Abstract
Poly(ADP-ribose)polymerase 1 (PARP1) is well characterized for its role in base excision repair (BER), where it is activated by and binds to DNA breaks and catalyzes the poly(ADP-ribosyl)ation of several substrates involved in DNA damage repair. Here we demonstrate that PARP1 associates with telomere repeat binding factor 2 (TRF2) and is capable of poly(ADP-ribosyl)ation of TRF2, which affects binding of TRF2 to telomeric DNA. Immunostaining of interphase cells or metaphase spreads shows that PARP1 is detected sporadically at normal telomeres, but it appears preferentially at eroded telomeres caused by telomerase deficiency or damaged telomeres induced by DNA-damaging reagents. Although PARP1 is dispensable in the capping of normal telomeres, Parp1 deficiency leads to an increase in chromosome end-to-end fusions or chromosome ends without detectable telomeric DNA in primary murine cells after induction of DNA damage. Our results suggest that upon DNA damage, PARP1 is recruited to damaged telomeres, where it can help protect telomeres against chromosome end-to-end fusions and genomic instability.
INTRODUCTION
Telomeres are structures at the ends of chromosomes that consist of repeats of noncoding TTAGGG sequences and telomere-associated proteins. Telomeres allow cells to distinguish natural chromosome ends from damaged DNA and protect chromosomes against degradation and fusion (reviewed in Greider and Blackburn, 1996). Telomere integrity in cells thus plays an essential role in the control of genomic stability. Uncapped telomeres, resulting from either loss of function of telomere-binding proteins or loss of telomeric repeats, directly associate with many DNA damage response proteins and/or induce a response similar to that observed for DNA breaks (Espejel et al., 2002a, 2002b, 2004; Karlseder et al., 2002, 2004; d'Adda di Fagagna et al., 2003; Takai et al., 2003; Hao et al., 2004; Tarsounas et al., 2004).
Several PARP family proteins associate with telomeres or telomerase (Smith et al., 1998; Kaminker et al., 2001; Cao et al., 2002; Sbodio et al., 2002; Dantzer et al., 2004; Liu et al., 2004). Tankyrases 1 and 2 can directly bind and poly(ADP-ribosyl)ate the telomere repeat binding factor 1 (TRF1) and affect its binding to telomeric DNA (Smith et al., 1998; Kaminker et al., 2001; Cook et al., 2002; Rippmann et al., 2002; Sbodio et al., 2002). Human tumor cells overexpressing a wild-type Tankyrase 1 promoted telomere elongation (Smith and de Lange, 2000; Cook et al., 2002). Poly(ADP-ribose) polymerase 2 (PARP2) interacts with TRF2 and regulates its telomeric DNA-binding activity through poly (ADP-ribosyl)ation. Primary Parp2-deficient mouse embryonic fibroblasts (MEFs) show normal telomere length and telomere capping but display an increase in chromosome ends lacking detectable telomeric DNA (Dantzer et al., 2004). We recently reported that Vault poly(ADP-ribose) polymerase (VPARP), a minor protein component of cytoplasmic vault particles, associates with telomerase activity in cell extracts; nevertheless, mice deficient for mVparp have normal telomerase activity, telomere length, and telomere capping (Liu et al., 2004).
PARP1 binds to and is activated by DNA breaks and catalyzes the poly(ADP-ribosyl)ation of several substrates involved in chromatin structure and DNA repair, favoring the recruitment of DNA repair proteins and their access at damaged sites (reviewed in Ame et al., 2004). Samper et al. (2001) previously reported that Parp1-deficient mice exhibited normal telomere length and chromosome end capping. In this report, we characterized the role of PARP1 at eroded telomeres in mammals and investigated the molecular mechanism of PARP1 in regulating telomeres. Our studies demonstrate that PARP1 interacts with TRF2 in human cell extract and its PARP activity affects the DNA-binding activity of TRF2. PARP1 is found sporadically at normal telomeres, whereas it appears preferentially at damaged telomeres in mammalian cells after induction of DNA damage or at short telomeres in telomerase-deficient murine embryonic stem (ES) cells. PARP1-TRF2 interaction helps PARP1 localize to sites of DNA strand breakages at telomeres. Loss of PARP1 function leads to telomere dysfunction and chromosomal abnormalities in primary MEFs exposed to DNA-damaging reagents. Our results suggest that PARP1 protects eroded telomeres and thus genomic stability through regulation of TRF2.
MATERIALS AND METHODS
Vectors
All the GST fusion constructs were previously published (Dantzer et al., 2004). A PCR fragment of pcDNA-FLAG-hTRF2 (a kind gift from Dr. Lea Harrington) was amplified using the primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGCTGGTGGTGGTGGTT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTAGTTCATGCCAAGTCTTT-3′, cloned into the pDONR 221 entry vector (Invitrogen) by Gateway technology and finally transferred to destination vectors with GST-tag (pDEST15, Invitrogen) or His-Tev-HA-tag (Wang et al., unpublished vector). The FLAG-CDC14B vector was published elsewhere (Cho et al., 2005).
Preparation of Primary mParp1 Null MEFs and mTert Null ES Cells
The generation of mParp1 null mice and mTert null ES cells were described elsewhere (de Murcia et al., 1997; Liu et al., 2000, 2002). MEFs were established from 13.5-d mouse embryos and were cultured in 3% oxygen (SANYO O2/CO2 incubator, MCO-18M).
Transfection and Nuclear Extracts
Empty vectors or vectors with cDNA of interest were transfected alone or cotransfected into human cells with Lipofectamine Plus Reagent (Invitrogen) or FuGene 6 (Roche) following the manufacturer's instructions. After 48 h of transfection, cells were harvested and lysed in Buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.6% NP-40, 1 mM DTT, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) and then centrifuged at 5000 rpm for 30 s at 4°C. To obtain nuclear lysates, the pelleted nuclei were resuspended and lysed in Buffer C (20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 1 mM PMSF) before clearing by centrifugation at 14,000 rpm for 2min at 4°C.
Protein Pulldown
A total of 500 μg of human 293T nuclear protein extracts were subjected to immunoprecipitation using Anti-FLAG M2 Affinity Gel (Sigma) or ImmunoPure Immobilized Protein A beads (Pierce, Rockford, IL) conjugated with either rabbit anti-PARP1 (Ame et al., 2001) or rabbit anti-TRF2 (508; van Steensel et al., 1998) antibodies in a buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM NaF, 5 mM EDTA, 0.1% NP-40, 1 mM DTT, 1 mM PMSF, and EDTA-free protease inhibitor cocktail tablet (Roche Diagnostics). Whole cell extracts from Cos-7 cells expressing GST or GST fusion proteins (Dantzer et al., 2004) were incubated with glutathione Sepharose 4B (Amersham) for 2 h at 4°C. The beads were washed four times in a wash buffer (50 mM Tris, pH 8.0, 200–500 mM NaCl, 0.25–0.5% NP-40, and 0.5 mM PMSF) before being subjected to SDS-PAGE and Western blot analysis. For DNA dependence interaction ethidium bromide (EtBr) was added to the lysates to a final concentration of 12.5 μg/ml, and lysates were incubated on ice for 30 min and then centrifuged at 15,000 rpm for 5 min. The supernatants were then used in pulldown experiments, keeping the same concentration of EtBr throughout the washes.
Purification of GST Fusion Proteins
BL21AI One Shot Escherichia coli (Invitrogen) cells were transformed with pDEST15 GST-hTRF2 and grown to an OD600 before inducing with 0.2% final concentration of l-arabinose (Sigma) for 2 h at 37°C. Cells were harvested by centrifugation, resuspended in NNTEE buffer, pH 8.0 (5 mM EGTA, 5 mM EDTA, 500 mM NaCl, 0.5%NP-40, 50 mM Tris, pH 7.6, 2 mM PMSF, and 5 mM DTT), and lysed by sonication. The lysate was cleared by centrifugation at 10,000 × g for 20 min at 4°C and incubated with glutathione Sepharose 4B (Amersham) for 1 h at 4°C. Beads were washed twice in NNTEE buffer followed by two washes in TEE buffer (5 mM EGTA, 5 mM EDTA, and 50 mM Tris, pH 8.0). The recombinant GST fusion proteins were eluted from the beads with 10 mM glutathione in 50 mM Tris (pH 7.5), dialyzed against phosphate-buffered saline (PBS) using Slide-A-Lyzer Dialysis cassette (Pierce), and then concentrated using a Centricon YM-50 column (Amicon).
Poly(ADP-ribosyl)ation Reaction and DNA-binding Assays
Heteromodification of GST Fusion Proteins by PARP1. The protocol for poly(ADP-ribosyl)ation of GST-hTRF2 by hPARP1 has been described elsewhere (Schreiber et al., 2002; Dantzer et al., 2004). In brief, GST pulldown assays were performed as described above, except that washes were done with HSB (20 mM Tris-HCl, pH 8, 500 mM NaCl, 0.5% NP-40, 0.5 mM PMSF). After a last wash with activity buffer (50 mM Tris-HCl, pH 8, 4 mM MgCl2, 0.3 mM DTT), the beads were pelleted and resuspended in activity buffer containing either 300 pmol or no hPARP1. The reaction was started by the addition of activity buffer containing DNase I–activated calf thymus DNA, 32P-labeled NAD, and TRF2. After 4 min at 25°C, the reaction was stopped by the addition of cold HSB on ice and beads were washed three times with HSB, before being analyzed by gel electrophoresis on 8% SDS-PAGE and autoradiography or by Western blot.
Electrophoretic Mobility Shift Assay. A 54-mer oligonucleotide (5′-GGCTGCTACCGGCACATCGTCCTAGCAAGGTTAGGGTTAGGGTTAGGGTTAGGG-3′) was annealed with the complimentary 48-mer oligonucleotide (3′-CCGACGATGGCCGTGTAGCAGGATCGTTCCAATCCCAATCCCAATCCC-5′) to produce a 3′ overhang duplex (Dantzer et al., 2004). 50 ng of the 3′ overhang telomeric DNA duplex were incubated with 5 μg of GST, GST-hTRF2, or 3U of hPARP1 (Trevigen) in binding buffer (20 mM HEPES, pH 7.9, 150 mM KCl, 0.1 mM EDTA, 1 mM DTT, and 500 μg/ml bovine serum albumin (BSA); Dantzer et al., 2004) for 30 min at room temperature in a final volume of 12 μl. For the formation of the TRF2-PARP1-DNA complexes, the telomeric DNA duplex was incubated with GST-hTRF2 for 20 min before incubation with the hPARP1 for 10 min under the same binding conditions. For PAR polymer synthesis, NAD+ (Fluka) was added to a final concentration of 400 μM. To inhibit PARP1 activity, 2 mM final concentration of 3-aminobenzamide (3-AB; Sigma) was added to the reaction. After addition of 3 μl of loading buffer (21% Ficoll in 60 mM HEPES, pH 7.9), the samples were electrophoresed in nondenaturing 4–20% Tris-Glycine Novex gels (Invitrogen) for 60–90 min at 150 Volts, stained with SYBR Green EMSA nucleic acid gel stain (Molecular Probes), and viewed under UV light.
Immobilized dsTelomeric DNA-binding Assay. Twenty picomoles of 5′-biotin labeled dsTelomeric DNA (Sigma Genosys) were bound to 150 μg of Dynabeads M-280 Streptavidin (DYNAL Biotech) by incubating in 2× B&W buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 2 M NaCl) for 15 min. The beads were then washed three times with 10 volumes of 1× B&W buffer followed by two washes with binding buffer (see above) to remove unbound/unlabeled DNA. Immobilized dsTelomeric DNA (100 ng; ∼2.8 pmol) was incubated with 7 μg GST-hTRF2 and/or 1.5 Units of hPARP1 in binding buffer. Assembly of the TRF2-PARP1-DNA complexes followed by the addition of NAD+ and/or 3-AB was as described for the electrophoretic mobility shift assay (see above). The beads were separated from the supernatant using a Magnetic Particle Concentrator Dynal MPC-S (DYNAL Biotech). The supernatant and the beads were subjected to SDS-PAGE and Western blot analysis.
Indirect Immunofluorescence
For induction of DNA damage, cells were treated with 5–10 mM hydrogen peroxide (H2O2) for 5–10 min before harvesting or with 5 Gy X-Ray 20 min before harvesting for immunofluorescence. Immunofluorescence was performed as previously described (Smith and de Lange, 1999). Briefly cells were fixed in cold methanol (Sigma) at –20°C for 10 min, permeabilized with 0.5% NP-40, and blocked in 1% BSA (IgG-free; Sigma). Cells were first immunostained with a rabbit anti-TRF2 antibody (1:100, Novus), rabbit anti-PARP2 antibody (1:50; Meder et al., 2005), or rabbit anti-GST antibody (1:2000, Upstate Biotechnology) overnight at 4°C followed by Alexa 488- or -594–labeled donkey anti-rabbit secondary antibody (1:500; Molecular Probes) for 1 h at 37°C and then immunostained with a mouse anti-PARP1 antibody (1:50 or 1:100, BD Bioscience) for 2 h at 37°C followed by Alexa 594– or 488–labeled donkey anti-mouse secondary antibody. Fluorescent images were acquired with a Leica SP2 laser scanning confocal microscope (Deerfield, IL) or a Zeiss axiophot fluorescence microscope (Thornwood, NY).
Telomere Fluorescence In Situ Hybridization and Immunofluorescence
Telomere fluorescence in situ hybridization and immunofluorescence (TEL-FISH) was performed according to previously published protocols (Smith and de Lange, 1999; Meeker et al., 2002; Hao et al., 2004) with some modifications. Cells were treated with 0.1 mg/ml Colcemid for 2–4 h. To induce DNA damage, cells were either treated with 5 mM H2O2 for 5–10 min before harvesting or exposed to 5 Gy X-Ray 20 min before harvesting by trypsinization. Cells were incubated in hypotonic RSB buffer (Hao et al., 2004) for 15 min at 37°C and then fixed in cold methanol at –20°C for 10 min. After adding one-fourth volume of glacial acetic acid, fixed cells were immediately dropped onto slides and air-dried briefly to obtain metaphase spreads. Fixed cells were washed with 0.25× PBS for 15 min, blocked in 1% BSA (IgG-free) for 30 min, and probed with a mouse anti-PARP1 antibody (1:50, BD Bioscience) or a rabbit anti-PARP1 antibody (1:5000; Ame et al., 2001) followed by Alexa 488– or 594–labeled secondary antibodies against the primary host (see Indirect Immunofluorescence). After acquiring PARP1 images, slides were washed with PBS for 15 min and then hybridized to a Cy-3–labeled (CCCTAA)3 PNA probe (see Telomeric Fluorescence In Situ Hybridization (Telomeric-FISH). Telomeric fluorescent images were acquired on a Zeiss axiophot fluorescence microscope then processed and merged with PARP1 images using Adobe Photoshop (San Jose, CA).
Telomeric Fluorescence In Situ Hybridization (Telomeric-FISH)
For examining chromosome abnormalities after DNA damage, primary MEFs were treated with 5 mM H2O2 or 2 mM methyl methanesulfonate (MMS) for 5–10 min or 5 Gy X-Ray. After treatment, cells were washed three times with warm PBS and then cultured in fresh medium for another 24 h after initial treatment. 0.1 μg/ml Colcemid was added 4 h before harvesting. Metaphase spread preparation and telomeric-FISH were performed as previously described (Blasco et al., 1997; Zijlmans et al., 1997). The Cy-3–labeled (CCCTAA)3 PNA (Applied Biosystems) was used as a probe. Metaphases were examined with a Zeiss axiophot fluorescence microscope.
RESULTS
PARP1 Interacts with TRF2
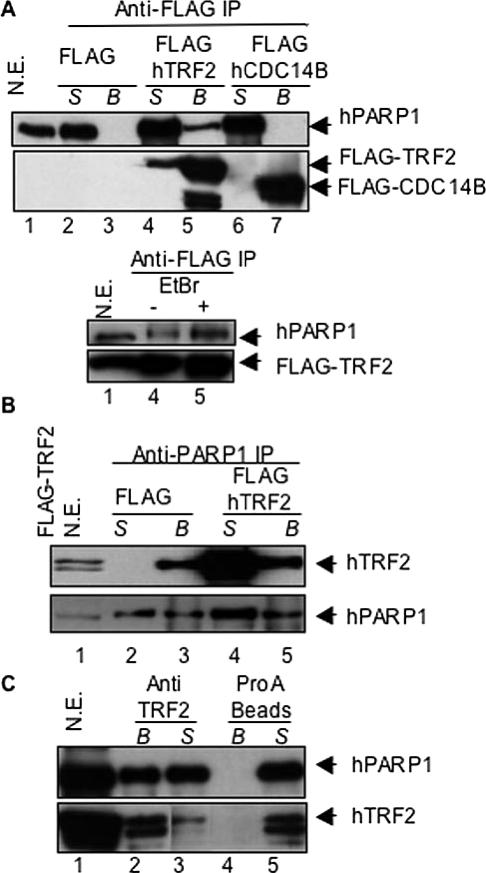
It has been shown that PARP2 interacts with TRF2 and its PARP activity regulates the DNA-binding activity of TRF2 (Dantzer et al., 2004). As PARP1 is a close relative of PARP2 (Ame et al., 2004), we hypothesize that PARP1 may share a similar role. To examine a possible interaction between PARP1 and TRF2 in vivo, human 293T cells were transiently transfected with FLAG-hTRF2, a FLAG-tagged unrelated nuclear protein (hCDC14B), or an empty vector. hPARP1 was present in the FLAG-hTRF2 immunoprecipitate (Figure 1A, top panel, lane 5), but absent from both FLAG-hCDC14B and vector control immunoprecipitates (lanes 3 and 7). Conversely, endogenous hTRF2 or FLAG-hTRF2 was detected in the hPARP1 immunoprecipitates of human 293T cells transfected with an empty vector or FLAG-hTRF2 (Figure 1B, top panel, lanes 3 and 5). To determine if this interaction occurred when both proteins were expressed at endogenous levels, human 293T nuclear extracts were subjected to immunoprecipitation with an anti-TRF2 antibody and analyzed for the presence of hPARP1. hPARP1 was detected in the hTRF2 immunoprecipitate, but not when protein A beads alone were used (Figure 1C, top panel, lanes 2 and 4). These results demonstrate that hPARP1 interacts with hTRF2 in human cell extracts in vivo.
Figure 1.
hPARP1 interacts with hTRF2 in vivo. (A) Nuclear extracts (N.E) of human 293T cells transfected with vector alone (lanes 2 and 3), FLAG-hTRF2 (lanes 4 and 5), or FLAG-hCDC14B (lanes 6 and 7) were subjected to immunoprecipitation (IP) with an anti-FLAG antibody. Supernatant (S) and beads (B) were examined by Western blot for coimmunoprecipitation of endogenous hPARP1 with a mouse anti-PARP1 antibody (BD Transduction) or for transfected FLAG proteins with a mouse anti-FLAG M2 antibody (Sigma). Bottom panel, pulldown experiments performed in the presence of 12.5 μg/ml ethidium bromide (EtBr). (B) Nuclear extracts of human 293T cells transfected with vector alone (lanes 2 and 3) or FLAG-hTRF2 (lanes 4 and 5) were subjected to immunoprecipitation with a rabbit anti-PARP1 antibody (Ame et al., 2001) and examined for the coimmunoprecipitation of endogenous or FLAG-hTRF2 with a mouse anti-TRF2 antibody (Upstate, top panel) or for endogenous hPARP1 (bottom panel). (C) Nuclear extracts of human 293T cells were subjected to immunoprecipitation with a rabbit anti-TRF2 antibody (van Steensel et al., 1998; lanes 2 and 3) or protein A beads alone (lanes 4 and 5) and examined for coimmunoprecipitation of endogenous hPARP1 (top panel) or endogenous hTRF2 (bottom panel). Forty micrograms of nuclear lysate were used as control (lane 1) in the experiments.
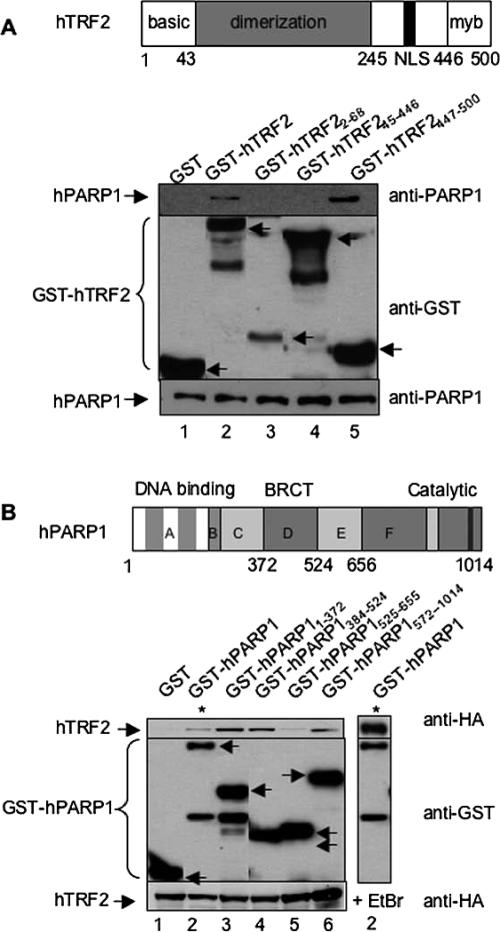
To map the domains in TRF2 that might be responsible for its interaction with PARP1, cell extracts from Cos-7 cells expressing GST, GST-hTRF2, or the GST-hTRF2 deletion mutants (basic domain, dimerization domain, or myb domain) were analyzed by GST pulldown and Western blot (Figure 2A). Coprecipitation of endogenous hPARP1 was achieved by either the full-length or the myb domain (aa 447–500) of hTRF2 (top panel, lanes 2 and 5). To map the PARP1 domains that might be involved in its interaction with TRF2, cell extracts from Cos-7 cells expressing His-Tev-HA-hTRF2 along with GST, GST-hPARP1, or GST-hPARP1 deletion mutants (encompassing the DNA-binding domain, the BRCT domain, or the catalytic domain) were analyzed by GST pulldown and Western blot (Figure 2B). Fusion proteins containing either the DNA-binding domain or the BRCT domain and to a lesser extent the PARP domain (top panel, lanes 3, 4, and 6, respectively) appeared to interact with His-Tev-HA hTFR2. The weaker interaction observed with full-length PARP1 is likely due to less expression of GST-hPARP-1 (top panel, lane 2). These results suggest that PARP1 interacts, mainly via its DNA-binding and BRCT domains, with the myb domain of TRF2.
Figure 2.
The Myb domain of hTRF2 interacts with hPARP1. (A) Interaction between GST-hTRF2 full-length or deletion constructs and endogenous hPARP1 in Cos-7 cells. Top panel, schematic representation of human TRF2. Bottom panel, GST, GST-hTRF2, and GST-hTRF2 deletion mutants expressed in Cos-7 cells were subjected to GST pulldown then examined by Western blot using, consecutively, mouse anti-hPARP1 (BD Transduction Laboratoriess, top) and rabbit anti-GST (Upstate, middle) antibodies. (B) Interaction between GST-hPARP1 full-length or deletion constructs and His-Tev-HA hTFR2 in Cos-7 cells. Top panel, schematic representation of human PARP1. Bottom panel, GST, GST-hPARP1 full-length, and GST-hPARP1 deletion mutants along with His-Tev-HA hTFR2 were expressed in Cos-7 cells. Cell lysates were subjected to GST pull-down in the absence (lanes 1–6) or presence (lane 7) of 12.5 μg/ml ethidium bromide and subsequently examined by Western blot using mouse anti-HA.11 (BAbCo, top), then rabbit anti-GST (middle) antibodies. To show the level of endogenous PARP1 or His-Tev-HA hTFR2 in each sample, 40 μg of whole cell lysates were subjected to Western blot with mouse anti-hPARP1 or mouse anti-HA antibodies, respectively (bottom). Arrows indicate the position of GST or GST fusion proteins. Bottom bands are protein degradation products.
Because both proteins independently bind to DNA, we examined if the coprecipitation of PARP1 and TRF2 was mediated by tethering to telomeric DNA. EtBr was added to the cell lysates and throughout the pulldown experiments to disrupt protein–DNA interactions. In the presence of 12.5 μg/ml EtBr the interactions between TRF2 and PARP1 remained intact (Figure 1A, bottom panel and Figure 2B, right panel) and are therefore DNA-independent.
PARP1 poly(ADP-ribosyl)ates the Dimerization Domain of TRF2, Influencing Its DNA-binding Activity
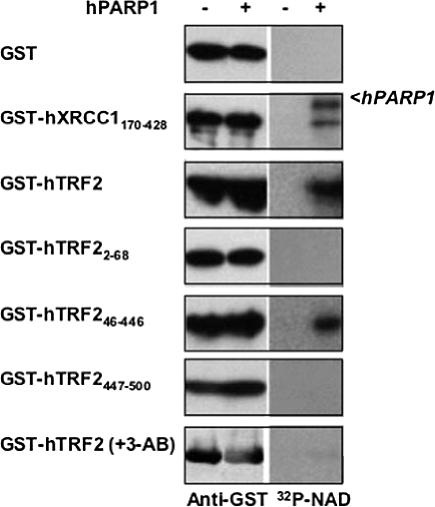
We next examined the ability of PARP1 to poly(ADP-ribosyl)ate TRF2. GST-hTRF2 or deletion mutants were expressed in Cos-1 cells and isolated by GST pulldown. The purified proteins were incubated with 32P-NAD (providing ADP-ribose) in the absence or presence of purified hPARP1. The resulting poly(ADP-ribosyl)ated proteins were visualized by autoradiography. In the presence of hPARP1, full-length hTRF2 and its dimerization domain (hTRF245–446) were poly(ADP-ribosyl)ated (Figure 3, right panel). The PARP inhibitor, 3-AB abolished poly(ADP-ribosyl)ation of hTRF2 by hPARP1, confirming that the radioactive signal detected was due to synthesis of poly(ADP-ribose) (PAR) polymers by hPARP1 at hTRF2 (bottom, right panel). Our results indicate that PARP1 poly(ADP-ribosyl)ates TRF2 at the dimerization domain.
Figure 3.
The dimerization domain of hTRF2 is poly(ADP-ribosyl)ated by PARP1. GST, GST-hXRCC170–428, GST-hTRF2, or deletion mutants were expressed in Cos1 cells, purified by GST pulldown, and incubated in a reaction buffer containing 32P-NAD and DNase I activated DNA with (+) or without (–) hPARP1 or the PARP inhibitor, 3-AB, as indicated. The fusion proteins were analyzed by Western blot (left panel) with rabbit anti-GST antibody (Upstate) and visualized by autoradiography (right panel). XRCC1170–428 was previously shown to be an acceptor protein for poly(ADP-ribosyl)ation by PARP1 (Masson et al., 1998) and used as a positive control.
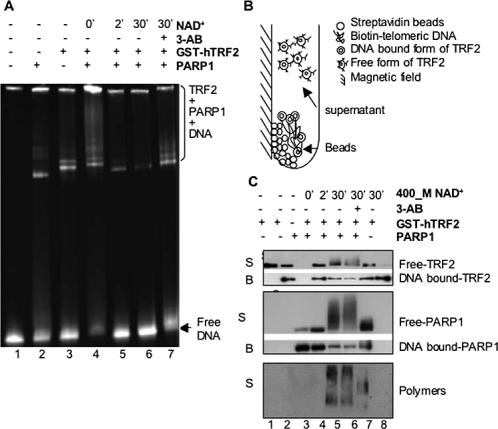
To test if poly(ADP-ribosyl)ation of TRF2 by PARP1 had an effect on the DNA-binding activity of TRF2, an electrophoretic mobility-shift assay was performed under conditions in which TRF2 and PARP1 bound to telomeric DNA. As shown in Figure 4A, both hPARP1 (lane 2) and GST-hTRF2 (lane 3) could form protein-telomeric DNA complexes, which increased with increasing concentrations of GST-hTRF2 or hPARP1 and were not due to unspecific binding through the GST tag, because no protein-DNA complexes were detected when telomeric DNA was incubated with GST protein (unpublished data). When GST-hTRF2 and hPARP1 were incubated together with telomeric DNA, a supershift occurred, accompanied by a concomitant decrease in free-DNA (lane 4 vs. lanes 2 and 3). When the PARP substrate NAD+ was added to initiate PAR polymer synthesis, the preassembled TRF2-PARP1-telomeric DNA complexes were greatly reduced into a progressively decreasing form of PARP1-telomeric DNA complex, accompanied by an increase in free telomeric DNA (lanes 5 and 6 vs. lane 4). However, when the PARP inhibitor 3-AB was added along with NAD+, some of the TRF2-PARP1-telomeric DNA complex was restored (lane 7 vs. lane 6), suggesting that poly-(ADP-ribosyl)ation has a negative effect on TRF2's DNA-binding activity.
Figure 4.
PARP1 negatively regulates TRF2's DNA-binding ability. (A) Electrophoretic mobility shift assay. The dsTelomeric DNA was incubated alone (lane 1) or with hPARP1 (lane 2), GST-hTRF2 (lane 3), or hPARP1 plus GST-hTRF2 (lanes 4–7). Some of the reactions were supplemented with NAD+ for various times in the presence or absence of 3-AB as shown (lanes 5–7). The positions of free DNA and covalent complexes are indicated. (B) Schematic representation of immobilized dsTelomeric DNA-binding assay, showing DNA-bound proteins in the immobilized beads and free poly(ADP-ribosyl)ated proteins in the supernatant. (C) Western blot analysis of the supernatant (S) and the beads (B) from an immobilized dsTelomeric DNA-binding assay using mouse anti-hTRF2 antibody (Upstate, top panel), mouse anti-PARP1 antibody (BD Transduction, middle panel), or rabbit anti-poly(ADP-ribose) antibody (BD Transduction, bottom panel). Streptavidin Dynabeads alone were incubated with GST-hTRF2 (lane 1) and biotin-dsTelomeric DNA-conjugated streptavidin Dynabeads were incubated with GST-hTRF2 (lane 2), hPARP1 (lane 3), or GST-hTRF2 plus hPARP1 (lanes 4–7) before addition of NAD+ and/or 3-AB as shown. Note that after addition of NAD+, free hTRF2 and hPARP1 are present in the supernatant as smearlike bands representing different degrees of poly(ADP-ribosyl)ation (lanes 5 and 6).
To confirm above results and also examine the effects of poly(ADP-ribosyl)ation at the protein level, we developed an immobilized DNA-binding assay coupled with Western blot analysis, which allowed direct comparison of free-versus DNA bound–TRF2 and PARP1 before and after poly(ADP-ribosyl)ation (Figure 4B). Biotin-labeled dsTelomeric DNA was first bound to magnetic streptavidin Dynabeads. The protein–DNA complexes were assembled on the beads and then treated with NAD+ and/or 3-AB. On the completion of reactions, the beads were separated from the supernatant by a magnetic field and both the beads and the supernatant were subjected to Western blot analysis to detect TRF2, PARP1, and their poly(ADP-ribosyl)ated forms (Figure 4C). When incubated together with Dynabeads alone (without DNA), GST-hTRF2 did not bind nonspecifically to the beads and remained in the supernatant as free protein (top panel, lane 1). After incubation with the telomeric DNA-conjugated beads, both GST-hTRF2 and hPARP1 were present mostly as DNA-bound proteins in the beads (top and middle panels, lanes 2–4). Although some free TRF2 and PARP1 could be detected in the supernatants, when less protein was used in the assay, free proteins could no longer be detected (unpublished data). When the preassembled PARP1-TRF2-biotin-telomeric DNA complexes were incubated with NAD+ for various times, there was a decrease in the DNA-bound proteins in the beads along with an increase in free poly(ADP-ribosyl)ated proteins in the supernatant (top and middle panels, lanes 5 and 6 vs. lane 4; poly(ADP-ribosyl)ated proteins are present as smear-like bands). When 3-AB was added along with NAD+, the presence of free proteins was reduced and some of the DNA bound-proteins were restored (top and middle panels, lane 7 vs. lanes 5 and 6). The TRF2's DNA-binding ability was unaffected by the presence of NAD+ (top panel, lane 8). Poly(ADP-ribosyl) ation of free TRF2 and PARP1 was further confirmed by detection of two clusters of smearlike signals using an anti-PAR antibody (bottom panel, lanes 5 and 6). The lower smear, starting at 75 kDa, corresponds to the poly(ADPribosyl)ated TRF2, whereas the higher smear, above 105 kDa, might contain both poly(ADP-ribosyl)ated PARP1 and TRF2. In the presence of 3-AB, these smears shifted toward a lower molecular weight, representing species of TRF2 and PARP1 with reduced degrees of poly(ADP-ribosyl)ation (bottom panel, lane 7 vs. lanes 5 and 6). These results demonstrate that poly(ADP-ribosyl)ated TRF2 was unable to bind to the biotin-telomeric DNA and was consequently released from the beads into the supernatant.
PARP1 Localizes at Telomeres, Preferentially in Cells Exposed to DNA-damaging Reagents, Which Partially Depends on Its Interaction with TRF2
We subsequently examined if PARP1 localized at telomeres, especially at telomeres with DNA strand breakages in vivo. As poly(ADP-ribosyl)ation is the first line of defense in response to DNA breaks (reviewed in Huber et al., 2004), we speculated that DNA damage at telomeres may immediately activate and recruit PARP1 to the site. To test these possibilities we examined and compared the frequency of PARP1 at telomeres of untreated cells versus cells exposed to DNA damaging reagents.
To determine if PARP1 was found at telomeres in vivo, we first examined the colocalization of PARP1 with TRF2 in interphase HeLa 1.2.11 cells (a subclone of HeLa) (Smith and de Lange, 1999) by indirect immunofluorescence. We found that fixation methods and permeabilization conditions affect PARP1's punctate nuclear staining necessary for the observation of its colocalization with TRF2 (unpublished data). We therefore used a previously published protocol in which cells are fixed in ice-cold methanol and then permeabilized with 0.5% NP-40 (Smith and de Lange, 1999). Colocalization of PARP1 with TRF2 was sporadic in untreated cells, but readily detected in cells exposed to DNA-damaging reagents (Figure 5). Similar results were found in other cell lines (e.g., HeLa and VA13), although VA13 cells showed a lesser degree of colocalization (unpublished data).
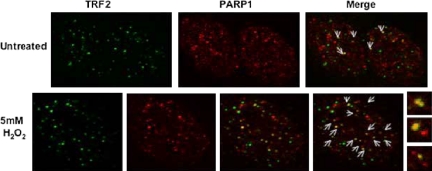
Figure 5.
PARP1 preferentially colocalizes with TRF2 after DNA damage. Double immunostaining of PARP1 and TRF2 in untreated and H2O2-treated HeLa 1.2.11 interphase cells. When compared with untreated cells, more PARP1 signals (in red) overlap with TRF2 signals (in green) in the treated cells. On the right, larger insets of the images show more clearly the signal overlaps in yellow (arrows). PARP1 antibody dilutions differ per treatment, 1:50 for untreated cells, and 1:100 for H2O2-treated cells.
Previously, Dantzer et al. (1999) reported that PARP2, another member of the PARP superfamily, did not colocalize with TRF2 in untreated telomerase-positive cells. PARP2 is closely related to PARP1. Its catalytic domain has the strongest resemblance to that of PARP1 with 69% similarity; however its DNA-binding domain differs from that of PARP1 and targets DNA gaps but not nicks. PARP2 and PARP1 also share common partners involved in the BER, kinetochore structure, and mitotic spindle checkpoint (reviewed in Ame et al., 2004). We thus examined if PARP2 would colocalize with TRF2 when DNA strand breakages occurred at telomeres. Unlike PARP1, the colocalization of PARP2 with TRF2 was rare in HeLa1.2.11 cells exposed to DNA-damaging reagents (Supplementary Figure S1).
Although we showed colocalization of PARP1 with TRF2 in interphase cells, such colocalization may not be necessarily at telomeres. Recent reports have indeed demonstrated that TRF2 associates with genomic double-strand breaks as an early response to DNA damage (Bradshaw et al., 2005; Tanaka et al., 2005). Therefore, we confirmed the localization of PARP1 at telomeres by the detection of PARP1 at chromosomal ends of HeLa 1.2.11 metaphase spreads. PARP1 signals were sporadically detected at chromosome interiors and ends of untreated metaphase spreads. In contrast, PARP1 signals were readily detectable at both telomeric and nontelomeric regions of chromosomes of nearly all the H2O2 or x-ray–treated metaphase spreads (Supplementary Figure S2). We corroborated that PARP1 was at telomeres, by showing the colocalization of PARP1 signals with telomeric DNA signals in HeLa 1.2.11 cells, primary MEFs and mouse ES cells using immunofluorescence in combination with TEL-FISH (Figures 6 and 7). Interestingly, PARP1 was also detected at chromosome ends without detectable telomeric DNA (telomere signal free ends or SFEs) in treated cells (Figures 6 and 7A). No punctatelike signals were found associated with the chromosomes of mParp1–/– MEFs, confirming the specificity of the PARP1 signals at the chromosomes (Figure 7D). The frequency of chromosome-associated PARP1 signals was higher in treated cells than in untreated cells (average number of PARP1 signals/chromosome: 0.932 in treated vs. 0.014 in untreated). A fraction of the PARP1 signals was seen at telomeres (average number of telomere-associated PARP1 signals/chromosome: 0.197 in treated vs. 0.002 in untreated cells), with some signals at SFEs (29.1% SFEs positive for PARP1 signals in treated cells; nearly all the chromosome ends had detectable telomeric DNA in untreated cells; Table 1). These results demonstrate that PARP1 rarely localizes to normal telomeres; however, the frequency of PARP1's telomeric associations increases after DNA damage in vivo.
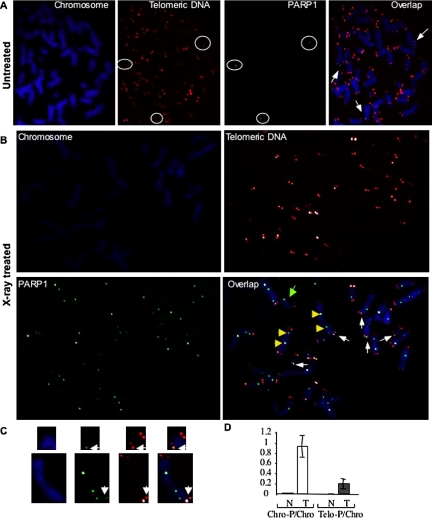
Figure 6.
PARP1 localizes at normal and damaged telomeres in human cells. TEL-FISH analysis of metaphase spreads of (A) untreated and (B) 5-Gy x-ray–treated HeLa 1.2.11 cells. More PARP1 signals (in green) were observed in the radiation-treated cells when compared with their untreated counterparts; these signals increasingly overlapped with telomeric DNA signals (in red) in the radiation-treated cells. PARP1 signals were also found at chromosome ends without detectable telomeric DNA signals or at internal regions of chromosomes of radiation-treated cells. Arrows, representative PARP1 signals at chromosome ends with (in white) or without (in green) detectable telomeric DNA signals or internal regions of chromosomes (in yellow). (C) Enlarged view of two chromosomes from treated cells showing overlap of PARP1 and telomeric DNA signals. (D) Bar graph comparing the average number of PARP1 signals/chromosome (Chro-P/Chro) and average number of telomere-associated PARP1 signals/chromosome (Tel-P/Chro) in both untreated (N) and treated (T)cells. The SD was calculated based on triplicate results.
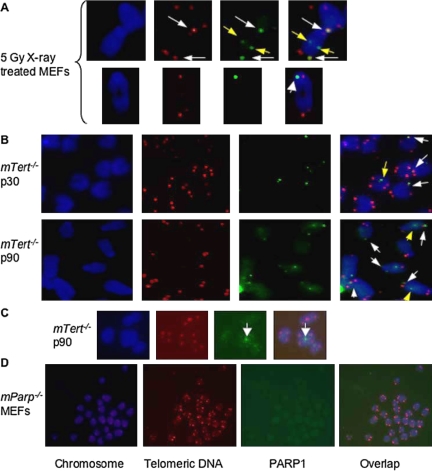
Figure 7.
PARP1 localizes at eroded telomeres induced by DNA damage or by loss of telomeric DNA in murine cells. TEL-FISH analysis of metaphase spreads of mTert-deficient ES cells with short telomeres and radiation-treated primary wild-type MEFs. PARP1 signals (in green) were observed at the chromosome ends with or without detectable telomeric DNA signals (in red) in (A) 5-Gy x-ray–treated MEFs or (B) late passages of telomerase-deficient mouse ES cells harboring short (passage 30) or critically short (passage 90) telomeres. (C) PARP1 signals found at chromosome end-to-end fusion sites without detectable telomeric DNA signals in mTert-deficient ES cells. (D) No punctate-like signals associated with the chromosomes of mParp1–/– MEFs. Arrows, representative PARP1 signals at chromosome ends (in white) or interiors (in yellow).
Table 1.
Frequencies of PARP1 signals at telomeres in untreated or radiation-treated HeLa1.2.11 cells
| Cell type | No. of chromosome-associated PARP1/no. of chromosomesa | No. of telomere-associated PARP1/no. of chromosomesb | No. of SFE-associated PARP1/no. of SFEsc |
|---|---|---|---|
| Untreated | 22/1523 (0.014) | 3/1523 (0.002) | 0/4 (0) |
| X-ray (5 Gy) | 1552/1666 (0.932)d | 329/1666 (0.197)d | 58/196 (30) |
Average number of PARP1 signals per chromosome. PARP1 signals include both chromosomal interiors and telomeres.
Average number of telomere-associated PARP1 signals per chromosome.
% SFEs positive for PARP1 signals in parentheses. Each chromosome is counted for four ends (for example, 1523 chromosomes of untreated cells have 6092 ends. Among those ends, four ends have SFEs).
p value between treated and untreated yielded a statistical difference of p < 0.01, using a chi-square test.
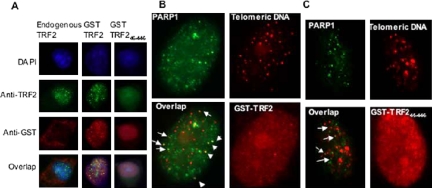
Although interactions between PARP1 and TRF2 are DNA independent in vivo (Figures 1 and 2), PARP1 is capable of binding telomeric DNA by itself in vitro (Figure 4). We therefore investigated if localization of PARP1 to damaged telomeres required its interaction with TRF2 in vivo. A TRF2 mutant (aa 45–446) with deletions of both basic and myb domains acts as a dominant-negative allele, disrupts the telomeric localization of endogenous TRF2 in vivo (van Steensel et al., 1998; Figure 8A) and is incapable of interacting with PARP1 (Figure 2). HeLa 1.2.11 cells transiently expressing GST-TRF2 or GST-TRF245–446 were treated with 10 mM H2O2 for 10 min and then examined for the localization of PARP1 at telomeres by TEL-FISH analysis. Although colocalization of PARP1 signals with telomeric DNA signals was observed in cells expressing either the wild-type or mutant GST-TRF2 fusion protein, it was less pronounced in the cells expressing GST-TRF245–446 (Figure 8, B and C, arrows). These results suggest that expression of a TRF2 dominant negative mutant interferes with the telomeric localization of PARP1.
Figure 8.
Effect of a TRF2 dominant negative mutant on the telomere binding of endogenous TRF2 or PARP1 in vivo. HeLa 1.2.11 cells were transiently transfected with GST-TRF2 or GST-TRF245–446 for 32 h. Before harvesting, cells were treated with 10 mM H2O2 for 10 min. (A) Double immunostaining of TRF2 (in green) and GST-TRF2 or GST-TRF245–446 (in red) in HeLa 1.2.11 interphase cells. The signature punctate staining of a functional TRF2 protein can be seen in untransfected cells (left panel), in cells transfected with GST-TRF2 (center panel), but not in cells transfected with a dominant negative mutant of TRF2, GST-TRF245–446 (right panel). (B and C) Representative TEL-FISH analysis of HeLa 1.2.11 interphase cells transiently expressing GST-TRF2 or GST-TRF245–446. Some PARP1 signals (in green) overlap with telomeric DNA signals (in red) (see arrows).
Critically Short Telomeres of Telomerase-deficient Murine ES Cells Recruit PARP1
Uncapped telomeres, caused by loss of telomeric repeats, directly associate with many DNA damage response proteins and induce a response similar to that observed with DNA breaks (d'Adda di Fagagna et al., 2003; Takai et al., 2003; Karlseder et al., 2004; Tarsounas et al., 2004). Hao et al. (2004) reported that γ-H2AX, a DNA damage response protein, localized to short telomeres in T-cells and fibroblasts of late generation telomerase RNA knockout mice. Together, these data suggest that critically short telomeres are likely to be recognized as DNA damage.
To test if critically short telomeres could also induce PARP1's localization to telomeres, we examined murine telomerase reverse transcriptase null (mTert–/–) ES cells by TEL-FISH analysis. We previously demonstrated that mTert–/– ES cells were telomerase deficient and underwent progressive loss of telomeric DNA during continuous passages in culture, and their chromosome ends eventually lost detectable telomeric DNA (Liu et al., 2000, 2002; Wang et al., 2005). A slight increase in PARP1 signals was observed at both telomeric and nontelomeric regions of chromosomes of wild-type (mTert+/+) ES cells during prolonged culture (average number of PARP1 signals/chromosome: 0.231 at passage 90 vs. 0.185 at passage 30). This effect was especially pronounced in mTert–/– ES cells with progressively shortened telomeres (0.923 at passage 90 vs. 0.3 at passage 30) (Supplementary Figure S3 and Table 2). PARP1 signals were detected at chromosome ends with short or critically short telomeres in mTert–/– ES cells (Figure 7B). At passage 90, there was a marked increase in telomere SFEs in the mTert–/– ES cells (% chromosome ends with SFEs: 12.8% at passage 90 vs. 0.2% at passage 30). Twenty-five percent of SFEs were positive for PARP1, whereas no SFEs were observed in the wild-type (mTert+/+) ES cells (Table 2). PARP1 was sometimes detected at chromosome end-to-end fusion sites without detectable telomeric DNA (Figure 7C). These results showed that murine ES cells maintained in long-term culture accumulate PARP1 and that in mTert–/– ES cells with critically short telomeres, there is an increase of PARP1 at telomeres.
Table 2.
Frequencies of PARP1 signals at telomeres in mTert-deficient mouse ES cells with short or critically short telomeres
| Cell type | No. of chromosome-associated PARP1/no. of chromosomesa | No. of telomere-associated PARP1/no. of chromosomesb | No. of SFE-associated PARP1/no. of SFEsc |
|---|---|---|---|
| Wild type | |||
| p30 | 187/1010 (0.185) | 15/1010 (0.0149) | 0/0 |
| p90 | 231/1002 (0.231) | 51/1002 (0.051) | 0/0 (0) |
| mTert-/-d | |||
| p30 | 313/1043 (0.3) | 78/1043 (0.075) | 5/9e |
| p90 | 946/1025 (0.923) | 315/1025 (0.307) | 131/526 (25)e |
a—c See footnotes in Table 1.
mTert-/- ES cells at passage (p) 30 or 90 have short or critically short telomeres, respectively (Liu et al., 2000, 2002; Wang et al., 2005).
Each chromosome is counted for four ends; 0.2% (9 SFEs/1043 × 4 chromosome ends) or 12.8% (526 SFEs/1025 × 4 chromosome ends) of chromosome ends have SFEs in mTert-/- ES cells at p30 or p90, respectively.
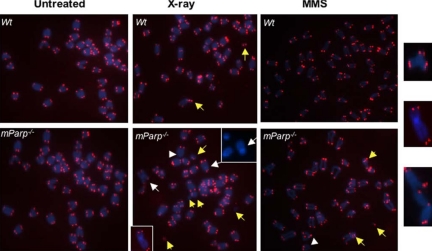
Parp1 Deficiency Leads to an Increase in Chromosome End-to-End Fusions and Telomere SFEs Induced by DNA Damage in Primary Mouse Embryonic Fibroblasts
So far, we demonstrated that PARP1 localized occasionally at normal telomeres but preferentially at telomeres upon induction of DNA damage or loss of telomeric DNA. These results suggest a role for PARP1 essentially at eroded telomeres. To examine the consequences of loss of PARP1 function in mammals, we examined the status of telomeres in proliferating primary wild-type and mParp1 null (mParp1–/–) MEFs. In agreement with previous studies (Samper et al., 2001), we found that primary mParp1–/– MEFs had normal telomeric DNA signals and did not exhibit the signature of telomere dysfunction (e.g., chromosome end-to-end fusions and SFEs; Figure 9, Table 3; unpublished data). Thus, PARP1 does not appear to have a major role in regulating normal telomeres.
Figure 9.
PARP1 deficiency leads to chromosome end-to-end fusions or telomere signal free ends in primary MEFs after DNA damage in vivo. Telomeric-FISH analysis of metaphase spreads of untreated, 5-Gy x-ray- or 2 mM MMS–treated early passage of primary wild-type (wt) or mParp–/– proliferating MEFs. Un-treated wild-type and mParp–/– MEFs showed normal telomeric DNA signals (in red) and chromosomes (in blue). Although chromosome breakages were detectable in wild-type MEFs 24 h after DNA damage, telomere signal free ends (arrowheads), chromosome or chromatid end-to-end fusions or telomere associations (white arrows, enlarged images at far-right), and chromosome breakages or frag-ments (yellow arrows) were found more frequently in x-ray– or MMS-treated mParp–/– MEFs.
Table 3.
Frequencies of chromosome abnormalities in untreated or radiation-treated wild-type and mParp1-deficient primary MEFs
| Cell type | End-to-end fusions/chromosomesa | SFEs/chromosomesa | Chromosome breakages/chromosomesa |
|---|---|---|---|
| Untreated | |||
| Wild type | 0/2034 (0) | 6/2034 (0.003) | 1/2034 (0) |
| mParp-/- | 2/2087 (0) | 8/2087 (0.004) | 17/2087 (0.008) |
| X-ray (5 Gy) | |||
| Wild type | 7/2050 (0.003) | 192/2050 (0.094) | 86/2050 (0.042) |
| mParp-/- | 44/2071 (0.021)d | 352/2071 (0.017)b | 269/2071 (0.13)b |
Average number of abnormal chromosome events/per chromosome.
p value between mParp-/- and wild type yielded a statistical difference of p < 0.01, using a chi-square test. The data were obtained from telomeric-FISH.
To elucidate the possible role of PARP1 at damaged telomeres, we challenged MEFs with DNA damaging reagents. Primary MEFs were briefly treated with 5 mM H2O2 or 2 mM MMS for 5–10 min or with 5 Gy x-ray and the culture medium was replaced before continuing culture. Twenty-four hours after exposure, metaphase spreads of the proliferating cells were examined for chromosome abnormalities. Although wild-type cells still showed some chromosome abnormalities, mParp1–/– MEFs had an increase in end-to-end fusions (average number of chromosome end-to-end fusions/chromosome: 0.021 in mParp1–/– vs. 0.003 in wild type), SFEs (average number of SFEs/chromosome: 0.17 in mParp1–/– vs. 0.094 in wild type), and chromosome fragments or breakages (average number of chromosomal breakages/chromosome: 0.13 in mParp1–/– vs. 0.042 in wild type; Figure 9 and Table 3; p < 0.01). It is worthy to point out that because mParp1–/– MEFs show accumulation of G2/M cells and cell death after DNA damage (Trucco et al., 1998), the frequency of chromosome abnormalities might have been underscored by our methods. Furthermore, we could not rule out that telomere SFEs may be the byproducts of chromosomal breakage. Nonetheless, these results demonstrate that primary mParp1–/– MEFs could not efficiently repair DNA strand breakages at telomeres and suggest a role for PARP1 in the capping and protection of damaged telomeres.
DISCUSSION
Functional Interaction of PARP1 with TRF2
In this report, we demonstrated that PARP1 interacts with TRF2 in human cell extracts and affects TRF2's telomeric DNA-binding ability. Therefore, the two solely described DNA damage dependent PARPs, PARP1 and PARP2, functionally interact with TRF2 (Dantzer et al., 1999 and this study).
The implication of the TRF2-PARP1 relationship in vivo was shown from the observation that PARP1 colocalized with TRF2 in mammalian cell lines. Moreover, such colocalization was rarely observed in untreated cells but increasingly in cells exposed to DNA damage reagents. Recent reports have demonstrated that TRF2 associates with genomic double-strand breaks as an early response to DNA damage (Bradshaw et al., 2005; Tanaka et al., 2005). Therefore, some of the PARP1-TRF2 signal overlaps observed may be at sites of genomic double-strand breaks and not exclusive to telomeres. Indeed, upon induction of DNA damage the overlap between PARP1 and telomeric DNA signals was less pronounced than the overlap between PARP1 and TRF2 signals as shown by TEL-FISH analysis. Nevertheless, we demonstrated that PARP1 signals overlap with telomeric DNA and that the frequency of such overlaps increased in cells that had loss of telomeric DNA or DNA strand breakages at telomeres. Moreover, PARP1 deficiency led to telomere dysfunction in murine cells exposed to DNA-damaging reagents. These results suggest a role for PARP1 especially at eroded telomeres.
PARP1 and PARP2 seem to share similar biochemical properties in regards to TRF2 (Ame et al., 2004; Dantzer et al., 2004 and this report). A previous report showed that PARP2 colocalized with TRF2 in a telomerase-negative ALT cell line (Dantzer et al., 2004). Here, we have shown that unlike PARP1, PARP2 rarely colocalizes with TRF2 in a telomerase-positive cell line, HeLa1.2.11, exposed to DNA-damaging reagents. Together, these data support a separate role for PARP2 in the regulation of telomeres maintained by telomerase-independent mechanisms. However, this phenotype could be cell type dependent. We are in the process of studying the possible role of PARP2 in the protection of eroded telomeres in mice.
Expression of a TRF2 dominant negative mutant in HeLa 1.2.11 cells abolished the PARP1-TRF2 interaction and partially disrupted PARP1's telomeric DNA localization. These data suggest that the PARP1-TRF2 interaction helps PARP1 to localize to sites of DNA strand breakages at telomeres. However, disrupting their interaction does not completely abolish PARP1's telomeric localization, indicating that PARP1 can localize to damaged telomeres through other means. PARP1 can bind the telomere-associated protein, TRF1 in vivo (Dantzer et al., unpublished data). In addition, PARP1 can directly bind telomeric DNA in vitro (Pion et al., 2003; Figure 4). Thus, upon DNA damage at telomeres, it is possible for PARP1 to bind telomeric DNA directly and/or interact with several telomeric associated proteins (e.g., TRF1, TRF2), which help recruit PARP1 to the sites of DNA strand breakages in telomeres.
Although the interaction between PARP1 and TRF2 is mediated by TRF2's myb domain, this domain is not required for poly(ADP-ribosyl)ation of TRF2, because the TRF2 deletion mutant containing only its dimerization domain is sufficient for PARP1 to modify. Thus, poly(ADP-ribosyl)ation of TRF2 may occur unaided by physical interaction between PARP1 and TRF2. Like PARP2, the PARP activity of PARP1 negatively affected TRF2's binding to telomeric DNA, as was evident by both the DNA bandshift assay and the immobilized dsTelomeric DNA-binding assay. This phenomenon suggests that a functional interaction between TRF2 and PARP1 will be of importance in instances in which the dissociation of TRF2 from telomeres is necessary, for example, at damaged telomeres (see below).
The Role of PARP1 at Eroded Telomeres as a Result of Gradual Loss of Telomeric DNA or DNA Strand Breakages
Uncapped telomeres generated by loss of telomeric repeats, associate with DNA damage response proteins (d'Adda di Fagagna et al., 2003; Hao et al., 2004; Tarsounas et al., 2004). Here we report that mTert–/– ES cells during prolonged growth in culture accumulate PARP1 at critically short telomeres, suggesting that uncapped telomeres are recognized as damaged DNA by PARP1. However, it has been reported that the absence of PARP1 does not increase the rate of telomere shortening or end-to-end fusions in response to gradual loss of telomeric DNA in telomerase-deficient MEFs (Espejel et al., 2004). This could be due to a gradual activation of PARP1 by the progressive loss of telomeric DNA in telomerase null mice may be complemented by other PARP family members and/or adaptive responses.
We have demonstrated that PARP1 localizes preferentially to damaged telomeres and loss of PARP1 function can result in telomere dysfunction (e.g., end-to-end fusions) in cells exposed to DNA-damaging reagents. Thus, telomeric DNA strand breaks may activate and recruit PARP1 to help repair the damaged telomeres by recruiting other DNA repair enzymes and/or by acting on the architecture of the damaged telomeres. Dantzer et al. (2004) previously reported that TRF2 copurified with XRCC1 (the scaffold protein that coordinates the enzymes in the BER pathway) and DNA polymerase β, implying they might participate in the repair of damaged telomeres. In mammals, telomeres normally exist in a t-loop structure stabilized by telomere repeat binding factors, especially TRF2 (Griffith et al., 1999; Baumann et al., 2002; Karlseder, 2003). The poly(ADP-ribosyl)ation of TRF2 upon DNA damage could favor the relaxation of the t-loop structure, and in turn, allow access of DNA damage response or repair machinery in the BER pathway (such as XRCC1 and DNA polymerase β) to the damaged telomeres. The synthesis of poly(ADP-ribose) at the damaged sites of telomeres may also trigger the immediate recruitment of these repair factors.
In summary, our data suggest that under genomic insult or telomere erosion conditions, PARP1 may be activated by and directly bind eroded telomeres or be recruited to eroded telomeres through its interaction with telomere-associated proteins, such as TRF2. Once there, PARP1 catalyzes the poly(ADP-ribosyl)ation of TRF2, disassociating it from telomeres and allowing access to the DNA damage repair machineries to repair eroded telomeres.
Supplementary Material
Acknowledgments
We sincerely thank Dr. Lea Harrington for her critical comments and many fruitful discussions during this study and for providing pcDNAFLAG-hTRF2 vector; Dr. Titia de Lange for kindly providing the rabbit anti-TRF2 antibody (508) and HeLa 1.2.11 cells, and Drs. Gilbert de Murcia, Katherine Friedman, and Hayes McDonald for critical reading of the manuscript. We acknowledge the support of the office of Biological and Environmental Research, U.S. Department of Energy under Contract DE-AC056-960R22464 with UT-Battelle LLC, the Laboratory of Directed Research and Development Program of Oak Ridge National Laboratory (3211-2049 and 3211-2050), managed by UT-Battelle, LLD, for the U.S. Department of Energy under contract No. DE-AC05-00OR22725, and Association pour la Recherche Contre le Cancer, Comité du Haut-Rhin de la Ligue Contre le Cancer, Electricité de France, Commissariat à l'Energie Atomique, Centre National de la Recherche Scientifique.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-07-0672) on January 25, 2006.
Abbreviations used: ES, embryonic stem; MEFs, mouse embryonic fibroblasts; PARP1, poly(ADP-ribose)polymerase 1; SFEs, telomere signal free ends; Tert, telomerase reverse transcriptase.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Ame, J. C., Schreiber, V., Fraulob, V., Dolle, P., de Murcia, G., and Niedergang, C. P. (2001). A bidirectional promoter connects the poly(ADP-ribose) polymerase 2 (PARP-2) gene to the gene for RNase P RNA structure and expression of the mouse PARP-2 gene. J. Biol. Chem. 276, 11092–11099. [DOI] [PubMed] [Google Scholar]
- Ame, J. C., Spenlehauer, C., and de Murcia, G. (2004). The PARP superfamily. Bioessays 26, 882–893. [DOI] [PubMed] [Google Scholar]
- Baumann, P., Podell, E., and Cech, T. R. (2002). Human Pot1 (protection of telomeres) protein: cytolocalization, gene structure, and alternative splicing. Mol. Cell Biol. 22, 8079–8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco, M. A., Lee, H. W., Hande, M. P., Samper, E., Lansdorp, P. M., DePinho, R. A., and Greider, C. W. (1997). Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34. [DOI] [PubMed] [Google Scholar]
- Bradshaw, P. S., Stavropoulos, D. J., and Meyn, M. S. (2005). Human telomeric protein TRF2 associates with genomic double-strand breaks as an early response to DNA damage. Nat. Genet. 37, 193–197. [DOI] [PubMed] [Google Scholar]
- Cao, Y., Li, H., Deb, S., and Liu, J. P. (2002). TERT regulates cell survival independent of telomerase enzymatic activity. Oncogene 21, 3130–3138. [DOI] [PubMed] [Google Scholar]
- Cho, H. P., Liu, Y., Gomez, M., Dunlap, J., Tyers, M., and Wang, Y. (2005). The dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol. Cell Biol. 25, 4541–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, B. D., Dynek, J. N., Chang, W., Shostak, G., and Smith, S. (2002). Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol. Cell Biol. 22, 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna, F., Reaper, P. M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N. P., and Jackson, S. P. (2003). A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198. [DOI] [PubMed] [Google Scholar]
- Dantzer, F., et al. (2004). Functional interaction between poly(ADP-Ribose) polymerase 2 (PARP-2) and TRF2, PARP activity negatively regulates TRF2. Mol. Cell Biol. 24, 1595–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer, F., Schreiber, V., Niedergang, C., Trucco, C., Flatter, E., De La Rubia, G., Oliver, J., Rolli, V., Menissier-de Murcia, J., and de Murcia, G. (1999). Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 81, 69–75. [DOI] [PubMed] [Google Scholar]
- de Murcia, J. M., et al. (1997). Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA 94, 7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel, S., Franco, S., Rodriguez-Perales, S., Bouffler, S. D., Cigudosa, J. C., and Blasco, M. A. (2002a). Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 21, 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel, S., Franco, S., Sgura, A., Gae, D., Bailey, S. M., Taccioli, G. E., and Blasco, M. A. (2002b). Functional interaction between DNA-PKcs and telomerase in telomere length maintenance. EMBO J. 21, 6275–6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel, S., Klatt, P., Menissier-de Murcia, J., Martin-Caballero, J., Flores, J. M., Taccioli, G., de Murcia, G., and Blasco, M. A. (2004). Impact of telomerase ablation on organismal viability, aging, and tumorigenesis in mice lacking the DNA repair proteins PARP-1, Ku86, or DNA-PKcs. J. Cell Biol. 167, 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider, C. W., and Blackburn, E. H. (1996). Telomeres, telomerase and cancer. Sci. Am. 274, 92–97. [DOI] [PubMed] [Google Scholar]
- Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H., and de Lange, T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503–514. [DOI] [PubMed] [Google Scholar]
- Hao, L. Y., Strong, M. A., and Greider, C. W. (2004). Phosphorylation of H2AX at short telomeres in T cells and fibroblasts. J. Biol. Chem. 279, 45148–45154. [DOI] [PubMed] [Google Scholar]
- Huber, A., Bai, P., de Murcia, J. M., and de Murcia, G. (2004). PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst) 3, 1103–1108. [DOI] [PubMed] [Google Scholar]
- Kaminker, P. G., Kim, S. H., Taylor, R. D., Zebarjadian, Y., Funk, W. D., Morin, G. B., Yaswen, P., and Campisi, J. (2001). TANK2, a new TRF1-associated poly(ADP-ribose) polymerase, causes rapid induction of cell death upon overexpression. J. Biol. Chem. 276, 35891–35899. [DOI] [PubMed] [Google Scholar]
- Karlseder, J. (2003). Telomere repeat binding factors: keeping the ends in check. Cancer Lett. 194, 189–197. [DOI] [PubMed] [Google Scholar]
- Karlseder, J., Hoke, K., Mirzoeva, O. K., Bakkenist, C., Kastan, M. B., Petrini, J. H., and de Lange, T. (2004). The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2, E240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder, J., Smogorzewska, A., and de Lange, T. (2002). Senescence induced by altered telomere state, not telomere loss. Science 295, 2446–2449. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Kha, H., Ungrin, M., Robinson, M. O., and Harrington, L. (2002). Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl. Acad. Sci. USA 99, 3597–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., et al. (2000). The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 10, 1459–1462. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Snow, B. E., Kickhoefer, V. A., Erdmann, N., Zhou, W., Wakeham, A., Gomez, M., Rome, L. H., and Harrington, L. (2004). Vault poly(ADP-ribose) polymerase is associated with mammalian telomerase and is dispensable for telomerase function and vault structure in vivo. Mol. Cell Biol. 24, 5314–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, M., Niedergang, C., Schreiber, V., Muller, S., Menissier-de Murcia, J., and de Murcia, G. (1998). XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell Biol. 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder, V. S., Boeglin, M., de Murcia, G., and Schreiber, V. (2005). PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J. Cell Sci. 118, 211–222. [DOI] [PubMed] [Google Scholar]
- Meeker, A. K., Gage, W. R., Hicks, J. L., Simon, I., Coffman, J. R., Platz, E. A., March, G. E., and De Marzo, A. M. (2002). Telomere length assessment in human archival tissues: combined telomere fluorescence in situ hybridization and immunostaining. Am. J. Pathol. 160, 1259–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pion, E., Bombarda, E., Stiegler, P., Ullmann, G. M., Mely, Y., de Murcia, G., and Gerard, D. (2003). Poly(ADP-ribose) polymerase-1 dimerizes at a 5′ recessed DNA end in vitro: a fluorescence study. Biochemistry 42, 12409–12417. [DOI] [PubMed] [Google Scholar]
- Rippmann, J. F., Damm, K., and Schnapp, A. (2002). Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J. Mol. Biol. 323, 217–224. [DOI] [PubMed] [Google Scholar]
- Samper, E., Goytisolo, F. A., Menissier-de Murcia, J., Gonzalez-Suarez, E., Cigudosa, J. C., de Murcia, G., and Blasco, M. A. (2001). Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerase-deficient mice and primary cells despite increased chromosomal instability. J. Cell Biol. 154, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbodio, J. I., Lodish, H. F., and Chi, N. W. (2002). Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1) and IRAP (insulin-responsive aminopeptidase). Biochem. J. 361, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber, V., Ame, J. C., Dolle, P., Schultz, I., Rinaldi, B., Fraulob, V., Menissier-de Murcia, J., and de Murcia, G. (2002). Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277, 23028–23036. [DOI] [PubMed] [Google Scholar]
- Smith, S., and de Lange, T. (1999). Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112 (Pt 21), 3649–3656. [DOI] [PubMed] [Google Scholar]
- Smith, S., and de Lange, T. (2000). Tankyrase promotes telomere elongation in human cells. Curr. Biol. 10, 1299–1302. [DOI] [PubMed] [Google Scholar]
- Smith, S., Giriat, I., Schmitt, A., and de Lange, T. (1998). Tankyrase, a poly-(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- Takai, H., Smogorzewska, A., and de Lange, T. (2003). DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., Mendonca, M. S., Bradshaw, P. S., Hoelz, D. J., Malkas, L. H., Meyn, M. S., and Gilley, D. (2005). DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proc. Natl. Acad. Sci. USA 102, 15539–15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas, M., Munoz, P., Claas, A., Smiraldo, P. G., Pittman, D. L., Blasco, M. A., and West, S. C. (2004). Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117, 337–347. [DOI] [PubMed] [Google Scholar]
- Trucco, C., Oliver, F. J., de Murcia, G., and Menissier-de Murcia, J. (1998). DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 26, 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel, B., Smogorzewska, A., and de Lange, T. (1998). TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Erdmann, N., Giannone, R. J., Wu, J., Gomez, M., and Liu, Y. (2005). An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc. Natl. Acad. Sci. USA 102, 10256–10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans, J. M., Martens, U. M., Poon, S. S., Raap, A. K., Tanke, H. J., Ward, R. K., and Lansdorp, P. M. (1997). Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. USA 94, 7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.