circumstance in order to dissect the function of the PDZ motif in regulating Rho GEF activity and cell migration.
MATERIALS AND METHODS
Cloning of Mouse Syx1 and Syx2 cDNA
The sequence of human KIAA0720 (NCBI accession number AB018263) was used for searching the NCBI mouse EST library. A putative full-length cDNA sequence was assembled in silico based on overlapping EST entries that spanned the entire coding sequence. Total RNA extracted (RNeasy, Qiagen, Chatsworth, CA) from mouse heart ECs was used for amplifying by reverse transcriptase PCR a 2.3-kb-long cDNA fragment corresponding to the central region of the Syx cDNA with primers derived from the assembled sequence (forward: GAAGAAAGCAAGGAGGAGACACA; reverse: CACAAAGTCTTGAAGGGAAG). The 5′ and 3′ ends of the fragment were extended (5′/3′ RACE, Roche, Indianapolis, IN) by RACE (5′ primer: CTTCTTCTTCATGGCTGGCAC; 3′ primer: GTCACTCCCACCAGCGAG) using the same total RNA pool. 5′ RACE generated a 0.6-kb fragment and 3′ RACE generated two 3′ fragments of 1.1 and 1.4 kb, corresponding to the Syx1 and Syx2 splice variant, respectively. The start codon was identified by comparison between human, mouse, and rat Syx genes and by GENESCAN (http://genes.mit.edu/GENSCAN.html) exon analysis. Each fragment was inserted separately into pCR2.1-TOPO (Invitrogen, Carlsbad, CA) by TA cloning and sequence-verified. The 0.6-kb fragment was subcloned into the plasmid containing the 2.3-kb fragment using ScaI (5′) and NotI (3′). The 3′ fragments of Syx1 and Syx2 were both extended by PCR (forward primer: GCTCCCCCACCATCCTTCGAAAGAGCAGCAACAGCCTCGACTCTGAGCACTGCACCTCAGATGGCTCCACGGAGAC; reverse primer: M13) into the 5′ direction so as to encompass a unique BstbI site present in the 3′ region of the 2.3-kb fragment. This site was then used to subclone each 3′ fragment into the plasmid containing the 0.6- and 2.3-kb fused fragments, producing full-length cDNAs of Syx1 and Syx2 reaching 3.8 and 4.1 kb, respectively. The cDNA sequences of Syx1 and Syx2 were deposited at NCBI with accession numbers AY605057 and AY605058, respectively.
DNA and RNA Constructs
Mouse Syx1 and Syx2 cDNA were amplified from their respective pCR2.1-TOPO constructs so as to avoid the start codon and the 5′ region and to facilitate subcloning into the target plasmids between the BspEI and XhoI sites (forward primer: GGTGATCACACGTAGTTTCCGGATGTAG; reverse primer: CCGCTCGAGATCTGGGACAGGACCGGGCGTCTC). Syx1 and Syx2 were subcloned into N-terminus fusion versions of vectors pEYFP, pEGFP, mRFP (all from BD Biosciences, San Diego, CA), and pcDNA4/HisMax (Invitrogen) between the BglII and BamHI sites. Syx1-specific siRNA (5′-CUCACUGCCUCGGAGGUAUGA-3′) was targeted to a 21-base-long sequence consisting of 12 bases at the 3′ end of exon 20 and 9 at the 5′ end of exon 21, thus preventing full hybridization to the Syx2 transcript. Syx2-specific siRNA was targeted to a 21-base-long sequence (5′-ACTCATGCCTGGGAGCATCGCATTT-3′) in the intron between exons 20 and 21, which is present only in the Syx2 transcript. Luciferase-targeted siRNA was used as negative control. Synectin-specific siRNA (5′-GCCGUACAGCUCCUUGACA-3′) was targeted to the 3′ region of the transcript. All siRNA oligos were synthesized and purchased from Qiagen.
Differential Reverse Transcriptase PCR of Syx1 and Syx2 mRNA
We took advantage of the splicing pattern of the Syx gene in order to design primers that amplified (Thermo-Script, Invitrogen) either 826- (human), 875- (rat), and 941 (mouse)-nt-long fragments from Syx1 mRNA, or 1087- (human), 1136- (rat), and 1202 (mouse)-nt-long fragments from Syx2 mRNA. The same forward and reverse primers were used for both Syx1 and Syx2 amplification. The forward primers (human: CTCTCAGCACTGTGCCTCAGATG; rat: CTCTCAGCACTGTGCCTCAGATG; mouse: GGGTGAAGAGAGCGGTACCTCAG) span the 3′ region of exon 20 and the 5′ region of exon 21. The reverse primers (human: GCTGTCTCTTGGTCAATGGCAC; rat: GGCAGCAGTGAAGCAGGTGC; mouse: GCGGTGAAGCAGGTGCTGTATC) correspond to the 3′ UTR. Because Syx2 mRNA contained an additional 3′ UTR fragment, its amplicon is longer by 261 bases (in all 3 species) than the amplicon generated from the Syx1 mRNA. All reverse transcriptase PCR results were verified by omitting the reverse transcriptase reaction.
The cell types probed by reverse transcriptase PCR for Syx1 and Syx2 expression were as follows: 1) cells of human origin: umbilical vein endothelial, arterial endothelial, retinal pigment epithelial, immortalized astrocytes, melanocytes, melanoma, osteosarcoma, glioma, glioblastoma multiforme, lymphoblastoma, breast cancer epidermal, and pheocromocytoma; 2) cells of rat origin: rat fat pad ECs (RFPECs), astrocytes, and gliosarcoma; and 3) cells of mouse origin: heart endothelial, lung endothelial, monocytes, helper T-lymphocyte, neuroblastoma, lymphoma, and lymphoblastoma.
Real-time PCR
mRNA purified as above was treated by DNase (Ambion, Austin, TX) in order to remove genomic DNA, reverse-transcribed (ThermoScript, Invitrogen) using 1 μg with random hexamers and amplified (7500 Real Time PCR System, Applied Biosystems, Foster City, CA) using primers and annealing temperatures listed in Table 1. Each sample contained 1 μl reverse-transcribed DNA, 15 μl 2× SYBR Green PCR Master Mix (Applied Biosystems), 1 μl forward and 1 μl reverse primer (5 pmol/μl), in a total volume of 30 μl. The cycling protocol consisted of 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C.
Table 1.
Sequences and annealing temperatures of primers used in real-time PCR experiments
| Gene | Forward primer | Reverse primer | Tm (°C)a | Amplicon length (bp) |
|---|---|---|---|---|
| Syx1 | CTCACTGCCTCGGAGGTCTG | GTCTCTTGGTCAATGGCACTCTT | 59.6,58.4 | 62 |
| Syx2 | CTGTGGTGGGCAGCAGTG | GACAGAGTCCTTCTTTGGCTGAC | 58.3,58.1 | 108 |
| β-Actin | TCAACACCCCAGCCATGTAC | GGTCAGGATCTTCATGAGGTAGTC | 57.9,56.9 | 203 |
Sequences were designed by Primer Express (Applied Biosystems) and synthesized by Invitrogen. The Syx1 and Syx2 forward primers span the 3′ region of exon 20 and the 5′ region of exon 21. The reverse primers correspond to the 3′ UTR.
Values are for forward and reverse primers, respectively
We calculated the amount of either Syx1 or Syx2 mRNA copy number, using β-actin as an internal control, according to the 2-ΔΔCT method (Livak and Schmittgen, 2001). The ratio between the mRNA copy numbers of Syx1 and Syx2 in each sample is given by 2-ΔΔCT, where CT is the threshold fractional cycle number. Syx1/Syx2 ratios represent the average of four amplification runs, calculated after subtracting the ratios of non-reverse-transcribed samples that were run in parallel. Validation experiments demonstrated that the amplification efficiencies of both Syx1, Syx2, and β-actin were approximately equal, as required for the 2-ΔΔCT method.
Cell Culture, Transfection, and FACS
RFPECs were grown in M199 with 10% fetal bovine serum (FBS) at 37°C/5% CO2. Mouse heart ECs from wild-type (WT) or synectin-/- mice (a gift of M. Simons, generated by disruption of the synectin gene with the gene trap method) were harvested and grown as described (Partovian et al., 2005). Glioblastoma multiforme (GBM) cell lines and telomerase-immortalized human astrocytes (both gift of M. Israel, Dartmouth) were grown in DMEM with 10% FBS.
RFPECs were transfected with Syx1 and Syx2 fused to visual fluorescent proteins (VFP, a generic term for all GFP derivatives) or with siRNA by FuGENE-6 (Roche) and by Targefect (Targeting Systems, San Diego, CA), respectively, according to the manufacturer's instructions. VFP-expressing cells were sorted (FACStar Plus, Becton Dickinson, Lincoln Park, NJ) into medium and high expresser populations by separating the top 7-9% and 2-4%, respectively, based on the fluorescence intensity of the fused VFP moieties. The medium expresser group was used in all experiments in order to avoid potential artifacts of supraphysiological expression levels.
Antibodies
Chicken polyclonal antibody to mouse Syx was raised and affinity purified (Aves Labs, Tigard, OR) using a peptide (QHRKLTLAQLYRIRTT) corresponding to residues 1046-1061, which are shared by Syx1 and Syx2. The corresponding sequences in rat Syx1 and Syx2 are identical. Rabbit affinity-purified antibody to synectin was produced as described (Dance et al., 2004) and used at 1 μg/ml for immunoblot and 10 μg/ml for immunofluorescence. Rabbit affinity-purified antibody to myosin VI was produced as described (Hasson and Mooseker, 1994) and used at 1 μg/ml for immunoblot. Antibodies for c-Jun N-terminus kinase (JNK) and phospho-JNK were from Cell Signaling Technology (Beverly, MA). Alexa-647-conjugated phalloidin was from Molecular Probes (Eugene, OR). α-VFP was purchased from BD Biosciences. Peroxidase-conjugated donkey anti-rabbit and donkey anti-chicken antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Phalloidin and all purchased antibodies were used at concentrations recommended by the manufacturers.
Confocal Microscopy
RFPECs or mouse heart ECs were plated on glass coverslips and processed as described (Horowitz et al., 2002). Images were acquired by laser-scanning confocal microscopy (Zeiss LSM 510 Meta, Thornwood, NY) with a ×63 objective. Plasma membrane (PM) targeting was quantified by counting the number of cells with PM-localized Syx1 or Syx2 in a total of 100 cells in each experiment.
Live Cell FRET Imaging
Subconfluent RFPECs were grown on fibronectin-coated (10 ng/ml, 1 h at 37°C) 35-mm plates with optical-quality 1-mm glass bottom (FluoroDish, World Precision Instruments, Sarasota, FL). Cells were cotransfected with the Raichu-RhoA FRET probe pRaichu-1237X (Yoshizaki et al., 2003) and with mRFP-Syx1 or mRFP-Syx2 constructs. Transfected cells were starved overnight in M199 medium (Invitrogen) supplemented with 0.25% FBS and 1% bovine serum albumin overnight. Cells selected for imaging had similar mRFP emission levels as judged by eye. A series of 50 images at 20-s intervals was acquired starting at 1 min after cell activation by platelet-derived growth factor (PDGF) or lysophosphatidic acid (LPA), so as to track the postactivation increase in the FRET response. Afterward, additional four series of 20 images at 20-s intervals were acquired every 6 min. Images of cell-free areas of the plate were acquired for each series and were subtracted from the raw images (ImagePro, Media Cybernetics, Silver Spring, MD).
Live cell FRET imaging was performed on an inverted microscope (Olympus IX71, Olympus, Melville, NY) using a plexiglass-enclosed stage kept at 37°C and 5% CO2. Images were acquired by cooled software-controlled (QED, Media Cybernetics) charge-coupled device camera (SensiCam, Cooke, Auburn Hills, MI). FRET capability was conferred by a dual beam splitter with CFP and YFP filters (HQ 465/30m and HQ 560/55m, respectively, Chroma, Brattleboro, VT). CFP was excited by a white light source (Lambda DG4, Sutter Instrument, Novato, CA) through a 438/24-nm filter (FF458-Ex01-25, Semrock) followed by a 20% neutral density filter and a 60× oil immersion objective (Olympus). Shutters were switched by a software-controlled driver (VMM-D3, Uniblitz). Exposure time was set to 0.5 s. After background subtraction, the FRET efficiency (the ratio of YFP/CFP emissions) was calculated by ImagePro (Madison, WI). Epifluorescence illumination was generated by a 100-W Xenon lamp (Olympus).
Cell Function Assays
Gap Closure. Assays were performed as described (Horowitz et al., 2002). Agonists were applied at the indicated concentrations throughout the assay. Cells were stimulated by LPA or by PDGF BB (Sigma), an optimal stimulator of RFPECs migration.
Transwell. Mouse heart ECs were loaded with 5 μg/ml calcein (Molecular Probes) by incubation for 2 h at 37°C in DMEM supplemented with 0.5% FBS. After detachment by dissociation solution (Cellstripper, Cellgro, Herndon, VA), 105 cells in 0.25 ml were seeded in Fluoroblock (BD Biosciences, pore size, 3 μm) inserts placed in a 24-well plate. A volume of 0.75 ml medium containing 5 ng/ml EGF (Sigma), an efficient stimulator of mouse heart EC migration, was placed in the bottom of the well. Cell migration was quantified by measuring calcein fluorescence (Synergy HT, using 485/20 and 530/25 nm filters; Bio-Tek, Winooski, VT) after a 4-h incubation at 37°C/5% CO2. Transwell was preferred over gap closure assay with mouse heart ECs because these cells did not form a well-defined advancing edge.
In Vitro Tube Formation. Twelve-well plates were coated with a ∼2-mm layer of Matrigel (BD Bioscience) according to the manufacturer's instructions. RFPECs, 105, were seeded per well in M199 with 0.5% FBS. Phase-contrast images of cells at the center of the wells were acquired 12 h after cell seeding.
RhoA Activity. RFPECs were starved for 24 h in M199 with 0.5% FBS and 0.5% bovine serum albumin. Cells treated by LPA (1 μM, 10 min at 37°C; Sigma), PDGF (50 ng/ml, 5 min; BD Biosciences), and either dimethyl sulfoxide (DMSO) or phosphate-buffered saline, respectively, were lysed and the RhoA activity was detected either by immunoblotting or by ELISA-based kits (Cytoskeleton, Denver, CO) according to the manufacturer's instructions. RhoA activity was quantified either by densitometry of the immunoblotted RhoA bands, or by measuring OD492 of the ELISA samples.
JNK Phosphorylation. Cells (104/well) were grown in a 96-well plate overnight at 37°C/5% CO2 and then starved for 24 h (0.25% fetal calf serum, 0.5% BSA in DMEM). Cells were treated by LPA (1 μM, 10 min at 37°C; Sigma) or by DMSO and processed for immunodetection of phospho-JNK (T183/Y185) and total JNK in each well by ELISA according to the manufacturers' instructions (CASE JNK kit, SupperArray, Bethesda, MD). Eight wells were used for each condition and cell type, four of which for the detection of phospho-JNK and four for detecting total JNK. Primary antibodies were detected by peroxidase-conjugated secondary antibody, followed by OD450 measurement (Multiscan MCC/340, Labsystems). OD450 value of each well was normalized by OD595 measured after colorimetric detection of the total protein in the well. JNK phosphorylation levels were presented as a ratio between the normalized OD450 corresponding to phospho-JNK and to total JNK.
Proliferation. GBM U87 cells, 2000 per well, were seeded in a 96-well plate and grown in DMEM supplemented with 10% FBS at 37°C/5% CO2. Cells were transfected by 10 pmol/well Syx2 or luciferase siRNA according to the manufacturer's (X-tremeGene; Roche) instructions. The initial size of the cell population was defined as a baseline value (day 0) and quantified by measuring the OD492 of a colorimetric reaction, following the manufacturer's instructions (CellTiter 96 AQueous One, Promega). Similar measurements were repeated with cells grown for additional 24, 48, and 72 h.
Immunoprecipitation
Confluent RFPECs were lysed in 0.5 ml RIPA buffer supplemented with a protease inhibitor cocktail (Complete, Roche), precleared by incubation with 20 μl protein G plus/protein A-agarose (Pierce, Rockford, IL) slurry for 1 h at 4°C, and incubated with 10 μg of the indicated antibody (or 2 μl of α-VFP) and 40 μl protein G plus/protein A-agarose for 4 h at 4°C. Beads were washed six times in lysis buffer and bound proteins were eluted by boiling in 40 μl Laemmli buffer for 5 min.
Statistical Analysis
Standard deviations are shown as bars in the histograms. Differences were considered statistically significant if p ≤ 0.05 by t test.
RESULTS
Syx Is Transcribed as Two Nearly Identical Splice Variants
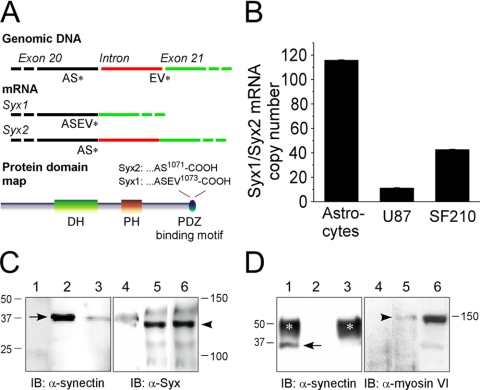
We found that the gene of the synectin-binding RhoA ex-change factor (Syx) initially identified by a yeast two-hybrid screen is transcribed as two splice variants. Strikingly, the amino acid sequences encoded by these variants differ only by two residues: one variant (Syx1) is 1073 amino acid long, whereas the other (Syx2) is shorter by two C-terminus residues (Figure 1A). Unlike the long splice variant, the C-terminus of the shorter one does not conform to the consensus sequence of type I PDZ-binding motif.
Figure 1.
Gene and mRNA structure, expression levels, and molecular interactions of Syx. (A) Structure of the Syx gene and mRNA near the 3′ region of the coding region and domain map of Syx. (B) Ratios between the mRNA copy numbers of Syx1 and Syx (Syx1/Syx2) were calculated according to the 2-ΔΔCT method (Livak and Schmittgen, 2001), using β-actin as an internal control. The Syx1/Syx2 ratios represent the average of four (±SD) amplification runs, calculated after subtracting the ratios of non-reverse-transcribed samples that were run in parallel. (C) Immunoprecipitation of synectin by anti (α)-Syx1 and vice versa. RFPEC lysates were immunoprecipitated (IP), resolved by SDS-PAGE and immunoblotted (IB) as indicated: lane 1, nonimmune chicken IgY; lane 2, RFPEC lysate; lane 3, anti-Syx IP; lane 4, ni rabbit IgG; lane 5, anti-synectin IP; lane 6, RFPEC lysate. Arrow, synectin band; arrowhead: Syx1 band. (D) VFP-Syx1 but not VFP-Syx2 immunoprecipitated synectin. Samples were processed as in C. Lane 1, anti-VFP IP from lysate of VFP-Syx1-expressing RFPECs; lane 2, nonimmune IgG IP from lysate of VFP-Syx1-expressing RFPECs; lane 3, anti-VFP IP from lysate of VFP-Syx2-expressing RFPECs. Anti-Syx immunoprecipitated myo6 only from VFP-Syx1-expressing RFPECs: lane 4, anti-VFP IP from lysate of VFP-Syx2-expressing RFPECs; lane 5, anti-VFP IP from lysate of VFP-Syx1-expressing RFPECs; lane 6, RFPEC lysate; arrow, synectin band; arrowhead, myo6 band.
The Syx gene is located on mouse chromosome 4. The difference between the transcripts of the two splice variants is produced by splicing out a 261 nucleotide (nt)-long intron located between exons 20 and 21 and between the codons of residues 1071 and 1072. The resulting transcript has a length of 3867 nt and codes for a 1073-residue-long protein (Figure 1A). When the above intron is not spliced out, transcription ends at a stop codon located in exon 20 immediately after the codon for residue 1071. The corresponding transcript is 4128 nt long.
We screened various cell types in order to detect the expression levels of Syx1 and Syx2. A total of 23 normal and tumor human, mouse, and rat cell types (see Materials and Methods for the full list) were probed by reverse transcriptase PCR. Syx2 expression was low or undetectable in all normal cells and most malignant cell lines, including RFPECs (unpublished data). The only cell type that expressed Syx2 at a significant level relative to Syx1 was GBM, the most common and most malignant primary brain tumor (Stark et al., 2005). The real time PCR-measured ratios between the mRNA copy numbers of Syx1 and Syx2 in the GBM cell lines U87 and SF210 were significantly lower than in human astrocytes (Figure 1B), which we considered to be a control normal cell type. Based on these ratios, the Syx2 expression level was 9 and 2.4% in U87 and SF210 cells, respectively, of the Syx1 expression level.
Because of the minimal difference between the sequences of Syx1 and Syx2, it was not possible to raise antibodies specific to each isoform. A similarity search with the Syx cDNA sequence revealed that the human (chromosome 1, as previously reported; De Toledo et al., 2001), rat (chromosome 5), and cow genomes also contain a Syx gene with the same structure as in the mouse.
Syx Interacts with Synectin and Myosin VI
Because Syx was initially detected as the prey of synectin in a yeast two-hybrid assay, we sought to confirm the interaction by coimmunoprecipitation. To begin with, we attempted to detect Syx itself in RFPEC lysate with an antibody raised against a 12-residue-long peptide corresponding to a sequence in the C-termini of both Syx1 and Syx2. The detected band migrated at ∼120 kDa, close to the calculated 119-kDa molecular weight of Syx1 (Figure 1C). A similar-sized band was previously detected in a Western immunoblot of rat hippocampus (Marx et al., 2005). Endogenous Syx was immunoprecipitated by synectin and vice versa (Figure 1C), confirming the yeast two-hybrid results and in agreement with the coimmunoprecipitation of Syx and synectin coexpressed in 293 cells (J. Henderson and J. Baraban, Johns Hopkins, personal communication). When we tagged the N-termini of Syx1 and Syx2 with VFP and used this tag for immunoprecipitation from VFP-Syx1 or VFP-Syx2-expressing RFPECs, only VFP-Syx1 coimmunoprecipitated synectin, whereas VFP-Syx2 did not (Figure 1D).
Synectin functions as an adaptor between proteins that bind to its PDZ domain and the actin-based molecular motor myosin VI (Hasson, 2003), which interacts with the C-terminus of synectin (Reed et al., 2005). Myosin VI was coimmunoprecipitated with VFP-Syx1 but not with VFP-Syx2 (Figure 1D), indicating that myosin VI associated with Syx1 in a synectin-dependent manner.
Syx Is Targeted in a Synectin-dependent Manner
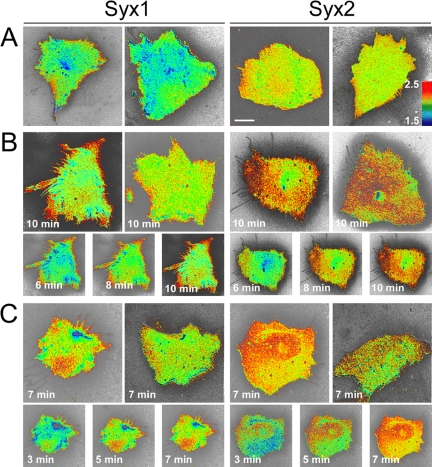
Given that PDZ adaptor proteins frequently regulate the targeting of their ligands (Sheng, 2001), we sought to compare the localization of Syx1 and Syx2. In quiescent RFPECs, endogenously expressed Syx, which is almost exclusively of the Syx1 isoform (real-time PCR, unpublished data), was localized in the cytoplasm (Figure 2A). LPA stimulation was accompanied by the appearance of a Syx1 perinuclear cluster, and by Syx1 targeting to the PM. In LPA-stimulated cells synectin collocated with Syx1 in both the perinuclear region and the PM, but there was no collocation of the two proteins in quiescent cells (Figure 2A). Similar to endogenous Syx1, VFP-Syx1 localization in quiescent RFPECs was cytoplasmic (Figure 2B). LPA stimulation was accompanied by an increase in the perinuclear concentration of both VFP-Syx1 and -Syx2. Only VFP-Syx1, however, was targeted to the PM and to PM-adjacent clusters, similar to endogenous Syx1 (Figure 2B). VFP-Syx1 was targeted to the PM in 54% of LPA-stimulated RFPECs, whereas VFP-Syx2 was targeted to the PM in 19% of RFPECs (Figure 3). These results suggest that the PDZ-binding motif is essential for the PM targeting of activated Syx.
Figure 2.
Localization of Syx1 and Syx2. (A) Localization of endogenous Syx and synectin in quiescent and LPA-stimulated (1 μM, 10 min) RFPECs (nuclear labeling is nonspecific). WT cells were fixed, permeabilized, and immunolabeled with the indicated antibodies, followed by fluorescently labeled anti-chicken IgY and anti-rabbit IgG. Syx and synectin collocated (arrows) in LPA-stimulated cells, but not in quiescent ones. Cells were fixed, permeabilized, and immunolabeled by anti-Syx, followed by fluorescently labeled anti-chicken IgY. (B) Localization of VFP-Syx1 or VFP-Syx2 in quiescent and LPA-stimulated RFPECs (fields are shown in triplicate; arrows point to VFP-Syx1 on the PM), which were transiently transfected by each of the two splice variants and fixed 24 h posttransfection. (C) Localization of endogenous Syx1 in WT and in synectin-/- mouse heart ECs. All images were acquired by confocal microscopy. Scale bars, 10 μm; 5 μm in insets.
Figure 3.
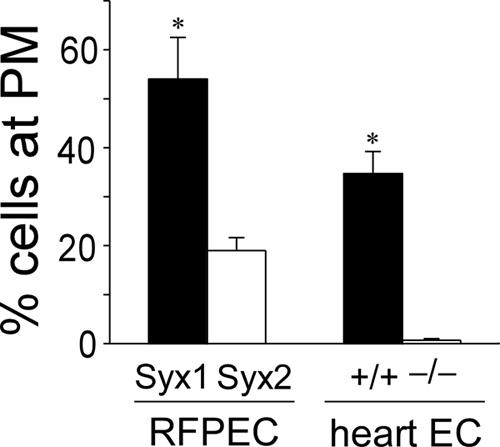
Quantification of PM targeting of Syx1 and Syx2. Percent of RFPECs imaged as in Figure 2B in which either VFP-Syx1 or VFP-Syx2 were targeted to the PM; percent of WT or synectin-/- mouse heart ECs imaged as in Figure 2C in which endogenous Syx1 was targeted to the PM. One hundred cells were counted in each of three different experiments with each cell type (±SD; * p < 0.05).
Conversely, we utilized heart ECs from synectin-/- mice in order to find if the absence of the adaptor protein alters Syx targeting. As in RFPECs, endogenous Syx in mouse heart ECs is predominantly of the Syx1 isoform (real-time PCR, unpublished data). In WT and synectin-/- quiescent heart ECs, Syx1 was distributed homogenously in the cytoplasm (Figure 2C). Upon LPA-stimulation a concentrated perinuclear Syx1 population appeared both in WT and synectin-/- heart ECs (Figure 2C), but PM targeting occurred only in the WT cells (Figure 2C). Syx1 was targeted to the PM in 35% of WT heart ECs, but virtually in none of the synectin-/- heart ECs (Figure 3). This result complements the evidence gathered from VFP-Syx1- and VFP-Syx2-expressing RFPECs for the dependence of PM targeting of Syx on its interaction with synectin.
Syx1 and Syx2 Produce Different Spatial Patterns of RhoA Activity
The differences between the localization of each Syx isoform may confer corresponding changes in the level and spatial distribution of RhoA activity. Initially, in order to assess the relative contribution of Syx1 to the overall RhoA activity level in RFPECs, we compared the results of RhoA assays of RFPECs transfected either by Syx1-specific or by control siRNA. The RhoA activation level in RFPECs transfected by Syx1-specific siRNA was reduced by ∼50% relative to RFPECs treated by control siRNA (Figure 4A), indicating that endogenous Syx1 is responsible for the activation of a major part of the total RhoA pool in RFPECs in response to LPA.
Figure 4.
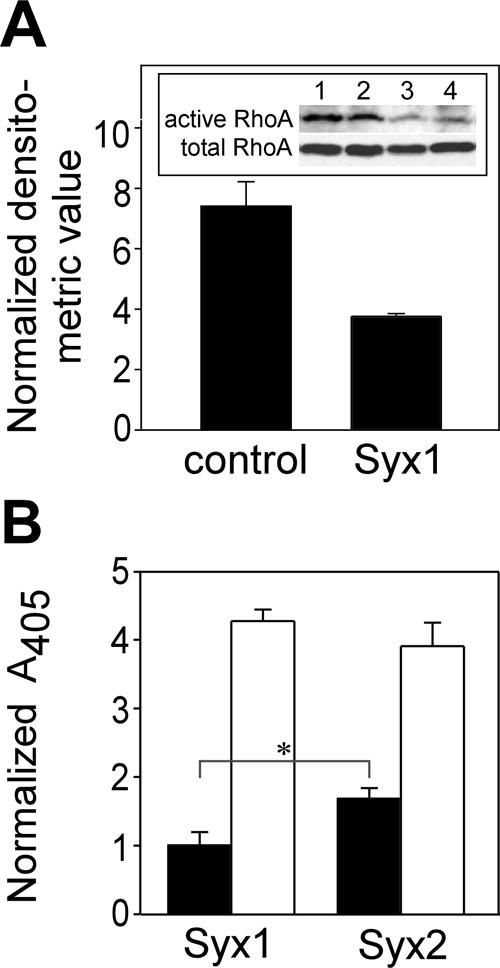
Contribution of Syx1 and Syx2 to global RhoA activity in RFPECs. (A) Relative contribution of Syx1 to RhoA activation. Immunoblots (in duplicate) of active RhoA and of total RhoA in lysates of WT RFPECs transfected by either Syx1-specific or control siRNA and activated by LPA. Means represent the densitometric values of the duplicate bands of active RhoA shown in A, normalized by the corresponding values of the total RhoA bands. (B) Global RhoA activity in quiescent (▪) and LPA-treated (1 μM, 10 min; □) RFPECs expressing each of the indicated Syx splice isoforms. Means represent ratios between OD492 of active and total RhoA of each sample after normalization by the mean of the quiescent Syx1 samples (n = 6, ±SD; * p < 0.05).
The global level of active RhoA in LPA-treated RFPECs expressing Syx1 was similar to that of Syx2-expressing cells (Figure 4B), indicating that Syx2 activates RhoA as efficiently as Syx1 in response to LPA. Surprisingly, however, the basal RhoA activity level was 70% higher in Syx2-expressing cells. The basal as well as the LPA-stimulated global levels of RhoA activity cannot account, therefore, for the higher migration rate of Syx1-versus Syx2-expressing cells.
We anticipated that the spatial patterns of RhoA activity may reflect the observed differences in the localization of each Syx isoform. For that purpose we imaged RFPECs cotransfected by either mRFP-Syx1 or mRFP-Syx2 and by a RhoA FRET probe that emits light in the YFP range only upon GTP binding to the RhoA moiety in the probe (Yoshizaki et al., 2003).
In unstimulated RFPECs expressing mRFP-Syx1, RhoA activity was low across the cells except for the PM (Figure 5A). In quiescent mRFP-Syx2-expressing cells, however, there was a higher level of active RhoA across the cell, likely reflecting the diffuse cytoplasmic distribution of Syx2 (see Figure 2B). On LPA stimulation, Syx1-expressing RFPECs exhibited highly increased RhoA activity along the PM (Figure 5B). Though the level of active cytoplasmic RhoA in Syx2-expressing RFPECs increased significantly relative to quiescent Syx2-expressing cells, the spatial pattern remained diffuse without the higher PM-associated activity seen in Syx1-expressing cells. PDGF treatment was accompanied by similar differences between the activation patterns of RhoA in Syx1-versus Syx2-expressing cells (Figure 5C).
Figure 5.
FRET-imaged RhoA activity in Syx1- and Syx2-expressing RFPECs. Duplicate FRET images of RFPECs expressing the Raichu-RhoA YFP-CFP probe and each of the indicated mRFP-Syx constructs. Cells were either quiescent (A) or stimulated by LPA (B, 1 μM, 10 min) or by PDGF (C, 50 ng/ml, 5 min). FRET efficiency is displayed as a thermal map corresponding to the scale shown in A. The smaller images in B and C show a time series corresponding to the larger image above on the left. Live cell FRET images were acquired as described in Materials and Methods. Scale bar, 10 μm.
Syx1 and Syx2 Have Contrasting Effects on EC Function
Because cell migration requires spatial and temporal coordination of GTPase activity (Fukata et al., 2003), the disparate patterns of active RhoA in Syx1-versus Syx2-expressing RFPECs could conceivably produce differences in the migration rates of each cell group.
Both unstimulated and PDGF- or LPA-treated RFPECs expressing Syx1 migrated threefold faster than vector-transfected control cells in gap closure assays (Figure 6A). Expression of Syx2, however, did not augment migration relative to vector-transfected cells. These results suggest that although the diffusely distributed Syx2 was less effective in supporting migration, it did not interfere with the function of endogenous Syx1.
Figure 6.
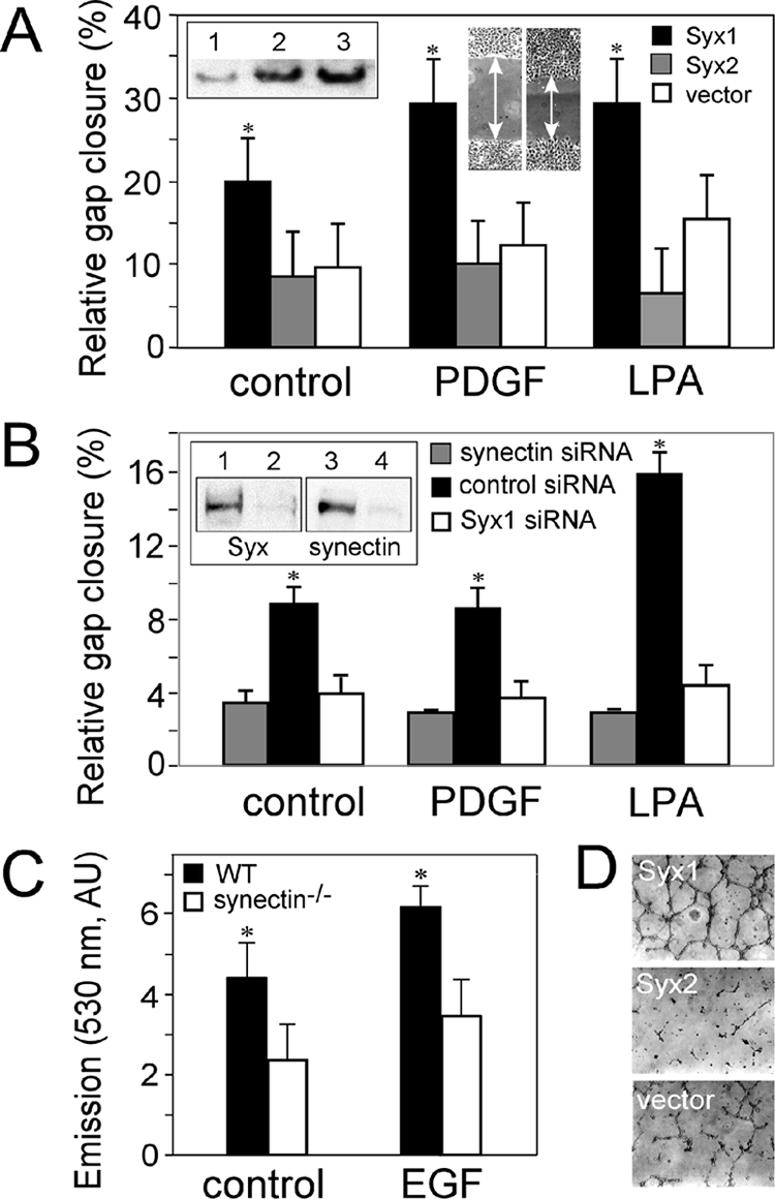
Functional assays of Syx1- and Syx2-expressing RFPECs. (A) Gap closure by quiescent, PDGF- (50 ng/ml) or LPA- (1 μM) treated RFPECs stably expressing VFP-Syx1, VFP-Syx2, or VFP alone. A confluent monolayer of serum-starved RFPECs was scratched before treatment, and the gap width was measured immediately before (W0) and after (W6) a 6-h incubation (ΔW = W0 - W6). Inset, immunoblot showing the total expression level of Syx in WT (lane 1), or of VFP-Syx2 (lane 2), and VFP-Syx1 (lane 3) in transfected RFPECs. (B) Gap closure by RFPECs transfected by each of the indicated siRNA species and treated as in A. Inset, immunoblot showing the expression levels of synectin and of Syx in RFPECs transfected by siRNA specific to synectin (lane 2), Syx1 (lane 4), or by control siRNA (lanes 1 and 3). (C) Migration rates of quiescent or EGF (5 ng/ml)-treated heart ECs from WT or from synectin-/- mice in Transwell assays, quantified by spectrometry of fluorescently labeled migrated cells at 530 nm, as described in Materials and Methods (AU, arbitrary units). (D) In vitro tube formation assays by RFPECs expressing the indicated constructs in the presence of 0.5% serum. RFPECs were seeded in Matrigel-coated plates and imaged 12 h later.
Because Syx1 interacts with synectin, we tested synectin's contribution to EC migration by transfecting Syx1-expressing RFPECs with synectin-specific siRNA. Silencing of synectin expression by more than 90% inhibited migration to an extent very similar to that of Syx1-specific siRNA (Figure 6B). We further investigated the involvement of synectin in cell migration by comparing the migration rates of ECs from hearts of WT mice to those of synectin-/- mice. ECs from WT mice migrated twice as fast as ECs of synectin-/- mice in response to EGF in a Transwell assay (Figure 6C). These results show that interaction with synectin is required for the contribution of Syx1 to cell migration.
In vitro tube formation or angiogenesis is a major functional marker of ECs and is regulated by RhoA (van Nieuw Amerongen et al., 2003). Similar to the effects on RFPEC migration, cells expressing Syx1 formed a continuous tubular network when grown on extracellular matrix substrate with 0.5% serum, whereas Syx2-expressing cells formed very few tubes, far less than the incomplete network formed by vector-transfected RFPECs (Figure 6D).
Syx2 Is Expressed in Brain Tumor Cells
Because we detected significant expression levels of Syx2 only in GBM cell lines, (Figure 1B), we hypothesized that this expression pattern is linked to tumorigenicity and that similar to Syx2-transfected RFPECs, the basal activity level of RhoA may be higher in GBM cells than in normal astrocytes. That, in turn, could contribute to the malignant transformation of these cells via transcription factors downstream of RhoA. Similar to Syx2-expressing RFPECs, the RhoA activity levels of LPA-treated astrocytes and GBM cells were comparable (Figure 7A), but the basal RhoA activity level was more than twofold higher in the two GBM cell lines we assayed. It appears, therefore, that even a relatively modest expression level of Syx2 (e.g., 9% of Syx1 expression level in U87 cells, Figure 1B) is sufficient to sustain elevated basal RhoA activity.
Figure 7.
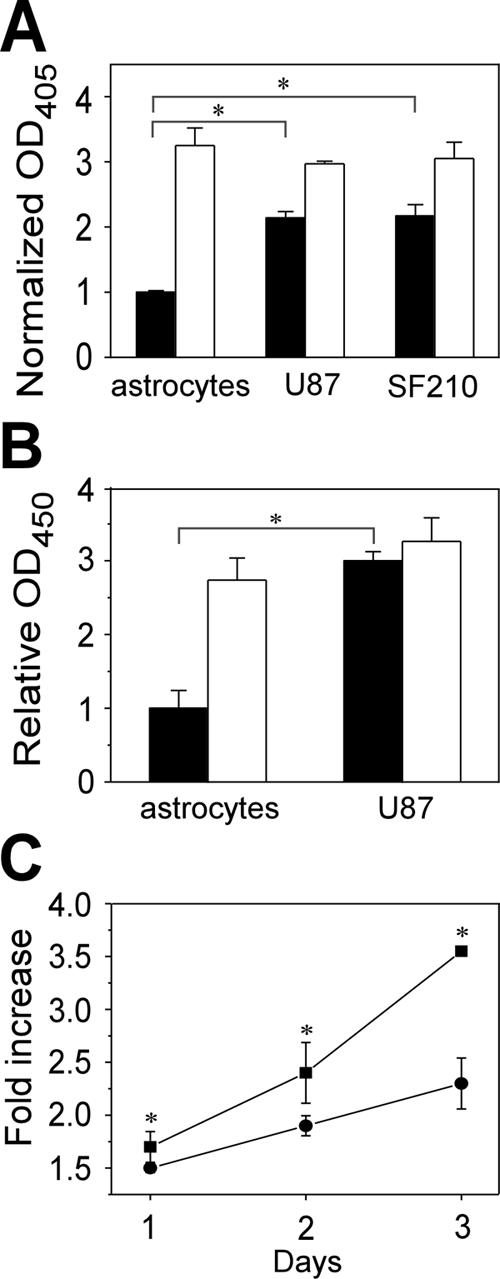
Basal RhoA and JNK activity, and proliferation rate in GBM cells are higher than in normal astrocytes. (A) Global RhoA activity levels in lysates of quiescent (▪) and LPA-treated (1 μM, 10 min; □) astrocytes and GBM cell lines U87 and SF210, assayed by ELISA, and measured by OD492. Means represent ratios between OD492 of active and total RhoA of each sample (see Materials and Methods) after normalization by the mean of the same ratios of quiescent cells (n = 4, ±SD; * p < 0.05). (B) Levels of phospho-JNK in quiescent (▪) and LPA-treated (1 μM, 10 min; □) normal astrocytes and U87 GBM cell line, assayed by ELISA, and presented as ratios between the OD450 of anti-phospho-JNK and the OD595 of anti-pan JNK. The absorbances were previously normalized by the sizes of the cell populations of each sample (n = 4, ±SD; * p < 0.05). (C) Proliferation rates of GBM cell line U87 treated by siRNA specific to Syx2 (•) or by control siRNA (▪), presented as ratios between the cell population size on each day and the initial population size (n = 4, ±SD; * p < 0.05).
RhoA regulates the expression of the c-Jun transcription factor through a pathway separate from RhoA-dependent regulation of the actin cytoskeleton. That pathway uses JNK (Marinissen et al., 2004), which is activated upon phosphorylation of Thr183 and Tyr185 (Derijard et al., 1994). Subsequently JNK phosphorylates transcription factors bound to the c-Jun promoter (Davis, 2000), thus increasing cell proliferation. JNK phosphorylation level was significantly higher in quiescent GBM U87 cells than in quiescent astrocytes and was not further increased by LPA treatment (Figure 7B), in agreement with the possibility that the higher level of basal RhoA activity in GBM cells resulted in up-regulation of downstream transcription factors.
U87 GBM cells transfected by Syx2-specific siRNA proliferated significantly slower than cells treated by control siRNA (Figure 7C), confirming that Syx2 expression is associated with the elevated growth rate typical of malignant cells.
DISCUSSION
Previously reported functions of PDZ motifs in Rho GEFs concerned mainly targeting, as in the case of Kalirin-7 that required the PDZ motif for positioning in dendritic spines. Mutant Kalirin-7 lacking this motif was diffusely distributed in the cytoplasm (Penzes et al., 2001), similar to Syx2. The dependence of Rac1 activation by Kalirin-7 on the engagement of its PDZ-binding motif was not clear cut, however, because PSD-95 down-regulated Rac1 activation by both WT and a Kalirin-7 mutant lacking the PDZ-binding motif. Similarly, conflicting results were reported regarding the dependence of the transforming activity of Net1 on its PDZ-binding motif (Qin et al., 2005), where a mutant lacking the PDZ-binding motif was less efficient in transforming 3T3 fibroblasts, but a splice variant with a PDZ-binding motif was similarly inefficient. A previous study where expression of a Syx1 human orthologue was shown to transform 3T3 fibroblasts used a partial clone (GEF720) lacking 160 N-terminus residues (De Toledo et al., 2001) but containing the PDZ-binding motif. This truncated version of Syx1 may have been disinhibited, however, because the N-terminus is required for the down-regulation of Syx1 (Marx et al., 2005). Therefore, this result does not necessarily reflect the transformation potential of full-length Syx1, which is likely to be lower than that of GEF720.
The failure of Syx2 expression to augment cell migration in the same manner as Syx1 suggests that the PDZ-binding motif and the interaction with synectin are required for the functional coupling of Syx to the actin cytoskeleton. A second possible consequence of the absence of a PDZ-binding motif and the diffuse distribution in the cytoplasm is reduced catalytic regulation, as indicated by the higher basal RhoA activity in Syx2-expressing RFPECs and in GBM cells. The lower regulation of Syx2 could result from better access to the cytoplasmic pool of RhoA or its inability to associate with other proteins required for regulating Syx2 itself and/or RhoA.
The association of myosin VI with the complex containing Syx1 and synectin probably links Syx1 to the actin cytoskeleton and could facilitate translocation of the complex in the retrograde direction along actin filaments (Wells et al., 1999). Because synectin is associated with endocytic vesicles (Aschenbrenner et al., 2003), the specific function of Syx1 may involve rearrangement of the actin cytoskeleton during inward vesicle traffic. In support of this possibility, we observed collocation between Syx1 and several markers of early endosomes (unpublished data).
Possibly the most intriguing question is the biological significance of the existence of almost identical Rho GEF splice variants. Because we were able to detect a significant level of Syx2 expression only in malignant cells, we hypothesize that the less tightly regulated Syx2, which gives rise to a sustained basal level of RhoA activity, up-regulates tumorigenic transcription factors. Association between tumorigenicity and Syx2 expression is suggested also by a previous study, which reported that synectin degradation mediated via its interaction with the viral oncoprotein E6, as well as RNAi silencing of synectin, increased cell proliferation (Favre-Bonvin et al., 2005). Together with the results presented here, this initial data set suggests a novel mechanism of malignant transformation that does not involve truncation of proto-oncogenic Rho GEF (Ron et al., 1989), but rather the expression of an unmodified and loosely targeted Rho GEF splice variant.
Acknowledgments
We thank Dr. M. Simons, Dartmouth Medical School, for synectin-/- mice; Dr. Michiyuki Matsuda, Osaka University, for FRET probes; Dr. Tama Hasson, University of California at San Diego, for synectin and myosin VI antibodies; Dr. Mark Israel and Nathan Watson, Dartmouth Medical School; for human astrocytes and GBM cells; and Dr. Brent Harris, Dartmouth Medical School, for rat astrocytes. We thank Drs. Michael Simons, Dartmouth Medical School; Jay Baraban, Johns Hopkins University; and Anne Blangy, Center for Research of Macromolecular Biochemistry, France, for sharing unpublished data. We thank Dr. Anna Salikhova, Dartmouth Medical School, for help with the Transwell assay.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-01-0002) on February 8, 2006.
Abbreviations used: AU, arbitrary units; EC, endothelial cell; FRET, fluorescence resonance energy transfer; GBM, glioblastoma multiforme; GEF, guanine exchange factor; IB, immunoblot; IP, immunoprecipitation; JNK, c-Jun N-terminus kinase; LPA, lysophosphatidic acid; PDZ, postsynaptic density 95, disk large, zona occludens-1; PM, plasma membrane; RFPEC, rat fat pad endothelial cell; VFP, visual fluorescent protein; WT, wild type.
References
- Aschenbrenner, L., Lee, T., and Hasson, T. (2003). Myo6 facilitates the translocation of endocytic vesicles from cell peripheries. Mol. Biol. Cell 14, 2728-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobe, G. C., Liu, X., Fang, S. J., How, T., and Lodish, H. F. (2001). A novel mechanism for regulating transforming growth factor beta (TGF-beta) signaling. Functional modulation of type III TGF-beta receptor expression through interaction with the PDZ domain protein, GIPC. J. Biol. Chem. 276, 39608-39617. [DOI] [PubMed] [Google Scholar]
- Dance, A. L., Miller, M., Seragaki, S., Aryal, P., White, B., Aschenbrenner, L., and Hasson, T. (2004). Regulation of myosin-VI targeting to endocytic compartments. Traffic 5, 798-813. [DOI] [PubMed] [Google Scholar]
- Davis, R. J. (2000). Signal transduction by the JNK group of MAP kinases. Cell 103, 239-252. [DOI] [PubMed] [Google Scholar]
- De Toledo, M., Coulon, V., Schmidt, S., Fort, P., and Blangy, A. (2001). The gene for a new brain specific RhoA exchange factor maps to the highly unstable chromosomal region 1p36.2-1p36.3. Oncogene 20, 7307-7317. [DOI] [PubMed] [Google Scholar]
- Derijard, B., Hibi, M., Wu, I. H., Barrett, T., Su, B., Deng, T., Karin, M., and Davis, R. J. (1994). JNK1, a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76, 1025-1037. [DOI] [PubMed] [Google Scholar]
- Erickson, J. W., and Cerione, R. A. (2004). Structural elements, mechanism, and evolutionary convergence of Rho protein-guanine nucleotide exchange factor complexes. Biochemistry 43, 837-842. [DOI] [PubMed] [Google Scholar]
- Favre-Bonvin, A., Reynaud, C., Kretz-Remy, C., and Jalinot, P. (2005). Human papillomavirus type 18 E6 protein binds the cellular PDZ protein TIP-2/GIPC, which is involved in transforming growth factor beta signaling and triggers its degradation by the proteasome. J. Virol. 79, 4229-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata, M., Nakagawa, M., and Kaibuchi, K. (2003). Roles of Rho-family GTPases in cell polarisation and directional migration. Curr. Opin. Cell Biol. 15, 590-597. [DOI] [PubMed] [Google Scholar]
- Gao, Y., Li, M., Chen, W., and Simons, M. (2000). Synectin, syndecan-4 cytoplasmic domain binding PDZ protein, inhibits cell migration. J. Cell Physiol. 184, 373-379. [DOI] [PubMed] [Google Scholar]
- Hasson, T. (2003). Myosin VI: two distinct roles in endocytosis. J. Cell Sci. 116, 3453-3461. [DOI] [PubMed] [Google Scholar]
- Hasson, T., and Mooseker, M. S. (1994). Porcine myosin-VI: characterization of a new mammalian unconventional myosin. J. Cell Biol. 127, 425-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz, A., Tkachenko, E., and Simons, M. (2002). Fibroblast growth factor-specific modulation of cellular response by syndecan-4. J. Cell Biol. 157, 715-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuno, R., Nagase, T., Waki, M., and Ohara, O. (2002). HUGE: a database for human large proteins identified in the Kazusa cDNA sequencing project. Nucleic Acids Res. 30, 166-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402-408. [DOI] [PubMed] [Google Scholar]
- Lou, X., Yano, H., Lee, F., Chao, M. V., and Farquhar, M. G. (2001). GIPC and GAIP form a complex with TrkA: a putative link between G protein and receptor tyrosine kinase pathways. Mol. Biol. Cell 12, 615-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinissen, M. J., Chiariello, M., Tanos, T., Bernard, O., Narumiya, S., and Gutkind, J. S. (2004). The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol. Cell 14, 29-41. [DOI] [PubMed] [Google Scholar]
- Marx, R., Henderson, J., Wang, J., and Baraban, J. M. (2005). Tech: a RhoA GEF selectively expressed in hippocampal and cortical neurons. J. Neurochem. 92, 850-858. [DOI] [PubMed] [Google Scholar]
- Park, E., Na, M., Choi, J., Kim, S., Lee, J. R., Yoon, J., Park, D., Sheng, M., and Kim, E. (2003). The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the βPIX guanine nucleotide exchange factor for Rac1 and Cdc42. J. Biol. Chem. 278, 19220-19229. [DOI] [PubMed] [Google Scholar]
- Partovian, C., Zhuang, Z., Moodie, K., Lin, M., Ouchi, N., Sessa, W. C., Walsh, K., and Simons, M. (2005). Protein kinase Cα activates eNOS and increases arterial blood flow in vivo. Circ. Res. 97, 482-487. [DOI] [PubMed] [Google Scholar]
- Penzes, P., Johnson, R. C., Sattler, R., Zhang, X., Huganir, R. L., Kambampati, V., Mains, R. E., and Eipper, B. A. (2001). The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29, 229-242. [DOI] [PubMed] [Google Scholar]
- Qin, H., Carr, H. S., Wu, X., Muallem, D., Tran, N. H., and Frost, J. A. (2005). Characterization of the biochemical and transforming properties of the neuroepithelial transforming protein 1. J. Biol. Chem. 280, 7603-7613. [DOI] [PubMed] [Google Scholar]
- Reed, B. C., Cefalu, C., Bellaire, B., Cardelli, J. A., Louis, T., Salamon, J., Bloecher, M. A., and Bunn, R. C. (2005). GLUT1CBP(TIP2/GIPC1) interactions with GLUT1 and Myosin VI: evidence supporting an adapter function for GLUT1CBP. Mol. Biol. Cell 16, 4183-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron, D., Graziani, G., Aaronson, S. A., and Eva, A. (1989). The N-terminal region of proto-dbl down regulates its transforming activity. Oncogene 4, 1067-1072. [PubMed] [Google Scholar]
- Rossman, K. L., Der, C. J., and Sondek, J. (2005). GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell. Biol. 6, 167-180. [DOI] [PubMed] [Google Scholar]
- Sheng, M. (2001). Molecular organization of the postsynaptic specialization. Proc. Natl. Acad. Sci. USA 98, 7058-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, J. T., Worthylake, D. K., Rossman, K. L., Betts, L., Pruitt, W. M., Siderovski, D. P., Der, C. J., and Sondek, J. (2002). Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat. Struct. Biol. 9, 468-475. [DOI] [PubMed] [Google Scholar]
- Songyang, Z., Fanning, A. S., Fu, C., Xu, J., Marfatia, S. M., Chishti, A. H., Crompton, A., Chan, A. C., Anderson, J. M., and Cantley, L. C. (1997). Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275, 73-77. [DOI] [PubMed] [Google Scholar]
- Stark, A. M., Nabavi, A., Mehdorn, H. M., and Blomer, U. (2005). Glioblastoma multiforme-report of 267 cases treated at a single institution. Surg. Neurol. 63, 162-169; discussion 169. [DOI] [PubMed] [Google Scholar]
- van Nieuw Amerongen, G. P., Koolwijk, P., Versteilen, A., and van Hinsbergh, V. W. (2003). Involvement of RhoA/Rho kinase signaling in VEGF-induced endothelial cell migration and angiogenesis in vitro. Arterioscler. Thromb. Vasc. Biol. 23, 211-217. [DOI] [PubMed] [Google Scholar]
- Wells, A. L., Lin, A. W., Chen, L. Q., Safer, D., Cain, S. M., Hasson, T., Carragher, B. O., Milligan, R. A., and Sweeney, H. L. (1999). Myosin VI is an actin-based motor that moves backwards. Nature 401, 505-508. [DOI] [PubMed] [Google Scholar]
- Yoshizaki, H., Ohba, Y., Kurokawa, K., Itoh, R. E., Nakamura, T., Mochizuki, N., Nagashima, K., and Matsuda, M. (2003). Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J. Cell Biol. 162, 223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Chittenden, T., and Simons, M. (2004). Characterization of synectin expression and promoter activity. Gene 342, 29-34. [DOI] [PubMed] [Google Scholar]