Abstract
In the yeast Saccharomyces cerevisiae, two similar phosphatidylinositol 3-kinase complexes (complexes I and II) function in distinct biological processes, complex I in autophagy and complex II in the vacuolar protein sorting via endosomes. Atg14p is only integrated into complex I, likely facilitating the function of complex I in autophagy. Deletion analysis of Atg14p revealed that N-terminal region containing the coiled-coil structures was essential and sufficient for autophagy. Atg14p localized to pre-autophagosomal structure (PAS) and vacuolar membranes, whereas Vps38p, a component specific to complex II, localized to endosomes and vacuolar membranes. Vps34p and Vps30p, components shared by the two complexes, localized to the PAS, vacuolar membranes, and several punctate structures that included endosomes. The localization of these components to the PAS was Atg14p dependent but not dependent on Vps38p. Conversely, localization of these proteins to endosomes required Vps38p but not Atg14p. Vps15p, regulatory subunit of the Vps34p complexes, localized to the PAS, vacuolar membranes, and punctate structures independent of both Atg14p and Vps38p. Together, these results indicate that complexes I and II function in distinct biological processes by localizing to specific compartments in a manner mediated by specific components of each complex, Atg14p and Vps38p, respectively.
INTRODUCTION
Membrane trafficking events should occur in a spatiotemporally regulated manner to deliver vesicles and their cargo proteins to the correct destination. The class III phosphatidylinositol 3-kinase (PI3-kinase; hereafter PI3-kinase indicates this class III kinase) participates in the regulation of membrane trafficking. PI3-kinase is a lipid kinase that specifically phosphorylates phosphatidylinositol at the D-3 position of the inositol ring to produce phosphoinositide 3-phosphate (PI3-P) (Schu et al., 1993; Stack and Emr, 1994). The generated PI3-P is recognized by downstream effectors; these effector molecules concentrate to a restricted area of the membrane where the subsequent membrane trafficking events occur (reviewed in Martin, 1998; Odorizzi et al., 2000). PI3-kinase is involved in membrane trafficking events, including endocytosis, the sorting of soluble vacuolar proteins, and autophagy (Schu et al., 1993; Munn and Riezman, 1994; Kihara et al., 2001). Currently, the mechanisms by which PI3-kinase mediates these diverse functions while maintaining the fidelity of each membrane-trafficking event remains poorly understood.
The involvement of PI3-kinase in membrane trafficking has been extensively studied in yeast. Vps34p, the only PI3-kinase encoded by yeast, is required for the sorting of nascent soluble vacuolar hydrolases, such as carboxypeptidase Y (CPY), via endosomes (Robinson et al., 1988; Schu et al., 1993). Yeast cells harboring point mutations in the Vps34p lipid kinase domain missort CPY into the secretory pathway. The function of Vps34p is dependent on the protein kinase activity of Vps15p, a serine/threonine kinase; therefore, Δvps15 cells display a similar phenotype to that seen for Δvps34 cells (Herman and Emr, 1990; Herman et al., 1991a, b; Stack et al., 1993, 1995). PI3-kinase is also involved in protein transport in mammalian cells. Proteins resident in lysosomes, an organelle functionally equivalent to the yeast vacuole, are erroneously secreted after treatment with PI3-kinase inhibitors (Brown et al., 1995; Davidson, 1995), suggesting a conserved function for PI3-kinases in protein sorting into the vacuole/lysosome in both yeast and mammalian cells. The human homologue of Vps15p associates with the Vps34p homologue in vivo (Volinia et al., 1995).
Autophagy is another major pathway delivering proteins into the vacuole/lysosome. Using this process, cells degrade large quantities of their cytoplasmic content to recycle the components in response to severe shortages of nutrients. This process is composed of multiple steps that are well conserved among eukaryotes. On induction of autophagy, the cytoplasmic contents are enclosed into a double membrane structure called an autophagosome. The autophagosome then fuses with a vacuole/lysosome; the inner membrane structures and packaged cytoplasmic macromolecules are degraded within this compartment (Baba et al., 1994; Mizushima et al., 2001; reviewed in Klionsky and Ohsumi, 1999). Microscopic analysis suggested that the majority of autophagy-related (Atg) proteins localize to a perivacuolar structure called pre-autophagosomal structure (PAS) at which autophagosomes are thought to originate (Suzuki et al., 2001). Because subcellular fractionation and electron microscopic studies have revealed that this poorly characterized structure does not contain vacuolar, endoplasmic reticulum (ER), Golgi, or late-endosomal markers, autophagosome formation likely involves a novel process that is distinct from the mechanisms governing the classical endomembrane system (reviewed in Noda et al., 2002). PI3-kinase also functions in the membrane dynamics of autophagy (Kiel et al., 1999). Δvps34 and Δvps15 mutant cells exhibited a complete loss of autophagic activity as seen for other autophagy-defective mutants (atg mutants) (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Kihara et al., 2001). Two additional genes essential for autophagy, ATG14 and ATG6/VPS30, are subunits of a PI3-kinase complex (Kihara et al., 2001). In addition, autophagy is suppressed in mammalian cells treated with PI3-kinase inhibitors (Petiot et al., 2000), confirming the conserved role of PI3-kinase in autophagy from yeast to mammals.
Although only one functional lipid kinase is encoded by yeast, we have previously identified two distinct PI3-kinase complexes, complexes I and II, in this organisms (Kihara et al., 2001). Complex I functions in autophagy, whereas complex II functions in vacuolar protein sorting. Both of these complexes contain three common subunits, Vps15p, Vps34p, and Vps30p. Vps15p tethers the complex to the membrane; although this protein is myristylated at its N terminus, membrane association of the complex is not dependent solely on myristoylation (Herman et al., 1991b). Vps34p is recruited to the membrane and activated by Vps15p (Stack et al., 1995). The protein kinase activity of Vps15p is essential for complex formation; the lipid kinase activity of Vps34p is not. Whereas the function of Vps30p within these PI3-kinase complexes is not well understood, the homologous protein Beclin 1 has also been implicate in autophagy in both animals and plants (Liang et al., 1999; Liu et al., 2005). In addition to the common subunits, each complex also contains a unique factor. Atg14p is integrated into complex I, whereas Vps38p is specific for complex II. These additional subunits may act as connecter molecules, bridging Vps30p and Vps34p to allow complex formation (Kihara et al., 2001). These two complexes function in distinct biological processes; disruption of ATG14 does not affect vacuolar protein sorting, whereas deletion of VPS38 does not affect autophagy (Kametaka et al., 1998; Kihara et al., 2001). The mechanisms determining the specificity of these PI3-kinase complex functions have not been examined. Because both complexes exhibit PI3-kinase activity (Kihara et al., 2001), we assume that the primary functions of the two complexes are the same, namely, the production of PI3-P. Additional factors are likely necessary to direct each PI3-kinase complex to function in distinct biological processes, despite generating a common primary product. Atg14p and Vps38p may be these key factors, both conferring upon each complex a specificity of function and connecting Vps30p and Vps34p.
In this study, we investigated the function of Atg14p by deletion analysis. We also monitored the intracellular localization of the components of individual PI3-kinase complexes. Based on these analyses, we demonstrated the function of Atg14p in autophagy. These results led to our proposal of a model detailing the mechanisms by which PI3-kinase complexes I and II acquire distinct functions.
MATERIALS AND METHODS
Yeast Strains and Media
The S. cerevisiae strains used in this study were derived from SEY6210 (Robinson et al., 1988) or BJ2168 (Yeast Genetic Stock Center, University of California, Berkeley, CA) strains, as specified in Table 1. We used standard methods and media for yeast manipulation (Kaiser et al., 1994). Δatg14::kanMX mutants were constructed by amplifying the region containing the disruption marker and the flanking sequence by PCR from genomic DNA prepared from the BY4741 Δatg14::kanMX strain. The ATG14 locus was replaced with the amplified fragment. Δvps38::HIS3, Δvps38::TRP1, and Δvps38::LEU2 mutants were constructed by amplifying the regions containing the disruption marker and the flanking sequences by PCR using pRS313, pRS314, and pRS315 (Sikorski and Hieter, 1989) as templates, respectively. These amplified disruption cassettes replaced VPS38. Disruption of ATG11 was performed as described previously (Kim et al., 2001). Green fluorescent protein (GFP) was integrated into the genomic DNA as follows. A region containing GFP (S65T), the ADH1 termination sequence, and the kanMX6 marker was amplified by PCR from pFA6a-GFP(S65T)-kanMX6 (Longtine et al., 1998) using primer sets containing the flanking regions of the target genes and then inserted directly into the chromosomes. Vps15-GFP, Vps30-GFP, and Vps34-GFP fusion proteins were functional for the maturation of API and growth at 37°C. API-RFP::kanMX cells were constructed as follows. Sequence encoding red fluorescent protein (RFP) (monomeric RFP [mRFP]; Campbell et al., 2002) was amplified by PCR from the RFP sequence on pRSETB vector obtained from Carlsberg Research Center (Copenhagen, Denmark), to have PacI and AscI sites at 5′ and 3′ end, respectively. Amplified fragment was ligated into the PacI/AscI site of pFA6a-GFP (S65T)-kanMX6 to replace the GFP sequence. From this plasmid, the region containing RFP, ADH1 termination sequence, and kanMX6 marker was amplified by PCR with primer sets containing flanking regions of the target genes. Amplified cassettes were inserted directly into the chromosome. API-RFP::natMX cells were constructed by replacing the kanMX marker with a PCR-amplified natMX fragment. All primer sequences are available upon request.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SEY6210 | MATα leu2 ura3 his3 trp1 lys2 suc2 | Robinson et al. (1988) |
| KVY55 | SEY6210 pho8::PHO8Δ60 | Kirisako et al. (1999) |
| YOK9 | KVY55 Δatg14::kanMX | This study |
| YOK359 | KVY55 Δatg11::LEU2 | This study |
| YOK360 | KVY55 Δatg11::LEU2 Δatg14::kanMX | This study |
| YOK135 | SEY6210 Δvps38::HIS3 | This study |
| GYS115 | SEY6210 Δatg14::LEU2 | Suzuki et al. (2001) |
| BJ2168 | MATa leu2 trp1 ura3 pep4-3 prb1-1122 prc1-407 | Yeast Genetic Stock Center |
| STY103 | BJ2168 Δatg14::kanMX | This study |
| YOK219 | SEY6210 Δatg14::LEU2 Δvps38::HIS3 | This study |
| YOK358 | KVY55 Δatg14::kanMX Δvps38::HIS3 | This study |
| YOK435 | MATa/α Δatg14::LEU2/Δatg14::LEU2 Δvps38::HIS3/Δvps38::HIS3 API-mDsRed::natMX/API | This study |
| YOK89 | MATa/α Δvps38::LEU2/VPS38-YEGFP::natMX API-mDsRed::kanMX/API | This study |
| YOK152 | MATa/α VPS34/VPS34-YEGFP::kanMX API-mDsRed::natMX/API | This study |
| YOK153 | YOK152 Δatg14::LEU2/Δatg14::LEU2 | This study |
| YOK154 | MATa/α VPS34/VPS34-YEGFP::kanMX API-mDsRed::kanMX/API Δvps38::TRP1/Δvps38::HIS3 | This study |
| YOK173 | YOK152 Δatg14::LEU2/Δatg14::LEU2 Δvps38::HIS3/Δvps38::TRP1 | This study |
| YOK179 | MATa/α VPS30/VPS30-YEGFP::kanMX API-mDsRed::natMX/API | This study |
| YOK180 | YOK179 Δatg14::LEU2/Δatg14::HIS3 | This study |
| YOK181 | MATa/α VPS30/VPS30-YEGFP::kanMX API-mDsRed::kanMX/API Δvps38::TRP1/Δvps38::HIS3 | This study |
| YOK172 | MATα VPS30-YEGFP::kanMX Δatg14::HIS3 Δvps38::TRP1 | This study |
| YOK190 | MATa/α VPS15-YEGFP::kanMX/VPS15-YEGFP::kanMX API-mDsRed::natMX/API | This study |
| YOK191 | YOK190 Δatg14::LEU2/Δatg14::LEU2 | This study |
| YOK192 | YOK190 Δvps38::TRP1/Δvps38::HIS3 | This study |
| YOK193 | YOK190 Δatg14::LEU2/Δatg14::LEU2 Δvps38::HIS3/Δvps38::TRP1 | This study |
Autophagy was induced by transferring the cells into nitrogen-depleted medium SD (–N), or S (–NC) medium in which both nitrogen and carbon have been depleted.
Plasmid Construction
The promoter region of ATG14, a hemagglutinin (HA)-GFP sequence, and the 3′ untranslated region of ATG14 were cloned in tandem into pRS316 to generate the pOK4 vector. Linker sequences (two repeats of the sequence encoding GlyGlyGlySer) were added before the coding sequences for HA and GFP. Sequences encoding the Atg14p deletion series were ligated in frame into the site between the promoter sequence and HA-GFP in pOK4. To construct multicopy plasmids overexpressing the variants, the KpnI/SacI fragment derived from these plasmids was ligated into the KpnI/SacI site of pRS426. VPS38-HA-GFP constructs were generated in the same manner. Successful plasmid construction was confirmed by sequencing. All primer sequences and plasmid maps are available upon request.
Estimation of Autophagic Activity
To quantify the extent of autophagy, we performed alkaline phosphatase (ALP) assays as described previously (Noda and Ohsumi, 1998). Cell viability in SD (–N) medium was measured by counting the number of dead cells stained with phloxine B (final concentration 2 μg/ml) that exhibited bright fluorescence (Onodera and Ohsumi, 2004). Maturation of aminopeptidase I (API) was estimated by immunoblotting with an anti-API antibody. The accumulation of autophagic bodies was examined by phase-contrast microscopy (model IX-71; Olympus, Tokyo, Japan). Images were acquired with MetaMorph software (Universal Imaging, Downingtown, PA).
Coimmunoprecipitation
Spheroplasts were lysed by osmotic shock and then solubilized for 30 min at 4°C in immunoprecipitation (IP) buffer (50 mM HEPES-NaOH, pH 8.0, 200 mM sorbitol, 150 mM NaCl, 10 mM 2-mercaptoethanol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF]), 40 μg/ml aprotinin, 10 μg/ml pepstatin A, 20 μg/ml leupeptin, 40 μg/ml benzamidin, and protease inhibitor cocktail (Complete, EDTA-free; Roche Diagnostics, Indianapolis, IN). After the removal of cell debris by centrifugation at 1500 × g for 5 min, samples were centrifuged at 100,000 × g for 1 h. Supernatants were incubated with anti-GFP (rabbit polyclonal antibody; Molecular Probes, Eugene, OR) or anti-Atg6 (Kihara et al., 2001) antibodies at 4°C for 2 h. After the addition of protein G-Sepharose beads, samples were incubated for an additional hour at 4°C. Beads were washed four times with IP buffer. Bound proteins were eluted with SDS sample buffer and separated by SDS-PAGE.
Immunoblotting
Immunoblotting was performed using anti-HA (HA-7; Sigma-Aldrich, St. Louis, MO), anti-API, anti-Vps15p, affinity-purified anti-Vps30p, and affinity-purified anti-Vps34p (Kihara et al., 2001) antibodies. Immunodetection used an ECL system (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) with a bioimaging analyzer (LAS1000; Fujifilm, Tokyo, Japan). To detect Atg14p variants and Vps15p, immunoreaction enhancer (Can Get Signal; Toyobo Engineering, Osaka, Japan) was used according to the manufacturer's instructions.
Subcellular Fractionation
Yeast spheroplasts were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 200 mM sorbitol, 1 mM PMSF, 40 μg/ml aprotinin, 10 μg/ml pepstatin A, 20 μg/ml leupeptin, 40 μg/ml benzamidin, and protease inhibitor cocktail) by extrusion through a polycarbonate 3-μm filter (Vida and Gerhardt, 1999). To remove cell debris, the filtrate was centrifuged at 500 × g for 5 min. The supernatant (total) was subsequently centrifuged at 13,000 × g for 15 min to generate a low-speed pellet (LSP) and low-speed supernatant. The supernatant was further centrifuged at 100,000 × g for 1 h to generate a high-speed pellet (HSP) and high-speed supernatant (HSS).
Microscopy
The intracellular localization of GFP-tagged proteins and RFP-tagged API was observed using inverted fluorescence microscopes (IX-71 and IX-81; Olympus) equipped with cooled charge-coupled device cameras (CoolSNAP HQ; Nippon Roper, Tokyo, Japan). Images were acquired using MetaMorph software (Universal Imaging) and processed using Adobe PhotoShop software (Adobe Systems, Mountain View, CA).
RESULTS
Coiled-Coils at the N-Terminal Half of Atg14p Are Essential for Autophagy
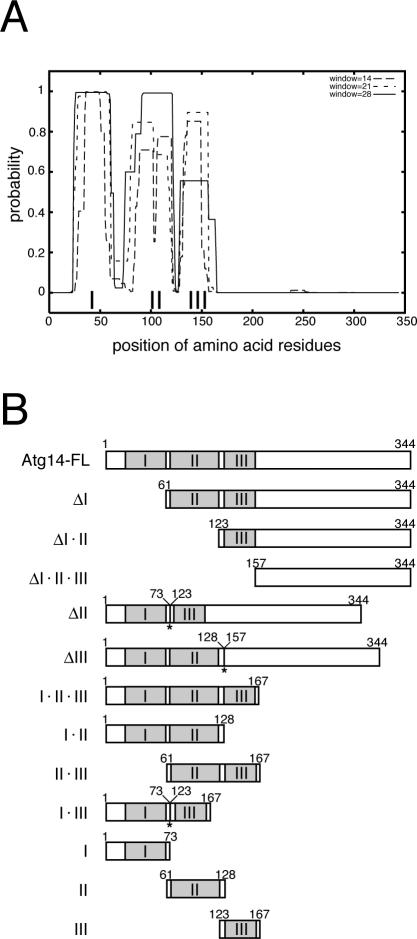
Structurally, Atg14p was predicted to possess three coiled-coil regions within the N-terminal half of the protein (Figure 1A). No prominent motifs could be identified within the C-terminal half of Atg14p. To determine which regions of Atg14p were critical, we performed deletion analysis of Atg14p based on these predictions. A deletion series for Atg14p lacking one to three of the coiled-coil regions or the C-terminal half were constructed and designated as follows: for example, full-length of Atg14p, Atg14p lacking coiled-coil I, and Atg14p containing the three coiled-coil regions but not the C-terminal half were termed Atg14-FL, Atg14-ΔI, and Atg14-I·II·III, respectively (Figure 1B). The C terminus of each variant was fused to an HA-GFP tag by a linker sequence (two repeats of GlyGlyGlySer), present before both the HA and GFP coding sequences. We confirmed that HA–GFP-tagged Atg14p was functional for autophagy (our unpublished data).
Figure 1.
Atg14p deletion series. (A) The presence of three coiled-coil regions (I, II, and III) were predicted within Atg14p (COILS; http://www.ch.embnet.org/software/COILS_form.html). (B) The deleted variants of Atg14p are shown schematically. HA-GFP, with a linker sequence (two repeats of GlyGlyGlySer) before both the HA and GFP sequences was fused to the C terminus of each variant. The coiled-coils are shown in gray. Two amino acid residues (SerSer, corresponding to the ligation site, XbaI/SpeI) were inserted at the position denoted by the asterisks.
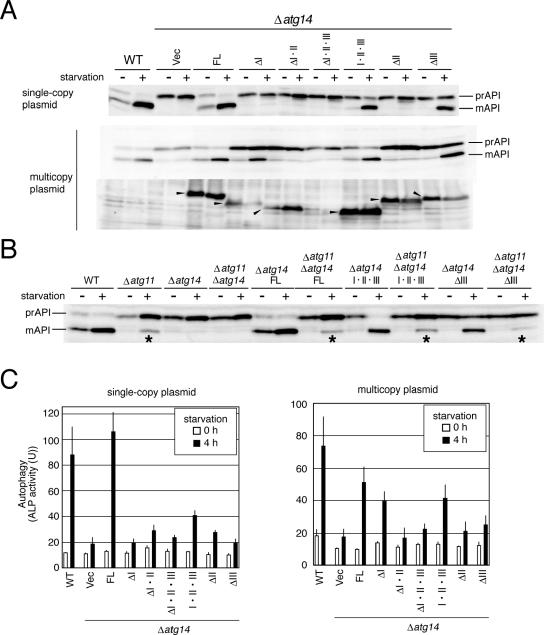
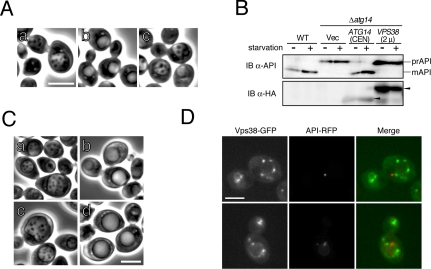
These variants were expressed in Δatg14 cells from single- or multicopy plasmids under the control of the ATG14 promoter. Protein functionality was assayed first by monitoring the maturation of proform of API, which is transported into vacuoles via the cytoplasm-to-vacuole targeting (Cvt) pathway or by autophagy under nutrition-rich and starvation conditions, respectively. The protein is then processed into a mature form within vacuoles. API maturation was enhanced, especially under starvation conditions, in cells expressing Atg14-FL, Atg14-I·II·III, or Atg14-ΔIII from single-copy plasmids (Figure 2A). API maturation was also restored in cells overexpressing Atg14-ΔI from a multicopy plasmid. None of the other variants were functional, even when expressed from multicopy plasmids. The amounts of the variants except Atg14-ΔI·II·III were at least at several times higher levels than that of endogenous Atg14p when expressed from multicopy plasmids, although only Atg14-FL and Atg14-I·II·III were detectable when expressed from single-copy plasmids (our unpublished data). We next assayed autophagy-dependent API-maturation in Δatg11 cells, which lacks one of the factors required for the Cvt pathway, but not for autophagy (Kim et al., 2001) (Figure 2B). Levels of autophagy-dependent API maturation was similar to that in Δatg11 cells, expressing endogenous Atg14p, in Δatg11Δatg14 cells expressing exogenous Atg14-FL, Atg14-I·II·III, or Atg14-ΔIII.
Figure 2.
Autophagic activity of cells expressing deleted variants of Atg14p. Cells grown in SD + CA + AdeTrp medium at 30°C were collected during logarithmic phase growth (starvation–) and at 4 h after transfer to SD (–N) medium (starvation+). Lysates were prepared by the alkaline-trichloroacetic acid (TCA) method (A and B) or using glass beads (C). (A) Maturation of API was monitored by immunoblotting with an anti-API antibody. The amount of each Atg14p variant (arrowheads) was estimated by immunoblotting with an anti-HA antibody (HA-7). (B) Autophagy-dependent API maturation was estimated in cells lacking ATG11. Each variant was expressed from a single-copy plasmid. The asterisks indicate autophagy-dependent API maturation. (C) Autophagic activity was estimated using an ALP assay. The means ± SD of results from triplicate measurements are shown.
Next, autophagic activity was estimated by measuring the activity of ALP, which was expressed in the cytosol as a truncated proform and delivered to and activated in vacuoles in an autophagy-dependent manner (Noda et al., 1995). After both single-copy and multicopy expression, autophagic activity in Atg14-I·II·III cells under starvation conditions was significantly higher than that in Δatg14 cells but lower than that in wild-type (WT) cells (Figure 2C). Results of ALP assays represent the activities of bulk and nonselective degradation of the cytoplasm via autophagy (Noda et al., 1995). It is a less sensitive system to detect partial autophagic activities than API maturation assay, which represents a selective transport activity of a large complex of API via both Cvt pathway and autophagy. The lower ALP activity in Atg14-I·II·III cells than in WT, in spite of the normal API maturation activity, may result from the difference in the sensitivity of the assays used. These results indicate that the N-terminal half of Atg14p containing the three coiled-coil structures is functional for autophagy, although it is not fully active. In contrast, the C-terminal half of Atg14p is not essential for autophagy.
Cells expressing Atg14-FL from a single-copy plasmid possessed higher autophagic activity under starvation conditions than WT cells. This is likely because of the higher expression of the protein from a single-copy plasmid than endogenous expression (our unpublished data), which suggests that the amount of Atg14p may be an important factor limiting the extent of autophagy. Overexpression of Atg14-FL from a multicopy plasmid, however, yielded lower autophagic activity than that seen in WT cells, suggesting that excess amount of Atg14p affects autophagy to some extent and that the proper amount of Atg14p is required for normal autophagy.
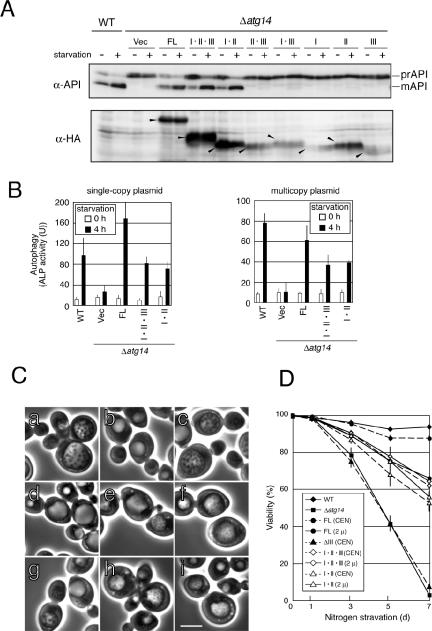
Because the three coiled-coil regions within the N-terminal half of Atg14p are essential for autophagy, whereas the C-terminal half is dispensable, we further dissected the N-terminal region containing the three coiled-coils. We examined cells expressing further deleted variants for autophagic activity. Atg14-I·II-expressing cells exhibited similar autophagic activity as those expressing Atg14-I·II·III (Figure 3, A and B), whereas none of the other further deleted variants exhibited any function to induce API maturation. Therefore, Atg14-I·II is the minimal region essential for autophagy.
Figure 3.
Minimal region essential for Atg14p function in autophagy. Cells grown in SD + CA + AdeTrp medium at 30°C were collected during logarithmic growth (starvation–) and 4 h after transfer to SD (–N) (starvation+) (A and B). Lysates were prepared by the alkaline-TCA method (A) or using glass beads (B). (A) Maturation of API was monitored by immunoblotting with an anti-API antibody. The amount of each Atg14p variant (arrowheads) expressed from a multicopy plasmid was estimated by immunoblotting with an anti-HA antibody (HA-7). (B) The extent of autophagy was estimated by ALP assay. The means ± SD of results from triplicate measurements are shown. (C) The accumulation of autophagic bodies was examined on the BJ2168 background, which lacks vacuolar proteases. Cells in logarithmic phase growth were transferred to S (–NC) medium. After 4 h of culture in S (–NC) medium, cells were observed by phase-contrast microscopy. WT cells bearing an empty vector (a) and Δatg14 cells bearing an empty vector (b) or expressing Atg14-FL from a single-copy plasmid (c), Atg14-ΔIII from a single-copy plasmid (d), Atg14-I·II·III from a single-copy plasmid (e), Atg14-I·II from a single-copy plasmid (f), Atg14-FL from a multicopy plasmid (g), Atg14-I·II·III from a multicopy plasmid (h), or Atg14-I·II from a multicopy plasmid (i) are shown. Bar, 5 μm. (D) Cell viability under nitrogen-starvation conditions was measured. Cells were grown to logarithmic phase in SD + CA + AdeTrp and then transferred to SD (–N) medium. Cell viability was determined by phloxine B staining, as described in Materials and Methods.
Cells expressing Atg14-I·II, Atg14-FL, or Atg14-I·II·III accumulated autophagic bodies under starvation conditions (Figure 3C). The autophagic bodies accumulated in cells expressing either Atg14-I·II·III or Atg14-I·II, however, were smaller than those seen in WT and Atg14-FL cells. This change in autophagosome size may be responsible for the reduction in autophagic activity monitored by ALP assay. Activity of PI3-kinase complex I might influence the size of the autophagosomes. Cells overexpressing Atg14-FL from a multicopy plasmid accumulated normal size autophagic bodies, although the population of cells accumulating autophagic bodies was reduced from that of WT cells (our unpublished data), which may explain the reduced autophagic activity measured.
Cells expressing Atg14-I·II·III and Atg14-I·II maintained significantly higher viabilities in nitrogen-starved medium than autophagy-defective cells such as Δatg14 and Atg14-ΔIII cells, although these levels could not reach those of WT and Atg14-FL (single-copy plasmid) cells. These results correlate well with the fact that these cells exhibited partial autophagic activity, monitored by ALP assay, and accumulated smaller autophagic bodies. Together, the N-terminal region containing coiled-coils I and II is essential and sufficient for autophagy.
Atg14p Binds to Vps30p and Vps34p at the Coiled-Coil I and II
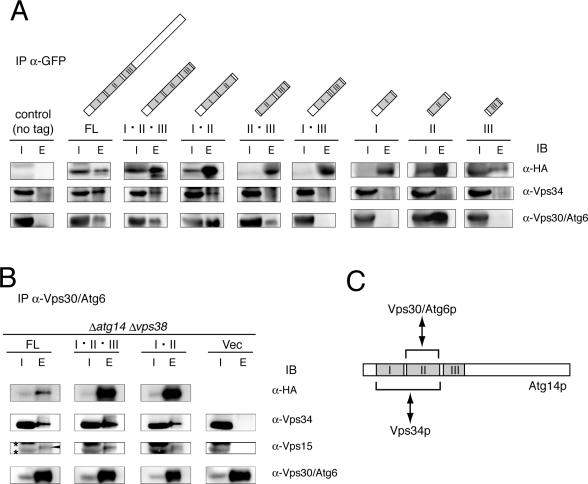
Because coiled-coil regions often mediate protein–protein interaction, we investigated the associations of Atg14p variants with other components of PI3-kinase complex I by coimmunoprecipitation. HA–GFP-tagged Atg14p variants were immunoprecipitated with an anti-GFP antibody; coimmunoprecipitates were examined by immunoblotting. In cells expressing Atg14-FL, Vps34p and Vps30p were coimmunoprecipitated (Figure 4A). Atg14-I·II·III and Atg14-I·II also interacted with Vps34p and Vps30p. The region containing only coiled-coil II was sufficient to interact with Vps30p.
Figure 4.
Interaction of Atg14p with Vps30p and Vps34p is mediated by coiled-coils I and II. Proteins were coimmunoprecipitated from the supernatants of Triton X-100-treated lysates using anti-GFP (A) and anti-Vps30 (B) antibodies. Isolated proteins (E, elution fraction) were resuspended in 1/10 (A) or 1/40 (B) volume of the starting supernatant fraction (I, input); equal (A) or 1.5× volume (B) was loaded onto SDS-PAGE. (A) Atg14p interacts with Vps30p and Vps34p at coiled-coil I and II regions. Atg14-I, Atg14-II, and Atg14-III were expressed from multicopy plasmids, and the other variants were expressed form single-copy plasmids. (B) Vps34p and Vps15p coimmunoprecipitated with Vps30p in a manner dependent on the variants of Atg14p expressed from single-copy plasmids in Δvps38Δatg14 cells. Arrow and asterisks indicate Vps15p and nonspecific bands, respectively. (C) In this schematic representation of Atg14p-mediated complex formation, the regions required for this interaction are indicated.
To determine whether Atg14p, Vps34p, and Vps30p were isolated as part of a single complex or as the combination of Atg14p-Vps34p and Atg14p-Vps30p complexes, we further attempted immunoprecipitation with an anti-Vps30 antibody. We used Δvps38 cells to exclude any potential interactions between Vps34p and Vps30p that were mediated by Vps38p. Vps34p coimmunoprecipitated with Vps30p in the presence of Atg14-FL, Atg14-I·II·III, or Atg14-I·II (Figure 4B). This interaction was completely dependent on the presence of an Atg14p variant, indicating that Vps30p, Atg14p, and Vps34p are integrated into a single complex within these cells. Vps15p was also coimmunoprecipitated with Vps30p in the presence of Atg14-FL, Atg14-I·II·III, or Atg14-I·II, indicating that all subunits of complex I form a complex in these cells.
Atg14p Localizes to the PAS and the Vacuolar Membrane
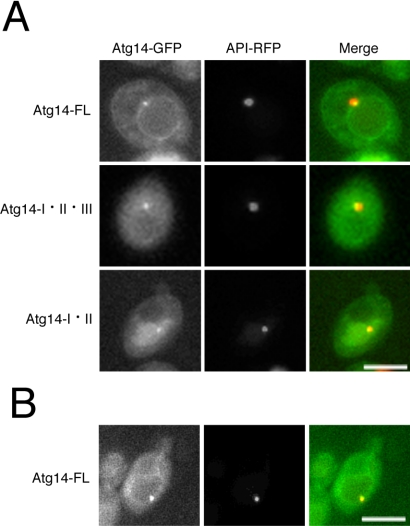
Atg14p is known to localize to the PAS (Strømhaug et al., 2004), which we confirmed by examining the colocalization of Atg14p with RFP-tagged API, a PAS marker. We also detected Atg14-FL, expressed from single-copy plasmid, on the vacuolar membrane (Figure 5A). Similarly, integrated Atg14-GFP localized to the PAS and vacuolar membrane (our unpublished data). Because Atg14p is unstable, making it easily degraded to undetectable levels in the absence of interactions with Vps30p and Vps34p (Kihara et al., 2001), Atg14p at these compartments is integrated into PI3-kinase complex I, as confirmed by immunoprecipitation experiments (Figure 4). Both Atg14-I·II·III and Atg14-I·II retained the ability to localize to the PAS, although localization to the vacuolar membrane was significantly reduced to nearly undetectable levels. It is unclear whether the reduced vacuolar fluorescence is the cause of the reduced autophagic activity. The localization pattern of Atg14-FL was not affected by the disruption of VPS38 (Figure 5B). The same was true in the case of Atg14-I·II·III and Atg14-I·II (our unpublished data).
Figure 5.
PAS-localization of Atg14p variants. (A) Atg14-FL, Atg14-I·II·III, and Atg14-I·II expressed from single-copy plasmids colocalized with API, a PAS marker. Vacuolar fluorescence was significantly reduced in cells expressing either Atg14-I·II·III or Atg14-I·II. (B) Atg14-FL expressed from a single copy-plasmid in Δatg14Δvps38 cells (YOK435). Bars, 5 μm.
Atg14-I·II, which interacts with both Vps30p and Vps34p, localized to the PAS and was able to function in autophagy (Table 2), confirming that this is the minimum essential for the function in autophagy. These results indicate that the critical functions of Atg14p are mediated by protein–protein interactions of coiled-coils I and II.
Table 2.
Summary of Atg14p deletion analysis
| Variant | Autophagy | PAS localization | Binding to Vps30p | Binding to Vps34p |
|---|---|---|---|---|
| Atg14-FL | + | + | + | + |
| Atg14-I·II·III | + | + | + | + |
| Atg14-I·II | + | + | + | + |
| Atg14-II·III | - | N.D. | + | - |
| Atg14-I·III | - | N.D. | - | - |
N.D., not determined.
Complexes I and II Localize to Distinct Compartments Dependent on Their Specific Components
Previous studies demonstrated that disruption of ATG14 did not affect vacuolar protein sorting and that disruption of VPS38 did not affect autophagic activity. These results indicate that complexes I and II function in distinct biological processes (Kametaka et al., 1998; Kihara et al., 2001). We have confirmed this concept by additional criteria. Autophagic bodies were accumulated normally in Δvps38 cells (Figure 6). Overexpression of Vps38-HA-GFP did not restore API maturation and the accumulation of autophagic bodies in Δatg14 cells, although it could reverse the defect in vacuolar protein sorting in Δvps38 cells. In conjunction with previous studies, these results clearly indicate that the functions of complexes I and II do not overlap.
Figure 6.
Vps38p and autophagy. (A) SEY6210 strain cells at logarithmic phase growth were transferred to S (–NC) medium in the presence of 1 mM PMSF. After 4 h of culture in S (–NC) medium, WT cells (a), Δatg14 cells (b), and Δvps38 cells (c) were observed by phase-contrast microscopy. (B) API maturation was monitored in Δatg14 cells overexpressing Vps38-HA-GFP. Cells, grown in SD + CA + AdeTrp medium at 30°C, were collected during logarithmic growth (starvation –) or 4 h after transfer to SD (–N) medium (starvation +). Lysates were subjected to immunoblotting with anti-API and anti-HA (HA-7) antibodies. (C) The accumulation of autophagic bodies was examined in Vps38-HA-GFP-overexpressing Δatg14 cells, on the BJ2168 background which lacks vacuolar proteases. WT cells (a), Δatg14 cells (b), Δatg14 cells expressing Atg14-FL from a single-copy plasmid (c), and Δatg14 cells expressing Vps38-HA-GFP from a multicopy plasmid (d) were transferred to S (–NC) medium. After 4 h of culture in S (–NC) medium, cells were observed by phase-contrast microscopy. (D) We investigated the localization of Vps38p. Diploid cells expressing chromosomal Vps38-GFP and API-RFP (YOK89) were cultured in SD + CA + AdeTrpUra medium and subjected to fluorescence microscopy. Two representative fields are shown. Bars, 5 μm.
We examined the specificity of PI3-kinase complex function by investigating the intracellular localization of each complex. As described above, Atg14p localized to the PAS and the vacuolar membrane (Figure 5). We next investigated the localization of Vps38p, the factor specific for complex II. Vps38p localized to the vacuolar membrane and a subset of punctate structures unique from the PAS (Figure 6). These punctate structures are endosomes, because integrated Vps38p is known to colocalize with an endosome marker, Snf7p (Huh et al., 2003). We have confirmed the colocalization of Vps38p with Snf7p (our unpublished data). To avoid confusion, fluorescent punctate structures will hereafter be described as “endosomes” when they are Vps38p positive or Vps38p dependent. In addition to the endosomes, Vps34p and Vps15p localized to additional punctate structures (see below) that are described as “dot structures” for convenience. Because Vps38p is essential for the formation of complex II (Kihara et al., 2001), the absence of detectable Vps38p at the PAS indicates that complex II does not accumulate at the PAS.
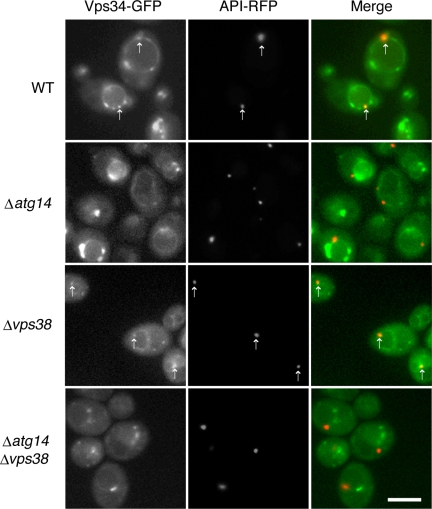
We also examined the localization of Vps34p, Vps30p, and Vps15p in WT, Δatg14, Δvps38, and Δatg14Δvps38 cells. Vps34p localized to the PAS, the vacuolar membrane, endosomes, and dot structures (Figure 7). The PAS localization of Vps34p was abolished in Δatg14 cells, indicating that targeting of this molecule is dependent on Atg14p. In contrast, Vps34p staining of endosomes, dot structures, and vacuolar membranes was retained in Δatg14 cells. In Δvps38 cells, a significant amount of Vps34p was dispersed throughout the cytoplasm. Vps34p was still detected at the PAS, the vacuolar membrane, and dot structures, indicating that localization of Vps34p to these structures does not require Vps38p. Vsp34p localization to dot structures and vacuolar membranes could be detected even in Δatg14Δvps38 cells, although the fluorescence from the vacuolar membranes was significantly reduced.
Figure 7.
Localization of Vps34p. Diploid cells expressing chromosomal Vps34-GFP and API-RFP were observed by fluorescence microscopy. The arrows indicate the structures to which Vps34-GFP and API-RFP colocalize. Bar, 5 μm.
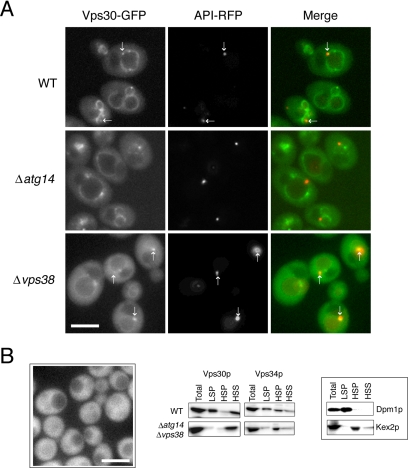
Vps30p was localized to the PAS, vacuolar membranes, and endosomes (Figure 8). As seen for Vps34p, Atg14p was required for the PAS localization of Vps30p but dispensable for the localization of Vps30p to endosomes. The PAS localization of Vps30p did not require Vps38p, because Vps30p remains localized to the PAS in Δvps38 cells. In contrast, Vps30p disappeared from endosomes, leaving the majority of Vps30p dispersed throughout the cytoplasm in Δvps38 cells, demonstrating an essential role for Vps38p in the endosomal localization of Vps30p. In Δatg14Δvps38 cells, Vps30p was distributed evenly throughout the cytoplasm with no apparent compartment. This result was consistent with the cell fractionation experiments, demonstrating that the majority of Vps30p was found in the postlysis supernatants of Δatg14Δvps38 cells (Figure 8B).
Figure 8.
Localization of Vps30p. (A) Diploid cells expressing chromosomal Vps30-GFP and API-RFP were observed by fluorescence microscopy. The arrows indicate the structures to which Vps30-GFP and API-RFP colocalize. (B) Haploid cells expressing chromosomal Vps30-GFP (left). Vps30-GFP was dispersed throughout the cytoplasm, which was confirmed by cell fractionation analysis of WT and Δatg14Δvps38 cells expressing an untagged Vps30p (right). Dpm1p and Kex2p were used as markers for the ER and late Golgi, respectively. Bars, 5 μm.
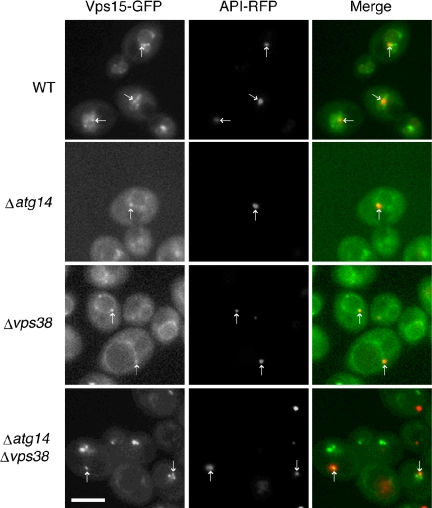
Similar to Vps34p, Vps15p localized to the PAS, the vacuolar membrane, endosomes, and dot structures (Figure 9). In contrast to the results seen for Vps34p and Vps30p, however, the PAS localization of Vps15p was Atg14p independent. In addition, Vps38p was not required for the localization of Vps15p to the PAS. In Δatg14Δvps38 cells, Vps15p remained localized to dot structures (see Discussion), vacuolar membranes, and the PAS, although the fluorescence from the vacuolar membranes was significantly reduced.
Figure 9.
Localization of Vps15p. Within the fluorescence images of diploid cells expressing chromosomal Vps15-GFP and API-RFP, the arrows indicate the structures at which Vps15-GFP and API-RFP colocalize. Bar, 5 μm.
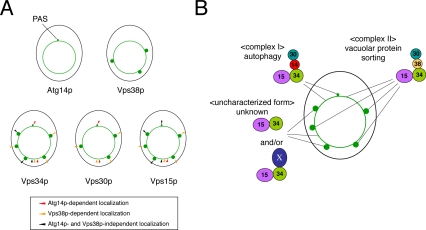
A summary of the microscopy is detailed in Figure 10. The PAS and vacuolar membranes were sites to which all components of complex I, including Atg14p, localized. PAS localization of Vps34p and Vps30p was Atg14p dependent but Vps38p independent. In contrast, endosomes and vacuolar membranes were the sites to which the components of complex II localized. Localization of these components to endosomes required Vps38p but not Atg14p. In addition to a shared distribution to vacuolar membranes, both complexes I and II had an additional specific site of intracellular localization, complex I to the PAS and complex II to endosomes. This localization was specified by the components exclusive to each complex. Vps15p and Vps34p also were observed on dot structures; this localization required neither Atg14p nor Vps38p.
Figure 10.
Distinct distribution of the two PI3-kinase complexes. (A) The intracellular localization of each component of the PI3-kinase complexes is shown in green, whereas the outline of the cell is detailed in black. (B) The distribution of the PI3-kinase complexes is summarized. “X” represents an unknown component(s).
DISCUSSION
In this study, we focused on Atg14p, a specific component of PI3-kinase complex I that is essential for autophagy. Analyses using a deletion series of Atg14p indicated that the region containing coiled-coils I and II within the N-terminal half were essential and sufficient for the function in autophagy. This region was minimum essential for the localization of Atg14p to the PAS. The two coiled-coil structures in this region were responsible for interactions with Vps34p and Vps30p to form PI3-kinase complex I. These results indicate that the primary function of Atg14p is mediated by protein–protein interactions of the coiled-coils.
PI3-kinase complex I is essential for autophagy but dispensable for vacuolar protein sorting. Conversely, complex II functions in the sorting of vacuolar proteins but is dispensable for autophagy. How this specificity of the function is determined has not been elucidated. In this study, we investigated the intracellular localization of the two complexes by microscopy, which provided several important discoveries. All the components of complex I were present at the PAS. The localization of Vps34p and Vps30p to the PAS was dependent on the expression of Atg14p, indicating an essential role for Atg14p in the accumulation of PI3-kinase complex I at the PAS. Atg14p, however, was not essential for the localization of Vps34p and Vps30p to vacuolar membranes and endosomes. Vps38p, a specific component of complex II, did not localize to the PAS, but localized to endosomes and vacuolar membranes. Vps38p was not required for the PAS localization of Vps34p, Vps30p, and Vps15p but was essential for their localization to endosomes. Therefore, Vps38p plays a crucial role in the association of complex II with endosomes. From these results, we concluded that only complex I can accumulate at the PAS, whereas only complex II is found at endosomes. These unique localization patterns are mediated by the specific components of each complex, Atg14p and Vps38p. Here, we propose a model that the distinct functions of PI3-kinase complexes I and II are acquired by the specific association of each complex to a distinct compartment that is mediated by the specific components.
Unlike Vps34p and Vps30p, Vps15p localized to the PAS in the absence of Atg14p (Figure 9). The mechanism by which Vps15p localizes to the PAS is currently unknown. This Atg14p-independent localization of Vps15p suggests a potential mechanism by which Atg14p can promote the accumulation of complex I at the PAS. Atg14p may recruit Vps34p and Vps30p to the PAS, forming a complex with preexisting Vps15p on the membrane. In this way, Atg14p would not have to bring a preformed complex containing Vps15p to the PAS. Alternatively, Atg14p could prevent the dissociation of complex I, stabilizing it at the PAS. In any case, the interaction between certain PAS-localized proteins and the complex I subunits may be critical.
All of the components of PI3-kinase complexes I and II localized to the vacuolar membrane. At present, it is not clear whether the PI3-kinase complexes on the vacuolar membrane are functional. Vesicles undergoing anterograde transport finally fuse with vacuoles. Similarly, autophagosomes finally fuse with vacuoles (Baba et al., 1994; Kirisako et al., 1999). Therefore, it remains possible that the presence of complexes I and II at vacuolar membranes simply represents the consequence of membrane-trafficking events. Fab1p, a phosphoinositide kinase that phospholylates PI3-P to generate phosphoinositide-3,5-diphosphate, is involved in the regulation of vacuolar morphology (Yamamoto et al., 1995; Bonangelino et al., 1997; Gary et al., 1998; Dove et al., 2002, 2004). Alternatively, PI3-kinase complexes at vacuolar membranes may function in the maintenance of vacuolar homeostasis in combination with Fab1p and its possible effectors. It will be interesting to examine the vacuolar membrane localization of PI3-kinase complex subunits in various mutants defective in anterograde or retrograde transport, retrieval of the vacuolar membrane components, and autophagy (Tsukada and Ohsumi, 1993; Yamamoto et al., 1995; Bryant et al., 1998; Gary et al., 1998; Dove et al., 2002, 2004).
Vps34p and Vps15p remain localized to dot structures and vacuolar membranes in Δatg14Δvps38 cells (Figures 7 and 9). These GFP-positive signals may represent Vps34p–Vps15p complex and/or a novel PI3-kinase complex containing Vps34p, Vps15p, and potentially novel undefined components. The existence of PI3-kinase complex(es) unique from both complexes I and II has been suggested (Kihara et al., 2001). Given that ∼80% of WT PI3-kinase activity is retained in Δvps30 cells and that Δvps34 and Δvps15 cells have additional phenotypes beyond that of Δvps30 cells (Robinson et al., 1988; Raymond et al., 1992; Kihara et al., 2001), the putative novel complex likely does not contain Vps30p. This result would be in good agreement with our microscopy and cell fractionation data demonstrating that Vps30p was dispersed throughout the cytosol and did not localize to a specific compartment in Δatg14Δvps38 cells (Figure 8).
To date, genes with significant homology with ATG14 and VPS38 have not been found in animals and plants. Given the conserved roles of class III PI3-kinase in autophagy and protein sorting to the vacuole/lysosome from yeast to mammals (Petiot et al., 2000; Brown et al., 1995; Davidson, 1995), it would be interesting to search the functional homologues of Atg14p and Vps38p that potentially function to connect Vps34p and Vps30p in mammals.
We demonstrated that PI3-kinase complex I accumulates at the PAS in a manner dependent on Atg14p. What is the function of PAS-localized complex I in autophagy? In mammals, wortmannin and LY294002, two inhibitors of PI3-kinase, inhibit autophagy (Petiot et al., 2000), suggesting that the lipid kinase activity of a Vps34p homologue is required for this process. We have confirmed that the lipid kinase activity of Vps34p is also essential for autophagy in yeast (our unpublished data). In a variety of biological processes, PI3-P marks specific membrane sites to which downstream molecules need to be recruited (Martin, 1998; Odorizzi et al., 2000). One possible role of PI3-kinase complex I in autophagy may be to produce PI3-P at the PAS to recruit PI3-P-binding molecules, which in turn recruit additional downstream molecules to the PAS. Atg14p is required for the localization of Atg2p, Atg5p, and Atg8p to the PAS (Shintani et al., 2001; Suzuki et al., 2001). Therefore, complex I may act primarily at a relatively early point in the autophagosome formation, potentially during formation and/or maintenance of the PAS through the production of PI3-P.
Acknowledgments
We thank Dr. Roger Y. Tsien for providing the plasmid of mRFP. We thank R. Ichikawa for technical support. We also thank the members of Ohsumi's laboratory for providing materials and constructive discussion. This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture of Japan.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-09-0841) on January 18, 2006.
Abbreviations used: ALP, alkaline phosphatase; API, aminopeptidase I; CPY, carboxypeptidase; PAS, preautophagosomal structure; PI3-kinase, phosphatidylinositol 3-kinase; PI3-P, phosphoinositide 3-phosphate; WT, wild-type.
References
- Baba, M., Takeshige, K., Baba, N., and Ohsumi, Y. (1994). Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino, C. J., Catlett, N. L., and Weisman, L. S. (1997). Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell. Biol. 17, 6847–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W. J., DeWald, D. B., Emr, S. D., Plutner, H., and Balch, W. E. (1995). Role for phosphatidylinositol 3-kinase in the sorting and transport of newly synthesized lysosomal enzymes in mammalian cells. J. Cell Biol. 130, 781–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, N. J., Piper, R. C., Weisman, L. S., and Stevens, T. H. (1998). Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J. Cell Biol. 142, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, H. W. (1995). Wortmannin causes mistargeting of procathepsin D. evidence for the involvement of a phosphatidylinositol 3-kinase in vesicular transport to lysosomes. J. Cell Biol. 130, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove, S. K., McEwen, R. K., Mayes, A., Hughes, D. C., Beggs, J. D., and Michell, R. H. (2002). Vac14 controls PtdIns(3,5)P(2) synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 12, 885–893. [DOI] [PubMed] [Google Scholar]
- Dove, S. K., et al. (2004). Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 23, 1922–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary, J. D., Wurmser, A. E., Bonangelino, C. J., Weisman, L. S., and Emr, S. D. (1998). Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, P. K., and Emr, S. D. (1990). Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 6742–6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, P. K., Stack, J. H., and Emr, S. D. (1991b). A genetic and structural analysis of the yeast Vps15 protein kinase: evidence for a direct role of Vps15p in vacuolar protein delivery. EMBO J. 10, 4049–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, P. K., Stack, J. H., DeModena, J. A., and Emr, S. D. (1991a). A novel protein kinase homologue essential for protein sorting to the yeast lysosome-like vacuole. Cell 64, 425–437. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Kaiser, C., Michaelis, S., and Mitchell, A. (1994). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Kametaka, S., Okano, T., Ohsumi, M., and Ohsumi, Y. (1998). Apg14p and Apg6p/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 273, 22284–22291. [DOI] [PubMed] [Google Scholar]
- Kiel, J.A.K.W., Rechinger, K. B., van der Klei, I. J., Salomons, F. A., Titorenko, V. I., and Veenhuis, M. (1999). The Hansenula polymorpha PDD1 gene product, essential for the selective degradation of peroxisomes, is a homologue of Saccharomyces cerevisiae Vps34p. Yeast 15, 741–754. [DOI] [PubMed] [Google Scholar]
- Kihara, A., Noda, T., Ishihara, N., and Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kamada, Y., Stromhaug, P. E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S. V., Ohsumi, Y., Dunn, W. A., Jr., and Klionsky, D. J. (2001). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., Noda, T., and Ohsumi, Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., and Ohsumi, Y. (1999). Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 1–32. [DOI] [PubMed] [Google Scholar]
- Liang, X. H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., and Levine, B. (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Czymmek, K., Talloczy, Z., Levine, B., and Dinesh-Kumar, S. P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Martin, T.F.J. (1998). Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 14, 231–264. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Yamamoto, A., Hatano, M., Kobayashi, Y., Kabeya, Y., Suzuki, K., Tokuhisa, T., Ohsumi, Y., and Yoshimori, T. (2001). Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn, A. L., and Riezman, H. (1994). Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J. Cell Biol. 127, 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Matsuura, A., Wada, Y., and Ohsumi, Y. (1995). Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126–132. [DOI] [PubMed] [Google Scholar]
- Noda, T., and Ohsumi, Y. (1998). Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966. [DOI] [PubMed] [Google Scholar]
- Noda, T., Suzuki, K., and Ohsumi, Y. (2002). Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 12, 231–235. [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., Babst, M., and Emr, S. D. (2000). Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25: 229–235. [DOI] [PubMed] [Google Scholar]
- Onodera, J., and Ohsumi, Y. (2004). Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J. Biol. Chem. 279, 16071–16076. [DOI] [PubMed] [Google Scholar]
- Petiot, A., Ogier-Denis, E., Blommaart, E. F., Meijer, A. J., and Codogno, P. (2000). Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275, 992–998. [DOI] [PubMed] [Google Scholar]
- Raymond, C. K., Howald-Stevenson, I., Vater, C. A., and Stevens, T. H. (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3, 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. S., Klionsky, D. J., Banta. L. M., and Emr, S. D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 8, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu, P. V., Takegawa, K., Fry, M. J., Stack, J. H., Waterfield, M. D., and Emr, S. D. (1993). Phosphatidylinositol 3–kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88–91. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., Suzuki, K., Kamada, Y., Noda, T., and Ohsumi, Y. (2001). Apg2p functions in autophagosome formation on the perivacuolar structure. J. Biol. Chem. 276, 30452–30460. [DOI] [PubMed] [Google Scholar]
- Stack, J. H., and Emr, S. D. (1994). Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J. Biol. Chem. 269, 31552–31562. [PubMed] [Google Scholar]
- Stack, J. H., DeWald, D. B., Takegawa, K., and Emr, S. D. (1995). Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3–kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack, J. H., Herman, P. K., Schu, P. V., and Emr, S. D. (1993). A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3–kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 12, 2195–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strømhaug, P. E., Reggiori, F., Guan. J., Wang, C. W., and Klionsky, D. J. (2004). Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 15, 3553–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm, M., Egner, R., Koch, B., Schlumpberger, M., Straub, M., Veenhuis, M., and Wolf, D. H. (1994). Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280. [DOI] [PubMed] [Google Scholar]
- Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. [DOI] [PubMed] [Google Scholar]
- Vida, T., and Gerhardt, B. (1999). A cell-free assay allows reconstitution of Vps33p-dependent transport to the yeast vacuole/lysosome. J. Cell Biol. 146, 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia, S., Dhand, R., Vanhaesebroeck, B., MacDougall, L. K., Stein, R., Zvelebil, M. J., Domin, J., Panaretou, C., and Waterfield, M. D. (1995). A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 14, 3339–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., DeWald, D. B., Boronenkov, I. V., Anderson, R. A., Emr, S. D., and Koshland, D. (1995). Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell. 6, 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]