Abstract
Slx1 and Slx4 are subunits of a structure-specific DNA endonuclease that is found in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other eukaryotic species. It is thought to initiate recombination events or process recombination structures that occur during the replication of the tandem repeats of the ribosomal DNA (rDNA) locus. Here, we present evidence that fission yeast Slx1-Slx4 initiates homologous recombination events in the rDNA repeats that are processed by a mechanism that requires Rad22 (Rad52 homologue) but not Rhp51 (Rad51 homologue). Slx1 is required to generate ∼50% of the spontaneous Rad22 DNA repair foci that occur in cycling cells. Most of these foci colocalize with the nucleolus, which contains the rDNA repeats. The increased fork pausing at the replication fork barriers in the rDNA repeats in a strain that lacks Rqh1 DNA helicase is further increased by expression of a dominant negative form of Slx1. These data suggest that Slx1-Slx4 cleaves paused replication forks in the rDNA, leading to Rad22-dependent homologous recombination that is used to maintain rDNA copy number.
INTRODUCTION
While replicating the genome, DNA polymerases will sometimes stall when they encounter DNA lesions, protein complexes bound to DNA, or nucleotide starvation. Stalled forks pose serious threats to genomic integrity because they are prone to collapse or rearrangement (McGlynn and Lloyd, 2002). In contrast, there are many examples of natural replication pause and termination sites in a variety of organisms (Rothstein et al., 2000). These sites are often called replication fork barriers (RFBs). It is somewhat paradoxical that in some cases these “programmed” RFBs are thought to play important roles in maintaining genome integrity.
The best-characterized RFBs occur in the ribosomal DNA genes (rDNA), which are present in long tandem repeats at one or a few chromosomal loci in most eukaryotic species, including yeasts and humans (Brewer and Fangman, 1988; Linskens and Huberman, 1988; Little et al., 1993; Gerber et al., 1997; Sanchez et al., 1998). The budding yeast Saccharomyces cerevisiae carries ∼150 copies of the rDNA genes. The RFB is located in the nontranscribed sequence (NTS) between the 3′ end of the 35S rRNA gene and the autonomously replicating sequence (ARS). The RFB inhibits replication fork progression in the direction opposite to rDNA transcription. This mechanism helps to prevent collisions between replication forks and the transcription machinery (Takeuchi et al., 2003). The protein fork blocking 1 (Fob1) binds DNA in the RFB region and is essential for RFB activity (Kobayashi and Horiuchi, 1996).
The size of the rDNA array in S. cerevisiae is not static. For example, it will contract upon inactivation of RNA polymerase I and expands after RNA polymerase I activity is restored (Kobayashi et al., 1998). Fob1 is required for the mechanisms that control the copy number of rDNA repeats (Kobayashi et al., 2001; Johzuka and Horiuchi, 2002). The current model for the regulation of the rDNA copy number involves a DNA endonuclease activity, which creates a broken fork structure that leaves a free DNA end at the RFB. The free DNA end starts a homologous recombination (HR) process by invading complementary sequences within the rDNA array. This may lead to expansion or contraction (or no net change) in the rDNA array depending on exactly where the HR event occurs. This mechanism seems to be dependent on Rad52 (Park et al., 1999; Johzuka and Horiuchi, 2002), which is known to be required for all HR pathways in S. cerevisiae (Paques and Haber, 1999).
The fission yeast Schizosaccharomyces pombe has ∼150 rDNA repeats separated in two clusters at both ends of chromosome III. As in budding yeast, one rDNA repeat consists of a transcription unit for the 35S rRNA precursor and a NTS, which contains an ARS element (ARS3001) and RFB (see Figure 5A) (Sanchez et al., 1998). However, unlike budding yeast, fission yeast has four closely spaced polar replication barriers named RFB1-3 (Ter1-3) and RFP4 (Krings and Bastia, 2004; Sanchez-Gorostiaga et al., 2004). Fork blockage at RFB1-3 requires the Swi1-Swi3 protein complex. Blockage at RFB2 and RFB3 additionally requires Reb1, a transcription termination protein that has specific binding sites at RFB2 and RFB3. Interestingly, RFB2 and RFB3 resemble the fork barriers present in the mouse rDNA repeats, which is regulated by TTF1 transcription termination factor (Lopez-estrano et al., 1998). In contrast, RFB1 is similar to the budding yeast RFB. RFB1 is bound by the switch-activating protein Sap1 to generate a polar replication fork arrest (Krings and Bastia, 2005; Mejia-Ramirez et al., 2005).
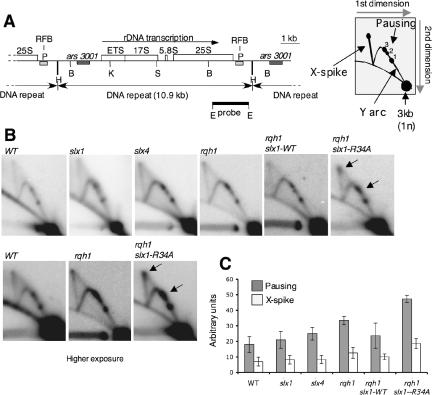
Figure 5.
Paused replication forks in the RFB of the rDNA accumulate in rqh1Δ slx1-R34A: leu1 cells. (A) Map of the rDNA repeats as reported previously (Sanchez et al., 1998) and diagram of the migration pattern of replication intermediate that can be detected by 2D-gel electrophoresis. The ARS3001 box indicates the origin region, and the RFB box indicates a pause site (Sanchez et al., 1998). The restriction enzyme sites are indicated (H, HindIII; B, BamHI; K, KpnI; S, SacI; and E, EcoRI). (B) 2D-gel analysis of rDNA RFB site in slx1Δ, slx4Δ, rqh1Δ, rqh1Δ slx1+ slx1+:LEU1, and rqh1Δ slx1+ slx1-R34A:LEU1 mutants. Genomic DNA samples were digested with BamHI and hybridized with the EcoRI-EcoRI probe. Three distinct pausing sites where detected as previously described (Sanchez-Gorostiaga et al., 2004). The slx1-R34A allele provokes accumulation of replication forks blocked at the RFB and the amassing of X structures. (C) Quantification of pausing (RFB1-3) and X-spike signals relative to 1N spot signal [((X-spike + Pausing)/1N spot)x1000]. Error bars show the SD of three independent experiments.
DNA helicases of the RecQ family play a central role in maintaining genome stability (Enomoto, 2001; Opresko et al., 2004). This family includes budding yeast Sgs1, fission yeast Rqh1, and human BLM, WRN, and RecQL proteins. Interestingly, Sgs1, Rqh1, and BLM form functional enzyme complexes with DNA topoisomerase III. Inactivation of BLM or WRN in humans leads to increased rates of sister-chromatid exchange or DNA rearrangements as well as increased sensitivity to certain types of DNA-damaging agents (Hickson et al., 2001; Wu and Hickson, 2003; Mankouri and Hickson, 2004). In budding and fission yeasts, loss of Sgs1/Rqh1 is associated with increased recombination, several forms of chromosome instability, and enhanced sensitivity to several types of genotoxic agents (Watt et al., 1995, 1996; Stewart et al., 1997; Laursen et al., 2003). Notably, the yeast mutants are hypersensitive to hydroxyurea (HU), which causes replication forks to stall through dNTP starvation. The mutants are also sensitive to agents such as methyl methanesulfonate (MMS) and UV light, which also cause fork stalling. This spectrum of sensitivity to genotoxic agents, together with a number of genetic and biochemical studies, have led to the idea that RecQ family helicases, acting together with TopIII, have central roles in the rescue of stalled forks through a nonrecombinogenic mechanism (Doe et al., 2000; Boddy et al., 2001; Cox, 2001; Hickson et al., 2001). They have been proposed to unwind the stalled fork that has regressed to form a Holliday junction-like structure known as a chicken foot (Opresko et al., 2004). In vitro studies have also shown that RecQ-like helicases and TopIII can act together to dissolve double Holliday junctions (Wu and Hickson, 2003). This activity might resolve DNA structures that arise from regressed or converged replication forks.
The slx1 and slx4 genes were first identified in a screen for genes that are essential in S. cerevisiae mutants that lack the Sgs1 DNA helicase (Mullen et al., 2001). Subsequent analysis showed that Slx1 and Slx4 form a protein complex that has structure-specific DNA endonuclease activity, with a preference for certain types of branched DNA structures (Fricke and Brill, 2003; Coulon et al., 2004). The Slx1 homologue in S. pombe was subsequently identified by virtue of its sequence similarity to S. cerevisiae Slx1, and the highly divergent Slx4 homologue in S. pombe was discovered through its physical association with Slx1 (Coulon et al., 2004). Both Slx1 and Slx4 were shown to be essential in the absence of Rqh1 in fission yeast. The functional conservation of S. pombe and S. cerevisiae Slx1-Slx4 complexes, and the presence of Slx1 homologues in other sequenced eukaryotic genomes, suggests that Slx1-Slx4 function is likely to be conserved in humans and other eukaryotes (Kaliraman and Brill, 2002; Fricke and Brill, 2003; Coulon et al., 2004). We found that Slx1-Slx4 complex partially purified from S. pombe is a structure-specific DNA endonuclease that introduces a single-strand cut in duplex DNA on the 3′ side of a double-strand/single-strand junction. Slx1 is likely the nuclease per se because it belongs to UvrC-Intron-Type (URI) family (Aravind and Koonin, 2001) and has weak endonuclease activity in the absence of Slx4 (Fricke and Brill, 2003). Interestingly, Slx4 has a SAF-A/B, Acinus and PIAS (SAP) DNA binding domain, indicating that it may act as an enhancer or recruiter of Slx1 nuclease activity (Aravind and Koonin, 2000). We showed that loss of either Slx1-Slx4 or Rqh1 leads to contraction of the rDNA repeats (Coulon et al., 2004), findings that were consistent with the analysis of Slx1-Slx4 and Sgs1 in S. cerevisiae (Kaliraman and Brill, 2002). These findings suggested that Slx1-Slx4 complex is dedicated to a specific DNA cleavage event that is involved in rDNA maintenance. We proposed a model in which the Slx1-Slx4 complex is required for a recombination pathway that maintains or expands the rDNA repeats, whereas Rqh1 is required in a parallel pathway that prevents the loss of rDNA copies. This model is consistent with the evidence that an sgs1-ts slx4Δ double mutant is unable to undergo S phase at restrictive temperature and arrests in late S or G2 phase (Kaliraman and Brill, 2002). Together, these observations suggest that in the absence of Sgs1 or Rqh1, the Slx1-Slx4 complex may be required to process DNA structures formed at the RFB to allow complete replication of the rDNA loci and proper chromosome segregation.
Although it is evident that Slx1 and Slx4 associate to form a functional endonuclease complex, several recent studies with S. cerevisiae have revealed that Slx4 has some Slx1-independent functions. In comparison to slx1Δ cells, slx4Δ cells were found to be more sensitive to DNA damage caused by MMS or the topoisomerase I poison camptothecin (Fricke and Brill, 2003; Deng et al., 2005; Flott and Rouse, 2005). In addition, Slx4 was found to have an Slx1-independent function in promoting the phosphorylation of Rtt107/Esc4 by a mechanism that requires the checkpoint kinase Mec1 (Roberts et al., 2006). It is unknown whether similar distinctions exist between Slx1 and Slx4 in S. pombe.
In this report, we investigate the hypothesis that Slx1-Slx4 cleaves DNA structures in the rDNA arrays that are processed by the HR machinery. We present evidence that Slx1-Sx4 cleaves DNA structures in the rDNA that are processed in a pathway that requires Rad52 (Rad22) but not Rad51 (Rhp51). These studies also indicate that the replication forks stalled at the RFB are the substrates of Slx1-Slx4 complex in vivo.
MATERIALS AND METHODS
General Techniques
S. pombe methods and media have been described in Moreno et al. (1991).
Strains and Plasmids
Strains used in this study are ura4-D18 and leu1-32 unless otherwise stated: PR109, wild-type (h-); PR110, wild-type (h+); PS2343, wild-type (h- smt0); SC3250, rqh1::ura4 (h-); SC3240, slx1::kanMx6 (h+); SC3243, slx4::kanMx6 (h-); TNM3317, rad22::LEU2 (h+); SC3463, slx1::kanMx6 rad22::LEU2 (h+); SC3464, slx1::kanMx6 (h- smt0); SC3465, rad22::LEU2 (h- smt0); SC3466, slx1::kanMx6 rad22::LEU2 (h- smt0); SC3467, slx4::kanMx6 (h- smt0); SC3468, slx4::kanMx6 rad22::LEU2 (h+); SC3469, slx4::kanMx6 rad22::LEU2 (h- smt0); EN3190, mus81::kanMx6 (h+); SC3470, mus81::kanMx6 rad22::LEU2 (h+); EN3220, rad22-YFP:kanMx6 (h+); SC3471, slx1::kanMx6 rad22-YFP:kanMx6; SC3472, rqh1::ura4 rad22-YFP:kanMx6; SC3473 mus81::kanMx6 rad22-YFP:kanMx6; SC3474, rad22-RFP:kanMx6 (h+); SC3475 slx1::kanMx6 rad22-RFP:kanMx6; SC3476, rqh1::ura4 rad22-RFP:kanM6; PS2388, rhp51::ura4 (h+); SC3477, slx1::kanMx6 rhp51::ura4 (h+); PS2345 rhp51::ura4 (h- smt0); SC3478, slx1::kanMx6 rhp51::ura4 (h- smt0), SC3479, slx4::kanMx6 rhp51::ura4 (h+); SC3480, rqh1::ura4 slx1wt-TAP:leu1 (h+); SC3481, rqh1::ura4 slx1R34A-TAP:leu1 (h-); SC3247, slx1::kanMx6 slx1wt-TAP: leu1; SC3482, slx1::kanMx6 slx1wt-TAP:leu1 slx4-13myc::kanMx6; SC3283, slx1::kanMx6 slx1R34A-TAP:leu1 slx4-13myc::kanMx6. Strains SC3474, SC3475, and SC3476 have been transformed with pTA57 (Ding et al., 2000).
Fluorescent Microscopy
Cells were grown at 25°C in Edinburgh minimal medium (EMM) with necessary supplements. Cell grown at 25°C have stronger yellow fluorescent protein (YFP) or red fluorescent protein (RFP) signal for Rad22 and weaker background fluorescence. Cells were concentrated by centrifugation and kept on ice before microscopy. Microscopy was performed with Nikon Eclipse E800 microscope equipped with a Photometrics Quantix charge-coupled device camera and YFP-green fluorescent protein (GFP)/RFP filter set. Images were acquired with IPlab Spectrum software (Signal Analytics, Vienna, VA). Quantification of Rad22-YFP foci has been performed at least three times and at least 400 cells were counted for each strain in each experiment. Identical quantification has been performed for Rad22-RFP foci. Nucleolus and nonnucleolus have been discriminated using plasmid pTA57 encoding for the Rrn5-GFP fusion protein (Ding et al., 2000). Rrn5-GFP binds to rDNA and is used as a marker of nucleolus. According to previous results, the bright spot of Rrn5-GFP signal marks the nucleolus because it does not overlap with 4,6-diamidino-2-phenylindole (DAPI) staining (see Figure 2A). A weaker diffuse Rrn5-GFP signal is found to be overlapped with DAPI staining in nonnucleolus compartment. This feature of Rrn5-GFP signal allowed us to distinguish Rad22-RFP foci located inside or outside the nucleolus. Cells were excluded when the Rrn5-GFP signal did not show clear distinction between nucleolus and nonnucleolus compartments or when categorization of Rad22-RFP was subjective.
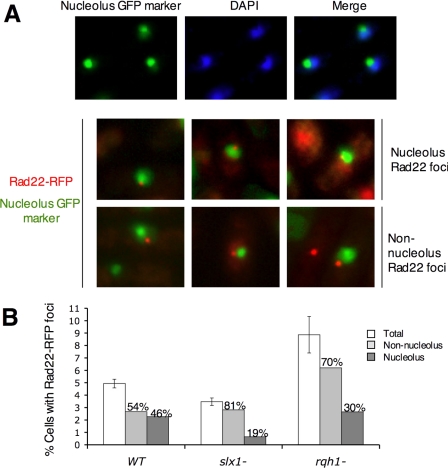
Figure 2.
Slx1-dependent Rad22 foci colocalize with the nucleolus. Cells that had genomic rad22-RFP and plasmid pTA57 encoding for the Rrn5-GFP fusion protein were grown in EMM medium at 25°C until mid-log phase. Rrn5-GFP binds to rDNA and is used in this study as a marker of nucleolus (Ding et al., 2000). (A) Top, costaining of Rrn5-GFP (green, nucleolus region) and DAPI staining (blue, nonnucleolus region). Rrn5-GFP and DAPI staining do not colocalize. Middle, three examples of Rad22-RFP foci (red) localized inside the nucleolus, judging from Rrn5-GFP localization. Bottom, three examples of Rad22-RFP foci (red) localized outside the nucleolus. (B) The location of Rad22-RFP foci relative to the Rrn5-GFP signal were quantified in wild-type, slx1Δ, and rqh1Δ strains. Error bars show the SD of three independent experiments. At least 1000 cells were scored for each strain in each experiment.
Two-dimensional (2D) Gel Electrophoresis
2D gel electrophoresis was performed as described previously (Noguchi et al., 2003). For the analysis of the RFB region, 5 μg of DNA was digested with 60 U of BamHI. Precipitated DNA was run on 0.4% agarose gel for the first dimension and a 1% agarose gel for the second dimension. Gels were transferred to Hybond-N+ membranes (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). After hybridization, radioactive signals were detected with a Storm 840 machine (GE Healthcare). The membranes were probed with the 1.35-kb EcoRI-EcoRI rDNA fragment.
RESULTS
Partial Suppression of rad22Δ by Inactivation of Slx1-Slx4
Rad22, the fission yeast homologue of Rad52, is thought to be required for most or all of the HR events in S. pombe (van den Bosch et al., 2001; Doe et al., 2004). This conclusion is supported by damage survival assays and genetic recombination measurements as well as by direct physical assays of recruitment of Rad22 to double-strand breaks (DSBs). With respect to the physical assays, chromatin immunoprecipitation analysis has shown that Rad22 binds near DSBs at the mating-type locus (Kim et al., 2000), and we have observed that Rad22-YFP is very rapidly localized at DSBs in live cells (Du et al., 2003). All of these data are consistent with the analysis of Rad52 in S. cerevisiae (Tsukamoto et al., 2003).
We hypothesized that Slx1-Slx4 complex might cleave stalled replication forks, leading to HR events that require Rad22. As an initial test of this model, we determined whether inactivation of Slx1 corrects the slow growth phenotype of rad22Δ mutants. The appropriate single and double mutants were created in an isogenic strain background with the h- smt0 mating-type locus. Serial dilutions of these strains were plated on yeast extract, glucose, and supplements (YES). This analysis showed that rad22Δ cells grew poorly relative to wild type (Figure 1A), as expected (Doe et al., 2004). The growth of slx1Δ cells was indistinguishable from wild type (Figure 1A). Interestingly, the growth of slx1Δ rad22Δ cells was better than rad22Δ cells (Figure 1A). The colony size of the double mutant did not match that of slx1Δ or wild type, but it was larger than the rad22Δ mutant. These results were confirmed by the analysis of the generation time of cells grown in liquid media (Figure 1C). These findings showed that inactivation of Slx1 partially alleviates the requirement for Rad22 for the normal robust growth of fission yeast cells.
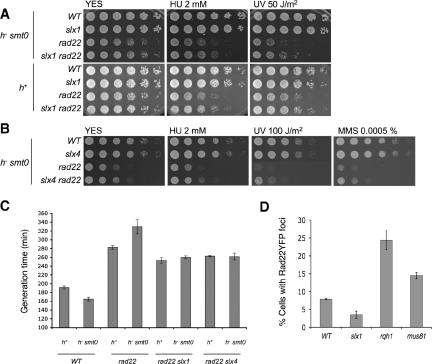
Figure 1.
Genetic interactions involving Slx1-Slx4 endonuclease and Rad22. (A) The slx1Δ mutation improves the growth of a rad22Δ mutant in both h- smt0 and h+ mating-types. Five-fold serial dilutions of S. pombe cells were plated on YES agar medium and incubated for 2-5 d at 30°C. YES plates were supplemented with the indicated amounts of HU or exposed to the indicated dose of UV. (B) The slx4Δ mutation improves the growth of a rad22Δ mutant in a h- smt0 background. (C) The slx1Δ and slx4Δ mutations increase the growth rate of a rad22Δ mutant. (D) Reduction of spontaneous Rad22-YFP foci in slx1Δ cells. Cells with a single copy of rad22-YFP were grown in EMM medium at 25°C until mid-log phase. Rad22-YFP foci were quantified in wild-type, slx1Δ, rqh1Δ, and mus81Δ mutants. Error bars show the SD of three independent experiments. At least 400 cells were counted for each strain in each experiment.
These studies were extended to include exposure of cells to genotoxic stress agents. HU arrests replication by causing dNTP starvation, whereas UV creates DNA lesions that if left unexcised can block the progression of replication forks. The slx1Δ cells were not noticeably hypersensitive to media containing 2 mM HU or exposure to 150 J/m2 UV (Figure 1A). In contrast, rad22Δ cells were very sensitive to both types of genotoxic stress (Figure 1A). The slx1Δ rad22Δ cells grew better than rad22Δ cells in HU-containing media (Figure 1A). The slx1Δ rad22Δ cells were likewise improved in their survival of UV (Figure 1A). The growth of slx1Δ rad22Δ cells did not match that of slx1Δ cells; therefore, we conclude that the slx1Δ mutation was not fully epistatic to rad22Δ, but the genotoxic agents seemed to accentuate the suppression of rad22Δ by slx1Δ. Perhaps the added demand of repairing damage caused by genotoxic agents accentuates the effect of relieving the need to repair breaks created by Slx1-Slx4.
These findings indicated that Slx1-Slx4 endonuclease initiates recombination events that are repaired by a Rad22-dependent recombination mechanism. To further test this hypothesis, we determined whether slx4Δ suppresses the rad22Δ mutation in an h- smt0 background. As was the case for the slx1Δ mutation, we observed that slx4Δ did not impair growth on YES media, but it did improve growth in a rad22Δ background (Figure 1B). Furthermore, in cells exposed to HU or UV, the growth of the slx4Δ rad22Δ strain was better than the rad22Δ strain (Figure 1B). In slx4Δ, we extended these studies to include MMS, another genotoxic agent that can cause DNA damage that interferes with replication fork progression. Consistent with the studies of the other genotoxic agents, we found that elimination of Slx4 partially suppressed the requirement for Rad22 for growth in media containing 0.005% MMS (Figure 1B).
All of the experiments described above were performed in isogenic strains with the h- smt0 mating-type locus. We exercised this precaution because we have observed that the mating type locus can affect the growth and genotoxic sensitivity of recombination mutants (our unpublished data). It is unknown why mating type has this effect. It might be connected to the mating-type specific distribution of recombination-promoting complex (RPC) containing Swi2 and Swi5 proteins at the silent mating-type region (Jia et al., 2004). We therefore repeated a subset of the studies involving slx1Δ in an h+ background. The poor growth phenotype caused by the rad22Δ mutation was not as severe as observed in the h- smt0 background (Figure 1A); nevertheless, the slx1Δ mutation was clearly able to partially suppress the poor growth and genotoxic sensitivity phenotypes caused by the rad22Δ mutation (Figure 1A). The analysis of generation times in liquid media confirmed the suppression of the rad22Δ poor growth phenotype in slx1Δ and slx4Δ strains in the h+ background (Figure 1C).
Reduction of Spontaneous Rad22 Foci in slx1Δ Cells
Rad22-YFP forms bright foci at the sites of DSBs created by the homing endonuclease and ionizing radiation (Du et al., 2003). In cultures of wild-type cells grown to log phase, ∼5-10% of cells have at least one spontaneous Rad22-YFP focus (2 or more spontaneous foci are rare). Most of these cells are in S phase or early G2 (Noguchi et al., 2003), indicating that foci arise from DNA damage or abnormal DNA structures that occur during DNA replication. To investigate whether some of these Rad22-YFP foci are formed as the result of the activity of Slx1-Slx4 endonuclease, we counted these foci in wild-type and slx1Δ cells. Particular care was taken to ensure that the strains were in the same state of growth (i.e., mid-log phase). Whereas in these studies ∼8% of the wild-type cells contained a Rad22-YFP focus, only ∼3.5% of the isogenic slx1Δ cells had a Rad22-YFP focus (Figure 1D). These findings suggest that Slx1-Slx4 endonuclease creates about one-half of the DNA structures that are processed by Rad22.
We also analyzed formation of spontaneous Rad22-YFP foci in mus81Δ and rqh1Δ cells. The Mus81-Eme1 endonuclease complex in S. pombe, and the analogous Mus81-Mms4 complex in S. cerevisiae, are essential for cell viability in the absence of Rqh1/Sgs1 DNA helicases (Boddy et al., 2000; Mullen et al., 2001). This property is shared with the Slx1-Slx4 endonucleases (Mullen et al., 2001; Coulon et al., 2004). The Mus81 structure-specific endonucleases have been proposed cleave a variety of DNA substrates in vivo, including relatively simple 3′-flaps, stalled replication forks, and Holliday junctions (Boddy et al., 2001; Kaliraman et al., 2001; Gaillard et al., 2003; Osman et al., 2003; Whitby et al., 2003). Cruciform structures that model regressed replication forks are particularly good in vitro substrates of the Mus81 complexes. In the mus81Δ culture, ∼15% of the cells had one or more Rad22-YFP foci (Figure 1D). This frequency of cells with Rad22-YFP foci was a significant increase above wild type. These data do not support a model in which Mus81-Eme1 complex has a prominent role in cleaving DNA structures that then lead to repair events that generate Rad22-YFP foci. The largest effect was observed with rqh1Δ cells, in which ∼25% of the cells had one or more Rad22-YFP foci (Figure 1D). These findings are consistent with the increased rates of recombination in rqh1Δ cells.
Nucleolar Localization of Slx1-dependent Rad22 Foci
Having observed that Slx1 is responsible for approximately one-half of the Rad22-YFP foci that arise spontaneously, we wished to determine whether the Slx1-dependent Rad22-YFP foci were occurring in the rDNA arrays. To answer this question we took advantage of the cytology of the nucleus. The nucleus of S. pombe is comprised of two hemispherical compartments. One hemisphere is enriched with RNA (nucleolus) and the other contains chromatin (stained by DAPI) that includes two protrusions of chromatin extending into the RNA-rich hemisphere. The protrusions of chromatin embedded in the nucleolus correspond to the location of rDNA arrays (Uzawa and Yanagida, 1992). Previous studies indicated that Slx1-Slx4 is specifically involved in the maintenance of rDNA arrays. If this model was correct, we would expect most of the Slx1-generated Rad22-YFP foci to occur in the DAPI-staining region of the nucleus that extends into the nucleolus. To test this hypothesis, we scored the location of Rad22-RFP foci while using a GFP fusion protein of Rrn5, an rDNA transcriptional activator, as a nucleolar marker (Figure 2A) (Ding et al., 2000). Quantification of Rad22-RFP foci was performed in wild-type, slx1Δ, and rqh1Δ cells transformed with the plasmid expressing the Rrn5-GFP fusion protein. The Rad22-RFP signal was substantially weaker than the Rad22-YFP signal, which explains why fewer spontaneous Rad22 foci were detected in this experiment (Figure 2B). Nevertheless, we measured a decrease in Rad22-RFP foci in slx1Δ cells and an increase in rqh1Δ cells. Both changes were statistically significant relative to each other and wild type. In wild-type cells, 46% of the Rad22-RFP foci overlapped with the Rrn5-GFP signal (Figure 2B), indicating that about one-half of the total Rad22 foci occur in the rDNA repeats. In contrast, ∼80% (n = 47) of the Rad22-RFP foci in slx1Δ cells did not overlap with the Rrn5-GFP signal. These findings indicated that most of the spontaneous Rad22-RFP foci that occur in the rDNA of wild-type cells are dependent on Slx1. Interestingly, the majority (∼70%; n = 40) of spontaneous Rad22-RFP foci in rqh1Δ cells were localized in the chromosomal region of the nucleus, a finding that that is consistent with Rqh1 functioning in both compartments of the nucleus.
Inactivation of Slx1 Does Not Suppress rhp51Δ
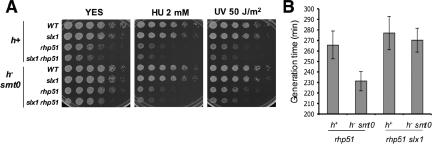
The studies described above indicated that Slx1-Slx4 is responsible for creating DNA breaks in the rDNA repeats that are repaired by a Rad22-dependent mechanism of HR. Studies with S. cerevisiae have shown that recombination intermediates in the rDNA repeats occur by a mechanism that requires Rad52 but not Rad51 (Zou and Rothstein, 1997). If such a situation exists in S. pombe, and Slx1-Slx4 primarily cleaves rDNA, it would be expected that elimination of Slx1-Slx4 activity should not suppress the phenotypes of mutants lacking Rhp51, which is the Rad51 homolog in fission yeast. Accordingly, we investigated the genetic interactions between slx1Δ and rhp51Δ mutations in both h+ and h- smt0 mating-type background. Serial dilution assays showed that rhp51Δ cells grew slower than wild type (Figure 3A). However, unlike the situation with rad22Δ, the slx1Δ mutation did not improve the growth of rhp51Δ cells. In fact, the slx1Δ rhp51Δ cells grew slightly slower than the rhp51Δ cells. These observations were confirmed by determination of generation times in liquid cultures (Figure 3B). This relationship was also maintained in cells exposed to HU or UV (Figure 3A). These data suggested that DNA cleaved by an Slx1-dependent mechanism is processed by an Rhp51-independent pathway.
Figure 3.
Genetic interactions involving Slx1-Slx4 endonuclease and Rhp51. The slx1Δ mutation does not improve the growth of rhp51Δ cells. (A) Single and double mutants in h+ or h- smt0 mating-type backgrounds were grown in YES media without exposure to genotoxic stress or after exposure to UV or in the presence of HU. (B) Generation times of rhp51Δ and slx1Δ rhp51Δ cells.
Expression of slx1-R34A Dominant Negative Allele in a rqh1Δ Strain Increases Stalled Forks in the rDNA
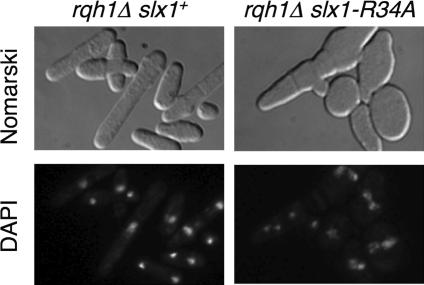
Our results supported a model in which Slx1-Slx4 endonuclease cleaves stalled forks in the rDNA array, generating DNA free ends that are repaired by a mechanism that requires Rad22 but not Rhp51. To specifically investigate the role of the endonuclease activity of Slx1-Slx4, we analyzed the slx1-R34A allele that encodes a mutant protein that is altered in the endonuclease catalytic site (URI domain) and is synthetic lethal with rqh1Δ (Coulon et al., 2004). We constructed a strain that has a plasmid containing slx1-R34A (with a TAP tag) integrated at the leu1 locus. This strain has no apparent phenotype (our unpublished data); however, when crossed into a rqh1Δ background, the resulting rqh1Δ slx1+ slx1-R34A:leu1 cells grew very poorly, forming small colonies with a plating efficiency that was reduced at least twofold relative to rqh1Δ cells. Swollen and multinucleated cells were detected in the culture (Figure 4). These phenotypes contrasted with a control strain that had a copy of slx1+ integrated at the leu1-32 locus (rqh1Δ slx1+ slx1+:leu1), which seemed to be identical to a rqh1Δ strain (Figure 4).
Figure 4.
Genetic interactions of rqh1Δ and slx1-R34A. Strains of the genotypes rqh1Δ slx1+:leu1 and rqh1Δ slx1-R34A:leu1 were grown in selective medium, washed three times with phosphate-buffered saline buffer, and DNA was stained with 1 μg/ml DAPI. Approximately 50% of rqh1Δ slx1-R34A:leu1 cells are swollen and contain multiple DAPI-staining nuclear structures, indicating a dominant negative effect of slx1-R34A.
From these results, we conclude that slx1-R34A has a dominant negative effect.
We used the rqh1Δ slx1+ slx1-R34A:leu1 strain and 2D-gel analysis of replication intermediates to investigate the role of Slx1-Slx4 endonuclease in processing stalled forks in the rDNA array. The 3-kb BamHI DNA fragment that encompasses RFB region of the rDNA was analyzed in chromosomal DNA samples taken from wild-type, slx1Δ, slx4Δ, rqh1Δ, rqh1Δ slx1+ slx1+:leu1, and rqh1Δ slx1+ slx1-R34A:leu1 strains (Figure 5, B and C). Y-arc and X-spike DNA structures were detected (Figure 5B). Y structures arise from moving replication forks within the BamHI fragment (Figure 5A). Pausing of replication forks at specific sites causes the accumulation of identical Y structures visible as distinct spots on the Y arc (Sanchez et al., 1998). X-shaped DNA structures are thought to arise from converged forks or recombination associated with DNA replication (Figure 5A).
Consistent with a recent study (Sanchez-Gorostiaga et al., 2004), we found that the RFB region in the S. pombe rDNA contains three pausing sites (RFB1, RFB2, and RFB3). Quantification of the RFB signals and X-shaped DNA structures as a percentage of the total replication and recombination intermediates revealed little difference between wild-type, slx1Δ, and slx4Δ strains (Figure 5, B and C). There was a moderate increase in both signals in the rqh1Δ strain. However, the largest effect was observed in the rqh1Δ slx1+ slx1-R34A:leu1 strain, which showed enhanced fork pausing and X-shaped structures relative to wild type, the rqh1Δ cells, and the matched rqh1Δ slx1+ slx1+:leu1 control strain (Figure 5, B and C). These effects of the mutant Slx1R34A-Slx4 complex are consistent with the model in which Slx1-Slx4 is directly involved in the maintenance of the rDNA arrays through processing of replication forks stalled at the RFBs.
DISCUSSION
In this study, we have analyzed the role of Slx1-Slx4 complex in initiating recombination events. Previous work with S. cerevisiae and S. pombe has shown that Slx1-Slx4 complex is a structure-specific DNA endonuclease that is essential for maintenance of the rDNA array in the absence of Sgs1/Rqh1 DNA helicase (Kaliraman and Brill, 2002; Fricke and Brill, 2003; Coulon et al., 2004). The studies in S. cerevisiae further indicated that the rDNA region cannot be fully replicated in cells that lack Slx1-Slx4 complex and Sgs1 (Kaliraman and Brill, 2002). In this article, we have attempted to provide deeper insight into the function of Slx1-Slx4 complex by determining whether its activity specifically provokes a requirement for HR proteins, as might be expected if it cleaves replication forks. We have found that Slx1-Slx4 complex indeed creates a need for Rad22, a protein that has a central role in HR. We found that elimination of Slx1-Slx4 activity improves the growth of rad22Δ mutants. Rad22 does not share this relationship with Mus81-Eme1 (Doe et al., 2004; our unpublished data), another structure-specific DNA endonuclease that is essential in the absence of Rqh1 and that has been proposed to cleave replication forks. Moreover, elimination of Slx1 does not suppress the growth defects of an rhp51Δ mutant, a finding that contrasts with the genetic interactions between rad22Δ and slx1Δ. These unique spectra of genetic interactions offer important clues about the function of Slx1-Slx4 endonuclease. The fact that slx1Δ and slx4Δ mutations partially suppress rad22Δ strongly suggests that Slx1-Slx4 endonuclease has a prorecombination activity in vivo.
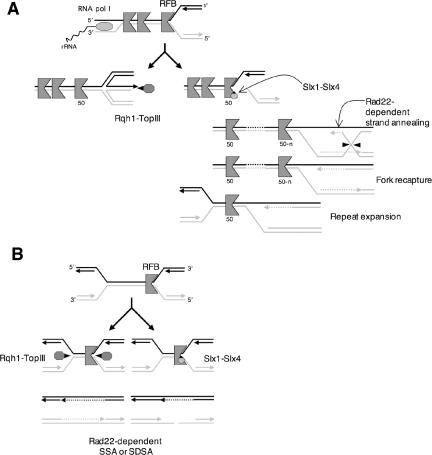
The data presented in this study suggest that the Slx1-Slx4 endonuclease complex creates DNA breaks that are processed by a pathway that requires Rad22 but is independent of Rhp51. Our data also suggest that the stalled replication forks at the RFB are the in vivo substrate of Slx1-Slx4 endonuclease. We propose a model of action of Slx1-Slx4 complex during replication of rDNA array (Figure 6). During S phase, the cleavage of stalled replication fork at RFB by Slx1-Slx4 may create a free DNA end. This DNA break may be processed by Rad22-dependent HR pathway (Figure 6A). This pathway may be analogous to the Rad52-dependent, Rad51-independent pathway of break-induced replication (BIR) described in S. cerevisiae (Malkova et al., 1996; Kraus et al., 2001). This mechanism might involve the activity of DNA helicases that unwind duplex DNA and thereby generate a substrate for Rad22-dependent strand annealing, possibly coupled with the presence of nicks or single strand gaps in the targeted rDNA repeat. Annealing of a free DNA end in an rDNA repeat located downstream of the initial Slx1-Slx4 cutting site restores replication fork and lead to expansion of rDNA repeats. How expansion/contraction of rDNA repeats is controlled is largely unknown, but a recent study in budding yeast showed that Sir2 in association with cohesin complex promotes equal sister-chromatid recombination in rDNA repeats preventing unequal rDNA rearrangements (Kobayashi et al., 2004). More recent work has shown that transcription from a noncoding bidirectional promoter within the rDNA spacer stimulates the dissociation of cohesin, permitting amplification of the rDNA repeats (Kobayashi and Ganley, 2005).
Figure 6.
Model for the action of Slx1-Slx4 complex on replication forks stalled at the RFB. Slx1-Slx4 cleaves replication forks stalled at RFB and the replication intermediates are processed by a Rad22-dependent pathway. (A) In S phase, cleavage of replication forks stalled at RFB by Slx1-Slx4 could trigger recombination events leading to expansion of rDNA repeats. DSBs generated by Slx1-Slx4 could be processed by a BIR-like mechanism. In contrast, Rqh1-TopIII could promote a nonrecombinogenic pathway by maintaining a chicken foot structure that arisen from a replication fork blocked at the RFB site. (B) During late S phase, a recombinogenic Slx1-Slx4-dependent pathway becomes essential in the absence of a nonrecombinogenic Rqh1-TopIII pathway to complete DNA replication.
Genetic evidence in S. cerevisiae and S. pombe showed that Slx1-Slx4 is essential in the absence of RecQ helicase (Mullen et al., 2001; Coulon et al., 2004). This synthetic lethality is not because of the loss of rDNA repeats but to a defect in completion of rDNA replication (Kaliraman and Brill, 2002; Coulon et al., 2004). In Escherichia coli, it has been shown that the concerted action of RecQ DNA helicase and topoisomerase III (TopIII) is able to promote single-strand DNA passage activity that can mediate decatenation of intertwined DNA molecules (Harmon et al., 1999, 2003). A related activity that dissolves two Holliday junctions has been described for human BLM helicase and TOPO III (Wu and Hickson, 2003). In our model (Figure 6A), we propose that Rqh1-TopIII could regress and maintain chicken foot structures that have arisen from replication forks blocked at an RFB site. This activity promotes a nonrecombinogenic pathway that is an alternative to an Slx1-Slx4-dependent pathway that can lead to rDNA rearrangements. It is also expected that converged forks produce intertwined DNA that can be decatenated by Sgs1- or Rqh1-TopIII complexes in yeast (Rothstein and Gangloff, 1995). In Figure 6B, we suggest a model in which Slx1-Slx4 becomes essential for resolution of converged fork at RFB due to the absence of Rqh1-TopIII. In this model, single cleavage of a stalled replication fork at RFB by Slx1-Slx4 can relax torsional forces and allow decatenation of parental DNA molecules in a pathway parallel to Rqh1-TopIII in late S phase. The replication of the noncleaved parental DNA molecule (black strands) is completed by gap filling with DNA polymerases, whereas replication of cleaved parental DNA molecule (gray strands) can be completed by single-strand annealing or synthesis-dependent strand annealing. We favor the single-cut model rather than simultaneous double-cut model proposed by Fricke and Brill (2003) because it has been shown that the structure of a stalled replication fork at RFB is different from the structure of traditional stalled replication fork due to DNA damage or torsional stress (Gruber et al., 2000). Slx1-Slx4 cleavage may be specific for a stalled replication fork at the RFB, but it is also possible that Slx1-Slx4 cleaves converged forks late in S phase as part of the termination of replication. Further studies will be required to test these models.
Current evidence indicates that in fission yeast the functions of Slx1 and Slx4 are confined to the heterodimeric Slx1-Slx4 endonuclease. For the purpose of this study, we have maintained this assumption and therefore have not carried out all of the experiments in parallel with both slx1Δ and slx4Δ mutations. However, it remains a formal possibility that either of the subunits have independent activities that are as yet undiscovered.
An important question that arises from these studies is whether Slx1-Slx4 complex cleaves stalled replication fork located outside of the rDNA arrays? In yeast, programmed pausing of replication fork is found in rDNA, mating-type locus, centromeric regions and tRNA genes. However, several types of data suggest that Slx1-Slx4 may be specific for the rDNA: 1) Slx1 is localized in rDNA (Coulon et al., 2004); 2) the sgs1-ts slx4 mutant has only a defect in replication of chromosome XII bearing the rDNA array in budding yeast at restrictive temperature (Kaliraman and Brill, 2002); and 3) in S. pombe, Slx1-Slx4 does not exhibit a mating-type switching defect, suggesting that a replication fork stalled at the mating-type locus is not a substrate for Slx1-Slx4 (our unpublished data). Although it seems that Slx1-Slx4 is specific for rDNA, the possibility that Slx1-Slx4 may also act at other sites in the genome cannot presently be excluded.
How does the Slx1-Slx4 complex work? Slx1 is a conserved protein through evolution, having clearly identifiable homologues throughout the eukaryotic kingdoms. These homologues share an endonuclease domain (URI domain) and a Ring-Finger domain. Related proteins in prokaryotic and archaeal species contain a URI domain but not a Ring-Finger domain (Aravind and Koonin, 2000). The Ring-Finger domain is thought to be involved in protein-protein interactions and in Slx1 is required for endonuclease activity (Fricke and Brill, 2003). ScSlx4 and SpSlx4 are very divergent proteins (Fricke and Brill, 2003; Coulon et al., 2004). However, both contain an SAP domain thought to promote protein-DNA interaction (Aravind and Koonin, 2000). A straightforward explanation is that Slx4 controls the Slx1 endonuclease activity and ensures substrate recognition through its SAP domain. In budding yeast, a large-scale genetic screen unveiled a synthetic lethal interaction between SLX4 and RAD50 (Tong et al., 2004) and supported the possibility that Slx4, but not Slx1, is involved in other pathways of tolerance of DNA damage. This hypothesis was supported by two recent publications that demonstrated that budding yeast Slx4 but not Slx1 is needed full resistance to camptothecin (Deng et al., 2005) and DNA alkylation damage (Flott and Rouse, 2005). Slx4 also has an Slx1-independent function in controlling the phosphorylation of Rtt107/Esc4, a protein involved in the maintenance of genome integrity (Roberts et al., 2006). These latest results unveil a multifaceted function of Slx4, whereas the role of Slx1 currently seems to be restricted to its endonuclease activity at the rDNA locus.
Acknowledgments
We thank Dr. Ding for the gift of plasmid pTA57 encoding for the Rrn5-GFP fusion protein. We thank all members of the Scripps Cell Cycle groups for encouragement and stimulating discussions, in particular Pierre-Henri Gaillard and Charly Chahwan. L.-L.D. is a fellow of the Leukemia and Lymphoma Society. T.M.N. was supported by the Damon Runyon Cancer Research Foundation Grant DRG-1565. This work was supported by Drexel University Start-up funds (to E. N.) and National Institutes of Health Grants CA-77325 and GM-59447 (to P. R.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1006) on February 8, 2006.
Abbreviations used: ARS, autonomously replicating sequence; 2D, two-dimensional; BIR, break-induced replication; DAPI, 4,6-diamidino-2-phenylindole; DSB, double-strand break; HJ, Holliday-Junction; HU, hydroxyurea; MMS, methyl methanesulfonate; RFB, replication fork barrier; WCE, whole cell extract.
References
- Aravind, L., and Koonin, E. V. (2000). SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25, 112-114. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E. V. (2001). Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 11, 1365-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M. N., Gaillard, P. H., McDonald, W. H., Shanahan, P., Yates, J. R., 3rd, and Russell, P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537-548. [DOI] [PubMed] [Google Scholar]
- Boddy, M. N., Lopez-Girona, A., Shanahan, P., Interthal, H., Heyer, W. D., and Russell, P. (2000). Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20, 8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, B. J., and Fangman, W. L. (1988). A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55, 637-643. [DOI] [PubMed] [Google Scholar]
- Coulon, S., Gaillard, P. H., Chahwan, C., McDonald, W. H., Yates, J. R., 3rd, and Russell, P. (2004). Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in Fission yeast. Mol. Biol. Cell 15, 71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, M. M. (2001). Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 35, 53-82. [DOI] [PubMed] [Google Scholar]
- Deng, C., Brown, J. A., You, D., and Brown, J. M. (2005). Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170, 591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D. Q., Tomita, Y., Yamamoto, A., Chikashige, Y., Haraguchi, T., and Hiraoka, Y. (2000). Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells 5, 169-190. [DOI] [PubMed] [Google Scholar]
- Doe, C. L., Dixon, J., Osman, F., and Whitby, M. C. (2000). Partial suppression of the fission yeast rqh1- phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19, 2751-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C. L., Osman, F., Dixon, J., and Whitby, M. C. (2004). DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 32, 5570-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L. L., Nakamura, T. M., Moser, B. A., and Russell, P. (2003). Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23, 6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto, T. (2001). Functions of RecQ family helicases: possible involvement of Bloom's and Werner's syndrome gene products in guarding genome integrity during DNA replication. J. Biochem. 129, 501-507. [DOI] [PubMed] [Google Scholar]
- Flott, S., and Rouse, J. (2005). Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem. J. 391, 325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke, W. M., and Brill, S. J. (2003). Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 17, 1768-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard, P. H., Noguchi, E., Shanahan, P., and Russell, P. (2003). The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell 12, 747-759. [DOI] [PubMed] [Google Scholar]
- Gerber, J. K., Gogel, E., Berger, C., Wallisch, M., Muller, F., Grummt, I., and Grummt, F. (1997). Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 90, 559-567. [DOI] [PubMed] [Google Scholar]
- Gruber, M., Wellinger, R. E., and Sogo, J. M. (2000). Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol. Cell. Biol. 20, 5777-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon, F. G., Brockman, J. P., and Kowalczykowski, S. C. (2003). RecQ helicase stimulates both DNA catenation and changes in DNA topology by topoisomerase III. J. Biol. Chem. 278, 42668-42678. [DOI] [PubMed] [Google Scholar]
- Harmon, F. G., DiGate, R. J., and Kowalczykowski, S. C. (1999). RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol. Cell 3, 611-620. [DOI] [PubMed] [Google Scholar]
- Hickson, I. D., Davies, S. L., Li, J. L., Levitt, N. C., Mohaghegh, P., North, P. S., and Wu, L. (2001). Role of the Bloom's syndrome helicase in maintenance of genome stability. Biochem. Soc. Trans. 29, 201-204. [DOI] [PubMed] [Google Scholar]
- Jia, S., Yamada, T., and Grewal, S. I. (2004). Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 119, 469-480. [DOI] [PubMed] [Google Scholar]
- Johzuka, K., and Horiuchi, T. (2002). Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7, 99-113. [DOI] [PubMed] [Google Scholar]
- Kaliraman, V., and Brill, S. J. (2002). Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet. 41, 389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman, V., Mullen, J. R., Fricke, W. M., Bastin-Shanower, S. A., and Brill, S. J. (2001). Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15, 2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, W. J., Lee, S., Park, M. S., Jang, Y. K., Kim, J. B., and Park, S. D. (2000). Rad22 protein, a rad52 homologue in Schizosaccharomyces pombe, binds to DNA double-strand breaks. J. Biol. Chem. 275, 35607-35611. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., and Ganley, A. R. (2005). Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309, 1581-1584. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Heck, D. J., Nomura, M., and Horiuchi, T. (1998). Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12, 3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., and Horiuchi, T. (1996). A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1, 465-474. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Horiuchi, T., Tongaonkar, P., Vu, L., and Nomura, M. (2004). SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117, 441-453. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Nomura, M., and Horiuchi, T. (2001). Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 136-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus, E., Leung, W. Y., and Haber, J. E. (2001). Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98, 8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings, G., and Bastia, D. (2004). swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 101, 14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings, G., and Bastia, D. (2005). Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J. Biol. Chem. 280, 39135-39142. [DOI] [PubMed] [Google Scholar]
- Laursen, L. V., Ampatzidou, E., Andersen, A. H., and Murray, J. M. (2003). Role for the fission yeast RecQ helicase in DNA repair in G2. Mol. Cell. Biol. 23, 3692-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens, M. H., and Huberman, J. A. (1988). Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, R. D., Platt, T. H., and Schildkraut, C. L. (1993). Initiation and termination of DNA replication in human rRNA genes. Mol. Cell. Biol. 13, 6600-6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-estrano, C., Schvartzman, J. B., Krimer, D. B., and Hernandez, P. (1998). Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J. Mol. Biol. 277, 249-256. [DOI] [PubMed] [Google Scholar]
- Malkova, A., Ivanov, E. L., and Haber, J. E. (1996). Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 93, 7131-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri, H. W., and Hickson, I. D. (2004). Understanding the roles of RecQ helicases in the maintenance of genome integrity and suppression of tumorigenesis. Biochem. Soc. Trans. 32, 957-958. [DOI] [PubMed] [Google Scholar]
- McGlynn, P., and Lloyd, R. G. (2002). Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 3, 859-870. [DOI] [PubMed] [Google Scholar]
- Mejia-Ramirez, E., Sanchez-Gorostiaga, A., Krimer, D. B., Schvartzman, J. B., and Hernandez, P. (2005). The mating type switch-activating protein Sap1 Is required for replication fork arrest at the rRNA genes of fission yeast. Mol. Cell. Biol. 25, 8755-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Mullen, J. R., Kaliraman, V., Ibrahim, S. S., and Brill, S. J. (2001). Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157, 103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, E., Noguchi, C., Du, L. L., and Russell, P. (2003). Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 23, 7861-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko, P. L., Cheng, W. H., and Bohr, V. A. (2004). Junction of RecQ helicase biochemistry and human disease. J. Biol. Chem. 279, 18099-18102. [DOI] [PubMed] [Google Scholar]
- Osman, F., Dixon, J., Doe, C. L., and Whitby, M. C. (2003). Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12, 761-774. [DOI] [PubMed] [Google Scholar]
- Paques, F., and Haber, J. E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, P. U., Defossez, P. A., and Guarente, L. (1999). Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 3848-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, T. M., Kobor, M. S., Bastin-Shanower, S. A., Ii, M., Horte, S. A., Gin, J. W., Emili, A., Rine, J., Brill, S. J., and Brown, G. W. (2006). Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol. Biol. Cell 17, 539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R., and Gangloff, S. (1995). Hyper-recombination and Bloom's syndrome: microbes again provide clues about cancer. Genome Res. 5, 421-426. [DOI] [PubMed] [Google Scholar]
- Rothstein, R., Michel, B., and Gangloff, S. (2000). Replication fork pausing and recombination or “gimme a break”. Genes Dev. 14, 1-10. [PubMed] [Google Scholar]
- Sanchez, J. A., Kim, S. M., and Huberman, J. A. (1998). Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp. Cell Res. 238, 220-230. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gorostiaga, A., Lopez-Estrano, C., Krimer, D. B., Schvartzman, J. B., and Hernandez, P. (2004). Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol. Cell. Biol. 24, 398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, E., Chapman, C., Al-Khodairy, F., Carr, A., and Enoch, T. (1997). rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16, 2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, Y., Horiuchi, T., and Kobayashi, T. (2003). Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17, 1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A. H., et al. (2004). Global mapping of the yeast genetic interaction network. Science 303, 808-813. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, M., Yamashita, K., Miyazaki, T., Shinohara, M., and Shinohara, A. (2003). The N-terminal DNA-binding domain of Rad52 promotes RAD51-independent recombination in Saccharomyces cerevisiae. Genetics 165, 1703-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa, S., and Yanagida, M. (1992). Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 101, 267-275. [DOI] [PubMed] [Google Scholar]
- van den Bosch, M., Vreeken, K., Zonneveld, J. B., Brandsma, J. A., Lombaerts, M., Murray, J. M., Lohman, P. H., and Pastink, A. (2001). Characterization of RAD52 homologs in the fission yeast Schizosaccharomyces pombe. Mutat. Res. 461, 311-323. [DOI] [PubMed] [Google Scholar]
- Watt, P. M., Hickson, I. D., Borts, R. H., and Louis, E. J. (1996). SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144, 935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt, P. M., Louis, E. J., Borts, R. H., and Hickson, I. D. (1995). Sgs 1, a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81, 253-260. [DOI] [PubMed] [Google Scholar]
- Whitby, M. C., Osman, F., and Dixon, J. (2003). Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 278, 6928-6935. [DOI] [PubMed] [Google Scholar]
- Wu, L., and Hickson, I. D. (2003). The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870-874. [DOI] [PubMed] [Google Scholar]
- Zou, H., and Rothstein, R. (1997). Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90, 87-96. [DOI] [PubMed] [Google Scholar]