Abstract
We have reported that extracts prepared from many human and mouse cell lines show little or no Sp2 DNA-binding activity and that Sp2 has little or no capacity to stimulate transcription of promoters that are activated by Sp1, Sp3, and Sp4. Using an array of chimeric Sp1/Sp2 proteins we showed further that Sp2 DNA-binding activity and trans-activation are each negatively regulated in mammalian cells. As part of an ongoing effort to study Sp2 function and regulation we characterized its subcellular localization in comparison with other Sp-family members in fixed and live cells. We report that 1) Sp2 localizes largely within subnuclear foci associated with the nuclear matrix, and 2) these foci are distinct from promyelocytic oncogenic domains and appear to be stable during an 18-h time course of observation. Deletion analyses identified a 37 amino acid sequence spanning the first zinc-“finger” that is sufficient to direct nuclear matrix association, and this region also encodes a bipartite nuclear localization sequence. A second nuclear matrix targeting sequence is encoded within the Sp2 trans-activation domain. We conclude that Sp2 preferentially associates with the nuclear matrix and speculate that this subcellular localization plays an important role in the regulation of Sp2 function.
INTRODUCTION
Sp/XKLF proteins are sequence-specific DNA-binding proteins that share a highly conserved, carboxy-terminal DNA-binding domain. This domain consists of three zinc-“fingers” of the Cys2-His2 class required for sequence-specific interactions with GC-rich promoter elements. Based on structural and sequence similarities, Sp proteins comprise one subfamily of Sp/XKLF proteins for which nine members (Sp1–Sp9) have been described. Sp1, Sp3, and Sp4 are the best characterized members of this subfamily, and each has been shown to regulate transcription of a constellation of genes including those governing cell cycle control, oncogenesis, and differentiation (Phillipsen and Suske, 1999; Suske, 1999; Black et al., 2001; Kaczynski et al., 2003; Chu and Ferro, 2005; Suske et al., 2005; Zhao and Meng, 2005). Sp proteins carry an amino-terminal trans-activation domain that has been divided into three subdomains, termed A, B, and C, based on their structural, biochemical, and functional properties. Domains A and B contain alternating serine/threonine- and glutamine-rich regions, whereas the C domain is characterized by a high proportion of charged amino acids. The glutamine-rich portion of each subdomain is believed to be required for trans-activation, whereas the function(s) of the serine/threonine-rich regions are less well understood. Although Sp proteins share structural and sequence similarities and each is widely expressed in mammalian cells, the often marked phenotypes of nullizygous animals suggest that each Sp protein is responsible for a unique set of cellular functions that cannot be supplanted by other members of the family (Supp et al., 1996; Marin et al., 1997; Bouwman et al., 2000; Harrison et al., 2000; Göllner et al., 2001a, 2001b; Nakashima et al., 2002; Bell et al., 2003).
Sp2 is a widely expressed protein that carries the least conserved DNA-binding domain among Sp-family members (Kingsley and Winoto, 1992; Phillipsen and Suske, 1999; Suske, 1999). We have reported that Sp2 binds poorly to a subset of DNA sequences bound by other family members and has little or no capacity to stimulate transcription of promoters that are potently activated by Sp1, Sp3, and Sp4 (Moorefield et al., 2004; Simmons and Horowitz, 2005). Using recombinant Sp2 protein prepared in insect cells and a PCR-assisted oligonucleotide selection protocol (“CASTing”), we established the consensus Sp2 DNA-binding sequence to be 5′-GGGCGGGAC-3′ and reported that recombinant Sp2 binds this sequence with high affinity in vitro (Moorefield et al., 2004). Yet, when consensus Sp2 DNA-binding sequences were incorporated within a well-characterized Sp-responsive promoter, dihydrofolate reductase (DHFR), only a modest increase in Sp2-mediated transcription was noted in transiently transfected mammalian cells. This modest increase in Sp2-mediated transcription remained 10–20-fold less than that induced by Sp1 or Sp3 in parallel assays. Using an array of chimeric proteins created by swapping analogous portions of Sp2 with those of Sp1, we showed further that Sp2 DNA-binding activity and trans-activation are each negatively regulated in mammalian cells (Moorefield et al., 2004). Indeed, surveying a wide variety of human and rodent cells, we were unable to detect endogenous Sp2 DNA-binding activity within nuclear extracts prepared from cells that express abundant amounts of Sp2. This latter finding stands in marked contrast with other Sp/XKLF-family members analyzed to date. Interestingly, a recent immunohistochemical analysis of primary human prostate tumors has indicated that Sp2 abundance is increased in late-stage tumors and further analyses have suggested that Sp2 overexpression may be oncogenic (Phan et al., 2004).
As part of an ongoing effort to study Sp2 function and regulation we characterized its subcellular localization in comparison with other Sp-family members in fixed and live cells. Herein we report that Sp2 is localized within discrete nuclear foci that are distinct from promyelocytic (PML) oncogenic domains (PODs) and that are associated with the nuclear matrix. Indeed, we report that the vast majority of endogenous and ectopically expressed Sp2 localizes to the nuclear matrix. Using time-lapse confocal microscopy we show that Sp2 nuclear foci are stable and virtually immobile during an 18-h time course of observation. In contrast to these results we report that 1) Sp3 is distributed diffusely throughout the nucleus and only a minority of Sp3 or Sp4 is associated with the nuclear matrix, and 2) the association of Sp1 with the nuclear matrix appears to be somewhat less frequent and tenacious than that of Sp2. To determine the portion of Sp2 required for nuclear matrix association, we examined the subcellular localization of a panel of partial Sp2-fusion proteins and in so doing identified a 37 amino acid sequence (amino acids 513–549) spanning the first zinc-finger and inter-finger linker of the Sp2 DNA-binding domain as being sufficient to direct nuclear matrix association. This portion of the Sp2 DNA-binding domain is the most divergent among Sp-family members perhaps accounting, at least in part, for its relatively unusual subnuclear distribution. We also report that a subset of this 37 amino acid region encodes a bipartite nuclear localization sequence and that an independent nuclear matrix targeting sequence (NMTS) is carried within the Sp2 trans-activation domain. We conclude from these studies that Sp2 preferentially associates with the nuclear matrix and speculate that this association may play an important role in the regulation of Sp2-mediated transcription.
MATERIALS AND METHODS
Cells and Antisera
HeLa and COS-1 cells were obtained from the Duke Comprehensive Cancer Center cell culture facility (Duke University Medical Center, Durham, NC) and cultured in DMEM (Invitrogen, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal bovine serum and 50 μg/ml Pipracil in humidified incubators under 5% CO2. Affinity-purified rabbit (sc-643, anti-Sp2 [K-20]; sc-20682, anti-Lamin B1 [H-90]; sc-805, anti-HA [Y-11]), mouse (sc-5621, anti-PML [PG-M3]), and goat antisera (sc-18555, anti-NuMA [N-20]) were obtained from Santa Cruz Biotechnology, (Santa Cruz, CA). Secondary antibodies (Alexa Fluor 488 goat anti-mouse, Alexa Fluor 594 goat anti-rabbit and rabbit anti-goat) were obtained from Molecular Probes (Eugene, OR).
Expression Constructs
The construction and properties of a mammalian expression construct carrying a full-length, HA-tagged human Sp2 cDNA have been described (pCMV4-Sp2/flu; Moorefield et al., 2004). An expression construct carrying an enhanced yellow fluorescent protein (EYFP)-Sp2 fusion was generated via the PCR using pCMV4-Sp2/flu as template and the following primers: 5′-GGGGGTACCGCCACCATGAGCGATCCACAGACCAGCATGGCTGCC-3′, and 5′-CCCGGATCCCTAGCTAGCGTAATCTGGAACATCGTATGGGTACAAGTTCTTCGTGACC-3′. The amplified cDNA was subsequently cloned at the KpnI and BamHI sites of pEYFP-C1 (Clontech, Mountain View, CA), creating pEYFP-Sp2. Deletion mutants were prepared via the PCR using pEYFP-Sp2 as template and Pfx platinum DNA polymerase (Invitrogen). Amino acids carried by each deletion mutant are as follows (amino acid numbers are correspond to GenBank accession number D28588): pEYFP-Sp2ΔA (amino acids 189–606), pEYFP-Sp2ΔAB (amino acids 390–606), pEYFP-Sp2ΔABC (amino acids 513–606), pEYFP-Sp2ΔZnIII (amino acids 1–582), and pEYFP-Sp2ΔZnII/III (amino acids 1–549). Primers used to create each deletion mutant were as follows: Sp2ΔA, 5′-GGGGGGTACCTGCGACTGCAGAATTCGAAGCTT G-3′, and 5′-GGGGGGTACCAGCGGGGCCAATGTGGTGAAGTTGACAGG-3′; Sp2ΔAB, 5′-GGGGGGTACCTGCGACTGCAGAATTCGAAGCTTG-3′, and 5′-GGGGGGTACCACCAGCAAAAAGCACTCAGCTGCAATTCTC-3′; Sp2ΔABC, 5′-GGGGGGTACCTGCGACTGCAGAATTCGAAGCTTG-3′, and 5′-GGGGGGTACCCAGGGCAAGAAGAAGCACGTTTGCCACATC-3′; Sp2ΔZnIII, 5′-CCCGCGGCCGCATACCCATACGATGTTCCAGATTACGCTAGCTAG-3′, and 5′-GGGGCGGCCGCCTGGGCGCACTCGAAGCGTTTGTCCCC-3′; Sp2ΔZnII/III, 5′-CCCGCGGCCGCATACCCATACGATGTTCCAGATTACGCTAGCTAG-3′, and 5′-GGGGCGGCCGCGACAAAGGGCCGCTCGCCAGTGTGC-3′. Deletion mutants Sp2ΔA, Sp2ΔAB, and Sp2ΔABC were digested with KpnI, Sp2ΔZnIII and Sp2ΔZnII/III were digested with NotI, and each was self-ligated. pEYFP-Sp2ZnI, (amino acids 520–549), was created via the PCR using the following primers 5′-GGGCTCGAGTGCCACATCCCCGACTGTGGCAAGACGTTCCG-3′, and 5′-CCCCCCGGGGACAAAGGGCCGCTCGCCAGTGTGCAGGCG-3′. The amplified cDNA fragment was subsequently cloned at the XmaI and XhoI sites of pEYFP-C1, generating pEYFP-Sp2ZnI. pEGFP-Sp2NLS-ZnI (amino acids 513–549) was created via the PCR using the following oligonucleotides 5′-GGGAGATCTCAGGGCAAGAAGAAGCACGTT-3′ and 5′-GGGGAATTCGACAAAGGGCCGCTCGCCAG-3′. The amplified cDNA fragment was subsequently cloned at the BglII and EcoRI sites of pEGFP-C1 (Clontech), generating pEGFP-Sp2NLS-ZnI. pEGFP-Sp2NLS (amino acids 513–519) was created via the PCR using the following primers 5′-AACGTGCTTCTTCTTGCCCTGA-3′ and 5′-GAATTCTGCAGTCGACGGTACCGCGG-3′. The amplified DNA was self-ligated, creating pEYFP-Sp2NLS. pEYFP-Sp2ABC (amino acids 1–519) was created via the PCR using the following primers 5′-GGGGGATCCACCGGATCTAGATAACTG-3′ and 5′-GGGGGATCCAACGTGCTTCTTCTTGCCC-3′. The amplified DNA was cleaved with BamHI and self-ligated, creating pEYFP-Sp2ABC. An EYFP-Sp1 expression construct was generated via the PCR using pCMV4-Sp1/flu (Udvadia et al., 1993) as template and the following primers: 5′-GGGCTCGAGGGATGGATGAAATGACAGCTGTGGTG-3′ and 5′-GGGGGATCCGCGCTAGCTAGCGTAATCTGGAAC-3′. The amplified cDNA fragment was subsequently cloned at the BamHI and XhoI sites of pEYFP-C1, creating pEYFP-Sp1. An EGFP-Sp3 expression construct was generated via the PCR using pCMV4-Sp3/flu (Kennett et al., 1997) as template and the following primers: 5′-GGGCTCGAGCCATGAACTCCGGGCCATCGCCGGG-3′ and 5′-GGGGAATTCCTAGCTAGCGTAATCTGGAACATCGTATGGGTACTC-3′. The amplified cDNA fragment was subsequently cloned at the EcoRI and XhoI sites of pEGFP-C1, creating pEGFP-Sp3. A plasmid carrying a human Sp4 cDNA, pBS-Sp4, was a generous gift from Dr. Richard Tsika (University of Missouri-Columbia, Columbia, MO). An HA-tagged Sp4 expression vector was created via the PCR using Deep Vent polymerase (New England Biolabs, Beverly, MA), pBS-Sp4 as template, and the following primers 5′-CAGATCTATGTATCCTTACGATGTGCCAGACTACGCTTCAGCAAAGATGAGCGATCAGAAGAAGGAGGAGG-3′ and 5′-GGTCAGAATTCTTCCATGTTGGTTGAAACATTGGG-3′. The amplified cDNA was subcloned in pCR-Blunt-II-Topo (Invitrogen), excised with XbaI and HindIII, and subcloned in pCMV4 (Andersson et al., 1989). The integrity of all expression constructs was confirmed by dideoxy-sequencing using Sequenase version 2.0 DNA polymerase (Amersham Life Science, Arlington Heights, IL). pECFP-PML and pECFP-Sp100 expression constructs were a generous gift of Dr. Roeland W. Dirks (Leiden University Medical Center, Leiden, The Netherlands) and have been described (Wiesmeijer et al., 2002).
In Situ Nuclear Matrix Preparation and Analysis
Proteins were localized in situ by direct and indirect immunofluorescence following the solubilization of nuclei and preparation of nuclear matrices essentially as described (Javed et al., 2000). Briefly, 2 × 105 COS-1 cells were plated on sterile glass coverslips in six-well plates and cultured at 37°C overnight. The following day cells were transfected with expression vectors (2 μg per well) using SuperFect transfection reagent (Qiagen, Valencia, CA) per the manufacturer's instructions and cultured for 24–48 h at 37°C. Plates were placed on ice and washed twice with ice-cold phosphate-buffered saline (PBS), and cells were solubilized in CSK buffer (100 mM NaCl, 10 mM PIPES, pH 6.8, 3 mM MgCl2, 1 mM EGTA, and 0.5% Triton X-100). After removal of CSK buffer, chromatin was digested at 30°C for 60 min via incubation of nuclei in digestion buffer (50 mM NaCl, 10 mM PIPES, pH 6.8, 3 mM MgCl2, 1 mM EGTA, and 0.5% Triton X-100). After removal of digestion buffer, nuclei were incubated for 10 min on ice in stop solution (digestion buffer containing 250 mM NH4SO4). Nuclei were subsequently fixed with 2% paraformaldehyde at room temperature for 15 min, washed with PBS, and stained with DAPI (4′,6-diamidino-2-phenylindole) in 0.5% Triton X-100/PBS. Coverslips were washed with PBS and then dH2O and mounted using Vectashield mounting media (Vector Labs, Burlingame, CA). Fluorescence was imaged using a Nikon TE-200 inverted epifluorescence microscope (Melville, NY) equipped with appropriate optics and filter blocks at a magnification of 100× under oil immersion. Results were recorded with a digital camera (SPOT, Jr.: Diagnostics Instruments, Sterling Heights, MI) and proprietary software using the manufacturer's instructions. Nuclear matrices prepared from mock-transfected cells were analyzed in parallel. Indirect immunofluorescence experiments performed on fixed, whole nuclei were performed as described (Moorefield et al., 2004; Spengler et al., 2005).
Time-Lapse Confocal Microscopy
For confocal microscopy 2–4 × 104 COS-1 cells were plated in 35-mm uncoated glass-bottom dishes (MatTek, Ashland, MA) and cultured overnight at 37°C. The following day cells were transfected with 3 μg of pEYFP-Sp2 using SuperFect transfection reagent (Qiagen) and cultured overnight at 37°C. Plates were transferred to a humidified microscope chamber (INC-2000 Incubator System; 20/20 Technologies, Wilmington, NC) heated to 37°C, and supplemented with 5% CO2. Nuclei were imaged using a Nikon Eclipse TE-2000E microscope configured with a 40× oil immersion lens (1.4 NA), YFP BP HYQ filter cube with a 535-nm bandpass filter, and C1 confocal workstation administered by EZ-C1 2.30 confocal laser scanning software. EYFP-positive cells were identified via wide-field epifluorescence and subsequently scanned into 35 Z-sections with an argon ion laser at 488 nm every 10 min for 18 h. Volume rendering software was applied to generate a composite image of all 35 Z-sections at each time point and then combined to generate a time-lapse video of the collected data.
Protein/DNA-binding Assays
Nuclear extracts were prepared from transfected and mock-transfected cells, and protein/DNA-binding (“gel-shift”) assays were performed precisely as described (Moorefield et al., 2004).
Nuclear Matrix Fractionation and Western Blotting
Cells were harvested and separated into cellular compartments essentially as described (Zaidi et al., 2001). Briefly, whole cell and fractionated cell extracts were prepared from COS-1 cells cultured on 100-mm dishes 48 h after transfection. For whole-cell lysates, cells were harvested in 300 μl Laemmli buffer (2% SDS, 2 M urea, 10% glycerol, 10 mM Tris-HCl, pH 6.8, 0.002% bromophenol blue, 10 mM DTT). Samples were immediately boiled, clarified, and stored at –80°C. For subcellular fractionation, cells were collected in ice-cold 1× PBS containing 1× Complete protease inhibitors (Roche, Indianapolis, IN). Cell pellets were resuspended in 300 μl CSK buffer containing 0.3 M sucrose, 1.6 mM ribonucleoside-vanadyl complex (New England Biolabs), and 1.2 mM AEBSF (4-(2-aminoethyl) benzenesulfonyl fluoride; Fisher Scientific, Suwanee, GA) for 10 min on ice. Nuclei were collected by centrifugation and supernatants containing soluble proteins (designated the soluble fraction) were stored at –80°C. Nuclei were subsequently extracted with 300 μl Digestion Buffer containing 400 U per ml RNase-free DNase I (Roche) for 30 min at room temperature. Digestion reactions were raised to 250 mM NH4SO4 using a 2 M stock solution and incubated for a further 5 min at room temperature. Extracted nuclei were centrifuged to separate chromatin-associated nuclear proteins from insoluble nuclear matrix (designated the nuclear matrix fraction), and each was boiled in 300 μl of Laemmli buffer. Equal volumes of each fraction were resolved on denaturing 8% polyacrylamide gels, and transferred to PVDF membranes (Millipore, Billerica, MA) using a semidry transfer apparatus. Membranes were blocked with 3% fat-free dried milk supplemented with 0.1% Tween-20, incubated with primary and secondary antisera (anti-Sp2 [K-20] or anti-HA [Y-11]; Santa Cruz Biotechnology), and antigen-antibody complexes were detected using a proprietary chemiluminescence kit according to the manufacturer's instructions (ECL; Amersham Pharmacia Biotech, Piscataway, NJ). Fractionated cell extracts prepared from transfected cells were obtained as follows. One day before transfection, 2 × 106 COS-1 cells were plated on 100-mm tissue culture dishes and incubated at 37°C. Each dish was transfected with 10 μg of a given Sp-expression vector using SuperFect transfection reagent (Qiagen) and cultured for 48 h at 37°C. Fractionated cell extracts prepared from transfected cells were obtained precisely as described above.
RESULTS
Sp-Family Members Differ in Their Subnuclear Localization
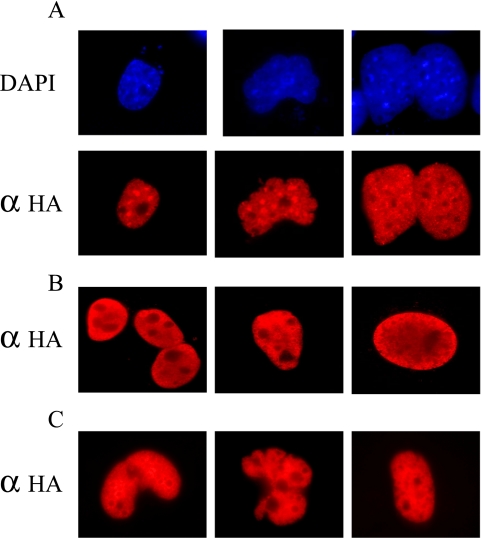
To compare the subcellular localization of Sp-family members, COS-1 cells were transiently transfected with expression vectors carrying human Sp1, Sp2, or Sp3 cDNAs linked to a carboxy-terminal epitope-tag derived from Influenza hemagglutinin. Western blotting indicated that equivalent amounts of ectopically expressed Sp proteins were produced in transfected cells (unpublished data and Moorefield et al., 2004). Transiently transfected cells were fixed with paraformaldehyde, challenged with a polyclonal rabbit anti-HA antiserum, and analyzed by indirect immunofluorescence 48 h after transfection. As expected all three Sp-family members localized exclusively to nonnucleolar portions of interphase nuclei (Figure 1). Yet, upon close inspection Sp protein-specific differences in nuclear localization were also noted. As shown in Figure 1A, in addition to generalized staining throughout transfected nuclei a significant fraction of ectopically expressed Sp2 appeared as punctate nuclear deposits resulting in nuclei with an overall “mottled” appearance. Ectopic expression of Sp1 led to a heterogeneous staining pattern that included small punctate deposits as well as nuclei featuring perinuclear and uniformly stained areas (Figure 1B). In contrast to these results, nuclei of Sp3-transfected cells revealed homogeneous, nonnucleolar staining (Figure 1C).
Figure 1.
Subcellular localization of Sp1, Sp2, and Sp3 in transiently transfected COS-1 cells. (A) COS-1 cells were transiently transfected with an expression vector encoding full-length, epitope-tagged human Sp2 and expressed protein was detected by indirect immunofluorescence using an anti-HA polyclonal antibody (αHA; Y-11) and Alexa Fluor 594 goat anti-rabbit secondary antibody. Nuclei were localized via DNA-staining (DAPI). (B and C) COS-1 cells expressing full-length, epitope-tagged human Sp1 (B) or Sp3 (C) were visualized as in A. Several representative nuclei are shown for cells transfected with each expression construct.
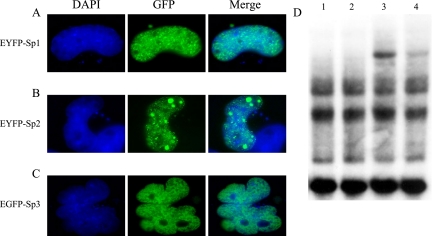
To confirm the immunolocalization results reported thus far and to ensure that these results were not artifacts of fixation, we prepared a series of fluorescent fusion proteins whose subcellular distribution could be evaluated in live cells. Human Sp1, Sp2, or Sp3 cDNAs were subcloned “in-frame” downstream of the coding regions of variants of green fluorescent protein (GFP) creating pEYFP-Sp1, pEYFP-Sp2, and pEGFP-Sp3, respectively. Each of these expression constructs were transiently transfected into COS-1 cells and the subnuclear distribution of resulting fusion proteins were examined in fixed and live cells. Consistent with results obtained by indirect immunofluorescence, each GFP-fusion protein exhibited a characteristic protein-specific subnuclear localization pattern in cells fixed with paraformaldehyde (Figure 2, A–C). In addition to diffuse staining throughout nonnucleolar portions of nuclei, the vast majority of cells expressing EYFP-Sp2 carried multiple punctate deposits of variable size (Figure 2B). EYFP-Sp1–transfected cells yielded numerous, relatively small punctate deposits as well as diffusely stained nuclei (Figure 2A), whereas nuclei of cells expressing EGFP-Sp3 were homogeneously stained (Figure 2C). Identical patterns of nuclear staining were also noted in paraformaldehyde-fixed HeLa and L929 cells as well as in live HeLa and COS-1 cells transfected with each expression construct (unpublished data and see Figure 4). This latter result indicates that Sp protein-specific staining patterns are not simply an artifact of cell fixation.
Figure 2.
Subcellular localization of EYFP-Sp1, EYFP-Sp2, and EGFP-Sp3, and characterization of EYFP-Sp2 DNA-binding activity. (A) COS-1 cells were transiently transfected with pEYFP-Sp1, EYFP-positive cells were identified via direct fluorescence microscopy (GFP), nuclei were localized via DNA staining (DAPI), and merged images are also shown (Merge). (B and C) COS-1 cells expressing EYFP-Sp2 (B), or EGFP-Sp3 (C) were analyzed as in A. (D) Whole cell extracts were prepared from mock-transfected COS-1 cells (lanes 1 and 2) as well as COS-1 cells transiently transfected with pEYFP-Sp2 (lanes 3 and 4), and extracts were incubated with a radiolabeled DHFR1* probe. Resulting protein/DNA complexes were resolved on a nondenaturing acrylamide gel. To confirm the identity of EYFP-Sp2/DNA complexes, anti-Sp2 antiserum (K-20) was added to lanes 2 and 4. An EYFP-Sp2/DNA complex is indicated by a closed arrowhead.
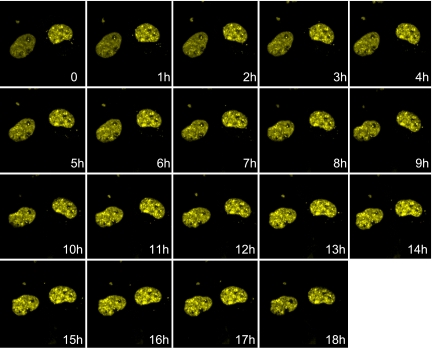
Figure 4.
Time-lapse confocal microscopy of EYFP-Sp2–expressing nuclei. COS-1 cells were transiently transfected with pEYFP-Sp2 and incubated overnight before analysis by confocal microscopy. Time-lapse images of 35 Z-sections were collected every 10 min for 18 h, and volume rendering software was used to derive a single composite image at each hourly time point.
Before performing additional subcellular localization experiments with GFP-fusion proteins, we wanted to determine if the fusion of GFP to the amino-termini of Sp proteins compromises their capacity to bind DNA. Thus, COS-1 cells were transiently transfected with an EYFP-Sp2 expression vector, nondenatured cell extracts were prepared, and these and control extracts were subsequently incubated with a radiolabeled oligonucleotide carrying a consensus Sp2-binding site (Moorefield et al., 2004). Protein/DNA-binding (“gel-shift”) reactions containing an extract prepared from EYFP-Sp2–expressing cells (lanes 3 and 4, Figure 2D) resulted in a modest amount of a novel protein/DNA complex not obtained in parallel reactions prepared with extracts from cells expressing EGFP alone (lanes 1 and 2, Figure 2D). To confirm that this novel protein/DNA complex results from the binding of EYFP-Sp2 to DNA, protein/DNA-binding assays were challenged with an Sp2-specific antibody. As expected, inclusion of anti-Sp2 antiserum led to a marked reduction in the abundance of this novel protein/DNA complex (lane 4, Figure 2D). Also as expected, inclusion of anti-Sp2 antiserum in control reactions did not lead to the depletion of protein/DNA complexes obtained in EGFP-expressing cells (lane 2, Figure 2D; Moorefield et al., 2004). We conclude from protein/DNA-binding assays that, at least in vitro, EYFP-Sp2 is competent to bind its cognate DNA-binding sequence. On the basis of the direct and indirect immunofluorescence data presented thus far, we also conclude that Sp1, Sp2, and Sp3 differ in their subnuclear distribution patterns. We presume that the distinct localization patterns exhibited by each Sp-family member reflect intrinsic functional differences and/or the influence of regulatory pathways governing their subnuclear localization.
Sp2 Subnuclear Foci Are Not Localized within PODs
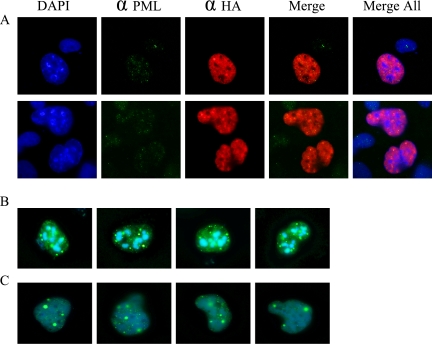
Given the unique distribution of Sp2 within nuclei, we reasoned that Sp2 might localize to nuclear subdomains termed PODs, nuclear domain 10 (ND10), or Kr-bodies (Maul et al., 1995, 2000; Doucas and Evans, 1996; Stein et al., 2000; Zhong et al., 2000; Negorev and Maul, 2001; Borden, 2002; Kießlich et al., 2002). PODs are subnuclear structures implicated in the regulation of cellular processes such as transcription and the response to DNA damage, and constituents such as PML and Sp100 appear as discrete nuclear foci or “speckles” in direct and indirect immunofluorescence studies. PML has also been shown to bind and negatively regulate Sp1-mediated trans-activation (Vallian et al., 1998). To determine if Sp2 is a POD constituent, two experiments were performed. First, COS-1 cells were transiently transfected with an epitope-tagged Sp2 expression vector and transfected cells were stained with anti-HA and anti-PML antibodies. Second, COS-1 cells were transfected with pEYFP-Sp2 and pECFP-PML or pEYFP-Sp2 and pECFP-Sp100 expression vectors, and the subnuclear distribution of ectopically expressed proteins was determined by direct immunofluorescence. Although it was difficult to detect endogenous PML in COS-1 cells, discrete subnuclear speckles were noted and superimposition of anti-HA and anti-PML images indicated that Sp2 and PML do not colocalize (Figure 3A). This conclusion was confirmed in transiently transfected cells receiving pEYFP-Sp2 and pECFP-PML or pECFP-Sp100. Foci of EYFP-Sp2 and ECFP-PML (Figure 3B) or EYFP-Sp2 and ECFP-Sp100 (Figure 3C) proteins localized to distinct nuclear subdomains in the vast majority of transfected cells analyzed. We conclude from these results that Sp2 nuclear foci do not colocalize with PODs and that the coexpression of Sp2 and PML or Sp100 does not recruit Sp2 to PODs.
Figure 3.
Sp2 subnuclear foci do not colocalize with PML or Sp100 within promyelocytic oncogenic domains. (A) COS-1 cells were transiently transfected with a mammalian expression vector (pCMV4-Sp2/flu) encoding a full length, epitope-tagged human Sp2 cDNA. Ectopically expressed Sp2 protein was detected by indirect immunofluorescence using an anti-HA polyclonal antiserum (αHA; Y-11) and Alexa Fluor 594 goat anti-rabbit secondary antibody, and endogenous PML was detected with anti-PML polyclonal antiserum (αPML; PG-M3) and Alexa Fluor 488 goat anti-mouse secondary antibody. Nuclei were detected by DNA-staining (DAPI). Superimposed images are as follows: anti-HA and anti-PML images (Merge) and anti-Sp2, anti-PML, and DAPI images (Merge All). (B and C) COS-1 cells were transiently cotransfected with expression vectors encoding (B) EYFP-Sp2 and ECFP-PML, or (C) EYFP-Sp2 and ECFP-Sp100, and ectopically expressed proteins were detected in situ. Several representative nuclei are shown for cells transfected with each pair of expression constructs.
Sp2 Nuclear Foci Are Relatively Stable and Immobile in Interphase Cells
The data reported thus far indicate that Sp2 localizes to discrete, subnuclear foci within interphase nuclei and that these foci are distinct from PODs. We reasoned that the localization of Sp2 within subnuclear domains might indicate that Sp2 performs its role as a regulator of gene expression within regional centers of transcriptional activity. We reasoned further that the location of such transcriptional centers might be dynamic, perhaps varying in size and/or position as a function of cell cycle progression. To monitor the subnuclear distribution of Sp2 in real time, we ectopically expressed EYFP-Sp2 in COS-1 cells and used time-lapse confocal microscopy to collect fluorescent images in 35 Z-sections every 10 min during an 18-h time course of observation. Although the movements of cells and whole nuclei were apparent during the 18 h of observation, few if any alterations were noted in the distribution or intensity of EYFP-Sp2 foci (Figure 4 and see movie in Supplementary Material). We conclude from these results that Sp2 nuclear foci are relatively stable and immobile in interphase cells.
Sp2 Is Associated with the Nuclear Matrix
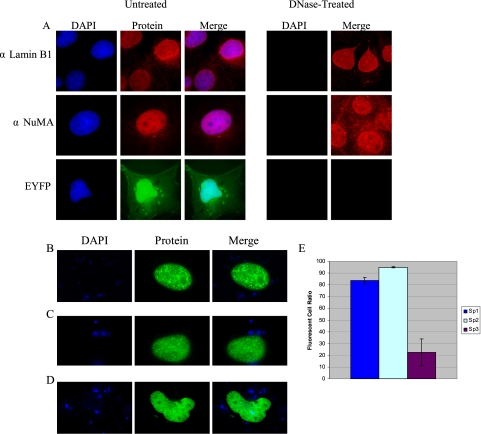
Given that Sp2 nuclear foci appeared to be stable and immobile in vivo, it became of interest to determine whether their subnuclear distribution reflected the association of Sp2 with chromatin or with components of the nuclear matrix. The latter possibility was particularly intriguing as a variety of transcription factors, including Sp1, have been reported to associate with the nuclear matrix (van Wijnen et al., 1993; Zeng et al., 1997; Javed et al., 2000; Parker et al., 2000; Zaidi et al., 2001; Dreuillet et al., 2002; Mattout-Drubezki and Gruenbaum, 2003; Djabali and Christiano, 2004; Seo et al., 2005). As a first step we verified that we could extract chromatin from nuclei in situ and identify previously characterized nuclear matrix-associated proteins. COS-1 cells cultured on glass coverslips were solubilized with Triton X-100, chromatin was removed by digestion with DNase I and extraction with 2 M ammonium sulfate, and resulting nuclear matrices were fixed with paraformaldehyde. Next, two well-characterized components of the nuclear matrix, Lamin B1 and NuMa, were visualized using polyclonal antisera and indirect immunofluorescence. As shown in Figure 5A, anti-Lamin B1 and anti-NuMA antibodies detected their respective antigens within the nuclei of untreated COS-1 cells as well as nuclei treated with DNase I. In contrast to these results, a protein that does not associate with the nuclear matrix, EYFP, was noted in all cell compartments of transfected COS-1 cells but was not detected within DNase I–treated nuclei prepared from transfected cells (Figure 5A).
Figure 5.
Detection of nuclear matrix-associated proteins in situ using direct and indirect fluorescence microscopy. (A) Endogenous lamins (αLamin B1; H-90) and NuMA (αNuMA; N-20) were detected in untreated COS-1 nuclei as well as DNase-treated nuclear matrices via indirect immunofluorescence using Alexa Fluor 594 goat anti-rabbit and Alexa Fluor 594 rabbit anti-goat secondary antibodies, respectively. EYFP was detected in untreated nuclei via direct fluorescence. Nuclei were localized via DNA-staining (DAPI), and superimposed images are also shown (Merge). (B–D) COS-1 cells were transfected with (B) pEYFP-Sp2, (C) pEYFP-Sp1, or (D) pEGFP-Sp3, nuclear matrices were prepared with DNase I, and labeled nuclei were identified in situ. (E) COS-1 cells were transfected with expression vectors encoding pEYFP-Sp1, pEYFP-Sp2, or pEGFP-Sp3, and the total numbers of fluorescent nuclei were enumerated in untreated cultures and in nuclear matrices prepared from cultures treated with DNase I. The graph represents the ratio of total fluorescent cells recovered in DNase I–treated cultures relative to the total number detected in untreated, transfected cultures. Each transfection experiment was repeated in triplicate to account for plate-to-plate variations in transfection efficiency. Error bars, SDs.
To determine whether Sp2 foci are associated with the nuclear matrix, COS-1 cells were transfected with pEYFP-Sp2 and nuclear matrices were prepared and analyzed by direct fluorescence microscopy. For comparison, parallel cell cultures were transfected with pEYFP-Sp1 or pEGFP-Sp3 expression vectors and analyzed similarly. As shown in Figure 5, B–D, each Sp-fusion protein was found in association with the nuclear matrix. However, significant differences in the numbers of resulting fluorescent nuclei were noted. To quantitate the efficiency with which each Sp-family member associates with the nuclear matrix, parallel cultures of COS-1 cells were transfected with expression vectors encoding each Sp-fusion protein and the total numbers of fluorescent nuclei were enumerated in cultures that were or were not treated with DNase I. This experiment was repeated at least three times for each Sp-family member to account for plate-to-plate differences in transfection efficiency, and the ratio of fluorescent cells recovered in DNase-treated cultures relative to those recovered in untreated cultures is plotted in Figure 5E. Consistent with the notion that Sp2 associates efficiently with components of the nuclear matrix, the vast majority (ca. 95%) of cells expressing EYFP-Sp2 gave rise to EYFP-Sp2–positive nuclear matrices. Sp1 appeared to be somewhat less efficient, as ∼82% of transfected COS-1 cells were found to express EYFP-Sp1 in association with the nuclear matrix. In marked contrast to these results, significantly fewer (ca. 22%) cells expressing EGFP-Sp3 gave rise to EGFP-Sp3–positive nuclear matrices. Results for Sp1 and Sp3 are entirely consistent with a recent report documenting the association of endogenous Sp1 and Sp3 with the nuclear matrix (He et al., 2005). It is worth noting that although EYFP-Sp1–positive nuclear matrices were recovered at relatively high frequency, these same nuclei exhibited considerably weaker staining than nuclei prepared from cells expressing EYFP-Sp2 or EGFP-Sp3. We presume that this difference in the intensity of nuclear staining reflects a relatively weaker association of Sp1 with the nuclear matrix.
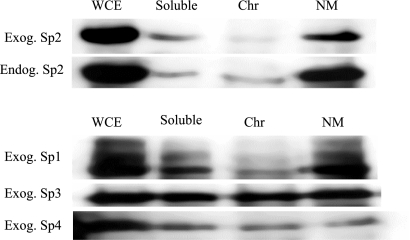
To corroborate results from in situ analyses, we examined the subcellular distribution of Sp-family members in fractionated cell extracts. Whole cell and fractionated extracts were prepared from mock transfected COS-1 cells as well as cells transiently-transfected with expression vectors encoding Sp1–4. Cell fractions containing soluble proteins, chromatin-associated proteins, and nuclear matrix–associated proteins were recovered, identical cell equivalents were resolved on denaturing polyacrylamide gels, and endogenous and exogenous Sp proteins were detected by Western blotting. Consistent with results obtained for in situ studies, the vast majority of endogenous and exogenous Sp2 was detected in fractions carrying components of the nuclear matrix (Figure 6). Only minor amounts of Sp2 were detected in soluble or chromatin-associated protein fractions. Similarly, the majority of ectopically expressed Sp1 was detected in association with the nuclear matrix although a significant amount of Sp1 protein was also detected within soluble protein fractions. In contrast to these results, and consistent with evidence from in situ analyses, Sp3 was detected uniformly in all subcellular fractions analyzed. Finally, Sp4 exhibited a localization pattern within subcellular fractions that was indistinguishable from that of Sp3. Taking results from cell fractionation experiments together with observations from indirect immunofluorescence and direct fluorescence assays, we conclude that Sp2 partitions largely to the nuclear matrix and that Sp-family members differentially associate with the nuclear matrix.
Figure 6.
Distribution of Sp-family members in fractionated cell extracts. Mock-transfected COS-1 cells and COS-1 cells transiently transfected with expression vectors encoding full-length human Sp1 (pCMV4-Sp1/flu), Sp2 (pCMV4-Sp2/flu), Sp3 (pCMV4-Sp3/flu), or Sp4 (pCMV4-Sp4/flu) were separated into cell fractions containing soluble proteins (Soluble), chromatin-associated proteins (Chr), and insoluble nuclear matrix–associated proteins (NM). Identical cell equivalents were resolved on denaturing polyacrylamide gels in parallel with denatured whole cell extracts (WCE) prepared from mock-transfected and transfected COS-1 cells. Endogenous Sp2 protein was identified with a rabbit polyclonal anti-Sp2 antiserum (Endog. Sp2; K-20), and exogenous Sp proteins were detected with a rabbit polyclonal anti-HA antiserum (Exog. Sp1, Exog. Sp2, Exog. Sp3, Exog. Sp4; Y-11).
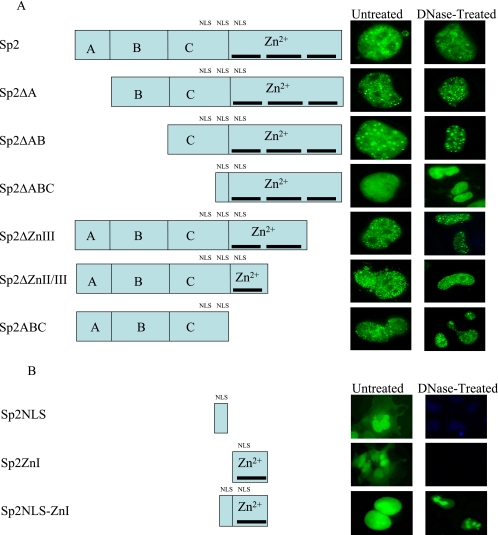
Two Independent Regions of Sp2 Specify Association with the Nuclear Matrix
Previous studies of nuclear matrix-associated transcription factors have identified 30–40 amino acid regions, each termed an NMTS, which are required for matrix association (Zeng et al., 1997; Parker et al., 2000; Zaidi et al., 2001; Djabali and Christiano, 2004; Seo et al., 2005). NMTS sequences identified to date exhibit little obvious sequence homology, and computer-assisted screens for NMTS-like sequences in Sp2 proved unsuccessful. Additional computer analyses, however, identified three putative nuclear localization signals (NLS), including one predicted bipartite signal, within portions of the Sp2 C and DNA-binding domains (Table 1 and Figure 7). To define amino acids required for the association of Sp2 with the nuclear matrix, a series of EYFP- and EGFP-Sp2 deletion mutants were prepared and each was examined for association with the nuclear matrix in situ. As shown in Figure 7A, deletion of individual subdomains (Sp2ΔA, Sp2ΔAB) or the entirety of the Sp2 trans-activation domain (Sp2ΔABC) resulted in EYFP-Sp2 fusion proteins that associated with nuclear matrices prepared from transiently transfected COS-1 cells. Interestingly, however, cells transfected with pEYFP-Sp2ΔABC exhibited homogeneous nuclear staining and lacked the subnuclear foci characteristic of Sp2-expressing cells. These results suggested that at least one NMTS mapped to the Sp2 DNA-binding domain and that amino acids within the C domain are likely to be required for nuclear speckling. To delimit amino acids within the Sp2 DNA-binding domain that specify nuclear matrix attachment, we created a series of EYFP-Sp2 fusion proteins lacking individual zinc-fingers. As shown in Figure 7A, removal of zinc-finger III (Sp2ΔZnIII) or zinc-fingers II and III (Sp2ΔZnII/III) did not effect the partitioning of EYFP-Sp2 to the nuclear matrix. Combined with results from deletions of the Sp2 trans-activation domain, these data suggested that one NMTS may reside within a 37 amino acid region encompassing zinc-finger I as well as the first inter-finger linker (amino acids 513–549) of the Sp2 DNA-binding domain. Yet these data did not rule out the possibility that one or more additional NMTS sequences reside within the Sp2 trans-activation domain, and thus an EYFP-Sp2 fusion was prepared that lacked the Sp2 DNA-binding domain (EYFP-Sp2ABC). Consistent with the notion that Sp2 may carry multiple NMTS sequences, COS-1 cells transfected with pEYFP-Sp2ABC exhibited characteristic subnuclear foci associated with the nuclear matrix (Figure 7A). On close inspection of the above EYFP-fusion constructs, however, we noted that pEYFP-Sp2ABC and pEYFP-Sp2ΔABC share seven amino acids (NH2-QGKKKHV-CO2H) at the extreme carboxy-terminal end of the Sp2 C domain that were predicted to span an NLS and/or a portion of a bipartite NLS. To rule out the unlikely possibility that these amino acids also functioned as an NMTS, COS-1 cells were transfected with a fusion construct (pEGFP-Sp2NLS) carrying these seven Sp2-derived amino acids. The distribution of EGFP-Sp2NLS in transfected cells was determined with and without treatment with DNase I in situ. Consistent with the contention that Sp2 carries multiple NMTS sequences, EGFP-Sp2NLS was distributed uniformly throughout transfected cells and did not associate with the nuclear matrix (Figure 7B).
Table 1.
Putative nuclear localization signals within human Sp2
| Amino acid sequence/positionsa | Sp2 region | NLS type |
|---|---|---|
| 489PGEKRRR495 | C domain | Monopartite |
| 515KKKH518 | C domain | Monopartite |
| 515KKK-HVCHIPDCG-KTFRK531 | C/DNA-binding domains | Bipartite |
A computer-assisted screen (http://psort.nibb.ac.jp/) of the human Sp2 amino acid sequence resulted in the identification of the putative nuclear localization sequences listed above. NLS classifications are based on consensus sequences derived previously (Hicks and Raikhel, 1995; Cokol et al., 2000).
Figure 7.
Identification of Sp2 regions required for association with the nuclear matrix. (A) The schematic diagram indicates the subdomain structure of Sp2, including the A, B, and C subdomains of the Sp2 trans-activation domain and the DNA-binding domain (Zn2+). Individual zinc-fingers within the DNA-binding domain are illustrated by a thick line. The approximate position of putative nuclear localization signals are also indicated (NLS). COS-1 cells were transiently transfected with expression vectors encoding EYFP-fusion proteins prepared with full-length human Sp2 or the indicated Sp2 deletion mutants. Fluorescent nuclei were detected via direct fluorescent microscopy in untreated cultures and nuclear matrices prepared from cultures treated with DNase I. Nuclei were localized via DNA-staining with DAPI and superimposed fluorescent images are presented in the right column. (B) COS-1 cells were transfected with an EGFP-fusion encoding the carboxy-terminal seven amino acids of the Sp2 C domain (Sp2NLS), an EYFP-fusion encoding a 30 amino acid region encompassing zinc-finger I and the inter-finger linker region of Sp2 (Sp2ZnI), or an EGFP-fusion encoding a 37 amino acid region carrying amino acids from both of these constructs (Sp2NLS-ZnI). Nuclei were localized via DNA-staining with DAPI and superimposed fluorescent images are presented in the rightmost column.
To more precisely delimit the NMTS present within the Sp2 DNA-binding domain, two further constructions carrying zinc-finger I were prepared. pEYFP-Sp2ZnI carries the 30 amino acids that encompass the first zinc-finger and inter-finger linker region, whereas pEGFP-Sp2NLS-ZnI carries these 30 amino acids as well as the seven Sp2 amino acids carried by pEGFP-Sp2NLS. A shown in Figure 7B, Sp2ZnI was distributed uniformly in transfected cells and did not associate with the nuclear matrix. Indeed, cells transiently transfected with this construction were indistinguishable from cells expressing EYFP alone. In contrast, the vast majority of cells expressing Sp2NLS-ZnI showed exclusively nuclear staining, and this EGFP-Sp2 fusion protein was associated with the nuclear matrix. We conclude that the 37 amino acids (amino acids 513–549) encompassing zinc-finger I and the first inter-finger linker of the Sp2 DNA-binding domain are sufficient to direct association with the nuclear matrix. We conclude further that the Sp2 trans-activation domain carries at least one additional NMTS. Finally, based on the differential subcellular localization of EYFP-Sp2ZnI, EGFP-Sp2NLS-ZnI, and EGFP-Sp2NLS we conclude that Sp2 likely carries a bipartite NLS that spans the junction of the C and DNA-binding domains.
DISCUSSION
Despite sharing structural similarities and a closely conserved DNA-binding domain, careful analyses have revealed that Sp-family members are each characterized by unique structural, biochemical, and functional attributes. Using a variety of molecular genetic and biochemical approaches our previous studies indicated that Sp2 is distinguished from other family members by a number of functional characteristics (Moorefield et al., 2004). For example, we have reported that Sp2 is a weak trans-activator in mammalian cells and little or no soluble Sp2 DNA-binding activity can be detected in many common human and rodent cell lines. Herein we extend these findings by reporting that Sp2 localizes within subnuclear foci that are distinct from PML oncogenic domains, and that the vast majority of endogenous and ectopically expressed Sp2 protein is associated with the nuclear matrix. Our comparative analyses also indicate that Sp-family members differ in their subnuclear localization and their association with the nuclear matrix.
The nuclear matrix is in part an intranuclear macromolecular scaffold composed of A/C- and B-type lamins, additional intermediate filament proteins and other structural proteins, as well as a variety of lamin-associated proteins. The nuclear matrix is also rich in ribonucleoproteins and a constellation of low-abundance proteins, including proteins responsible for chromatin modification, DNA replication and repair, transcription, and RNA splicing (van Wijnen et al., 1993; Stein et al., 2000; Nickerson, 2001; Mattout-Drubezki and Gruenbaum, 2003). A variety of sequence-specific DNA-binding proteins, such as steroid hormone receptors, SatB1, ATF, OCT-1, Pit-1, Runx/Cbfα/AML, and AP-1, have been shown to be associated with the nuclear matrix. Depending on cell growth/differentiation status, a number of matrix-associated transcription factors have been shown to also partition into soluble, nonmatrix-associated cellular fractions, suggesting that these proteins may shuttle between subnuclear compartments. Based on these observations, it has been postulated that the nuclear matrix may serve as a reservoir for the temporary localization of a subset of factors that may under appropriate circumstances be recruited to regulate gene expression at distal sites (Stein et al., 2000). For such factors association with the nuclear matrix may sequester and/or negatively regulate their DNA-binding activity, thus rendering them functionally inert. Other sequence-specific DNA-binding proteins that associate with the nuclear matrix form stable, active complexes that direct transcription within the context of discrete subnuclear foci. Such foci are believed to represent centers for the transcription of genes residing within specific chromatin loops that are themselves bound to the nuclear matrix via AT-rich matrix-attachment regions (MARs).
We report herein that the vast majority of Sp2 localizes to nuclear foci that are distinct from PML oncogenic domains and that are associated with the nuclear matrix. Such foci appear to be stable within nuclei, as little or no change in their location, size, or abundance, was noted during an 18-h time course of observation using confocal microscopy. Although such foci appear stable, it is worth pointing out that our time-lapse studies cannot rule out the possibility that Sp2 may dynamically shuttle between matrix-associated foci and the nucleoplasm. This possibility appears unlikely, however, as biochemical fractionation experiments revealed that only minimal amounts of endogenous or exogenous Sp2 were detectable in soluble or chromatin-associated cell compartments. Given the lack of endogenous Sp2 DNA-binding activity in soluble extracts, does nuclear matrix-associated Sp2 regulate gene expression? Although we can only speculate on this point, several alternatives are worth consideration. First, matrix-associated Sp2 may be competent to bind DNA and participate in the regulation of transcription within localized centers of gene expression. We previously identified a consensus Sp2 DNA-binding sequence (5′-GGGCGGGAC-3′) and reported that recombinant Sp2 binds this sequence with high-affinity (Kd = 225 pM; Moorefield et al., 2004). Given these data, it is certainly plausible that Sp2 functions as a sequence-specific DNA-binding protein in mammalian cells, perhaps regulating the expression of GC-rich promoters tethered to the nuclear matrix. Second, matrix-associated Sp2 may regulate gene expression indirectly via the sequestration of other regulators of gene expression. Consistent with this possibility, we have previously characterized an Sp2-associated protein that binds the B region of the Sp2 trans-activation domain but not the trans-activation domains of Sp1 or Sp3 (Moorefield et al., 2004). Should this as yet unidentified soluble protein play a role in the activation or repression of gene expression, then its effective concentration might be diminished by interactions with matrix-associated Sp2. We have also reported that Sp2 binds Nkx3.1, a homeodomain-containing transcription factor and tumor-suppressor gene product, and can neutralize Nkx3.1 DNA-binding activity in vitro (Simmons and Horowitz, 2005). Third, it is worth considering the possibility that matrix-associated Sp2 may have functions that are related only distantly to gene expression. For example, Sp2 may play a role in the anchoring of GC-rich chromosome regions to the nuclear matrix. Although this possibility may appear remote based on the well-established activities of other Sp proteins, the unique subcellular localization and functional properties of Sp2 suggest that its role in cell physiology may be quite different from other family members. Finally, it is certainly possible that matrix-associated Sp2 is functionally inert and awaits recruitment into an active, soluble form after stimulation by one or more signal transduction pathways. In this regard, it is worth mentioning that the stimulation of human and rodent cell lines with a variety of agonists has not as yet yielded evidence of increased soluble Sp2 DNA-binding activity (K. S. Moorfield and J. M. Horowitz, unpublished observations).
Our comparative studies have revealed that Sp-family members differ in their patterns of subcellular localization. Similar conclusions have recently been reported after a study of endogenous Sp1 and Sp3 localization in MCF-7 cells (He et al., 2005). Similar to Sp2, Sp1 localizes within subnuclear foci in fixed and live cells and the majority of Sp1 was detected within nuclear matrix fractions. It is also worth noting, however, that greater amounts of Sp1 were detected in soluble cell fractions than was obtained for Sp2, and Sp1 subnuclear foci are generally smaller in size than Sp2 foci. In marked contrast to the subcellular localization patterns of Sp1 and Sp2, Sp3 and Sp4 partition quite differently. In fixed and live cells, Sp3 did not localize to subnuclear foci and instead was detected diffusely and uniformly within nonnucleolar nuclear domains. Sp3 was detected in association with only a minority of nuclear matrices prepared in situ, and upon cell fractionation Sp3 and Sp4 were found in equivalent abundance within soluble, chromatin-associated and nuclear matrix compartments. In summary, we conclude that Sp-family members differ in their subnuclear localization and in their association with components of the nuclear matrix. Interestingly, van Wijnen et al. (1993) reported that a protein capable of binding specifically to a consensus Sp-binding site could be eluted from nuclear matrix preparations. Although this protein was not identified definitively by these workers, our results are consistent with the proposition that this DNA-binding activity was indeed Sp1 given its constitutive DNA-binding activity and its preferential partitioning to the nuclear matrix.
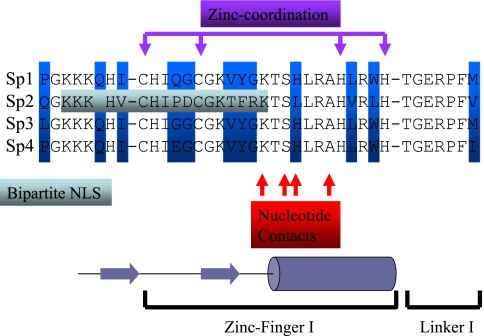
To delimit amino acid sequences that are necessary and sufficient to target Sp2 to the nuclear matrix, we created a series of EYFP- and EGFP-Sp2 deletion mutants and determined if their association with nuclei is dependent on the presence of chromatin. Deletion of the majority of the Sp2 trans-activation domain (EYFP-Sp2ΔABC) resulted in an EYFP-fusion protein that 1) localized exclusively to the nucleus, 2) associated with the nuclear matrix, and 3) did not give rise to subnuclear foci. These results indicate that at least one NLS and one NMTS reside within the carboxy-terminal 94 amino acids of Sp2 and that sequences within the Sp2 trans-activation domain are likely required for the formation of subnuclear foci. The subcellular localization of additional deletion mutants indicated that an NMTS is encoded within a 37 amino acid region that spans the junction of the C and DNA-binding domains, encompassing zinc-finger I and the first inter-finger linker region of Sp2 (Figure 8). Our data also strongly suggest that a bipartite NLS (amino acids 513–531) is encoded by a subset of amino acids within the Sp2 DNA-binding domain that function as an NMTS. Interestingly, the amino acids that comprise this bipartite NLS 1) are predicted to be part of a solvent-exposed, beta-pleated sheet structure that precedes amino acids required for nucleotide contacts and 2) are poorly conserved among other Sp-family members (Figure 8; Elrdod-Erickson et al., 1996; Phillipsen and Suske, 1999; Suske, 1999; Kaczynski et al., 2003). Thus, this substrate for nuclear translocation is likely to be another unique feature of Sp2. Alignment of the Sp2 NMTS with analogous portions of Sp1, Sp3, and Sp4 reveal that one-third (12/37) of Sp2 amino acids differ and that Sp2 diverges from the sequence of Sp1, Sp3, and Sp4 at identical amino acid positions (Figure 8). We presume that these sequence differences account, at least in part, for the subcellular localization patterns of each Sp-family member and impact the tenacity with which each associates with the nuclear matrix. Moreover, we postulate that these intrafamily differences in subcellular localization and nuclear matrix attachment reflect the specific roles of individual Sp-family members in cell physiology.
Figure 8.
Alignment of amino acids that comprise the NMTS within the Sp2 DNA-binding domain with analogous sequences of Sp1, Sp3, and Sp4. Amino acid differences within this region are highlighted in blue, and Sp2 amino acids that form a bipartite NLS are highlighted in gray. Amino acids responsible for zinc coordination and nucleotide recognition are indicated, and amino acids forming the beta sheet (arrows) and alpha helix (cylinder) that comprise the zinc-finger are indicated schematically below.
Although we have delimited an NMTS to a region that spans the Sp2 C and DNA-binding domains, our deletion studies also indicate that sequences outside this region collaborate in the association of Sp2 with the nuclear matrix. Based on the nuclear localization and matrix attachment of EYFP-Sp2ABC, at least one NLS and one NMTS must also reside within the Sp2 trans-activation domain. To our knowledge these data are the first to define independent domains capable of targeting a transcription factor to the nuclear matrix. Interestingly, and in apparent accord with these findings, we have reported that the Sp2 trans-activation and DNA-binding domains can also act independently to negatively regulate soluble DNA-binding activity and trans-activation by a series of chimeric Sp1/Sp2 proteins (Moorefield et al., 2004). Thus, it would appear that Sp2 carries distinct domains that conspire to tether it to the nuclear matrix and the association of a single Sp2 domain with the nuclear matrix can dominantly-interfere with Sp1 function in cis. Should Sp proteins regulate transcription when associated with the nuclear matrix, the functional differences exhibited by Sp1/Sp2 chimeras imply that Sp1 and Sp2 may be tethered to distinct targets.
Supplementary Material
Acknowledgments
We are grateful to Drs. R. W. Dirks and R. Tsika and for their generous contribution of reagents and to Drs. K. Zaidi and G. S. Stein for expert technical advice. We are also grateful to members of the Horowitz laboratory for reviewing this manuscript before submission. This work was supported by National Institutes of Health Grants CA105313 and GM065405, and training grants from the National Institute of Environmental Health Sciences (ES07046) and the National Science Foundation (IGERT 9987555).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1063) on February 8, 2006.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Andersson, S., Davis, D. N., Dahlback, H., Jornvall, H., and Russell, D. W. (1989). Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 264, 8222–8229. [PubMed] [Google Scholar]
- Bell, S. M., Schreiner, C. M., Waclaw, R. R., Campbell, K., Potter, S. S., and Scott, W. J. (2003). Sp8 is crucial for limb outgrowth and neuropore closure. Proc. Natl. Acad. Sci. USA 100, 12195–12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, A. R., Black, J. D., and Azizkhan-Clifford, J. (2001). Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188, 143–160. [DOI] [PubMed] [Google Scholar]
- Borden, K. L. B. (2002). Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell Biol. 22, 5259–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman, P., Göllner, H., Elsäaaer, H. -P., Eckhoff, G., Karis, A., Grosveld, F., Philipsen, S., and Suske, G. (2000). Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 19, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, S. and Ferro, T. J. (2005). Sp1, regulation of gene expression by phosphorylation. Gene 348, 1–11. [DOI] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djabali, K. and Christiano, A. M. (2004). Hairless contains a novel nuclear matrix targeting signal and associates with histone deacetylase 3 in nuclear speckles. Differentiation 72, 410–418. [DOI] [PubMed] [Google Scholar]
- Doucas, V. and Evans, R. M. (1996). The PML nuclear compartment and cancer. Biochem. Biophys. Acta 1288, M25–M29. [DOI] [PubMed] [Google Scholar]
- Dreuillet, C., Tillit, J., Kress, M., and Ernoult-Lange, M. (2002). In vivo and in vitro interaction between human transcription factor MOK2 and nuclear lamin A/C. Nucleic Acids Res. 30, 4634–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrdod-Erickson, M., Rould, M. A., Nekludova, L., and Pabo, C. O. (1996). Zif268 protein-DNA complex refined at 1.6 Å: a model system for understanding zinc-finger-DNA interactions. Structure 4, 1171–1180. [DOI] [PubMed] [Google Scholar]
- Göllner, H. et al. (2001a). Complex phenotype of mice homozygous for a null mutation in the Sp4 transcription factor gene. Genes Cells 6, 689–697. [DOI] [PubMed] [Google Scholar]
- Göllner, H., Dani, C., Phillips, B., Philipsen, S., and Suske, G. (2001b). Impaired ossification in mice lacking the transcription factor Sp3. Mech. Dev. 106, 77–83. [DOI] [PubMed] [Google Scholar]
- Harrison, S. M., Houzelstein, D., Dunwoodie, S. L., and Beddington, R. S. P. (2000). Sp5, a new member of the Sp1 family, is dynamically expressed during development and genetically interacts with Brachyury. Dev. Biol. 227, 358–372. [DOI] [PubMed] [Google Scholar]
- He, S., Sun, J. -M., Li, L., and Davie, J. R. (2005). Differential intranuclear organization of transcription factors Sp1 and Sp3. Mol. Biol. Cell 16, 4073–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, G. R., and Raikhel, N. V. (1995). Protein import into the nucleus: an integrated view. Annu. Rev. Cell Dev. Biol. 11, 155–188. [DOI] [PubMed] [Google Scholar]
- Javed, A. et al.. (2000). Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J. Cell Sci. 113, 2221–2223. [DOI] [PubMed] [Google Scholar]
- Kaczynski, J., Cook, T., and Urrutia, R. (2003). Sp1- and Krüppel-like transcription factors. Genome Biol. 4, 206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennett, S. B., Udvadia, A. J., and Horowitz, J. M. (1997). Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res. 25, 3110–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kießlich, A., von Mikecz, A., and Hemmerich, P. (2002). Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J. Struct. Biol. 140, 167–179. [DOI] [PubMed] [Google Scholar]
- Kingsley, C., and Winoto, A. (1992). Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol. Cell. Biol. 12, 4251–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin, M., Karis, A., Visser, P., Grosveld, F., and Philipsen, S. (1997). Transcription factor Sp1 is essential for early embryonic development but dispensible for cell growth and differentiation. Cell 89, 619–628. [DOI] [PubMed] [Google Scholar]
- Mattout-Drubezki, A., and Gruenbaum, Y. (2003). Dynamic interactions of nuclear lamina proteins with chromatin and transcriptional machinery. Cell Mol. Life Sci. 60, 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul, G. G., Negorev, D., Bell, P., and Ishov, A. M. (2000). Properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129, 278–287. [DOI] [PubMed] [Google Scholar]
- Maul, G. G., Yu, E., Ishov, A. M., and Epstein, A. L. (1995). Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell. Biochem. 59, 498–513. [DOI] [PubMed] [Google Scholar]
- Moorefield, K. S., Fry, S. J., and Horowitz, J. M. (2004). Sp2 DNA binding activity and trans-activation are negatively regulated in mammalian cells. J. Biol. Chem. 279, 13911–13924. [DOI] [PubMed] [Google Scholar]
- Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J. M., Behringer, R. R., and de Crombrugghe, B. (2002). The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29. [DOI] [PubMed] [Google Scholar]
- Negorev, D. and Maul, G. G. (2001). Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20, 7234–7242. [DOI] [PubMed] [Google Scholar]
- Nickerson, J. A. Experimental observations of a nuclear matrix. (2001). J. Cell Sci. 114, 463–474. [DOI] [PubMed] [Google Scholar]
- Parker, G. E., Sandoval, R. M., Feister, H. A., Bidwell, J. P., and Rhodes, S. J. (2000). The homeodomain coordinates nuclear entry of the Lhx3 neuroendocrine transcription factor and association with the nuclear matrix. J. Biol. Chem. 275, 23891–23898. [DOI] [PubMed] [Google Scholar]
- Phan, D., Cheng, C. -J., Galfione, M., Vakar-Lopez, F., Tunstead, J., Thompson, N. E., Burgess, R. R., Najjar, S. M., Yu-Lee, L. -Y., and Lin, S. -H. (2004). Identification of Sp2 as a transcriptional repressor of carcinoembryonic antigen-related cell adhesion molecule 1 in tumorigenesis. Cancer Res. 64, 3072–3078. [DOI] [PubMed] [Google Scholar]
- Phillipsen, S. and Suske, G. (1999). A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J., Lozano, M. M., and Dudley, J. P. (2005). Nuclear matrix binding regulates SATB1-mediated transcriptional repression. J. Biol. Chem. 280, 24600–24609. [DOI] [PubMed] [Google Scholar]
- Simmons, S. O., and Horowitz, J. M. (2005). Nkx3.1 binds and negatively regulates the transcriptional activity of Sp-family members in prostate-derived cells. (2006). Biochem. J. 393, 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler, M. L., Kennett, S. B., Moorefield, K. S., Simmons, S. O., Brattain, M. G., and Horowitz, J. M. (2005). Sumoylation of internally initiated Sp3 isoforms regulates transcriptional repression via a Trichostatin A-insensitive mechanism. Cell Signal. 17, 153–166. [DOI] [PubMed] [Google Scholar]
- Stein, G. S., van Wijnen, A. J., Stein, J. L., Lian, J. B., Montecino, M., Choi, J. -Y., Zaidi, S. K., and Javed, A. (2000). Intranuclear trafficking of transcription factors: implications for biological control. J. Cell Sci. 113, 2527–2533. [DOI] [PubMed] [Google Scholar]
- Supp, D. M., Witte, D. P., Branford, W. W., Smith, E. P., and Potter, S. S. (1996). Sp4, a member of the Sp1-family of zinc finger transcription factors, is required for normal murine growth, viability, and male fertility. Dev. Biol. 176, 284–299. [DOI] [PubMed] [Google Scholar]
- Suske, G. (1999). The Sp-family of transcription factors. Gene 238, 291–300. [DOI] [PubMed] [Google Scholar]
- Suske, G., Bruford, E., and Philipsen, S. (2005). Mammalian SP/KLF transcription factors: bring in the family. Genomics 85, 551–556. [DOI] [PubMed] [Google Scholar]
- Udvadia, A. J., Rogers, K. T., Higgins, P. D. R., Murata, Y., Martin, K. H., Humphrey, P. A., and Horowitz, J. M. (1993). Sp-1 binds promoter elements regulated by the Rb protein and Sp-1-mediated transcription is stimulated by Rb co-expression. Proc. Natl. Acad. Sci. USA 90, 3265–3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallian, S., Chin, K. -V., and Chang, K. -S. (1998). The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell Biol. 18, 7147–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen, A. J., Bidwell, J. P., Fey, E. G., Penman, S., Lian, J. B., Stein, J. L., and Stein, G. S. (1993). Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP-1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochem. 32, 8397–8402. [DOI] [PubMed] [Google Scholar]
- Wiesmeijer, K., Molenaar, C., Bekeer, I. M. L. A., Tanke, H. J., and Dirks, R. W. (2002). Mobile foci of Sp100 do not contain PML: PML bodies are immobile but PML and Sp100 proteins are not. J. Struct. Biol. 140, 180–188. [DOI] [PubMed] [Google Scholar]
- Zaidi, S. K., Javed, A., Choi, J. -Y., van Wijnen, A. J., Stein, J. L., Lian, J. B., and Stein, G. S. (2001). A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J. Cell Sci. 114, 3093–3102. [DOI] [PubMed] [Google Scholar]
- Zeng, C. et al. (1997). Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-α transcription factors. Proc. Natl. Acad. Sci. USA 94, 6746–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, C., and Meng, A. (2005). Sp1-like transcription factors are regulators of embryonic development in vertebrates. Dev. Growth Differ. 47, 201–211. [DOI] [PubMed] [Google Scholar]
- Zhong, S., Salomoni, P., and Pandolfi, P. P. (2000). The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2, E85–E90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.