Abstract
Actin interacting protein 1 (Aip1p) and cofilin cooperate to disassemble actin filaments in vitro and are thought to promote rapid turnover of actin networks in vivo. The precise method by which Aip1p participates in these activities has not been defined, although severing and barbed-end capping of actin filaments have been proposed. To better describe the mechanisms and biological consequences of Aip1p activities, we undertook an extensive mutagenesis of AIP1 aimed at disrupting and mapping Aip1p interactions. Site-directed mutagenesis suggested that Aip1p has two actin binding sites, the primary actin binding site lies on the edge of its N-terminal β-propeller and a secondary actin binding site lies in a comparable location on its C-terminal β-propeller. Random mutagenesis followed by screening for separation of function mutants led to the identification of several mutants specifically defective for interacting with cofilin but still able to interact with actin. These mutants suggested that cofilin binds across the cleft between the two propeller domains, leaving the actin binding sites exposed and flanking the cofilin binding site. Biochemical, genetic, and cell biological analyses confirmed that the actin binding- and cofilin binding-specific mutants are functionally defective, whereas the genetic analyses further suggested a role for Aip1p in an early, internalization step of endocytosis. A complementary, unbiased molecular modeling approach was used to derive putative structures for the Aip1p-cofilin complex, the most stable of which is completely consistent with the mutagenesis data. We theorize that Aip1p-severing activity may involve simultaneous binding to two actin subunits with cofilin wedged between the two actin binding sites of the N- and C-terminal propeller domains.
INTRODUCTION
Actin cytoskeleton dynamics requires precise and adaptable modes of regulation to carry out a multitude of essential cellular activities. Throughout the cell, actin interacting proteins work cooperatively to conduct cytoskeletal functions with a complexity that is only beginning to be understood. Many cellular processes, including those as basic as cell division and growth, demand the rapid turnover of actin networks, of which filament disassembly is a major rate-limiting factor. Actin interacting protein 1 (Aip1p) and cofilin are two actin binding proteins that function in concert to promote the rapid disassembly of actin filaments. Biochemical assays show that cofilin promotes depolymerization of actin filaments by accelerating pointed-end filament disassembly (Carlier et al., 1997; Lappalainen and Drubin, 1997) and by a severing activity that is very weak at physiological pH (Maciver et al., 1991; Ichetovkin et al., 2000). Aip1p dramatically enhances the depolymerization activity of cofilin-decorated actin filaments and is suspected to do so by assisting cofilin-induced severing and/or by capping the barbed ends of actin filaments (Okada et al., 1999; Rodal et al., 1999; Okada et al., 2002; Balcer et al., 2003; Mohri et al., 2004; Ono et al., 2004).
In Saccharomyces cerevisiae, an aip1 null mutant strain is viable and has subtle defects in actin organization, including apparent excessive actin assembly in actin cortical patches (Rodal et al., 1999). However, synthetic lethal interactions occur when an AIP1 deletion is combined with specific cofilin alleles (Iida and Yahara, 1999; Rodal et al., 1999), confirming a biological role for Aip1p in yeast. Furthermore, partial mislocalization of cofilin from cortical patches to actin cables has been observed in an aip1Δ yeast strain (Rodal et al., 1999). Aip1p is evolutionarily conserved and has been identified in a number of organisms as an important regulator of cytoskeletal dynamics. Xenopus Aip1p (XAip1) localizes to the cell cortex, cytoplasm, and nuclei (Okada et al., 1999). RNA interference (RNAi) inhibition of Aip1p in Drosophila S2 cells leads to accumulation of cortical F-actin and cell surface morphology defects (Rogers et al., 2003). In Caenorhabditis elegans, the loss of Aip1p (UNC-78) results in disorganized assembly of actin filaments in body wall muscle (Ono, 2001). A Dictyostelium null mutant shows impairments in chromosome segregation reliability, motility, endocytosis, and cytokinesis (Konzok et al., 1999; Gerisch et al., 2004). Last, in Arabidopsis, RNAi-induced reductions in Aip1p expression correlate with reduced leaf and plant size that at the lowest expression levels render the plant inviable (Ketelaar et al., 2004). This broad range of phenotypic defects suggests a central role for Aip1p in actin network organization and dynamics.
Aip1p consists of two contiguous seven-bladed β-propeller domains, each made up of seven WD-repeats (Figure 1A; Voegtli et al., 2003). The propellers are positioned at an angle to one another such that one surface of the molecule is concave, whereas the other is convex, and we refer to these as the front and back of the molecule, respectively. Aip1p was first identified in S. cerevisiae through its two-hybrid interaction with actin (Amberg et al., 1995). Subsequent two-hybrid analysis identified a physical interaction between Aip1p and cofilin (Rodal et al., 1999), although biochemical and two-hybrid analyses suggested that the interaction is stabilized by actin (Rodal et al., 1999). Aip1p was also found to be functionally related to cofilin because of its ability to suppress a cof1 temperature-sensitive mutant upon overexpression (Iida and Yahara, 1999). Aip1p increases filament disassembly at low stoichiometry, but this is dependent upon cofilin decoration of the filament and is optimal when the ratio of cofilin to actin is 1:1 (Rodal et al., 1999). Binding of Aip1p to cofilin-decorated actin filaments occurs laterally along the length of the filament and at filament ends (Okada et al., 1999). This “end-capping” of the actin filament has been suggested to assist cofilin by preventing elongation and reannealing of severed filaments (Okada et al., 1999). However, tethered actin filament assays conducted in the presence of cofilin, Aip1p, and filament-capping proteins favor an alternate proposal first presented by Rodal et al. (1999) in which Aip1p enhances the weak severing ability of cofilin (Ono et al., 2004). Therefore, additional evidence is needed to elucidate the mechanism of Aip1p's ability to enhance filament disassembly, in particular, the contributions of severing versus capping.
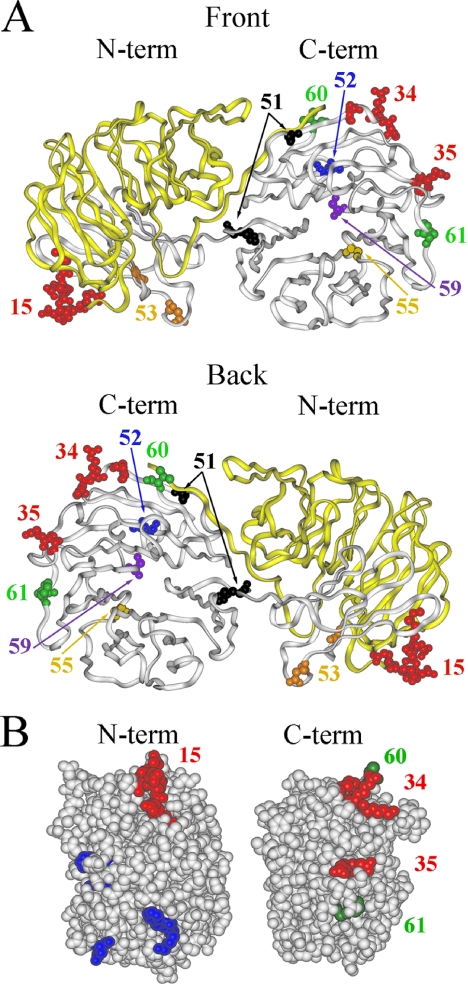
Figure 1.
Aip1p mutagenesis reveals actin and cofilin binding footprints on Aip1p. (A) Aip1p is shown from a front (top) and back (bottom) view with residues changed by aip1 mutant alleles highlighted. Residues involved in the actin interaction are highlighted in red. The region expressed by the aip1-56 truncation mutant, which interacts with actin but not cofilin, is colored in yellow. Cofilin-specific alleles are shown in orange, black, blue, purple, and brown. (B) Sides views of the Aip1p N-terminal (left) and C-terminal (right) propellers depict analogous actin binding domains. Red residues represent loss-of-function cluster charged-to-alanine mutations involved in the actin interaction. Green residues represent randomly selected gain-in-function mutants that apparently increased the Aip1p-actin two-hybrid interaction. Blue residues represent the S. cerevisiae equivalents of site-directed mutants generated in C. elegans AIP1 (UNC-78) that are defective for actin filament disassembly in vitro (Mohri et al., 2004).
To understand how Aip1p binding may alter the actin filament, there is a need to further define the molecular interactions within the Aip1p-cofilin-actin complex. Electron cryomicroscopy of cofilin-decorated filaments revealed that cofilin simultaneously binds two longitudinally adjacent actin monomers (McGough et al., 1997). The cofilin interaction sites on actin are predicted to be between domains 1 and 3 for the upper actin subunit and on domains 2 and 1 of the lower subunit (McGough et al., 1997). Yeast two-hybrid analyses confirm that charged amino acid residues in subdomain 3 of actin are essential for the cofilin interaction and also indicate that Aip1p binds subdomain 4 of actin, in a cofilin-dependent manner (Amberg et al., 1995; Rodal et al., 1999). This suggests that cofilin binding, by altering actin filament structure, creates a conformational change conducive to the Aip1p interaction. Cofilin contains two actin binding sites: the first site is essential for all actin interactions and is predicted to bind domains 1 and 3 of actin, whereas the second is essential for the F-actin interactions and is predicted to bind domains 1 and 2 of actin (Lappalainen and Drubin, 1997).
On cofilin decoration of F-actin, the longitudinal contacts between subdomains 1 and 2 of adjacent actin monomers are destabilized (Bobkov et al., 2002; Galkin et al., 2003). Similar loss of longitudinal contacts have been observed at the pointed ends of undecorated actin filaments, suggesting that cofilin binding maintains a preexisting destabilized conformation of the actin filament (Galkin et al., 2003). This is frequently associated with a 4-5°/subunit reduction in the actin filament twist (McGough et al., 1997). We hypothesize that this conformation may be optimal for Aip1p binding along the length of the filament, further destabilizing filaments through an intensified structural distortion that leads to enhanced filament breakage. After filament cleavage, Aip1p may remain bound to the newly formed filament barbed end, where it could potentially assist in barbed-end regulation.
To further unravel the molecular details of interactions within the Aip1p-cofilin-actin complex, we have undertaken a genetic dissection of AIP1 aimed at defining functional sites on Aip1p. This analysis confirmed the presence of a strong actin binding site on the N-terminal propeller of Aip1p (Mohri et al., 2004) but suggested an additional site of contact for actin on the analogous region of the C-terminal propeller. Separation-of-function mutants define a possible site of interaction for cofilin between the two actin binding sites of Aip1p bridging the two β-propeller domains. These data are completely consistent with an independently derived molecular model for the Aip1p-cofilin interaction. In addition, biochemical, genetic, and cytological approaches were used to further define the role of Aip1p within S. cerevisiae, while also attempting to address the filament severing versus filament capping debate. Our data link Aip1p to a discrete step in the process of endocytosis and for the first time indicate a physiological role for Aip1p in S. cerevisiae.
MATERIALS AND METHODS
Yeast Two-Hybrid Analysis of Site-directed and Random aip1 Mutants
Plasmid pAIP6 encoding a fusion of the GAL4 activation domain (AD) to AIP1 (Amberg et al., 1995) in vector pACT (Durfee et al., 1993) was the template in PCR reactions using Advantage DNA Polymerase (Clontech, Mountain View, CA). Site-directed mutants for cluster-charge-to-alanine scan alleles were created by overlap extension fusion PCR with external primers 2H1 (5′-TGATGAAGATACCCCACC-3′) and 2H5 (5′-ACAGTTGAAGTGAACTTGCG-3′) and internal primers specific to the mutant allele. The resulting PCR product was cotransformed with double-digested (BamHI and EcoRI) plasmid pACTII (gift of S. Elledge, Howard Hughes Medical Institute, Center for Genetics and Genomics, Boston, MA) by lithium acetate (Rose et al., 1989) into yeast strain Y187 (Table 1; Durfee et al., 1993), and the gap repair transformants were selected on SC-Leu plate medium. The transformants were analyzed for their abilities to interact with actin and cofilin by yeast two-hybrid analysis (Fields and Song, 1989). Briefly, Y187 cells carrying mutant aip1-AD fusion plasmids were spotted on SC-Leu and allowed to grow overnight. These were then replica plated to duplicate YPD plates, which were overlaid with a lawn of yeast strain Y190 transformed with plasmid pDAb7 encoding a fusion of ACT1 to the GAL4 DNA binding domain (Amberg et al., 1995) or plasmid pTY1 encoding a fusion of COF1 to the GAL4 DNA binding domain. The cells were allowed to mate for 1 d after which diploids were selected on SC media lacking tryptophan and leucine. The diploids were then replica plated to SD medium plus 10 μg/ml adenine and 25, 50, or 100 mM 3,5-aminotriazole. All mutants were rescued and amplified in Escherichia coli by standard methods (Rose et al., 1989).
Table 1.
S. cerevisiae strains
| Name | Genotype | Source |
|---|---|---|
| FY23×86 | MATa/α ura3-52/ura3-52 leu2Δ1/leu2Δ1 trp1Δ63/TRP1 HIS3/his3Δ200 | Rodal et al. (1999) |
| Y187 | MATα gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 GAL–lacZ | Bai and Elledge (1996) |
| Y190 | MATagal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 | Bai and Elledge (1996) |
| URA3::GAL–lacZ LYS2::GAL–HIS3cyhr | ||
| LGY2×3 | MATa/α aip1Δ::URA3/aip1Δ::URA3 ura3-52/ura3-52 | Rodal et al. (1999) |
| leu2Δ1/leu2Δ1 trp1D63/TRP1 HIS3/his3Δ200 | ||
| DDY1253 | MATα ura3-52 his3Δ200 lys2-801 cof1-4 | Lappalainen et al. (1997) |
| DDY1264 | MATα ura3-52 his3Δ200 lys2-801 cof1-19 | Lappalainen et al. (1997) |
| MCY9 | MATaaip1-15:G418r ura3-52 leu2Δ1 his3Δ200 | |
| MCY10 | MATα aip1-15:G418r ura3-52 leu2Δ1 trp1Δ63 | |
| MCY13 | MATaaip1-55:G418r ura3-52 leu2Δ1 his3Δ200 | |
| MCY14 | MATα aip1-55:G418r ura3-52 leu2Δ1 trp1Δ63 | |
| MCY17 | MATaaip1-59:G418r ura3-52 leu2Δ1 trp1Δ63 | |
| MCY18 | MATα aip1-59:G418r ura3-52 leu2Δ1 his3Δ200 | |
| MCY21 | MATaaip1-34/35:G418r ura3-52 leu2Δ1 trp1Δ63 | |
| MCY22 | MATα aip1-34/35:G418r ura3-52 leu2Δ1 his3Δ200 | |
| MCY25 | MATaaip1-60:G418r ura3-52 leu2Δ1 trp1Δ63 | |
| MCY26 | MATα aip1-60:G418r ura3-52 leu2Δ1 his3Δ200 | |
| MCY27 | MATacof1-19:LEU2 aip1-60:G418r ura3-52 leu2Δ1 trp1Δ63 | |
| MCY60 | MATα aip1Δ::NAT ura3Δ0 leuΔ0 his3Δ1 lys2Δ0 mfa1Δ::PMFA1-spHIS5 |
Random PCR mutagenesis used a similar method, except Taq polymerase was substituted. Successful transformants were screened for an ability to interact with actin, but not cofilin, or vice versa. Mutants showing such a defect were rescued, retested to confirm their specific defects, and submitted for sequencing. Images of aip1p mutants were generated using Insight II Version 2000 (Molecular Simulations, San Diego, CA). Coordinates for Aip1p were retrieved from the Collaboratory for Structural Bioinformatics Protein Data Bank (PDB file 1PI6).
Genomic Integration of aip1 Mutants
To generate strains carrying mutated aip1 genes, aip1-x:G418r cassettes were created by cloning appropriate mutation-containing aip1 fragments out of the two-hybrid vectors and into the plasmid pMC60 and then excising the aip1-x:G418r cassettes with SalI and KpnI; pMC60 contains the aip1-x:G418r cassette cloned into pBluescript II KS (+/-) (Stratagene, La Jolla, CA). An aip1-x:G418r cassette was inserted into the aip1Δ::URA3 genomic locus of diploid strain LGY2xLGY3 by homologous recombination. Successful transformants were selected on YPD + G418 media. Strains were confirmed by sequencing PCR products generated using external primers LGo-Aip1-1 (5′-AATACTAGCTATTGCTTTCCG-3′) and MCo-Aip1-130 (5′-AGTCTTTTCCTTACCCAT-3′) on genomic DNA templates.
Synthetic Sick/Lethal Testing of aip1 Mutants
For aip1 × cof1-4 analyses, aip1 mutant strains (MAT a aip1-x:G418r ura3-52 trp1Δ) were crossed to DDY1253. Diploids were selected on SC-leu + G418 media, sporulated for 4 to 6 d at 25°C, dissected, and analyzed for synthetic viability or growth defects.
For the aip1Δ × endocytosis-related gene deletion screen, yeast strain MCY60 was hand-crossed against 86 different gene deletion mutants from the EUROSCARF collection (MAT a geneΔ::G418r ura3-52 leu2Δ1 met15Δ his3Δ). Diploids were selected on YPD + G418 + Natr and then sporulated for 4 to 6 d at 25°C and dissected. Dissected tetrads were plated onto YPD, allowed to grow overnight at 30°C, and then replica plated to YPD + G418 (30°C) and YPD + Nat (30°C), to identify colonies in which both deletion alleles cosegregated. When double deletion mutant colonies remained viable, they were struck on YPD for single colonies and plated at 30 and 37°C to inspect for growth defects.
EUROSCARF deletion alleles that showed synthetic defects with an aipΔ were also tested against our array of aip1 mutant strains (MAT a aip1-x:G418r ura3-52 trp1Δ). Because both mutations were marked with G418r, PD versus NPD tetrads were used to identify synthetic growth defects at 30 and 37°C.
Yeast Immunofluorescence
Immunofluorescence was performed by standard protocols using a methanol/acetone fixation (Amberg et al., 2005). Affinity-purified anti-Aip1p antibody (primary) was used at a dilution of 1:100. Affinity-purified rabbit anti-cofilin antibody (primary) was used at 1:100 (Rodal et al., 1999). Fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Cappel, Costa Mesa, CA; ICN Biochemicals) was used at 1:1000.
Rhodamine-Phalloidin Staining
Staining of the actin cytoskeleton was performed using a standard protocol (Amberg et al., 2005). Briefly, a yeast cell culture grown to 2 × 107 was subjected to incubation with electron microscopy-grade formaldehyde at a final concentration of 4%, washed with phosphate-buffered saline (PBS), and treated with rhodamine-labeled phalloidin (1:10 dilution of 6.6 μM in methanol). After washing again with PBS, cells were suspended in mounting solution and viewed by fluorescence microscopy.
Protein Purification
Yeast actin was purified by a modified DNaseI affinity purification procedure (Kron et al., 1992). Briefly, 200 mg of DNase I (Roche Diagnostics, Indianapolis, IN) was coupled to 5 g of swelled Sepharose 4B (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. Then, 3 ml of DNase I-Sepharose was loaded into a 2.5 × 10-cm Econo-column (Bio-Rad, Hercules, CA), the column was washed with: 15 ml of 10% formamide in G-buffer (10 mM Tris, pH 7.5, 0.5 mM ATP, 0.2 mM dithiothreitol [DTT], and 0.2 mM CaCl2), 15 ml of G-buffer + 0.2 M NH4Cl, and 15 ml of G-buffer. Four liters of yeast strain FY23 × 86 was grown to a density of ∼2 × 107 cells/ml, pelleted in four tubes, and each resuspended in 40 ml of G-buffer (+ATP, DTT, phenylmethylsulfonyl fluoride, and Calbiochem protease inhibitor cocktail). Each was passed once through a French Press set at 1200 psig. The lysate was clarified in a Beckman JA-20 rotor at 12,000 rpm for 30 min at 4°C and then in a Beckman Ti50.2 rotor at 40,000 rpm for 120 min at 4°C. The supernatant was loaded in equal volumes onto two DNase I columns at a flow rate of ∼1-2 ml/min. Each column was washed with 15 ml of G-buffer + 10% deionized formamide, 14 ml of G-buffer + 0.2 M NH4Cl, and 15 ml of G-buffer. The actin was eluted with 15 ml of G-buffer + 50% deionized formamide. Contaminating formamide was removed by dialyzing overnight in 1 liter of G-buffer (0.05 μM ATP). Samples were concentrated in a Micron-10 device (Amicon, Billerica, MA), clarified by ultracentrifugation in a Beckman TLA100 rotor at 90,000 rpm for 20 min at 4°C, snap frozen, and stored at -80°C until use. Bradford assays were used to determine protein concentration.
Yeast cofilin was expressed in E. coli DH5α cells as a glutathione S-transferase (GST) fusion protein under the control of the Plac promoter. GST-cofilin was purified as described previously (Lappalainen and Drubin, 1997) but with the following alterations. Cells were grown to stationary phase in 1 liter of Luria broth + ampicillin. PBS was substituted with Tris-buffered saline (TBS) (50 mM Tris, pH 7.5, and 100 mM NaCl) in all relevant steps. Before binding to glutathione-agarose resin, Triton X-100 was mixed with the lysate to a 1% final concentration and then was spun in a Beckman centrifuge using a JA-20 rotor at 10,000 rpm for 20 min. The supernatant was extracted, added to 1.5 ml of a 50% resin slurry, and rocked for 90 min at room temperature. After washing 4 × 10 ml of ice-cold TBS, thrombin was added to the beads (3 ml of TBS, 10.4 μl of 0.5 M CaCl2, and 50 μl of thrombin at 1 U/μl) and incubated at room temperature for 3 h. After cleavage, the supernatant was collected and the resin was washed with 2 × 2 ml of TBS, washes were added to the supernatant. This was dialyzed overnight against 50 mM Tris, pH 7.5, at 4°C. Samples were further purified by fast-performance liquid chromatography (Bio-Rad) on a UNO-Q1 column and washed at 1 ml/min with 50 mM Tris, pH 7.5 and then eluted with a linear KCl gradient (100-400 mM) in 50 mM Tris, pH 7.5. Peak fractions were dialyzed overnight against 10 mM Tris, pH 7.5, and 50 mM NaCl for 5 h at 4°C, concentrated (Microcon YM-10 columns, Millipore, Bedford, MA), confirmed by Western assay, and stored at -80°C.
Yeast Aip1p was expressed in yeast cells as a GST fusion protein under the control of a galactose-inducible promoter [pEG(KT)]. Cells were inoculated into 1 liter of SC-Ura-Leu + 3% glycerol + 1% EtOH + 0.1% glucose and grown for 20-24 h at 30°C. Galactose was added to a 2% final concentration and induction continued for 10 h. GST-Aip1p purification proceeded as described for GST-cofilin, with the following alterations. Before binding to glutathione-agarose resin, cell lysate (with no Triton X-100) was pelleted in a Beckman centrifuge using a JA-20 rotor at 12 krpm for 30 min. The supernatant was then spun again in a Beckman ultracentrifuge using a 70 Ti rotor at 50,000 rpm for 50 min.
Profilin was purified from yeast strain FY23 × 86 as described previously (Haarer et al., 1990). Profilin was stored at 4°C on ice and used within 10 d of purification.
Molecular Docking
The coordinates of Aip1p and cofilin were obtained from the Protein Data Bank (PDB file 1PI6 and 1CFY, respectively). The Aip1p structure was missing a surface loop (544-549) that was first rebuilt using PLOP (Jacobson et al., 2004). Using the molecular dynamics package NAMD (Kale, 1999), simulations of both complexes were performed at 300 K using an NPT ensemble with CHARMM27 force-field and explicit solvent (TIP3P water). The molecular dynamics simulations were each comprised of energy minimization, followed by heating to 300 K at intervals of 75 K, equilibration for 500 ps, and finally a production run of 5 ns for Aip1p and 10 ns for cofilin. Representative conformations of each structure were extracted from the trajectory for subsequent use in molecular docking studies. Because both proteins are held rigid during the docking simulation, this step allows us to capture some of the backbone and side chain fluctuations in the proteins. Because cofilin is the smaller protein and exhibited significantly more dynamics than Aip1p, we selected Aip1p structures every 500 ps (total of 10 structures), whereas for cofilin we selected structures every 10 ns, resulting in 1000 conformations. Five docking trajectories were carried out with each combination of Aip1p and cofilin structures using AutoDock 3.0 (Morris et al., 1998). The resulting 50,000 docking predictions were clustered based on a root-mean-square deviation of 10 Å, and the structures exhibiting the best clustering were selected as candidate structures. This resulted in a total of six likely docked conformations with one conformation that showed particularly good correspondence with existing mutagenesis data.
RESULTS
Identification of the Actin Interaction Sites on Aip1p
We performed a cluster charged-to-alanine scan (Wertman et al., 1992) to introduce surface mutations into AIP1 to identify potential sites of actin interaction. This technique neutralized charged clusters of amino acid residues on the surface of Aip1p and was thus expected to disrupt points of electrostatic interaction between Aip1p and its binding partners (actin and cofilin). Each of 34 aip1 mutant alleles (aip1-1 to aip1-33) was tested against actin by yeast two-hybrid analysis (Table 2). Just one allele, aip1-15, showed a defect in actin binding, and this was only partial (Figure 2A). aip1-15 neutralizes a prominent patch of negatively charged residues (I245A, E246A, D247A, D248A, Q249A, E250A) within blade five of the N-terminal β-propeller (Figure 1A).
Table 2.
AIP1 charge-to-alanine scan alleles
| Allele | Amino acid changes |
|---|---|
| 1 | K7A, E8A |
| 2 | Q17A, R18A, N19A |
| 3 | D47A, D48A, D50A |
| 4 | D85A, E86A, K89A |
| 5 | D98A, K99A, E100A, N102A |
| 6 | K109A, E111A, Q113A |
| 7 | E127A, R129A, R130A |
| 8 | E136A, R138A, D139A, N140A |
| 9 | N151A, E155A |
| 10 | K169A, Q170A, R172A |
| 11 | Q161A, R162A |
| 12 | K194A |
| 13 | D228A, R229A, K230A |
| 14 | D235A, K237A |
| 15 | I245A, E246A, D247A, D248A, Q249A, E250A |
| 16 | D263A, Q265A, K266A |
| 17 | D293A, K294A, Q295A, Q296A |
| 18 | N299A, Q300A, Q301A |
| 19 | D329A, E330A, K333A |
| 20 | N339A, K340A |
| 21 | D356A, R358A |
| 22 | K285A, Q288A, K289A |
| 23 | Q369A, D370A |
| 24 | K382A, Q384A, E385A |
| 25 | N415A, N416A, D417A |
| 26 | E464A, E465A, N467A |
| 27 | D516A, Q518A, R520A, E521A |
| 28A | E543A, K544A |
| 28B | N547A, E548A, E549A, E550A |
| 29 | I551A, E552A, E553A, D554A |
| 30 | D562A, N564A |
| 31 | K575A, K578A |
| 32 | N588A, N589A |
| 33 | K608A, R609A, N611A |
| 34 | K571A, R572A, K575A |
| 35 | D516A, E521A |
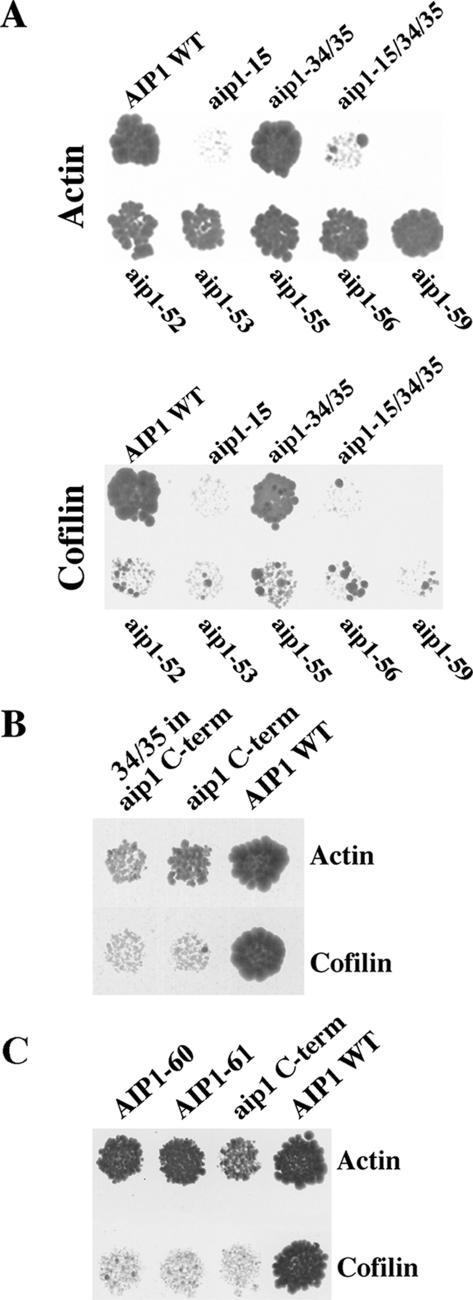
Figure 2.
Two-hybrid analysis of aip1 mutants. (A) Yeast two-hybrid interactions between mutants of Aip1p and actin (top) or cofilin (bottom) were measured based on activation of expression of the HIS3 reporter (growth on minimal media containing 100 mM 3-AT [top] and 50 mM 3-AT [bottom]). (B) The C-terminal propeller of aip1p (aip1 C-term) interacts by yeast two-hybrid with actin but not with cofilin (10 mM 3-AT). This interaction is severely impaired by addition of the aip1-34/35 mutations. (C) Two spontaneously generated mutants on the aip1 C-term (aip1-60 and aip1-61) increase the two-hybrid interaction with actin.
The inability of the charged-to-alanine scan to identify mutants that resulted in a complete loss of actin binding suggested either that the actin interacting site is too large to effectively disrupt with the neutralization of a single charged cluster or that more than one actin binding site exists on Aip1p. Taking into consideration the possibility that the double propeller structure of Aip1p could be the result of a gene duplication, we tested the C-terminal β-propeller (residues 337-615) for the ability to interact with actin in the two-hybrid system. As shown in Figure 2B, the C-terminal propeller of Aip1p demonstrates a weak two-hybrid interaction with actin.
Despite low protein sequence homology between the two propellers, we chose to test whether the observed functional homology (actin binding) corresponds to an actin interacting site located within corresponding regions of each propeller. Therefore, two charged clusters that had not been specifically targeted previously were identified at a region of the C-terminal propeller that is located in a structurally and sequentially similar location to the analogous region of the N-terminal propeller where the aip1-15 mutation is located. Each was converted to a new charged-to-alanine allele. These two alleles, aip1-34 (K571A, R572A, K575A) and aip1-35 (D516A, E521A) lie on the sixth and fifth blades, respectively, of the C-terminal propeller (Table 2 and Figure 1, A and B). When tested in the context of full-length Aip1p for a two-hybrid interaction with actin, neither of the mutants alone or in combination showed a defect (Figure 2A). However, when the aip1-15 mutant was tested in combination with the aip1-34 and aip1-35 mutants, the actin interaction was eliminated (Figure 2A). Furthermore, when the aip1-34 and aip1-35 mutations were combined in the context of the C-terminal propeller alone (residues 337-615), the weak actin interaction of this domain was markedly impaired (Figure 2B). These data suggest that two analogous actin binding sites are located on Aip1p: one on each β-propeller (Figure 1, A and B).
Confirmation of Two Independent Actin Interaction Sites
Additional mutants have been recovered that strengthen the argument that each of the two propellers contains an actin binding site. In each case, a random mutagenesis technique was implemented. First, the AIP1 gene was mutated through a PCR reaction in which a low fidelity Taq polymerase was used to incorporate random mutations into the gene. These mutated PCR products were introduced by recombination (Ma et al., 1987) into a two-hybrid DNA activation domain vector and differential interaction screening was used to isolate separation-of-function mutants that had the ability to interact with actin but not cofilin in two-hybrid assays. From this we recovered aip1-56, which replaces Trp261 with a stop codon, ending translation of Aip1p ∼80% into the N-terminal propeller. Thus, an N-terminal domain was identified that binds actin independent of the C-terminal propeller (Figure 1A in yellow and Figure 2A).
A slightly different strategy was used to confirm the site of actin interaction on the C-terminal propeller. The C-terminal propeller domain (expressing amino acids 337-615), which shows a weak two-hybrid interaction with actin and none with cofilin, was subjected to low-fidelity PCR as described above and screened in the two-hybrid system for gain-in-function mutations that display an apparent increased actin interaction. Two mutants were identified that increased activation of the two-hybrid reporters when tested against actin; sequencing of aip1-60 and aip1-61 revealed mutations Glu615Lys and Phe481Leu, respectively. These residues map very close and on either side of the aip1-34 and aip1-35 mutations, supporting the presence of an actin interacting site on the C-terminal propeller that is consistent with the location suggested by the two-hybrid results with the aip1-34 and aip1-35 mutants (Figures 1, A and B, and 2C; the final two C-terminal amino acid residues of Aip1p were not resolved in this crystal structure, so aip1-60, shown at residue 613, is actually closer to aip1-34 than this illustration suggests).
Differential Interaction Screening Identifies the Cofilin Interaction Site on Aip1p
To identify the cofilin binding site on Aip1p, random mutagenesis and differential interaction screening, as described above, were used to genetically dissect the ability of Aip1p to interact with actin and cofilin by two-hybrid. Mutants were selected based on an ability to interact well with actin at high levels of 3-AT inhibition of the His3p reporter but not with cofilin, even at low levels of 3-AT inhibition. By screening for isolates that did not lose their actin interacting activity, we targeted mutants that presumably maintain sound structural integrity, thus limiting their defects to the cofilin interaction site.
Three separate single mutations were found that were able to separate the actin and cofilin interactions: aip1-52 (Thr558Pro), aip1-55 (Gly449Asp), and aip1-59 (Ser492Leu) (Figures 1A and 2A). These mutations lay toward the center of the C-terminal propeller, between blades 5 and 6, 3 and 4, and 4 and 5, respectively, and suggest the existence of a cofilin interaction site on the front surface of the C-terminal propeller. A fourth mutant allele of the same phenotype, aip1-51, includes a combination of two mutations, H338P and A579T (our unpublished data). In addition, the mutant aip1-53 contains two mutations within the N-terminal propeller, Gly271Glu and Asp293Asn, which result in the loss of cofilin binding as well (Figures 1A and 2A). The unique aspect of this mutant is that it is the only allele that alters residues on the N-terminal propeller of Aip1p, with both mutations falling on blade 6. The necessity of the N-terminal propeller for the cofilin interaction is also supported by our preceding observation that the C-terminal propeller domain (residues 337-615), which has a weak two-hybrid interaction with actin, cannot interact with cofilin (as was also true for our gain-in-function C-terminal propeller mutations; Figure 2B). Therefore, these data collectively suggest that although cofilin largely contacts the C-terminal propeller of Aip1p, interactions with the N-terminal propeller are also essential to stabilize the interaction. All mutants and respective phenotypes addressed in this article are summarized in Table 3.
Table 3.
Summary of the phenotypic analyses of aip1 and cof1 mutants
| Two hybrid
|
Localization
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Allele | Mutations | Cell morphology | With actin | With cofilin | Aip1p | Cofilin | Actin organization | Biochemistry |
| AIP1 | WT | +++ | +++ | Cortical patches | Cortical patches | ++++ | Normal | |
| aip1Δ | WT | na | na | na | Catches and cables | +++ | na | |
| aip1-15 | 1245A, E246A, D247A, D248A, Q249A, E250A | WT | + | + | Cortical patches | Patches and cables | +++ | F-Actin depolym. defects |
| aip1-34/35 | K571A, R572A, K575A/D516A, E521A | 1% Elongated buds | +++ | +++ | Cortical patches | Patches and cables | +++ | Normal |
| aip1-15/34/35 | See above | nd | – | – | nd | nd | nd | Normal |
| aip1-51 | H338P, A579T | nd | ++ | – | nd | nd | nd | nd |
| aip1-52 | T558P | nd | ++ | – | nd | nd | nd | nd |
| aip1-53 | G271E, D293N | nd | ++ | – | nd | nd | nd | nd |
| aip1-55 | G449D | WT | ++ | – | Cortical patches | Patches and cables | +++ | Normal |
| aip1-56 | W261Stop | nd | ++ | – | nd | nd | nd | nd |
| aip1-59 | S492L | 1% Elongated buds | ++ | – | Cytosolic | Patches and cables | +++ | Normal |
| aip1 C-term | G337-E615 | nd | ± | – | nd | nd | nd | nd |
| aip1-60 | G337-E615, E615K | WT | + | – | Cortical patches | Cortical patches | ++++ | Normal |
| aip1-61 | G337-E615, F481L | nd | + | – | nd | nd | nd | nd |
| With actin | With Aip1p | Aip1p | Cofilin | |||||
| COF1 | WT | +++ | +++ | Cortical patches | Cortical patches | ++++ | Normal | |
| cof1-4 | S4A | Enlarged | +++ | – | Cortical patches | Cortical patches | + | Capping defect |
| cof1-19 | R109A, R110A | Slightly enlarged | ++ | ++ | Cytosolic | Cortical patches | ++ | Severing and capping defects |
| cof1-13 | E59A, D61A | WT | +++ | – | Cortical patches | Cortical patches | ++++ | nd |
| cof1-22 | E134A, R135A, R138A | nd | ++ | – | Cortical patches | Cortical patches | – | F-Actin depolym. |
depolym., depolymerization; na, not applicable; nd, no data; +++, WT growth; ++, moderate growth; +, weak growth; ±, very weak growth; –, no growth.
Molecular Docking of Aip1p and Cofilin
Molecular docking studies using Aip1p and cofilin resulted in a predicted complex between these two proteins. The complex shows impressive complementarity between the two protein surfaces considering that this is the result of docking simulations where both proteins were held rigid (Figure 3A). We have identified eight potential salt bridges that could be formed between the two proteins in this model and the residues involved are listed (cofilin residue-Aip1p residue): D123/E126-K533, E77-K89, D106-R18, D91-K410, R135-D585, R80-E111, K20-E136, and K82-D85. This model corresponds very well to the binding footprint predicted by our differential interaction screening, which identified Aip1p's cofilin binding domain (Figure 3B). In addition, biochemical evidence shows that cof1-19p is defective for Aip1p-induced actin filament disassembly (this work) and the mutated residues of this allele (R109, R110) are at the predicted Aip1p-cofilin interface. We also compared the docking results to three previously created mutant cofilin alleles, cof1-4 (S4A), cof1-13 (E59A, D61A), and cof1-22 (E134A, R135A, R138A) (Lappalainen and Drubin, 1997), which have been shown to disrupt the interaction with Aip1p but not actin in the yeast two-hybrid system (Rodal et al., 1999). The cof1-4 and cof1-22 mutations lie at the predicted interface of the Aip1p-cofilin interaction in agreement with our model, whereas the cof1-13 mutations face the opposite side of the cofilin molecule (Figure 3B). Although the cof1-13 allele does not fit well with our docking model, this discrepancy may be explained by the fact that the mutated residues lie directly above several basic residues that are at the binding interface and may be disrupting the interaction through conformational/allosteric changes. Furthermore, based on the spacial disparity among the three mutant cofilin alleles implicated in the Aip1p interaction, it does not seem possible that a single molecular docking model can satisfy all three constraints.
Figure 3.
Molecular docking of the Aip1p-cofilin complex. (A) The predicted protein complex between Aip1p (green) and cofilin (orange) as the result of molecular docking studies. The contacting interfaces of the two proteins are rendered as a transparent surface to illustrate their complementarities. (B) The specific residues found to affect Aip1p-cofilin binding have been labeled and rendered as spheres. Yellow residues represent mutations that disrupt cofilin's two-hybrid interaction with Aip1p but not actin. Purple residues represent mutations that disrupt Aip1p's two-hybrid interaction with cofilin but not actin. This figure was created using VMD (Humphrey et al., 1996).
Biochemical Analysis of Actin Filament Disassembly by aip1p Mutants
The following aip1p mutants that showed a defect by two-hybrid analysis were overexpressed in yeast as GST fusions and purified: aip1-15p, aip1-34/35p, aip1-55p, aip1-59p, aip1-60p (as a full-length version) as well as Aip1p WT. All aip1p mutants were then cleaved from GST by a thrombin digest, with the exception of aip1-15p, which we were unable to cleave. Our findings as well as those documented by Mohri et al. (2004) confirm that Aip1p-GST performs equally well as Aip1p in control disassembly assays. Each aip1p mutant was used in a severing assay, as described by Rodal et al. (1999). Although we are not positive that severing is the primary function being measured, we feel very strongly that this is the case and thus refer to these disassembly assays as severing assays. Briefly, 2.5 μM F-actin was combined with cofilin and Aip1p at a ratio of 20:20:1, respectively, and allowed to incubate at room temperature for 10 min. Samples were centrifuged at high speed (360,000 × g), and the efficiency of actin filament disassembly was measured by observing the pellet and supernatant fractions by SDS-PAGE. All of our severing assays showed that the aip1p mutants bound normally to the cofilin-decorated actin filaments, as observed by the amount of aip1p that pelleted with the remaining F-actin (our unpublished data). This indicates that none of the mutants were defective for F-actin binding in vitro. Only aip1-15p had a detectable severing defect, with a decrease in efficiency of ∼50% (Figure 4, lane 3 versus lane 4). Based on our data, the N-terminal propeller's interaction with actin is far stronger in the two-hybrid system than the C-terminal propeller (Figure 2, A and B). These two-hybrid findings coupled with our biochemical data suggest that the Aip1p N-terminal propeller has a greater role than the C-terminal propeller for in vitro actin filament disassembly.
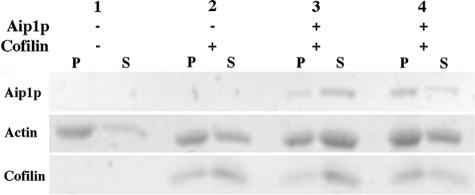
Figure 4.
Biochemical analysis reveals a severing defect for the aip1-15 mutation. Polymerized actin filaments (2.5 μM) were incubated with or without cofilin and GST-Aip1p (lane 3) or GST-aip1-15p (lane 4) for 10 min. The ratios of actin/cofilin and actin/Aip1p were 1:1 and 20:1, respectively. After high-speed centrifugation (360,000 × g), the extent of filament disassembly was determined based on the amount of actin that remained in the pellet fraction (F-actin) versus the supernatant fraction (G-actin), as detected by SDS-PAGE and SYPRO Ruby staining.
Biochemical Capping Analysis of Aip1p and Cof1p
We also intended to test our aip1p mutants using a biochemical actin filament-capping assay (Balcer et al., 2003). In this assay, F-actin is combined with profilin, cofilin, and Aip1p at a 20:40:4:1 ratio and allowed to incubate at room temperature for 20 min. Samples are then spun down at high speed (360,000 × g), and actin disassembly is assessed based on the amount of F-actin in the pellet versus the amount of actin in the supernatant. Theoretically, the cofilin concentration is too low for Aip1p to effectively bind and sever filaments (Rodal et al., 1999), but it should allow Aip1p to bind to barbed ends. As monomers dissociate from pointed ends, they form a dimeric complex with profilin, preventing them from rejoining at filament pointed ends but not at filament barbed ends. If Aip1p caps the barbed end of the actin filaments, the profilin-bound actin becomes sequestered, leading to a net depolymerization of the actin filaments from the pointed end.
We have observed a small but repeatable defect for cof1-19p in this assay, which is consistent with previous findings (Balcer et al., 2003). In addition, cof1-4p was found to have an even more severe defect (Figure 5 A, lanes 5-7). We also noticed that cof1-19p but not cof1-4 is defective in our severing assay (Figure 5A, lanes 2-4). Interestingly, when we repeated the capping assay using the cofilin mutants with and without Aip1p, the amount of actin disassembly looked nearly identical, indicating that in our hands this assay is not measuring Aip1p-related effects (compare Figure 5A, lanes 5-7 with Figure 5B, lanes 4-6). In an attempt to achieve Aip1p-dependent filament disassembly, we increased the actin concentration twofold (4 μM) and increased the profilin concentration 1.5-fold (6 μM), while leaving the cofilin and Aip1p concentrations the same (0.4 and 0.1 μM, respectively), thus increasing the actin:profilin:cofilin:Aip1p ratios to 40:60:4:1. The result was that disassembly no longer occurred without Aip1p, and subsequent addition of Aip1p to the reaction resulted in moderate filament disassembly (our unpublished data). However, we cannot be confident that this assay is in fact measuring barbed end capping rather than severing by Aip1p. Although the small amount of cofilin used in this experiment should not allow extensive Aip1p-induced severing, the cooperative nature of cofilin binding to filaments is likely to saturate short stretches of actin filaments (McGough et al., 1997; Ressad et al., 1998; Pope et al., 2000), creating ideal sites for Aip1p severing. We propose two scenarios of how minimal Aip1p-induced severing could lead to extensive disassembly of filaments in this assay. The first is that this assay is not measuring barbedend regulation but is instead measuring the cumulative effects of cofilin-enhanced pointed-end disassembly plus monomer sequestration by profilin, which has been reported under some conditions (Carlsson et al., 1977). Aip1p-enhanced severing would create an increased number of short filaments that would not pellet during centrifugation, while also increasing the number of pointed ends from which filaments could disassemble, allowing profilin to more rapidly sequester the pool of actin in a monomeric state. Therefore, the cofilin mutant defects observed in this assay could simple be attributed to a decreased enhancement of pointed-end disassembly. The second explanation is that cofilin is having a profilin-dependent barbed-end gating effect, in which it blocks profilin-bound monomers from rejoining filaments at the barbed end. Severing by Aip1p would enhance the number of cofilin-decorated barbed ends, thus enhancing filament disassembly. Our F-actin plus profilin alone control sample (Figure 5B, lane 3) shows significant filament disassembly, most likely because of actin monomer sequestration by molar excess of profilin. In lanes 7-9, the profilin concentration is reduced to 25% of the previous level and filament disassembly is markedly reduced, compared with lanes 4-6. It is not clear whether this effect is strictly related to decreased monomer sequestration or whether this supports the idea of a cofilin-dependent barbed-end gating effect, because there are fewer profilin-bound actin monomers in these samples. The very limited degree to which cofilin alone is able to disassemble actin filaments hints at a synergistic effect with profilin, which would favor the gating hypothesis. In other words, even the total loss of the limited filament disassembly caused by cofilin alone (Figure 5B, compare lanes 1 and 2) does not seem as though it would be sufficient to account for the differences between the wild-type and mutant cofilins when they are combined with profilin (Figure 5B, lanes 4-6).
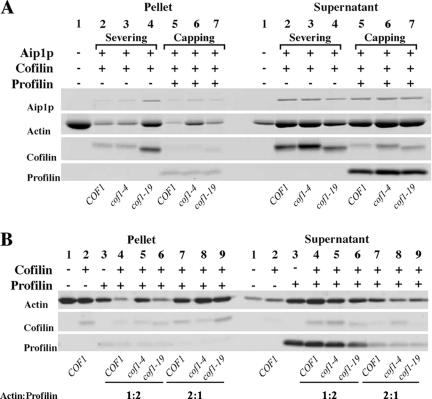
Figure 5.
Biochemical capping assays demonstrate that cofilin-specific disassembly defects are Aip1p independent. In all cases, actin filament disassembly was measured by high-speed centrifugation (360,000 × g) followed by SDS-PAGE and SYPRO Ruby staining to compare the relative amounts of actin in the pellet (F-actin) versus actin in the supernatant (G-actin). (A) The cof1p mutants were tested for actin filament-severing and -capping defects in the presence of Aip1p. Polymerized actin filaments (2 μM) were incubated with or without cofilin, Aip1p, and profilin (where applicable). In lanes 2-4 (severing assay), the ratios of actin/cofilin and actin/Aip1p were 1:1 and 20:1, respectively. In lanes 5-7 (capping assay), the ratios of actin/cofilin, actin/Aip1p, and actin/profilin added were 5:1, 20:1, and 1:2, respectively. Lanes 2 and 5 contained wild-type cofilin. Lanes 3 and 6 contained cof1-4p. Lanes 4 and 7 contained cof1-19p. These data show that cof1-4p and cof1-19p have different defects depending on the method used to measure actin filament dynamics. (B) cof1p mutants were tested for actin filament-capping defects without Aip1p to determine whether the defects observed in 6A are Aip1p dependent. Polymerized actin filaments (2 μM) were incubated with or without cofilin and profilin. The actin/cofilin ratio is 5:1. The ratios for actin/profilin added were 1:2 (lanes 4-6) or 2:1 (lanes 7-9). Lanes 4 and 7 contained wild-type cofilin. Lanes 5 and 8 contained cof1-4p. Lanes 6 and 9 contained cof1-19p. These data show that under these conditions differences in assembly at the barbed end are Aip1p independent but cofilin dependent and differentially effected by cof1-4p versus cof1-19p. cof1-4p is more defective in the capping assay than cof1-19p, whereas cof1-19p is more defective than cof1-4p for Aip1p-induced severing.
In summary, cof1p-associated defects in the capping assay persist in the absence of Aip1p and are thus unrelated to barbed-end capping by Aip1p. In addition, Aip1p-dependent filament disassembly was only achieved at increased actin concentrations, and this disassembly does not differentiate between actin filament severing and capping.
Genetic Analysis of aip1p Mutants
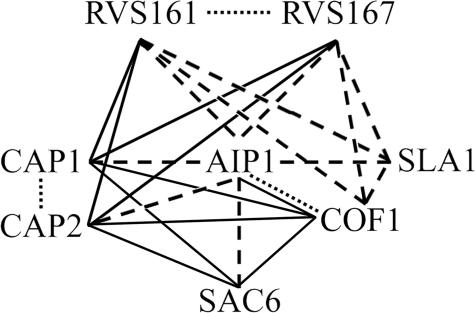
Despite the significant insights that have been made regarding Aip1p's ability to induce disassembly of cofilin-decorated actin filaments, the physiological role for these activities of Aip1p in yeast cortical actin patches has yet to be defined, and the relevance of Aip1p's reported filament-severing versus filament-capping activities remain a topic of debate. To begin to address these issues, we conducted a genetic screen in which we crossed an aip1Δ strain by a subarray of strains from the EUROSCARF nonessential gene deletion collection in S. cerevisiae. Each member of the subarray (listed in Table 4) was deleted for a gene whose product is known to encode a protein that localizes to cortical actin patches or is known to be involved in endocytosis (Balakrishnan et al., 2005). Because aip1Δ strains have fairly normal growth rates (Rodal et al., 1999), our goal was to identify synthetic growth defects in the double mutant strains, which would imply that the deleted subarray protein and Aip1p act in a common pathway or activity in the cell. Yeast strain MCY60 (aip1Δ::NATr) was mated to each of 74 deletion strains (geneΔ::G418r), and diploids were selected, sporulated, tetrads were dissected, and genotyping was conducted to verify that any observed phenotypic abnormalities cosegregated with a double gene-deletion genotype. Of the 74 knockout alleles tested, only six were found to exhibit a synthetic slow-growth phenotype with the aip1Δ allele at 30°C: sac6Δ, sla1Δ, rvs161Δ, rvs167Δ, cap1Δ, and cap2Δ. We have previously reported the interactions between aip1Δ and sac6Δ, sla1Δ, cap1Δ, and cap2Δ (Rodal et al., 1999). However, this is the first exhaustive test for genetic interactions between all genes encoding components of the cortical patch and AIP1. The failure to see interactions between aip1Δ and the other 68 cortical patch/endocytosis genes tested suggests that Aip1p does not directly cooperate or act in concert with these other proteins or that some of these genes have redundant functions with one another, making it difficult to uncover aip1Δ interactions. As for the six genes that did show an interaction with aip1Δ, all are involved in actin-related roles in the cell, are bona fide actin binding proteins, and are specifically linked to the vesicle internalization step of endocytosis (Engqvist-Goldstein and Drubin, 2003; Kaksonen et al., 2005). This is not surprising, because cortical actin patches have recently been proven to be early endocytic structures (Kaksonen et al., 2003; Huckaba et al., 2004; Rodal et al., 2005). The significance of these interactions is bolstered by the fact that Rvs161p and Rvs167p dimerize and are partially redundant for function (Lombardi and Riezman, 2001), whereas Cap1p and Cap2p dimerize to form the α/β heterodimeric actin filament barbed-end capping protein (Amatruda et al., 1990). Furthermore, these genes display extensive physical and genetic interactions among themselves (Balakrishnan et al., 2005; Figure 6). The synthetic genetic interactions between aip1Δ and the six cortical patch/endocytosis genes suggest an important physiological role for Aip1p in yeast cortical actin patches and the early steps of endocytosis. Also in support of these findings, aip1 null mutants in Dictyostelium demonstrate reduced rates of endocytosis (Konzok et al., 1999). Additional assays will be necessary to determine the precise activities in which Aip1p is involved.
Table 4.
Genes tested for genetic interactions with AIP1
| Gene name | ORF | Gene name | ORF | Gene name | ORF |
|---|---|---|---|---|---|
| LSB 6 | YJL100w | ENT2 | YLR206w | SRV2 | YNL138w |
| DNM1 | YLL001w | SLA1 | YBL007c | TPM1 | YNL079c |
| ENT4 | YLL038c | EDE1 | YBL047c | YPT53 | YNL093w |
| APP2/GYL1 | YMR192w | EDS1 | YBR033w | INP52 | YNL106c |
| INP53 | YOR109w | LSB5 | YCL034w | HUA1 | YGR268c |
| SHE4/DIM1 | YOR035c | ENT1 | YDL161w | YAP1802 | YGR241c |
| CLA4 | YNL298w | RVS161 | YCR009c | TVP18 | YMR071c |
| MON2 | YNL297c | RGD1 | YBR260c | YCK3 | YER123w |
| YCK2 | YNL154c | AKR1 | YDR264c | CRN1 | YLR429w |
| VPS21 | YOR089c | BRE4 | YDL231c | LSB4/YSC84 | YHR016c |
| HUA2 | YOR284w | SAC6 | YDR129c | NPL6 | YMR091c |
| SCP1 | YOR367w | SWA2 | YDR320c | AIP1 | YMR092c |
| YHL017w | YHL017w | PKH1 | YDR490c | SVP26 | YHR181w |
| INP51 | YIL002c | MST27 | YGL051w | SKM1 | YOL113w |
| CAP2 | YIL034c | RVS167 | YDR388w | MYO3 | YKL129c |
| TPM2 | YIL138c | GTS1 | YGL181w | SNO1 | YMR095c |
| BZZ1 | YHR114w | CHC1 | YGL206c | MYO5 | YMR109w |
| YIR003w | YIR003w | LSB1 | YGR136w | LSB 3 | YFR024c-A |
| YCK1 | YHR135c | YAP1801 | YHR161c | VRP1 | YLR337c |
| PRK1 | YIL095w | CLC1 | YGR167w | ARK1 | YNL020c |
| DID4 | YKL002w | TWF1 | YGR080w | YPT52 | YKR014c |
| CAP1 | YKL007w | BBC1/MTI1 | YJL020c | APL4 | YPR029c |
| APP1 | YNL094w | ARC18 | YLR370c | LSB2/PIN3 | YPR154w |
| END3 | YNL084c | SYP1 | YCR030c | BSP1 | YPR171w |
| SMY2 | YBR172c | ABP1 | YCR088w |
ORF, open reading frame. Bold text indicates a gene that shares a genetic interaction with AIP1.
Figure 6.
Physical and genetic interactions among cortical patch-localized genes and gene products. Dotted lines represent physical interactions between gene products. Dashed lines represent synthetic sick genetic interactions between null alleles or site-directed mutant alleles (cofilin). Solid lines represent synthetic lethal genetic interactions. Data obtained from the S. cerevisiae Genome Database (Balakrishnan, 2005) and this work.
Genetic Interactions With aip1 Mutants Confirm Active Sites on the Aip1p Surface
To examine the relevance of Aip1p interactions to in vivo function, we crossed five aip1 mutant strains, aip1-15, aip1-34/35, aip1-55, aip1-59, and aip1-60 to sac6Δ, sla1Δ, rvs161Δ, rvs167Δ, cap1Δ, cap2Δ, and cof1-4 strains to identify in vivo synthetic growth defects (Table 5). Interestingly, unlike with the aip1Δ allele, these double mutant strains had to be grown at 37°C rather than 30°C to detect any growth defects, indicating that our aip1p mutants are at least partially functional at 30°C. Only when crossed against the cof1-4 strains did the aip1 mutants mimic the phenotype of an aip1Δ allele at 30°C. We were unable to examine aip1 crosses with sac6Δ or sla1Δ at 37°C, because both genes are essential at this temperature. aip1-60, which carries a gain-in-function mutation that enhances two-hybrid interaction with actin (see above; Figure 2B), was also tested in the context of full-length Aip1p and demonstrated a slow growth phenotype with cof1-4 (Table 5).
Table 5.
Summary of genetic interactions uncovered in this study
| AIP1 | aip1Δ | aip1-15 | aip1-34/35 | aip1-55 | aip1-59 | aip1-60 | cof1-4 | cof1-19 | |
|---|---|---|---|---|---|---|---|---|---|
| 30°C | |||||||||
| sac6Δ | +++ | ++ | +++ | +++ | +++ | +++ | ++++ | + | ++ |
| sla1Δ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | + | +++ |
| rvs161Δ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++ | ++++ |
| rvs167Δ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++ | ++++ |
| cap1Δ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | – | + |
| cap2Δ | ++++ | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | – | ++ |
| cof1-4 | ++++ | – | – | – | – | – | ++ | na | na |
| cof1-19 | ++++ | ++++ | nt | nt | nt | nt | ++++ | na | na |
| 37°C | |||||||||
| sac6Δ | – | – | – | – | – | – | – | – | – |
| sla1Δ | – | – | – | – | – | – | – | – | – |
| rvs161Δ | +++ | + | + | ++ | +++ | + | ++++ | – | ++ |
| rvs167Δ | ++ | – | + | + | + | + | ++++ | + | ++ |
| cap1Δ | +++ | + | ++ | ++ | + | + | ++++ | – | – |
| cap2Δ | +++ | + | ++ | ++ | + | + | ++++ | – | – |
| cof1-4 | +++ | – | – | – | – | – | ++ | na | na |
| cof1-19 | ++++ | ++++ | nt | nt | nt | nt | ++++ | na | na |
nt, not tested; na, not applicable; ++++, WT growth; +++, moderate growth; ++, slow growth; +, very slow growth; –, dead.
Each of the four loss-of-function aip1p mutants tested in vivo showed significant synthetic growth defects when combined with the endocytosis-related gene deletion strains (Table 5). In many cases, the intensity of these defects varied depending on the deletion strain being tested. Only one cross, aip1-55 × rvs161Δ, failed to show a genetic interaction. As a whole, these findings show that both individual actin binding sites and the cofilin binding site are necessary for proper Aip1p activity in vivo.
If Aip1p is truly a barbed-end capping protein as reported previously (Okada et al., 2002; Balcer et al., 2003), we would expect the aip1 mutants to have more serious synthetic defects with the capΔ alleles, because they would be expected to be redundant in function. Interestingly, the actin binding site-specific aip1 alleles are more defective with the rvs161/167 deletions than they are with the cap1/cap2 deletions, suggesting a greater functional involvement of Aip1p and possibly Aip1p-facilitated severing with the RVS proteins than with the Cap proteins.
The cof1-4 and cof1-19 mutant alleles were also tested in combination with each aip1 mutant allele and the six endocytosis-related gene deletions (Table 5). In all cases, cof1-4 was more sick than cof1-19, suggesting that cof1-4 defects which seem to be related to on/off rates at filament ends are of greater functional significance during this stage of endocytosis than the Aip1p-associated severing defect of cof1-19.
In addition to the synthetic growth defects describe above, haploid strains carrying each of the aip1 mutants displayed various defects in cell morphology, actin organization, aip1p localization, and/or cofilin localization (Table 3 and Supplemental Figures S1-S3).
DISCUSSION
Mutagenesis and Computerized Docking Studies Predict a Model for the Actin-Cofilin-Aip1p Ternary Complex
Consistent with previously reported results (Mohri et al., 2004), we have identified an actin interaction site on Aip1p that seems to centralize along the outer rim of the N-terminal β-propeller. An incomplete loss of Aip1p function upon mutagenesis of this region led us to hypothesize that a second actin interacting site may exist on the C-terminal propeller, located analogously to the actin binding site of the N-terminal propeller. Several layers of genetic data support this proposition. First, a truncation mutant of Aip1p expressing only the C-terminal propeller shows a weak but consistent actin interaction in two-hybrid assays. Second, the suspected region for the actin interaction on this propeller was confirmed using site-directed loss-of-function mutations (alleles 34 and 35) as well as unbiased gain-in-function mutations (alleles 60 and 61). Third, when aip1-34/35 and aip1-60 mutants were integrated into the yeast genome in the context of the full-length Aip1p gene, both showed synthetic growth defects when combined with the cof1-4 allele. aip1-34/35 is also synthetic sick with a group of genes encoding endocytosis-related proteins.
Our random mutagenesis approach also identified a number of aip1 mutants that are able to interact with actin but not cofilin in the two-hybrid system, revealing a cofilin-binding footprint along the front, concave surface of the molecule with essential contact points on the N- and C-terminal propellers. Each cofilin-specific mutant tested was also shown to have synthetic growth defects with cof1-4 and genes encoding cortical patch proteins.
The active sites on Aip1p that we have identified are highly consistent with a molecular model acquired through independently conducted computerized docking studies of the Aip1p-cofilin complex. The cofilin-specific Aip1p mutations map extremely well to the Aip1p surface area that is buried by cofilin in the model. In agreement, cofilin mutants that have lost their ability to interact with Aip1p by two-hybrid (Rodal et al., 1999) are mostly consistent with the Aip1p binding interface on cofilin predicted by our model. Also of great significance is the fact that the cofilin residues mutated in the cof1-19 (R109, R110) allele, which we have demonstrated to be deficient in vitro for Aip1p-induced actin filament severing, are at the predicted molecular interface with Aip1p. In addition, this model indicates that the regions of cofilin and Aip1p thought to be involved in actin binding are available and reasonably positioned for these interactions to occur. The actin and cofilin binding footprints seem to indicate a simple model for lateral actin filament binding by Aip1p in which it straddles cofilin while potentially binding two adjacent actin monomers. Observations made by Mohri et al. (2004) support this model, because they showed that a C. elegans aip1p (unc-78) mutant capable of binding but not disassembling cofilin-decorated actin filaments reaches its filament binding saturation at a 2:1 actin-to-Aip1p ratio. Since actin has only one known Aip1p binding site and each Aip1p molecule has two actin binding sites, these data are consistent with the proposal that one molecule of Aip1p binds per two actin subunits within a filament. Furthermore, lateral filament binding by Aip1p has also been demonstrated by electron microscopy (Okada et al., 2002). Still, it remains unclear how Aip1p interacts with actin as a part of this complex. Specifically, we do not know whether both actin interacting sites bind to the actin filament simultaneously or whether one is used over the other under varying circumstances. Our experimental data suggest that both sites are important for optimal Aip1p function. In addition, how this leads to filament disassembly will require further investigation. We predict that Aip1p binding to cofilin-decorated actin filaments produces a torque that enhances the filament twist maintained by cofilin, leading to filament severing. Alternately, Aip1p may push cofilin deeper between subunits, wedging the filament apart by creating a steric hindrance between longitudinally associated actin monomers. In either case, we expect that significant conformational changes of the ternary complex occur immediately after Aip1p binding, which may cause difficulties in precisely predicting how this complex fits together. Our mutagenesis and modeling data coupled with the demonstrated ability of Aip1p to enhance the disassembly of cofilin-bound actin filaments (Rodal et al., 1999), and visual assays of tethered actin filaments treated with cofilin and Aip1p (Ono et al., 2004), strongly implicate Aip1p as an actin filament severing protein.
The inability of our aip1p mutants, with the exception of GST-aip1-15p, to show defects in a severing assay, although perplexing, is consistent with data from Mohri et al. (2004). Using C. elegans Aip1p (UNC-78), they were able to isolate N-terminal but not C-terminal propeller mutants (of four tested) that were defective in actin filament disassembly. Perhaps Aip1p-induced severing of actin filaments is so robust that more extreme mutational changes on the C-terminal propeller are necessary to obtain a biochemical defect. Consistent with this idea, we found that even while using minimal time periods of incubation and attempting to slow the rate of disassembly by conducting the experiments at 4°C, our severing reactions still proceeded to completion.
Is Aip1p a Capping Protein?
The biochemical activities of Aip1p from multiple sources (S. cerevisiae, X. laevis, and C. elegans) have been extensively tested in vitro (Okada et al., 1999, 2002; Rodal et al., 1999; Balcer et al., 2003; Mohri and Ono, 2003; Mohri et al., 2004; Ono et al., 2004). In actin pelleting assays, S. cerevisiae Aip1p very strongly induces the disassembly of cofilin-bound actin filaments, whereas C. elegans Aip1p (UNC-78) does so moderately, and X. laevis Aip1p (XAip1) does so very weakly (Okada et al., 1999; Rodal et al., 1999; Mohri and Ono, 2003). Although these disparities may be specific to the organism from which the Aip1p is obtained, it is possible that the weaker activities observed for UNC-78 and XAip1 have resulted from the use of rabbit skeletal muscle actin in the disassembly assays, whereas yeast Aip1p was tested using yeast actin. The ability of Aip1p to enhance severing of cofilin-decorated actin filaments has been convincingly documented (Okada et al., 1999; Ono et al., 2004). In addition, some findings indicate that Aip1p is able to cap the barbed ends of actin filaments, preventing filament elongation and thus enhancing disassembly (Okada et al., 2002; Balcer et al., 2003). Previous actin filament elongation assays using a range of XAip1 concentrations combined with cofilin provided persuasive evidence that XAip1 may exert a barbedend capping activity (Okada et al., 2002). However, we have reservations regarding the biological relevance of this result because of the variety of sources from which reagents were obtained: rabbit muscle actin, chicken cofilin, and XAip1. In addition, an experiment suggesting that XAip1 is functionally redundant with the barbed-end capping gelsolin-actin complex used an actin/cofilin ratio of 27.5:1, which is far too low of a cofilin concentration for any expected Aip1p-associated disassembly and is thus inconclusive (Rodal et al., 1999; Okada et al., 2002). Visual assays of tethered actin filaments tested with UNC-78 were not able to confirm barbed-end capping of actin filaments by Aip1p at the concentrations tested and instead favored filament severing (Ono et al., 2004).
For S. cerevisiae Aip1p, we have demonstrated that capping assay defects associated with cof1-4p and cof1-19p persist in the absence of Aip1p, suggesting that Aip1p is not involved in the barbed-end activity that this assay was thought to measure. When we performed the capping assays using increased actin concentrations and lower respective profilin/cofilin/Aip1p ratios, Aip1p became necessary for filament disassembly, resulting in a moderate shift of actin from the pellet to the supernatant fraction. However, we are not convinced that these results represent barbed-end capping as opposed to severing for reasons described previously. Therefore, additional studies are necessary to further investigate barbed-end capping and to investigate the nature of possible concentration and/or species specificities associated with this activity.
Interestingly, in our actin disassembly assays we observed that cof1-4p is more defective in the capping assay while cof1-19p is more defective in the severing assay, indicating that cofilin is playing a different role in each of these two assays. Cofilin's role in the capping assay seems to be profilin related, although it is not clear whether enhanced actin filament disassembly is caused by additive profilin/cofilin effects or synergistic effects caused by profilactin-specific barbed-end gating by cofilin. Approximations for the concentrations of actin and profilin in yeast are 60 and 20 μM, respectively (Magdolen et al., 1993). Presuming that the F-actin/G-actin ratio in yeast is at least 1:1, as has been stated for other organisms (Korn, 1982; Hug et al., 1995), then the G-actin/profilin ratio would be expected to be ∼3:2. Therefore, because the majority of actin monomers within the cell could be profilin bound, cofilin gating of barbed ends is a feasible hypothesis for a novel mode of barbed-end regulation.
If Aip1p were not involved in barbed-end capping, that would leave filament severing as Aip1p's only recognized role in the cell. Therefore, it seems paradoxical that an aip1Δ strain is SL with cof1-4, which may be defective in barbedend regulation but not in severing assays, whereas it is not SL with cof1-19, which shows considerable Aip1p-induced severing defects. Our proposed potential barbed-end regulating activity for cofilin, if verified, might explain this observation. It may be that a primary role of Aip1p is to create cofilin-decorated barbed ends through severing. On the other hand, Aip1p severing would also create more cofilin-decorated pointed ends, potentially enhancing filament disassembly by subunit release from this end of the filament. Furthermore, cofilin-bound filament ends could differ drastically from bare ends in terms of growth kinetics and in the ability to interact with other filament end-associated proteins, such as capping proteins, formins, and the Arp2/3 complex.
Aip1p capping and profilactin gating by cofilin could both occur at actin filament barbed ends, with local protein concentrations being a deciding factor as to which activity prevails. Therefore, additional visual and kinetic biochemical assays are necessary to conclusively differentiate between severing, capping, and gating of actin filaments. If Aip1p is involved with the regulation of filament barbed ends, it would likely act in a cooperative manner with the filament severing activity: Aip1p induces actin filament severing at its site of lateral filament binding, then remains bound to the newly created barbed end where it may participate in blocking subunit addition. Thus, severing could serve as a way for Aip1p to self-target to filament barbed ends. Electron microscopy in conjunction with molecular docking studies may provide details regarding the precise mechanisms by which Aip1p induces cofilin-mediated actin filament disassembly.
Genetic Interactions Reveal That Aip1p Plays a Role in Endocytosis in Yeast
During the process of endocytosis in yeast, Rvs161p and Rvs167p have been implicated in linking actin to the neck of the invaginated vesicle, where all three proteins contribute to vesicular scission (Schmidt et al., 1999; Baggett and Wendland, 2001; Jeng and Welch, 2001). Sla1p is known to be involved with organization and assembly of actin networks within cortical patches and a sla1Δ strain has large, flattened cortical patches and endocytic defects (Rodal et al., 1999; Warren et al., 2002). Sac6p (fimbrin) is an actin filament bundling protein that forms parallel cross-links between actin filaments (Bretscher, 1981). Cap1p and Cap2p form a heterodimeric complex that caps the barbed ends of actin filaments (Amatruda et al., 1992). All six of these proteins have been directly linked to vesicle internalization in yeast (Kaksonen et al., 2005). The slow growth phenotypes observed when aip1Δ is combined with these gene deletions but not with dozens of other deletions of cortical patch genes point to a direct role for Aip1p in endocytic vesicle internalization.
Cytological Observation of aip1 Mutant Strains Expose In Vivo Defects
Consistent with our genetic findings, we were able to visually observe morphological aberrations within our aip1 mutant strains using fluorescent microscopy. Actin organizational defects at least partially similar to an aip1Δ strain were seen in all mutant strains except for aip1-60 (Supplemental Figure S1). Interestingly, aip1-34/35 and aip1-59 also displayed elongated buds in 1-2% of the cells observed. We have never observed this novel phenotype in WT or aip1Δ strains, suggesting that the presence of impaired aip1p has the potential to be more functionally detrimental to the cell than the elimination of Aip1p all together. This is likely the result of some partial obstruction of Aip1p and/or cofilin activity that leads to an imbalance in actin filament growth dynamics.
Immunofluorescence of aip1p in the mutant strains also showed deviations from normal, ranging from a slight decrease to a total loss of cortical patch localization for the loss of function mutants, whereas aip1-60p demonstrated an enhanced localization at cortical patches (Supplemental Figure S2). Only aip1-34/35p did not have any observable mislocalization.
Cofilin localization was also performed in our aip1p mutant strains (Supplemental Figure S3). In an aip1Δ strain, cofilin partially mislocalizes to the cytoplasm where it exists in a filamentous state, presumably decorating cellular actin cables. Of the mutants tested, only aip1-60 did not at least partially mimic this phenotype. However, aip1-60 did enhance the cytoplasmic cofilin localization of the cof1-19 strain. The accumulation of cofilin-decorated cables in the cytoplasm of aip1 and cof1 strains suggest that the roles played by these proteins are not limited to actin cortical patches and that they likely act in cytoplasmic cable disassembly, perhaps by clearing excess filaments out of the cytoplasm. Furthermore, the fact that our mutants partially imitate an aip1Δ phenotype bolsters the physiological relevance of the active sites that we have identified on Aip1p.
The complexities of the cooperative activities conducted by Aip1p, cofilin, and possibly profilin extend beyond these proteins to include regulatory factors as well. Because of the intensely robust ability that Aip1p and cofilin have to disassemble actin filaments, as demonstrated by in vitro biochemical assays, cellular mechanisms must exist that prevent these proteins from uncontrollably shredding filaments in vivo. Were this not the case, Aip1p and cofilin would not colocalize with actin filaments in cortical patches. Thus, considerable progress remains to be made in further elucidating the precise activities of Aip1p and cofilin in vivo and in evaluating how these functions are regulated and balanced to enhance the dynamics of actin networks.
Supplementary Material
Acknowledgments
We thank B. Goode, K. Okada, S. Ono, and K. Mohri for sharing unpublished data and constructive suggestions. We also thank T. Duncan for assistance with molecular modeling programs to create figures and all members of the Amberg laboratory for helpful input. This research was supported by National Institutes of Health Grants GM-067246 (to D. S.) and GM-56189 (to D.C.A.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-10-0956) on January 18, 2006.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Amatruda, J. F., Cannon, J. F., Tatchell, K., Hug, C., and Cooper, J. A. (1990). Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature 344, 352-354. [DOI] [PubMed] [Google Scholar]
- Amatruda, J. F., Gattermeir, D. J., Karpova, T. S., and Cooper, J. A. (1992). Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution and polarized secretion in yeast. J. Cell Biol. 119, 1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D. C., Basart, E., and Botstein, D. (1995). Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2, 28-35. [DOI] [PubMed] [Google Scholar]
- Amberg, D. C., Burke, D. J., and Strathern, J. N. (2005). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Baggett, J. J., and Wendland, B. (2001). Clathrin function in yeast endocytosis. Traffic 2, 297-302. [DOI] [PubMed] [Google Scholar]
- Balakrishnan, R., et al. (2005). Saccharomyces Genome Database. http://www.yeastgenome.org/ (accessed 23 December 2005).
- Balcer, H. I., Goodman, A. L., Rodal, A. A., Smith, E., Kugler, J., Heuser, J. E., and Goode, B. L. (2003). Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13, 2159-2169. [DOI] [PubMed] [Google Scholar]
- Bobkov, A. A., Muhlrad, A., Kokabi, K., Vorobiev, S., Almo, S. C., and Reisler, E. (2002). Structural effects of cofilin on longitudinal contacts in F-actin. J. Mol. Biol. 323, 739-750. [DOI] [PubMed] [Google Scholar]
- Bretscher, A. (1981). Fimbrin is a cytoskeletal protein that crosslinks F-actin in vitro. Proc. Natl. Acad. Sci. USA 78, 6849-6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier, M. F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G. X., Hong, Y., Chua, N. H., and Pantaloni, D. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson, L., Nystrom, L. E., Sundkvist, I., Markey, F., and Lindberg, U. (1977). Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 115, 465-483. [DOI] [PubMed] [Google Scholar]
- Durfee, T., Becherer, K., Chen, P.-L., Yeh, S.-H., Yang, Y., Kilburn, A.E., Lee, W.-H., and Elledge, S. J. (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7, 555-569. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E., and Drubin, D. G. (2003). Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19, 287-332. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340, 245-246. [DOI] [PubMed] [Google Scholar]
- Galkin, V. E., Orlova, A., VanLoock, M. S., Shvetsov, A., Reisler, E., and Egelman, E. H. (2003). ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J. Cell Biol. 163, 1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch, G., Faix, J., Kohler, J., and Muller-Taubenberger, A. (2004). Actin-binding proteins required for reliable chromosome segregation in mitosis. Cell Motil. Cytoskeleton 57, 18-25. [DOI] [PubMed] [Google Scholar]
- Haarer, B. K., Lillie, S. H., Adams, A.E.M., Magdolen, V., Bandlow, W., and Brown, S. S. (1990). Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J. Cell Biol. 110, 105-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba, T. M., Gay, A. C., Pantalena, L. F., Yang, H. C., and Pon, L. A. (2004). Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 167, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug, C., Jay, P. Y., Reddy, I., McNally, J. G., Bridgman, P. C., Elson, E. L., and Cooper, J. A. (1995). Capping protein levels influence actin assembly and cell motility in Dictyostelium. Cell 81, 591-600. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., and Schulten, K. (1996). VMD: visual molecular dynamics. J. Mol. Graph 14, 27-28, 33-38. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., Han, J., Pang, K. M., Knecht, D. A., and Condeelis, J. S. (2000). Actin filaments are severed by both native and recombinant Dictyostelium cofilin but to different extents. Cell Motil. Cytoskeleton 45, 293-306. [DOI] [PubMed] [Google Scholar]
- Iida, K., and Yahara, I. (1999). Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes to Cells 4, 21-32. [DOI] [PubMed] [Google Scholar]
- Jacobson, M. P., Pincus, D. L., Rapp, C. S., Day, T. J., Honig, B., Shaw, D. E., and Friesner, R. A. (2004). A hierarchical approach to all-atom protein loop prediction. Proteins 55, 351-367. [DOI] [PubMed] [Google Scholar]
- Jeng, R. L., and Welch, M. D. (2001). Cytoskeleton: actin and endocytosis-no longer the weakest link. Curr. Biol. 11, R691-R694. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D. G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, 475-487. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Toret, C. P., and Drubin, D. G. (2005). A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123, 305-320. [DOI] [PubMed] [Google Scholar]
- Kale, L.E.A. (1999). NAMD2: greater scalability for parallel molecular dynamics. J. Comput. Phys. 151, 283-312. [Google Scholar]
- Ketelaar, T., Allwood, E. G., Anthony, R., Voigt, B., Menzel, D., and Hussey, P. J. (2004). The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14, 145-149. [DOI] [PubMed] [Google Scholar]
- Konzok, A., Weber, I., Simmeth, E., Hacker, U., Maniak, M., and Muller-Taubenberger, A. (1999). DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis, and motility. J. Cell Biol. 146, 453-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn, E. D. (1982). Actin polymerization and its regulation by proteins from nonmuscle cells. Physiol. Rev. 62, 672-737. [DOI] [PubMed] [Google Scholar]
- Kron, S. J., Drubin, D. G., Botstein, D., and Spudich, J. A. (1992). Yeast actin filaments display ATP-dependent sliding movement over surfaces coated with rabbit muscle myosin. Proc. Natl. Acad. Sci. USA 89, 4466-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen, P., and Drubin, D. G. (1997). Cofilin promotes rapid actin filament turnover in vivo. Nature 388, 78-82. [DOI] [PubMed] [Google Scholar]
- Lombardi, R., and Riezman, H. (2001). Rvs161p and Rvs167p, the two yeast amphiphysin homologs, function together in vivo. J. Biol. Chem. 276, 6016-6022. [DOI] [PubMed] [Google Scholar]
- Ma, H., Kunes, S., Schatz, P. J., and Botstein, D. (1987). Plasmid construction by homologous recombination in yeast. Gene 58, 201-216. [DOI] [PubMed] [Google Scholar]
- Maciver, S. K., Zot, H. G., and Pollard, T. D. (1991). Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 115, 1611-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdolen, V., Drubin, D. G., Mages, G., and Bandlow, W. (1993). High levels of profilin suppress the lethality caused by overproduction of actin in yeast cells. FEBS Lett. 316, 41-47. [DOI] [PubMed] [Google Scholar]
- McGough, A., Pope, B., Chiu, W., and Weeds, A. (1997). Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 138, 771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri, K., and Ono, S. (2003). Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 116, 4107-4118. [DOI] [PubMed] [Google Scholar]
- Mohri, K., Vorobiev, S., Fedorov, A. A., Almo, S. C., and Ono, S. (2004). Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279, 31697-31707. [DOI] [PubMed] [Google Scholar]
- Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., and Olson, A. J. (1998). Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 19, 1639-1662. [Google Scholar]
- Okada, K., Blanchoin, L., Abe, H., Chen, H., Pollard, T. D., and Bamburg, J. R. (2002). Xenopus actin interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 277, 43011-43016. [DOI] [PubMed] [Google Scholar]
- Okada, K., Obinata, T., and Abe, H. (1999). XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J. Cell Sci. 112, 1553-1565. [DOI] [PubMed] [Google Scholar]
- Ono, S. (2001). The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152, 1313-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., Mohri, K., and Ono, K. (2004). Microscopic evidence that actin-interacting protein 1 actively disassembles actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279, 14207-14212. [DOI] [PubMed] [Google Scholar]
- Pope, B. J., Gonsior, S. M., Yeoh, S., McGough, A., and Weeds, A. G. (2000). Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. J. Mol. Biol. 298, 649-661. [DOI] [PubMed] [Google Scholar]
- Ressad, F., Didry, D., Xia, G. X., Hong, Y., Chua, N. H., Pantaloni, D., and Carlier, M. F. (1998). Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J. Biol. Chem. 273, 20894-20902. [DOI] [PubMed] [Google Scholar]
- Rodal, A. A., Kozubowski, L., Goode, B. L., Drubin, D. G., and Hartwig, J. H. (2005). Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell 16, 372-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal, A. A., Tetreault, J. W., Lappalainen, P., Drubin, D. G., and Amberg, D. C. (1999). Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145, 1251-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S. L., Wiedemann, U., Stuurman, N., and Vale, R. D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., Winston, F., and Hieter, P. (1989). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schmidt, A., Wolde, M., Thiele, C., Fest, W., Kratzin, H., Podtelejnikov, A. V., Witke, W., Huttner, W. B., and Soling, H. D. (1999). Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature 401, 133-141. [DOI] [PubMed] [Google Scholar]
- Voegtli, W. C., Madrona, A. Y., and Wilson, D. K. (2003). The structure of Aip1p, a WD repeat protein that regulates cofilin-mediated actin depolymerization. J. Biol. Chem. 278, 34373-34379. [DOI] [PubMed] [Google Scholar]
- Warren, D. T., Andrews, P. D., Gourlay, C. W., and Ayscough, K. R. (2002). Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J. Cell Sci. 115, 1703-1715. [DOI] [PubMed] [Google Scholar]
- Wertman, K. F., Drubin, D. G., and Botstein, D. (1992). Systematic mutational analysis of the yeast ACT1 gene. Genetics 132, 337-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.