Abstract
Survivin is a component of the chromosomal passenger complex (CPC) that plays a role in maintenance of an active spindle checkpoint and in cytokinesis. To study whether these different functions can be attributed to distinct domains within the Survivin protein, we complemented Survivin-depleted cells with a variety of point- and deletion-mutants of Survivin. We show that an intact baculovirus IAP repeat (BIR) domain is required for proper spindle checkpoint functioning, but dispensable for cytokinesis. In line with this, mutants lacking an intact BIR domain localized normally to the central spindle, but their localization to inner centromeres was severely perturbed. Consequently, these mutants failed to recruit Aurora B, Borealin/Dasra B, and BubR1 to centromeres and kinetochores, but they had retained the ability to recruit Aurora B and Borealin/Dasra B to the midzone and midbody. Thus, the C terminus of Survivin is sufficient for central spindle localization and execution of cytokinesis, but the additional presence of a functional BIR domain is essential for centromere targeting and spindle checkpoint function. Importantly, our data show that the function of the CPC at the centromere can be separated from its function at the central spindle and that execution of cytokinesis does not require prior concentration of the CPC at centromeres.
INTRODUCTION
Aneuploidy is a hallmark of many types of cancers and is thought to contribute to carcinogenesis (Rajagopalan and Lengauer, 2004). Therefore, the generation of aneuploid progeny should be avoided during cell division. Aneuploidy can arise as a consequence of chromosome missegration and/or of a defect in the coordination of chromosome segregation and initiation of cytokinesis (Storchova and Pellman, 2004). In mitosis, the spindle checkpoint prevents anaphase onset until all sister-chromatids are properly attached to the mitotic spindle, thereby contributing to the maintenance of a stable diploid genome (Rieder et al., 1995; Shah and Cleveland, 2000; Musacchio and Hardwick, 2002). The Mad (Mad1, Mad2, BubR1/Mad3) (Li and Murray, 1991; Li and Benezra, 1996; Taylor et al., 1998) and Bub (Bub1-3) (Hoyt et al., 1991) proteins form the heart of the spindle checkpoint by inhibiting Cdc20, an accessory subunit of the anaphase promoting complex/cyclosome (Hwang et al., 1998; Kim et al., 1998; Sudakin et al., 2001; Tang et al., 2001). However, it was recently shown that also a number of chromosomal passenger proteins play an important auxiliary role in the spindle checkpoint (Biggins and Murray, 2001; Lens and Medema, 2003).
In human cells, the chromosome passenger proteins INner CENtromere Protein (INCENP) (Cooke et al., 1987), Aurora B (Bischoff et al., 1998), Borealin/Dasra B (Gassmann et al., 2004; Sampath et al., 2004), and Survivin (Ambrosini et al., 1997) exist in a complex termed the chromosomal passenger complex (CPC) (Vagnarelli and Earnshaw, 2004). Together, they localize to inner centromeres from G2 until the metaphase-to-anaphase transition and then translocate to the spindle midzone during anaphase and eventually localize to the midbody during telophase (Adams et al., 2001; Vagnarelli and Earnshaw, 2004). RNA interference (RNAi) knockdown experiments in U2OS and HeLa cells showed that these proteins are mutually dependent on each other for binding to the centromere (Carvalho et al., 2003; Ditchfield et al., 2003; Honda et al., 2003; Lens et al., 2003; Gassmann et al., 2004).
For one of the chromosome passenger proteins, Survivin, not only a mitotic regulatory function but also an apoptosis inhibitory function was proposed based on overexpression experiments with mutant Survivin proteins. Overexpression of a Survivin point mutant in which the baculovirus IAP repeat (BIR) domain was disrupted (SurvivinC84A) resulted both in cell division defects and cell death of HeLa cells (Li et al., 1998, 1999). In contrast, overexpression of a Survivin mutant that could no longer be phosphorylated by cyclin B/cdc2 (SurvivinT34A) did not result in cell division defects but induced apoptosis, suggesting that Survivin could play a role as an apoptosis inhibitor and as a regulator of cell division (O'Connor et al., 2000). Importantly, RNAi-mediated knockdown of Survivin in human cells clearly resulted in different mitotic defects, but it did not induce rapid cell death as would be expected from knockdown of an apoptosis inhibitor (Carvalho et al., 2003; Lens et al., 2003). These types of experiments not only confirmed the evolutionary conserved roles of Survivin in chromosome alignment and cytokinesis (Fraser et al., 1999; Li et al., 2000; Speliotes et al., 2000) but also revealed that, together with the other passenger proteins, this protein is required to maintain spindle checkpoint activity in particular when cells are challenged with drugs that interfere with the generation of tension between paired sister kinetochores (e.g., paclitaxel and monastrol) (Carvalho et al., 2003; Ditchfield et al., 2003; Hauf et al., 2003; Lens et al., 2003). Unlike Mad2 and BubR1, Survivin and the other passengers do not directly inhibit the APC/C, yet they enable the cell to communicate lack of tension back to the attached microtubules (Lens and Medema, 2003). To understand how Survivin is able to coordinate its various functions during mitosis, we complemented Survivin knockdown cells with different point- and deletion mutants of Survivin. Using this approach, we were able to uncouple Survivin's function in the spindle checkpoint from its role during cytokinesis.
MATERIALS AND METHODS
Plasmids
pSuper-Survivin, full-length Survivin1-142 and a mutant that is nonresponsive to our RNAi-targeting vector have all been described previously (Lens et al., 2003). The T34A and C84A point mutations were introduced into the RNAi-insensitive mutant of Survivin by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and appropriate primers. The 1-89 aa and 89-142 aa deletion mutants of Survivin were generated by PCR by using specific primers harboring either a C-terminal vesicular stomatitis virus (VSV)- or FLAG-tag and the RNAi-insensitive Survivin1-142 as template. PCR products were digested with EcoRI and XhoI and cloned into pc-DNA3. Generation of a siRNA-targeting vector for Borealin/Dasra B and an RNAi-resistant Borealin construct has been described previously (Vader et al., 2006). Borealin1-141 and Borealin142-280 deletion mutants were generated by PCR using appropriate primers and the RNAi-resistant Borealin cDNA as a template. The PCR products were cloned into pCR3 containing an N-terminal VSV epitope tag. Correctly mutated constructs were confirmed by sequence analysis. The pBabe-puro vector (Brummelkamp et al., 2002), spectrin-GFP (Kalejta et al., 1997), histone H2B-GFP, (Kanda et al., 1998), histone H2B-diHcRed (gift from Dr. J. Ellenberg, EMBL, Heidelberg, Germany) and INCENP-GFP (Vader et al., 2006) expression plasmids have all been described previously.
Antibodies and Dyes
Mouse anti-MPM-2 and rabbit anti-phospho-CENP-A (Ser7) were from Upstate Biotechnology (Charlottesville, VA), rabbit anti-Aurora B and anti-CENP-A were from Abcam (Cambridge, United Kingdom), mouse anti-Aurora B was from BD Transduction Laboratories (Lexington, KY), rabbit anti-Aurora A was from Cell Signaling Technology (Beverly, MA), and Survivin was from R&D Systems (Minneapolis, MN). Mouse anti-FLAG (M2) and mouse anti-VSV were obtained from Sigma-Aldrich (St. Louis, MO), and sheep anti-BubR1 was a kind gift from Dr. S. Taylor (University of Manchester, Manchester, United Kingdom) (Taylor et al., 2001). Rabbit anti-Dasra B was a generous gift from Dr. H. Funabiki (Rockefeller University, New York, NY) (Sampath et al., 2004). Cy5-conjugated donkey anti-mouse was from Jackson ImmunoResearch Laboratories (West Grove, PA). Goat anti-rabbit/Alexa568, goat anti-rabbit/Alexa647, and goat anti-mouse/Alexa568 or Alexa 647 conjugates were from Molecular Probes (Eugene, OR). Peroxidase-conjugated goat anti-rabbit and goat anti-mouse antibodies were from Dako Cytomation Denmark (Glostrup, Denmark). Propidium iodide (PI), thymidine, and paclitaxel were from Sigma-Aldrich.
Cell Culture, Transfection, and Synchronization
U2OS osteosarcoma cells and human embryonic kidney (HEK)293 cells were grown in DMEM supplemented with 6% fetal calf serum and antibiotics. Transfections were performed by the standard calcium phosphate transfection protocol. Where indicated, the cells were synchronized at the G1/S transition by addition of 2.5 mM thymidine directly after washing away the calcium-phosphate precipitate. Cells were maintained in thymidine for 24 h, after which they were released from the block either in the presence or absence of taxol (paclitaxel; 1 μM). Eighteen hours after release, cells were harvested and analyzed by flow cytometry or immunofluorescence. Alternatively, transfected cells were monitored by time-lapse analysis.
Time-Lapse Analysis
U2OS cells were plated on 35-mm glass-bottom culture dishes (Willco-dish, Amsterdam, The Netherlands) and transfected the next day with 1 μg of the pSuper plasmids and 1 μg of the indicated expression plasmids in combination with 0.1 μg of H2B-GFP. Cells were synchronized with thymidine for 24 h and followed by time-lapse microscopy starting 10 h after release from the thymidine block. For life imaging, dishes were transferred to a heated stage (37°C) on a Zeiss Axiovert 200M microscope equipped with a 0.55 numerical aperture (N.A.) condenser and a 40× Achroplan objective (0.60 N.A.). Twelve bits differential interference contrast (DIC) and fluorescence (100-ms exposures) images were captured every 1 to 5 min by using a Photometrics CoolSNAP HQ charged-coupled device camera set at gain 1.0 (Scientific, Tucson, AZ) and appropriate filter cubes (Chroma Technology, Brattleboro, VT) to select specific fluorescence. Images were processed using MetaMorph software (Universal Imaging, Downingtown, PA).
Flow Cytometry
Cells were grown in 10-cm2 dishes and transfected with GFP-spectrin (1 μg), in combination with pSuper or pS-Survivin (10 μg each), and expression plasmids encoding FLAG-tagged versions of the indicated Survivin mutants (1 μg for Survivin1-142, 10 μg for the mutant proteins). Sixty hours after transfection, cells were harvested and fixed in ice-cold 70% ethanol. The fixed cells were washed once with phosphate-buffered saline (PBS) containing 0.05% Tween 20 and then incubated with anti-MPM-2 monoclonal antibody (mAb) diluted in PBS containing 0.05% Tween 20 and 2% bovine serum albumin (BSA) to specifically stain mitotic cells. Finally, cells were stained with a secondary Cy5-conjugated goat-anti-mouse antibody and counterstained with propidium iodide. MPM-2 positivity of the GFP-positive cells was analyzed using flow cytometry (CellQuest; BD Biosciences, San Jose, CA) as described previously (Smits et al., 2000).
Immunofluorescence
Cells were grown on glass coverslips in 10-cm2 dishes, transfected the next day with GFP-spectrin (1 μg), in combination with pSuper or pS-Survivin (10 μg each) and expression plasmids encoding VSV-tagged versions of the indicated Survivin mutants (1 μg for Survivin1-142, 5-10 μg for the mutant proteins), and synchronized using thymidine. Fourteen hours after thymidine release, coverslips were removed from the culture dish and carefully washed twice with PBS. Cells were then fixed for 5 min in 4% paraformaldehyde plus 0.2% (wt/vol) sucrose at room temperature and subsequently washed with PBS. Cells were then permeabilized for 5 min with 0.5% Triton X-100 (in 20 mM Tris-Cl, pH 7.4, 50 mM NaCl, 300 mM sucrose, and 3 mM MgCl2), blocked in PBS plus 3% BSA, and subsequently incubated with the appropriate primary/secondary antibody combinations diluted in PBS/0.01% Tween 20 and 2% BSA as described previously (Pines, 1997). Confocal fluorescence images were obtained on a Leica TCS NT (Leica Microsystems, Heidelberg, Germany) confocal system, equipped with an Ar/Kr laser. Images were taken using a 63× 1.3. N.A. objective. Possible bleed-through of the different fluorochromes, which could give rise to false positive colocalization of the signals, was avoided by careful selection of the imaging conditions. Standard filter combination(s) and Kalman averaging was used.
Western Blotting and Immunoprecipitation
Cells were transfected with pBabe-puro (1 μg) in combination with pSuper or pS-Survivin (10 μg each) and expression plasmids encoding FLAG-tagged versions of the indicated Survivin mutants (1 μg for Survivin1-142, 10 μg for the mutant proteins). Twenty-four hours after transfection, puromycin was added, and the viable cells were harvested 36 h later. Cells were lysed in RIPA (1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 0.15 M NaCl, 20 mM Tris-Cl, pH 7.4, and 2 mM EDTA) buffer containing protease inhibitors (Complete; Roche Diagnostics, Mannheim, Germany) on ice, and lysates were cleared by centrifugation. Equal amounts of protein were loaded on SDS-PAGE and subsequently subjected to Western blotting. For immunoprecipitation, cells were lysed in E1A-buffer supplemented with protease inhibitors (Complete) for 30 min at 4°C (Smits et al., 2000). GFP-tagged proteins were immunoprecipitated with 3 μg of α-GFP polyclonal antibody (pAb) precoupled to protein G-Sepharose (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
RESULTS
Complementation of Survivin Knockdown Cells Identified Survivin Mutants with Differential Rescue Capacity
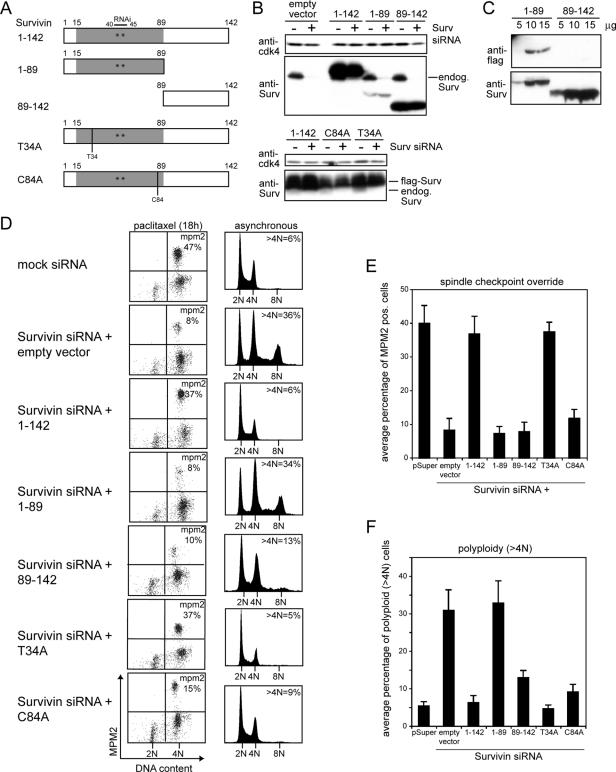
Using an RNAi-complementation approach, we aimed to separate the various mitotic functions of Survivin. To this end, we knocked down endogenous Survivin in U2OS cells by using vector-driven siRNA (pS-Survivin) and replaced the endogenous protein with exogenous epitope-tagged Survivin proteins whose cDNA contained two silent mutations in the RNAi targeting sequence, rendering these proteins insensitive to RNA interference by our Survivin siRNA (Lens et al., 2003). Because the C terminus of Survivin contains a potential protein–protein-interacting coiled-coil domain (Chantalat et al., 2000; Muchmore et al., 2000; Verdecia et al., 2000), a deletion mutant was generated that either lacked the C terminus but harbored the BIR domain (Survivin1-89) or a deletion mutant that only contained the C terminus (Survivin89-142). In addition, we mutated the potential cyclin B-cdc2 phosphorylation site threonine 34 (T34) and the zinc-chelating cysteine 84 of the BIR domain (C84) into alanine (Figure 1A). Together with amino acids C57, C60, and H77, cysteine 84 forms the zinc finger motif that stabilizes most of the BIR domain (Chantalat et al., 2000; Muchmore et al., 2000; Verdecia et al., 2000). Both point mutants have been described to act as dominant negative mutants (Li et al., 1998; O'Connor et al., 2000).
Figure 1.
Survivin RNAi complementation approach identifies two mutants that can rescue the cytokinesis defect but not the spindle checkpoint override of Survivin-depleted cells. (A) Schematic representation of the Survivin deletion and point mutants. Gray, BIR domain; white, C-terminal coiled coil. Stars indicate the position of the silent mutations in the RNAi targeting region. (B and C) Protein expression of the Survivin mutants in the presence or absence of Survivin siRNA. Western blots were probed with anti-Survivin pAb (B and C, bottom) or with anti-FLAG mAb (C, top) to detect only the exogenously expressed proteins. Anti-cdk4 was used as loading control (B). (D) Functional complementation by the mutant Survivin proteins. U2OS cells were cotransfected with mock siRNA or Survivin siRNA, the indicated Survivin plasmids and spectrin-GFP. Cells were either synchronized with thymidine and released for 18 h in the presence of paclitaxel (left) or left to grow asynchronously (right) Sixty hours after transfection cells were harvested, fixed, and stained with anti-MPM-2 mAb in combination with PI (left) or with PI only (right), and DNA profiles were analyzed in the GFP+ cell population. (E and F) Average percentages (±SEM) of mitotic, MPM-2–positive cells (E), and polyploid, >4N cells (F) of six independent experiments.
First, expression of these mutant proteins was confirmed by Western blotting (Figure 1, B and C). When blots were probed with an anti-Survivin pAb, both the endogenous and ectopically expressed Survivin proteins could be visualized. Using this antibody, the Survivin1-89 mutant seemed to be less well expressed than ectopic Survivin1-142 and the Survivin89-142 mutant (Figure 1B, top). However, when we transfected different amounts of the Survivin1-89 and Survivin89-142 expression plasmids and probed Western blots with an anti-FLAG mAb, we found that the Survivin1-89 mutant was better expressed than the Survivin89-142 mutant (Figure 1C, anti-FLAG blot) but less well detected by the anti-Survivin pAb (Figure 1C, anti-Survivin blot). These blots show that the anti-Survivin pAb is more sensitive than the anti-FLAG mAb and that the anti-Survivin pAb, predominantly recognizes epitopes residing within the C terminus of Survivin. Finally, both SurvivinT34A and SurvivinC84A were expressed to a slightly lesser extent than full-length Survivin (Survivin1-142) (Figure 1B, bottom).
To test the functionality of these different mutants, U2OS cells were cotransfected with Survivin siRNA, expression plasmids encoding the various proteins and GFP-spectrin as transfection marker. The override of a paclitaxel-induced mitotic arrest was used as measure for an impaired spindle checkpoint. Polyploidy (>4N DNA content) in asynchronous growing cells and tetraploidy (4N DNA content) in synchronized cells (not shown) were taken as a read out for defective cytokinesis (Lens et al., 2003). When synchronized cells were released from a thymidine block in the presence of paclitaxel, cells lacking Survivin failed to remain arrested in mitosis, resulting in a low mitotic index (Figure 1D, left, and E; Carvalho et al., 2003; Lens et al., 2003). Interestingly, only Survivin1-142 and SurvivinT34A were able to restore the spindle checkpoint defect, because cells transfected with these expression plasmids arrested in mitosis upon paclitaxel treatment. However, neither Survivin1-89, nor Survivin89-142 or SurvivinC84A were able to rescue the spindle checkpoint defect (Figure 1D, left, and E). The low mitotic indexes were not because of a G1/S arrest, because all the transfected cell populations accumulated with a 4N DNA content after thymidine release in the presence of paclitaxel (Figure 1D, left). Although ineffective in restoring the spindle checkpoint defect of Survivin-depleted cells, SurvivinC84A and Survivin89-142 were capable of reverting polyploidization induced by Survivin depletion. Whereas SurvivinC84A rescued the cytokinesis failure to a similar extent as the wild-type protein (Survivin1-142) and SurvivinT34A, Survivin89-142 rescued this failure somewhat less efficiently because a fraction of the reconstituted cells accumulated with a 4N DNA content (Figure 1D, right). Survivin1-89, in contrast, was totally incapable of reverting the cytokinesis defect (Figure 1D, right, and F). Thus, whereas Survivin1-142 and SurvivinT34A can functionally complement for endogenous Survivin, the mutant that lacks the C terminus (Survivin1-89) is completely inert. Interestingly, Survivin89-142 and SurvivinC84A can only rescue from polyploidization, but they are incapable of restoring a paclitaxel-induced mitotic delay.
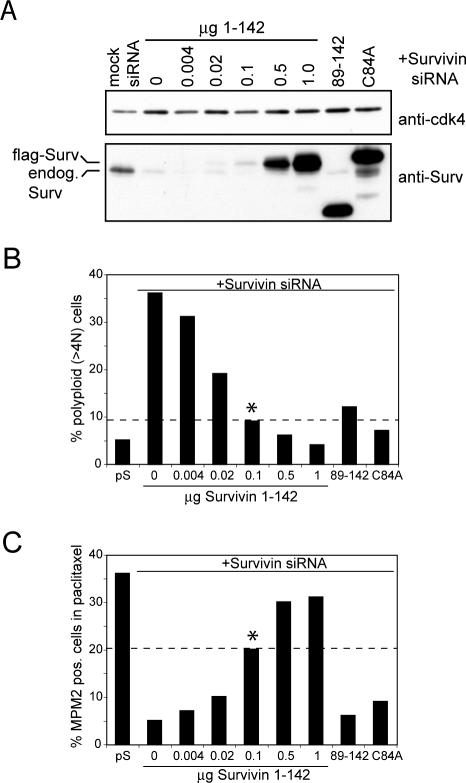
Low Expression Levels of Survivin89-142 and SurvivinC84A Do Not Explain Their Inability to Restore the Spindle Checkpoint Defect
The inability of Survivin89-142 and SurvivinC84A to restore the spindle checkpoint defect of Survivin-depleted cells could be explained by low expression levels of these proteins. In other words, low levels of Survivin might be sufficient to rescue cytokinesis, whereas higher levels of Survivin might be required to sustain the spindle checkpoint. To investigate this possibility, we cotransfected U2OS cells with Survivin siRNA and different concentrations of the Survivin1-142 expression plasmid. If indeed higher Survivin levels are needed to rescue the spindle checkpoint defect than the cytokinesis defect, it is expected that at a certain level of Survivin1-142 this difference becomes apparent. Titration of Survivin1-142 resulted in a concomitant decrease in rescue potential of the cytokinesis (Figure 2B) and spindle checkpoint defect (Figure 2C). The lowest concentration of Survivin1-142 that rescued the cytokinesis failure to a comparable level as the Survivin89-142 and SurvivinC84A mutants was 0.1 μg (Figure 2B). Yet, at this concentration, Survivin1-142 still significantly restored spindle checkpoint function (Figure 2C). We were unable to find a concentration at which Survivin1-142 mimics the differential rescue effect of Survivin89-142 and SurvivinC84A. These findings therefore suggest that the differential rescue capacity of Survivin89-142 and SurvivinC84A is not because of reduced expression levels but because of a functionally different behavior: the C-terminal α-helical coiled coil of Survivin is necessary and sufficient to restore the cytokinesis defect of Survivin-depleted cells, but the additional presence of a functional BIR domain is necessary to restore the spindle checkpoint defect.
Figure 2.
The inability of the Survivin89-142 and SurvivinC84A mutants to rescue the spindle checkpoint defect induced by Survivin depletion is not because of low expression levels. U2OS cells were cotransfected with mock siRNA or Survivin siRNA together with pBabe-puro and 10 μg of Survivin89-142, 10 μg of SurvivinC84A, or titrated amounts of Survivin1-142. (A) Puromycin-selected cells were lysed and subjected to SDS-PAGE and Western blotting. A degradation product of the overexpressed SurvivinC84A migrates at the size of endogenous Survivin. (B) DNA content of asynchronous growing GFP+ cells. (C) Mitotic index (%MPM-2–positive cells) of synchronized cells released in the presence of paclitaxel. Dashed line is for comparison of the percentage of polyploid (>4N) and mitotic cells complemented with 0.1 μg of Survivin1-142 (*) to cells complemented with SurvivinC84A and Survivin89-142.
Survivin89-142 and SurvivinC84A Colocalize with Aurora B and INCENP at the Midzone and Midbody but Fail to Maintain High Levels of These Proteins at the Centromeres
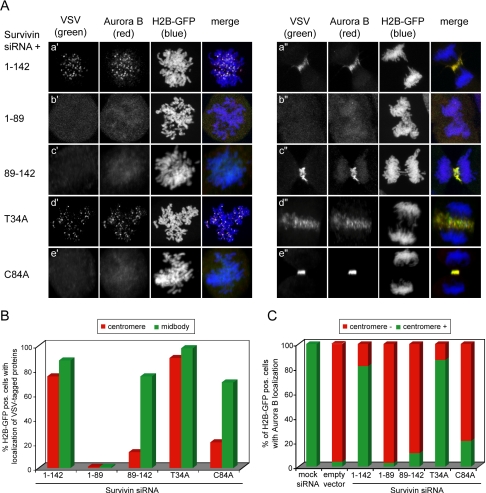
By titrating Survivin1-142, it is expected that Survivin levels will become limiting both at the centromeres and the central spindle, and this is most likely why the capacity to rescue the spindle checkpoint defect and the cytokinesis failure were equally affected. It was next asked whether the difference in rescue potential of the Survivin mutants could be explained by differences in subcellular localization. As expected, VSV-tagged Survivin1-142 localized as the endogenous protein: it was tethered to the centromeres during (pro)metaphase and to the midzone and midbody during anaphase and telophase, respectively (Figure 3Aa′ + a″, VSV columns, and B, and Supplemental Figure 1). The SurvivinT34A mutant that functionally behaves as the Survivin1-142 protein, also localized correctly during mitosis (Figure 3Ad′ + d″, VSV columns, and B). Importantly, Aurora B, which is mislocalized in Survivin-depleted cells (Carvalho et al., 2003; Honda et al., 2003; Lens et al., 2003), colocalized with exogenous Survivin1-142 and SurvivinT34A at the centromeres and midzone in Survivin knockdown cells (Figure 3Aa′ + a″, d′ + d″, Aurora B columns, and C).
Figure 3.
Colocalization of VSV-tagged Survivin proteins and endogenous Aurora B in Survivin-depleted cells. U2OS cells were grown on glass coverslips and cotransfected with Survivin siRNA, plasmids expressing VSV-tagged versions of the indicated Survivin proteins and with H2B-GFP to visualize the DNA. Cells were fixed in paraformaldehyde 14 h after release from a thymidine block. (A) The exogenously expressed proteins were detected with mouse anti-VSV mAb (columns labeled VSV) and endogenous Aurora B with rabbit anti-Aurora B pAb (columns labeled Aurora B). The left panel shows prometaphases, and the right panel shows early and late anaphases. (B) Percentage of transfected (H2B-GFP pos.) cells with centromeric (red) or midbody (green) localization of the VSV-tagged Survivin proteins. For each mutant,50 prometaphases and 50 telophases were scored. In the Survivin1-89 mutant, only 10 aberrant ana/telophases could be scored because the majority of these cells did not align their chromosomes nor underwent anaphase. (C) Percentage of transfected cells with centromeric localization of endogenous Aurora B. For each Survivin mutant, 75 prometaphases were scored.
Localization of the other mutant proteins correlated very well with their functional behavior. In line with its lack of rescue activity, Survivin1-89 did not localize to the centromeres or to the midzone but was found dispersed throughout the cytoplasm (Figure 3Ab′ + b″, VSV columns, and B). In line with this, Survivin1-89 also completely failed to relocalize Aurora B to centromeres and central spindle (Figure 3Ab′ + b″, Aurora B column, and C). Interestingly, Survivin89-142 and SurvivinC84A, incapable of rescuing the spindle checkpoint defect but capable of rescuing the cytokinesis failure, were hardly detectable on centromeres but clearly detectable on midzone and midbody (Figure 3Ac′ + c″, e′ + e″, VSV column, and B, and Supplemental Figure 1). In 13% of Survivin89-142 and 21% of the SurvivinC84A-reconstituted cells, we could detect these mutants on the centromeres but always at very low levels (Figures 3B and 6Bd′), implying that these mutants are able to interact with centromeres but that they have a severely reduced affinity for this chromosome structure. Importantly, in Survivin89-142 and SurvivinC84A-reconstituted cells, Aurora B colocalized with these mutant proteins on the midzone and midbody, but it was hardly detectable on the centromeres (Figure 3Ac′ + c″, e′ + e″, Aurora B column, and C). In line with the reduced centromeric localization of Aurora B in the Survivin1-89, Survivin89-142, and SurvivinC84A-reconstituted cells phosphorylation of the Aurora B substrate, CENP-A was also severely impaired in these cells (Supplemental Figure 2, A and B) (Zeitlin et al., 2001; Lampson and Kapoor, 2005).
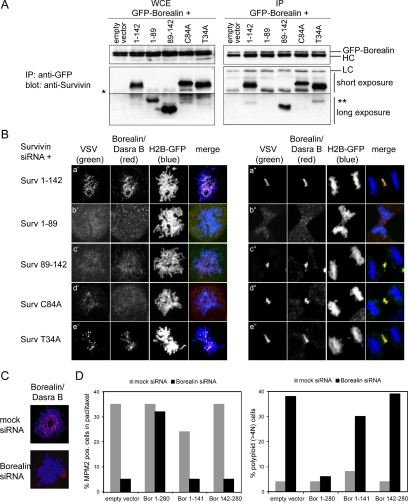
Figure 6.
Borealin/Dasra B interaction and localization with wild-type and mutant Survivin proteins and reconstitution of Borealin-depleted cells with Borealin/Dasra B truncation mutants. (A) HEK293 cells were cotransfected with GFP-tagged Borealin and the indicated Survivin mutants. Immunoprecipitations (IPs) were performed with anti-GFP pAb, and Western blots were probed with anti-Survivin pAb. Left, whole cell extracts (WCE). Right, immunoprecipitated proteins (IP). GPF-tagged Borealin runs just above the heavy chain (HC) and SurvC84A migrates slower than Surv1-142 and SurvT34A. Asterisk (*) is endogenous Survivin, and double asterisk (**) is a degradation product of the exogenously expressed protein. (B) Confocal images of U2OS cells cotransfected with Survivin siRNA, H2B-GFP, and the expression plasmids of the indicated VSV-tagged Survivin mutants. Endogenous Borealin/Dasra B was visualized with an anti-Dasra B pAb and the Survivin mutants with an anti-VSV mAb. (C) U2OS cells, grown on coverslips, were cotransfected with mock siRNA or Borealin siRNA and H2B-GFP as transfection marker. Fourteen hours after thymidine release, coverslips were fixed and stained with anti-Dasra B pAb. Borealin/Dasra (red) and H2B-GFP (blue). (D) U2OS cells were cotransfected with spectrin-GFP, Borealin siRNA and the indicated Borealin cDNAs. Left, cells were synchronized with thymidine and released in the presence of paclitaxel for 18 h. Mitotic index was determined by combined MPM2/PI staining and FACS analysis. Right, cells were grown asynchronously, harvested 60 h after transfection, stained with PI, and DNA content was determined by FACS analysis.
Finally, to further confirm that centromere and central spindle localization of the CPC is uncoupled in Survivin89-142-reconstituted cells, we imaged the localization of INCENP-GFP in living cells (Vader et al., 2006). In Survivin1-142-reconstituted cells (Figure 4A), INCENP-GFP was found both on the chromatin and clearly concentrated at the centromeres in (pro)metaphase. In anaphase, the centromeric localization was progressively lost and midzone localization became apparent. In Survivin89-142 and SurvivinC84A-reconstituted cells (Figure 4B; not shown), INCENP-GFP was found weakly dispersed on chromatin but failed to concentrate at centromeres. This weak chromatin localization was also found in Survivin-depleted cells (Figure 4C). However, in the Survivin89-142-reconstituted cells INCENP-GFP was able to localize to the midzone and midbody during anaphase and telophase, respectively. Thus, absence of a functional BIR domain affects the centromere localization of Survivin, Aurora B, and INCENP but not their central spindle localization and implicates that previous concentration at the centromere is not a prerequisite for midzone and midbody localization.
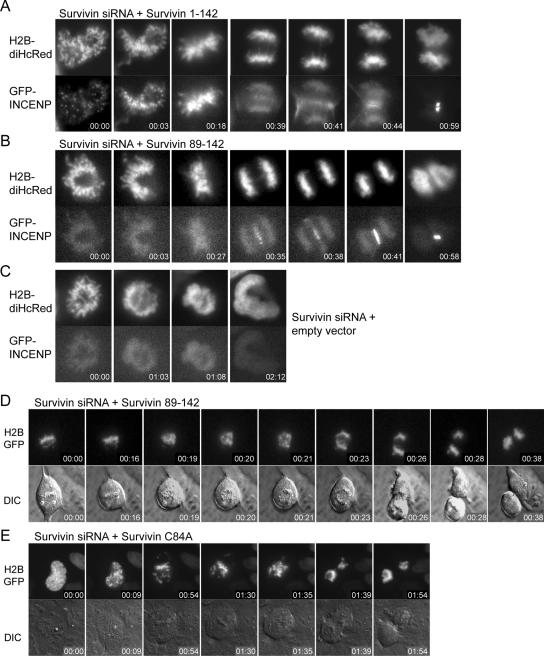
Figure 4.
INCENP-GFP localization, chromosome segregation, and cytokinesis in living Survivin1-142, Survivin89-142, and SurvivinC84A complemented cells. U2OS cells were grown in glass-bottom wells and cotransfected with Survivin siRNA and Survivin1-142 (A) Survivin89-142 (B and D), empty plasmid (C), or SurvivinC84A (E). To visualize CPC behavior, INCENP-GFP and H2B-diHcRed were cotransfected (A–C); in the other panels, only H2B-GFP was cotransfected (D and E). Cells were mounted on a time-lapse microscope 10 h after release from a thymidine block. Fluorescent and DIC images were captured every 1–5 min. Imaging was started at the onset of nuclear envelope breakdown in D and in prometaphase for A–C and E. To better visualize the localization of INCENP-GFP, enlargements of the images are shown in A–C. (D and E) Both the Survivin89-142 (3/8) and SurvivinC84A (3/9) reconstituted cells partially align their chromosomes. However, even in the situation where chromosome alignment was incomplete, cells start to segregate their sister chromatids, enter anaphase, and undergo cytokinesis (D and E). Note that in two or eight Survivin89-142- and five of nine SurvivinC84A-reconstituted cells alignment was normal. Time is hours:minutes.
Chromosome Behavior and Cytokinesis in Survivin Knockdown Cells Complemented with Survivin89-142 and SurvivinC84A
To further strengthen our observation that Survivin knockdown cells complemented with Survivin89-142 or SurvivinC84A indeed undergo cytokinesis, we cotransfected these cells with H2B-GFP and performed live imaging of cells in which the mitotic spindle was not disturbed by paclitaxel. The DIC images clearly show that cells expressing either mutant can undergo cytokinesis (Figure 4, D and E, bottom). Interestingly, when monitoring chromosome behavior, we found that in three of nine SurvivinC84A-complemented cells and three of eight Survivin89-142-complemented cells chromosome alignment was impaired, yet these cells entered anaphase and performed cytokinesis, indicating premature silencing of the spindle checkpoint (Figure 4, E and D, top, and Table 1). Also, after fixation we frequently found lagging chromosomes and anaphase bridges in the Survivin89-142 and SurvivinC84A-reconstituted cell populations (Figure 3Ac″ and Supplemental Figure 1B), reflecting the incapacity of these mutants to restore Survivin function at the centromere. Whereas mitotic exit in the presence of misaligned chromosomes is reminiscent of Survivin knockdown cells, anaphase and cytokinesis are not (Figure 4C; Lens et al., 2003). The phenotype of the reconstituted cells is in line with the fluorescence-activated cell sorting (FACS) data, where we found a reduction in the percentage of polyploid cells best explained by restoration of the cytokinesis defect, but an override of a paclitaxel-induced mitotic delay because of an unrestored spindle checkpoint defect. This thus confirms that Survivin89-142 and SurvivinC84A can indeed complement for the cytokinesis defect observed after Survivin depletion.
Table 1.
Summary of live cell imaging experiments
| Survivin siRNA+
|
|||||
|---|---|---|---|---|---|
| Category of defect: | Mock siRNA (n = 9) | Empty vector (n = 9) | Surv1-142 (n = 10) | SurvC84A (n = 9) | Surv89-142 (n = 8) |
| Aligned chromosomes and successful cytokinesis (“normal” mitosis) | 100 | 22 | 90 | 56 | 24 |
| Misaligned chromosomes and no cytokinesis | 0 | 78 | 10 | 11 | 38 |
| Misaligned chromosomes and successful cytokinesis | 0 | 0 | 0 | 33 | 38 |
Values are percentage of transfected U2OS cells with chromosome alignment and cytokinesis defects after reconstitution with Survivin mutants.
Importantly, in 24% of the Survivin89-142-reconstituted and 56% of the SurvivinC84A-complemented cells we found that chromosome alignment was normal (Table 1 and Figure 4B). Because Aurora B is important for the establishment of bipolar attachments of the microtubules from the mitotic spindle to the sister chromatids (Tanaka et al., 2002; Lampson et al., 2004), the most likely explanation for this result is that low levels of Aurora B protein and kinase activity at the centromeres, as occasionally observed in the Survivin89-142 and SurvivinC84A-expressing cells (Figure 3C and Supplemental Figure 2, A and B), are sufficient for correcting maloriented microtubules in an unperturbed mitosis but may fail to do so when normal spindle formation is perturbed by drugs. These data thus imply that higher levels of Survivin and Aurora B are needed at the centromere to sustain a mitotic delay after paclitaxel treatment than to resolve occasional maloriented kinetochore–microtubule attachments in an unperturbed mitosis.
Survivin89-142 and SurvivinC84A Are Unable to Retain High Levels of BubR1 at Kinetochores
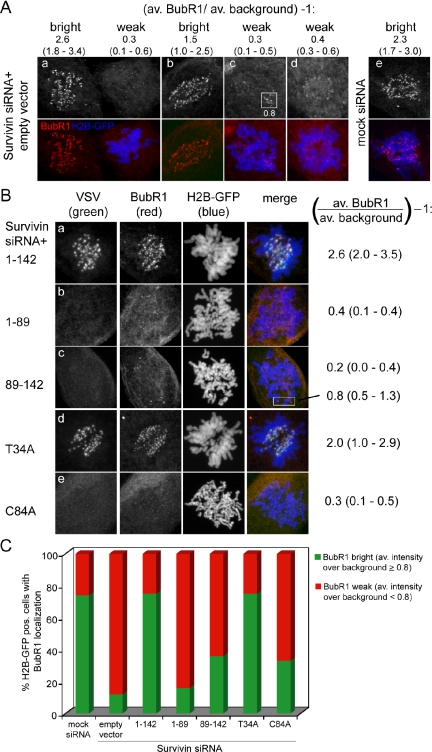
To further understand why Survivin knockdown cells complemented with Survivin89-142 and SurvivinC84A are unable to restore a mitotic arrest after paclitaxel treatment, we checked kinetochore localization of BubR1. As shown previously, kinetochore localization of BubR1 is greatly reduced in cells lacking Survivin, even when treated with paclitaxel (Carvalho et al., 2003; Lens et al., 2003) (Figure 5A). Importantly, the proteins that could rescue the spindle checkpoint override (Survivin1-142 and SurvivinT34A) were also able to localize BubR1 to the kinetochores (Figure 5Ba,d and C). In contrast, for the mutants that failed to restore a paclitaxel-induced mitotic delay (e.g., Survivin1-89, Survivin89-142, and SurvivinC84A) we found that the BubR1 intensity levels at the majority of kinetochores were very low (<0.8 over background) (Figure 5Bb,c,e and C). Thus, the inability of the latter mutants to maintain high levels of BubR1 at the kinetochores correlates very well with their incapacity to restore the spindle checkpoint defect in Survivin-depleted cells.
Figure 5.
BubR1 localization in Survivin knockdown cells reconstituted with wild-type and mutant Survivin proteins. U2OS cells were grown on glass coverslips and cotransfected with Survivin siRNA, plasmids expressing VSV-tagged versions of the indicated Survivin proteins and with H2B-GFP to visualize the DNA. Cells were fixed in paraformaldehyde 14 h after release from a thymidine block. (A) Examples of transfected (H2B-GFP pos., blue) prometaphases without BubR1 kinetochore localization and nontransfected (H2B-GFP neg.) prometaphases with BubR1 kinetochore localization (red) within one coverslip. Using the in-built area measurement module of the MetaMorph software, at 20 different positions per cell, background and kinetochore pixel intensities were measured. For each cell the average kinetochore pixel intensity was divided by the average background pixel intensity and subtracted with 1, to obtain the average BubR1 intensity levels over background. The lowest and highest BubR1 kinetochore intensities of that particular cell are indicated between parentheses. (B) The exogenously expressed proteins were detected with mouse anti-VSV mAb (columns labeled VSV) and endogenous BubR1 with sheep anti-human BubR1 pAb (columns labeled BubR1). For each transfection BubR1 levels of nontransfected cells were comparable with those in A. The indicated average BubR1 kinetochore pixel intensity (lowest - highest intensity) was calculated as in A. (C) Percentage of transfected cells with bright (average pixel intensity over background ≥0.8) or weak (average pixel intensity over background <0.8) kinetochore localization of endogenous BubR1. For each Survivin mutant, 100 prometaphases were scored.
The C Terminus of Survivin Interacts with Borealin/Dasra B and Can Localize the Protein to the Midbody
Recently, Borealin/Dasra B was identified as a novel interaction partner of Survivin (Gassmann et al., 2004; Sampath et al., 2004). An N-terminal fragment of Borealin (aa 1–141) was found to interact with Survivin and when overexpressed, this N-terminal fragment displaced Survivin and other passenger proteins from the centromeres but not from the central spindle (Gassmann et al., 2004). This observation implied an important function of Borealin/Dasra B in centromere targeting of the chromosome passenger holocomplex. Yet, the observation that a single point mutation in the BIR domain of Survivin affected localization and function of Survivin and Aurora B at the centromere but not at the central spindle also suggests a function for the Survivin BIR domain in centromere targeting of the CPC. For CSC-1 (the functional homologue of Borealin/Dasra B in Caenorhabditis elegans), it was shown that mutation of C83 in the Survivin homologue BIR-1 abolished the interaction between CSC-1 and BIR-1. Similar to C84 in Survivin, C83 of BIR-1 is predicted to chelate zinc, which is important for stabilization of its BIR domain (Romano et al., 2003). We therefore first investigated whether disruption of the Survivin BIR domain also affected the interaction between human Survivin and Borealin/Dasra B. GFP-tagged Borealin was coexpressed with the various Survivin mutants in HEK293 cells and subsequently immunoprecipitated from these cells. Interestingly, Borealin/Dasra B interacted with Survivin1-142, SurvivinT34A and with SurvivinC84A (Figure 6A). Moreover, the C terminus of Survivin and not the N-terminal BIR domain was able to interact with Borealin/Dasra B, although less efficiently as Survivin1-142 (Figure 6A). Importantly, whereas Survivin1-142 and SurvivinT34A restored localization of endogenous Borealin/Dasra B to both centromeres and central spindles, SurvivinC84A and Survivin89-142 could restore localization of endogenous Borealin/Dasra B to the midzone/midbody but not to the centromere (Figure 6Bc′ + c″, d′ + d″), suggesting that the Survivin–BIR domain contains essential cues for centromere localization of the CPC. Finally, when we complemented Borealin-depleted cells (Figure 6C) with the N-terminal fragment of Borealin/Dasra B (Bor1-141) we found that, despite its capacity to localize to the central spindle (Supplemental Figure 3, A and Bb″; Gassmann et al., 2004), it did not have the functional capacity to revert the cytokinesis defect due to Borealin/Dasra B depletion (Figure 6D).
DISCUSSION
Using an RNAi complementation approach, we were able to discriminate distinct domains in Survivin that are involved in localization of the chromosomal passenger complex to different subcellular structures and that play a role in separate functions of Survivin. We found that the C-terminal domain of Survivin (aa 89-142) was capable of localizing to the midzone and midbody and of restoring the cytokinesis defect induced by Survivin depletion. However, this mutant had a reduced ability to localize to the centromeres and to retain high levels of Aurora B, INCENP, Borealin/Dasra B, and BubR1 at these sites. As a consequence, Survivin89-142 was unable to restore a spindle checkpoint defect induced by the microtubule-stabilizing drug paclitaxel. Interestingly, the SurvivinC84A mutant behaved in a similar manner as the Survivin89-142 mutant. By mutating C84 into alanine, the architecture of the BIR domain is disturbed (Chantalat et al., 2000; Muchmore et al., 2000; Verdecia et al., 2000), and this most likely explains why the SurvivinC84A mutant mimics the rescue effect of the Survivin89-142 deletion mutant. Not surprisingly, complete deletion of the BIR domain has a more dramatic effect on the localization and function of Survivin than perturbation of the BIR by a single point mutation. Clearly, the add-back experiments with these mutants show that an intact BIR domain is essential for proper centromere localization and spindle checkpoint function. Yet, the BIR domain by itself is not sufficient because expression of Survivin1-89 could not restore centromere localization and spindle checkpoint function in Survivin-depleted cells. Moreover, because SurvivinC84A and Survivin89-142 fail to recruit the CPC to the centromeres, but do allow execution of cytokinesis, these data indicate that concentration of the CPC at centromeres is not a requirement for cytokinesis, as had been proposed previously.
SurvivinC84A has been described to act as a dominant negative protein that could induce apoptosis and cell division defects by displacing wild-type, endogenous Survivin from polymerized microtubules (Li et al., 1998, 1999). Similar to Skoufias et al. (2000), mere overexpression of SurvivinC84A in U2OS cells did not result in any cell division defects in our hands (Lens and Rodriguez, unpublished data). More importantly, when localization of this mutant was monitored in the presence of endogenous Survivin, it was found to localize diffusely throughout the mitotic cell. The typical midzone and midbody localization of SurvivinC84A became only apparent when expression of endogenous Survivin was suppressed by siRNA (Lens, unpublished observation). Its incapacity to displace endogenous Survivin from centromeres and the central spindle may explain why this typical localization pattern of SurvivinC84A was missed previously (Skoufias et al., 2000) and why in our hands overexpression of this mutant does not have any dominant negative effects. However, by combining mutational analysis with RNAi complementation, we have revealed a novel function for the BIR domain; it is essential for centromere localization and spindle checkpoint function of Survivin.
Overexpression experiments with SurvivinT34A suggested that also this mutant could act in a dominant negative manner (O'Connor et al., 2000; Grossman et al., 2001). The fact that this mutant can localize to all the expected sites during cell division, even in the presence of endogenous Survivin (Lens, unpublished observation), suggests that SurvivinT34A is able to compete with the localization of endogenous Survivin and that in principle it could act as a dominant negative protein. Yet, overexpression of this mutant did not affect the cell cycle or cell survival (Lens and Rodriguez, unpublished data). More importantly, this mutant was as effective as the wild-type protein in restoring all the cell cycle defects induced by Survivin depletion, indicating that CDK-dependent phosphorylation of T34 is not essential for Survivin function during mitosis.
Survivin-depleted cells are unable to sustain a mitotic delay induced by treatment with paclitaxel (Carvalho et al., 2003; Lens et al., 2003). The Survivin89-142 and SurvivinC84A mutants, although capable of restoring cytokinesis, could not restore this mitotic delay in Survivin-depleted cells most likely because these mutants did not fully reestablish Aurora B and BubR1 localization at the centromeres and kinetochores, respectively. Aurora B is thought to influence spindle checkpoint activity by destabilizing kinetochore–microtubule attachments that do not create tension (e.g., that are nonbipolar) (Biggins and Murray, 2001; Tanaka et al., 2002; Lampson et al., 2004). The resulting unattached kinetochore most likely (re)recruits Mad2 that will inhibit the APC/C (Zhou et al., 2002; Lens and Medema, 2003). In addition, sustained kinetochore localization of BubR1 seems to be Survivin/Aurora B-dependent (Carvalho et al., 2003; Ditchfield et al., 2003; Lens et al., 2003). Interestingly, when mitotic progression was studied in the absence of paclitaxel, in 38% of the Survivin89-142- and 33% of the SurvivinC84A-reconstituted cells chromosome congression was impaired. Still, in 24% of the Survivin89-142- and 56% of the SurvivinC84A-reconstituted cells chromosome alignment was apparently normal. This could mean that the low protein levels and kinase activity of centromeric Aurora B that we sometimes observed in Survivin89-142- and SurvivinC84A-reconstituted cells is sufficient to reorientate an occasional misattached microtubule in an unperturbed mitosis but is not sufficient to sustain a mitotic delay after microtubule stabilization induced by paclitaxel. Moreover, the inability of the Survivin89-142 and SurvivinC84A mutants to restore normal levels of BubR1 at the kinetochores may also contribute to their incapacity to restore a paclitaxel-induced mitotic delay. Interestingly, these data also suggest that a gradual reduction of the levels of the CPC at the centromeres results in a gradual loss of checkpoint functionality, and as such (minor) changes in the level of CPC components could potentially lead to mild checkpoint defects.
By combining mutational analysis and siRNA complementation, we were able to uncouple Survivin's centromere localization and spindle checkpoint function from its central spindle localization and cytokinesis function. Interestingly, in Drosophila secondary spermatocytes, cytokinesis can take place in the absence of chromosomes and importantly, Aurora B was found to be normally localized at the midzone (Bucciarelli et al., 2003). This is in line with our findings and suggests that centromeric concentration of the CPC is not a prerequisite for its accumulation at the central spindle. Similarly, overexpression of an N-terminal fragment of Borealin (aa 1-141) in Borealin-proficient cells displaced the other passenger proteins from the centromeres but did not affect their midbody localization (Gassmann et al., 2004). Interestingly, when this N-terminal Borealin fragment was expressed in Borealin-depleted cells, it was also able to localize to the central spindle, but not to the centromeres. Thus, the Borealin N terminus seems to localize in a similar manner in Borealin-depleted cells as the Survivin C terminus in Survivin-depleted cells. Nonetheless, the Borealin N terminus could not efficiently rescue the cytokinesis defect of Borealin-depleted cells. Whereas full-length Borealin can directly interact with INCENP, the Borealin N-terminal fragment cannot (Gassmann et al., 2004). Moreover, we found Borealin/Dasra B to be important for an efficient interaction between Survivin and INCENP, and Survivin to be capable of localizing the CPC in the absence of Borealin/Dasra B when directly fused to INCENP (Vader et al., 2006). Thus, in Survivin-depleted cells reconstituted with Survivin89-142, endogenous full-length Borealin/Dasra B will be able to interact with both the Survivin mutant as well as with INCENP/Aurora B. In contrast, in Borealin-depleted cells reconstituted with Borealin1-141, Borealin/Dasra B may be able to interact with endogenous Survivin but cannot make the connection with INCENP/Aurora B. Together with Survivin, this Borealin mutant could therefore localize to the central spindle but is most likely unable to relocalize the other passengers to this structure, hence its incapacity to functionally complement the cytokinesis defect.
With respect to the centromere localization, we propose that the Survivin BIR domain is the portion of the CPC that interacts with the centromere. A single point mutation in the BIR domain of Survivin is sufficient to disturb its centromere localization but does not abolish its interaction with Borealin/Dasra B. Thus, unlike BIR-1 in C. elegans (Romano et al., 2003), the BIR domain of mammalian Survivin is not essential for Borealin/Dasra B interaction. Again, Borealin/Dasra B may help in connecting centromere-bound Survivin to INCENP and Aurora B, and the entire complex may subsequently stabilize the Survivin–centromere interaction. By binding to Survivin but failing to make the connection with INCENP and Aurora B, Borealin1-141 may thus prevent stabilization of the Survivin–centromere interaction.
Our combined findings imply that Survivin is crucial for the spatial control of the chromosomal passenger complex. By interfering with its localization during mitosis, typical CPC-associated functions, such as sustaining a mitotic arrest in the absence of bipolar spindle attachments and execution of cytokinesis, can be uncoupled. Further understanding on how the different domains within Survivin are able to dictate its localization will provide insight into the spatiotemporal regulation of the CPC and its role in maintaining a stable genome.
Supplementary Material
Acknowledgments
We thank Dr. J. Laoukili for critical reading of the manuscript and helpful discussions and Drs. S. Taylor and H. Funabiki for the generous gift of antibodies. This work is supported by Dutch Cancer Society Grant NKI 2002–2764.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0727) on January 25, 2006.
Abbreviations used: BIR, baculovirus IAP repeat; CPC, chromosomal passenger complex.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adams, R. R., Carmena, M., and Earnshaw, W. C. (2001). Chromosomal passengers and the (Aurora) ABCs of mitosis. Trends Cell Biol. 11, 49-54. [DOI] [PubMed] [Google Scholar]
- Ambrosini, G., Adida, C., and Altieri, D. C. (1997). A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat. Med. 3, 917-921. [DOI] [PubMed] [Google Scholar]
- Biggins, S., and Murray, A. W. (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15, 3118-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, J. R., et al. (1998). A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17, 3052-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T. R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- Bucciarelli, E., Giansanti, M. G., Bonaccorsi, S., and Gatti, M. (2003). Spindle assembly and cytokinesis in the absence of chromosomes during Drosophila male meiosis. J. Cell Biol. 160, 993-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, A., Carmena, M., Sambade, C., Earnshaw, W. C., and Wheatley, S. P. (2003). Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 116, 2987-2998. [DOI] [PubMed] [Google Scholar]
- Chantalat, L., Skoufias, D. A., Kleman, J. P., Jung, B., Dideberg, O., and Margolis, R. L. (2000). Crystal structure of human survivin reveals a bow tie-shaped dimer with two unusual alpha-helical extensions. Mol. Cell 6, 183-189. [PubMed] [Google Scholar]
- Cooke, C. A., Heck, M. M., and Earnshaw, W. C. (1987). The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J. Cell Biol. 105, 2053-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield, C., Johnson, V. L., Tighe, A., Ellston, R., Haworth, C., Johnson, T., Mortlock, A., Keen, N., and Taylor, S. S. (2003). Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161, 267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, A. G., James, C., Evan, G. I., and Hengartner, M. O. (1999). Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr. Biol. 9, 292-301. [DOI] [PubMed] [Google Scholar]
- Gassmann, R., Carvalho, A., Henzing, A. J., Ruchaud, S., Hudson, D. F., Honda, R., Nigg, E. A., Gerloff, D. L., and Earnshaw, W. C. (2004). Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166, 179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, D., Kim, P. J., Schechner, J. S., and Altieri, D. C. (2001). Inhibition of melanoma tumor growth in vivo by survivin targeting. Proc. Natl. Acad. Sci. USA 98, 635-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf, S., Cole, R. W., LaTerra, S., Zimmer, C., Schnapp, G., Walter, R., Heckel, A., van Meel, J., Rieder, C. L., and Peters, J. M. (2003). The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, R., Korner, R., and Nigg, E. A. (2003). Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 14, 3325-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M. A., Totis, L., and Roberts, B. T. (1991). S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66, 507-517. [DOI] [PubMed] [Google Scholar]
- Hwang, L. H., Lau, L. F., Smith, D. L., Mistrot, C. A., Hardwick, K. G., Hwang, E. S., Amon, A., and Murray, A. W. (1998). Budding yeast Cdc 20, a target of the spindle checkpoint. Science 279, 1041-1044. [DOI] [PubMed] [Google Scholar]
- Kalejta, R. F., Shenk, T., and Beavis, A. J. (1997). Use of a membrane-localized green fluorescent protein allows simultaneous identification of transfected cells and cell cycle analysis by flow cytometry. Cytometry 29, 286-291. [DOI] [PubMed] [Google Scholar]
- Kanda, T., Sullivan, K. F., and Wahl, G. M. (1998). Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8, 377-385. [DOI] [PubMed] [Google Scholar]
- Kim, S. H., Lin, D. P., Matsumoto, S., Kitazono, A., and Matsumoto, T. (1998). Fission yeast Slp 1, an effector of the Mad2-dependent spindle checkpoint. Science 279, 1045-1047. [DOI] [PubMed] [Google Scholar]
- Lampson, M. A., and Kapoor, T. M. (2005). The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7, 93-98. [DOI] [PubMed] [Google Scholar]
- Lampson, M. A., Renduchitala, K., Khodjakov, A., and Kapoor, T. M. (2004). Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. [DOI] [PubMed]
- Lens, S. M., and Medema, R. H. (2003). The survivin/Aurora B complex: its role in coordinating tension and attachment. Cell Cycle 2, 507-510. [DOI] [PubMed] [Google Scholar]
- Lens, S. M., Wolthuis, R. M., Klompmaker, R., Kauw, J., Agami, R., Brummelkamp, T., Kops, G., and Medema, R. H. (2003). Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22, 2934-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Ackermann, E. J., Bennett, C. F., Rothermel, A. L., Plescia, J., Tognin, S., Villa, A., Marchisio, P. C., and Altieri, D. C. (1999). Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat. Cell Biol. 1, 461-466. [DOI] [PubMed] [Google Scholar]
- Li, F., Ambrosini, G., Chu, E. Y., Plescia, J., Tognin, S., Marchisio, P. C., and Altieri, D. C. (1998). Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396, 580-584. [DOI] [PubMed] [Google Scholar]
- Li, F., Flanary, P. L., Altieri, D. C., and Dohlman, H. G. (2000). Cell division regulation by BIR1, a member of the inhibitor of apoptosis family in yeast. J. Biol. Chem. 275, 6707-6711. [DOI] [PubMed] [Google Scholar]
- Li, R., and Murray, A. W. (1991). Feedback control of mitosis in budding yeast. Cell 66, 519-531. [DOI] [PubMed] [Google Scholar]
- Li, Y., and Benezra, R. (1996). Identification of a human mitotic checkpoint gene: hsMAD2. Science 274, 246-248. [DOI] [PubMed] [Google Scholar]
- Muchmore, S. W., Chen, J., Jakob, C., Zakula, D., Matayoshi, E. D., Wu, W., Zhang, H., Li, F., Ng, S. C., and Altieri, D. C. (2000). Crystal structure and mutagenic analysis of the inhibitor-of-apoptosis protein survivin. Mol. Cell 6, 173-182. [PubMed] [Google Scholar]
- Musacchio, A., and Hardwick, K. G. (2002). The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell. Biol. 3, 731-741. [DOI] [PubMed] [Google Scholar]
- O'Connor, D. S., Grossman, D., Plescia, J., Li, F., Zhang, H., Villa, A., Tognin, S., Marchisio, P. C., and Altieri, D. C. (2000). Regulation of apoptosis at cell division by p34cdc2 phosphorylation of survivin. Proc. Natl. Acad. Sci. USA 97, 13103-13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines, J. (1997). Localization of cell cycle regulators by immunofluorescence. Methods Enzymol. 283, 99-113. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, H., and Lengauer, C. (2004). Aneuploidy and cancer. Nature 432, 338-341. [DOI] [PubMed] [Google Scholar]
- Rieder, C. L., Cole, R. W., Khodjakov, A., and Sluder, G. (1995). The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130, 941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, A., Guse, A., Krascenicova, I., Schnabel, H., Schnabel, R., and Glotzer, M. (2003). CSC-1, a subunit of the Aurora B kinase complex that binds to the survivin-like protein BIR-1 and the incenp-like protein ICP-1. J. Cell Biol. 161, 229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath, S. C., Ohi, R., Leismann, O., Salic, A., Pozniakovski, A., and Funabiki, H. (2004). The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118, 187-202. [DOI] [PubMed] [Google Scholar]
- Shah, J. V., and Cleveland, D. W. (2000). Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell 103, 997-1000. [DOI] [PubMed] [Google Scholar]
- Skoufias, D. A., Mollinari, C., Lacroix, F. B., and Margolis, R. L. (2000). Human survivin is a kinetochore-associated passenger protein. J. Cell Biol. 151, 1575-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, V. A., Klompmaker, R., Arnaud, L., Rijksen, G., Nigg, E. A., and Medema, R. H. (2000). Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2, 672-676. [DOI] [PubMed] [Google Scholar]
- Speliotes, E. K., Uren, A., Vaux, D., and Horvitz, H. R. (2000). The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell 6, 211-223. [DOI] [PubMed] [Google Scholar]
- Storchova, Z., and Pellman, D. (2004). From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell. Biol. 5, 45-54. [DOI] [PubMed] [Google Scholar]
- Sudakin, V., Chan, G. K., and Yen, T. J. (2001). Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154, 925-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T. U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M. J., and Nasmyth, K. (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317-329. [DOI] [PubMed] [Google Scholar]
- Tang, Z., Bharadwaj, R., Li, B., and Yu, H. (2001). Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell 1, 227-237. [DOI] [PubMed] [Google Scholar]
- Taylor, S. S., Ha, E., and McKeon, F. (1998). The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, S. S., Hussein, D., Wang, Y., Elderkin, S., and Morrow, C. J. (2001). Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114, 4385-4395. [DOI] [PubMed] [Google Scholar]
- Vader, G., Kauw, J. J., Medema, R. H., and Lens, S. M. (2006). Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 7, 85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli, P., and Earnshaw, W. C. (2004). Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma 113, 211-222. [DOI] [PubMed] [Google Scholar]
- Verdecia, M. A., Huang, H., Dutil, E., Kaiser, D. A., Hunter, T., and Noel, J. P. (2000). Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat. Struct. Biol. 7, 602-608. [DOI] [PubMed] [Google Scholar]
- Zeitlin, S. G., Shelby, R. D., and Sullivan, K. F. (2001). CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Yao, J., and Joshi, H. C. (2002). Attachment and tension in the spindle assembly checkpoint. J. Cell Sci. 115, 3547-3555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.