Abstract
Hypoxia-inducible factor 1 (HIF-1) is controlled through stability regulation of its alpha subunit, which is expressed under hypoxia but degraded under normoxia. Degradation of HIF-1α requires association of the von Hippel Lindau protein (pVHL) to provoke ubiquitination followed by proteasomal digestion. Besides hypoxia, nitric oxide (NO) stabilizes HIF-1α under normoxia but destabilizes the protein under hypoxia. To understand the role of NO under hypoxia we made use of pVHL-deficient renal carcinoma cells (RCC4) that show a high steady state HIF-1α expression under normoxia. Exposing RCC4 cells to hypoxia in combination with the NO donor DETA-NO (2,2′-(hydroxynitrosohydrazono) bis-ethanimine), but not hypoxia or DETA-NO alone, decreased HIF-1α protein and attenuated HIF-1 transactivation. Mechanistically, we noticed a role of calpain because calpain inhibitors reversed HIF-1α degradation. Furthermore, chelating intracellular calcium attenuated HIF-1α destruction by hypoxia/DETA-NO, whereas a calcium increase was sufficient to lower the amount of HIF-1α even under normoxia. An active role of calpain in lowering HIF-1α amount was also evident in pVHL-containing human embryonic kidney cells when the calcium pump inhibitor thapsigargin reduced HIF-1α that was stabilized by the prolyl hydroxylase inhibitor dimethyloxalylglycine (DMOG). We conclude that calcium contributes to HIF-1α destruction involving the calpain system.
INTRODUCTION
The transcription factor hypoxia-inducible factor-1 (HIF-1) constitutes a central component in coordinating adaptive responses toward low oxygen availability, i.e., hypoxia. HIF-1 is a heterodimer composed of the 120-kDa HIF-1α subunit and the 91-94-kDa HIF-1β subunit (Semenza and Wang, 1992; Wang and Semenza, 1995). Although HIF-1β is constitutively expressed, oxygen facilitates continues destruction of HIF-1α via the 26S proteasome. This requires polyubiquitination by an E3-ubiquitin ligase complex that contains the von Hippel Lindau protein (pVHL; Kaelin, 2002; Bruick, 2003; Huang and Bunn, 2003; Pugh and Ratcliffe, 2003; Semenza, 2003). Ubiquitination and binding of pVHL to HIF-1α demands hydroxylation of Pro564 and/or Pro402 within the oxygen-dependent degradation domain (ODD) of HIF-1α (Ivan et al., 2001; Jaakkola et al., 2001; Masson et al., 2001). Hydroxylation is mediated by prolyl hydroxylases, also known as PH domain-containing enzymes (PHD, i.e., PHD1 to PHD4; Bruick and McKnight, 2001; Epstein et al., 2001; Oehme et al., 2002). In addition to regulating HIF-1α protein stability, oxygen affects the transcriptional activity of HIF-1, by regulating hydroxylation of a critical Asn803 residue within the C-terminal transactivation domain (CTAD) of HIF-1α (Lando et al., 2002). The asparagine hydroxylase known as FIH (factor inhibiting HIF; Mahon et al., 2001; Hewitson et al., 2002) renders CTAD unable to bind the coactivator p300/CBP. Thus, hypoxia attenuates Pro564/402 as well as Asn803 modifications, provoking protein stabilization and coactivator recruitment, which concomitantly results in HIF-1 transactivation.
Signals other than hypoxia such as nitric oxide (NO) and/or reactive nitrogen intermediates (RNI) participate in hypoxic signaling (Kimura et al., 2000; Palmer et al., 2000; Sandau et al., 2001; Thomas et al., 2004). Under normoxia, RNI evoke HIF-1α stabilization and formation of the active HIF-1 dimer to mimic a hypoxic response. These observations are compatible with the notion that RNI block PHD-activity. Thus, attenuating proline hydroxylation of HIF-1α hinders association with pVHL with the consequence of protein accumulation due to decreased ubiquitination/proteasomal degradation (Metzen et al., 2003). Opposite results are observed when RNI are generated under hypoxic conditions. RNI attenuate hypoxia-induced HIF-1α accumulation, HIF-1 DNA-binding and HIF-1 transcriptional activation (Liu et al., 1998; Sogawa et al., 1998; Huang et al., 1999). Although mechanistic details remained unclear it has been proposed that NO, derived from sodium nitroprusside, enhanced the interaction between HIF-1α and pVHL in vitro through reactivation of PHD activity (Wang et al., 2002). More recently it was proposed that NO, by blocking cytochrome c oxidase, leaves more oxygen available for PHD to regain its activity under hypoxic conditions (Hagen et al., 2003). Alternatively, reactive oxygen species including ONOO– may contribute to destabilize HIF-1α under hypoxia by mechanisms yet to be identified (Agani et al., 2002; Wellman et al., 2004).
We approached the question of RNI action in destabilizing HIF-1α by using renal cell carcinoma (RCC4) cells which are pVHL deficient and therefore permanently express HIF-1α and show active HIF-1. In RCC4 cells the combination of hypoxia and the NO donor DETA-NO lowered HIF-1α expression and HIF-1 activity by activating the calpain system without affecting transcription and translation of HIF-1α. The role of calpain in HIF-1α destruction was verified by applying calpain inhibitors, showing calcium dependencies and observing a direct binding of calpain to HIF-1α. Moreover, we provide evidence that calpain operates in pVHL-containing human embryonic kidney (HEK293) cells as well. We conclude that calcium and calpains participate in regulating HIF-1α expression and thus modulate responses toward hypoxia.
MATERIALS AND METHODS
Materials
Medium and supplements were purchased from PAA (Linz, Austria). Fetal calf serum (FCS) was from Biochrom (Berlin, Germany). S-nitrosoglutathione (GSNO) was synthesized as described (Hart, 1997). 2,2′-(hydroxynitrosohydrazono) bis-ethanimine (DETA-NO), NG-nitro-l-arginine methylester (l-NAME), cycloheximide (CHX), Z-Leu-Leu-Leu-al (MG132), lactacystin, epoxomicin, LPS, calpain inhibitor I (N-acetyl-Leu-Leu-Norleu-al, ALLN), calpain inhibitor II (N-Acetyl-l-leucyl-l-leucyl-l-methioninal, ALLM), ionomycin, thapsigargin, and anti-actin antibody were ordered from Sigma (Schnelldorf, Germany). Dimethyloxalylglycine (DMOG) was bought from Frontier Scientific (Lancashire, United Kingdom). BAPTA-AM (1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid-acetomethylester) was purchased from Molecular Probes (Leiden, Netherlands). Calpastatin peptide and human calpain-1 were from Calbiochem (Bad Soden, Germany). Interferon γ (IFNγ) and protease inhibitor cocktail came from Roche (Mannheim, Germany). Nitrocellulose membrane, ECL detection system and horseradish peroxidase (HRP)-labeled anti-mouse or anti-rabbit secondary antibodies were delivered by Amersham Biosciences (Freiburg, Germany). Anti-HIF-1α antibody and Clontech Advantage RT-for-PCR kit were purchased from Becton Dickinson (Heidelberg, Germany). Anti-IκBα antibody was bought from Santa Cruz (Heidelberg, Germany), the anti-HA monoclonal antibody came from CRP (Denver, CO), anti-calpain-1 antibody was delivered by Biomol (Hamburg, Germany) and anti-mouse-DynaBeads were purchased from Invitrogen (Karlsruhe, Germany). Primers were ordered from MWG-Biotech (Ebersberg, Germany). The plasmid pHA-HIF-1α encoding HA-tagged-HIF-1α was kindly provided by Dr. P. J. Ratcliffe (Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, United Kingdom). The plasmid pGLEPOHRE harboring three erythropoietin hypoxia-responsive elements (HRE) was from Dr. T. Kietzmann (University of Kaiserslautern, Kaiserslautern, Germany). Reporter plasmid cap-Luc and luciferase activity assay kit were supplied by Promega (Mannheim, Germany).
Cell Culture
Renal cell carcinoma (RCC4), RCC4 cells with pVHL being reintroduced (RCC4/VHL) and human embryonic kidney 293 (HEK293) cells were cultured in DMEM with 4.5 g/l glucose, supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 10% FCS. Cells were kept at 37°C in a humidified atmosphere with 5% CO2. For hypoxic exposure, cells were incubated at 0.5% O2 in a hypoxia workstation (Ruskinn Technology, Leeds, United Kingdom).
Western Blotting
RCC4 cells, 5 × 105, were seeded in 6-cm dishes 1 d before experiments. After treatments, cells were scraped off, lysed in 150 μl buffer A (50 mM Tris/HCl, 150 mM NaCl, 5 mM EDTA, 0.5% Nonidet-40, protease inhibitor cocktail, pH 7.5), and sonicated. After centrifugation (15000 × g, 15 min) the protein content was determined in the supernatants by a protein assay kit (Bio-Rad, Munich, Germany) and 80 μg protein was added to the same volume of 2× SDS-PAGE sample buffer (125 mM Tris/HCl, 2% SDS, 10% glycerin, 1 mM DTT, 0002% bromophenol blue, pH 6.9) and boiled for 5 min. Proteins were resolved on 10% SDS-polyacrylamide gels and blotted onto nitrocellulose membranes. Nonspecific binding sites were blocked with 5% (wt/vol) defatted milk powder in TTBS (50 mM Tris/HCl, 140 mM NaCl, 0.05% Tween-20, pH 7.2) for 1 h. The HIF-1α, actin and IκBα antibodies (1:1000 in 1% milk/TTBS) were added and incubated overnight at 4°C. Afterward, nitrocellulose membranes were washed three times for 5 min each with TTBS. Blots were then incubated with goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with HRP (1:2000 in 1% milk/TTBS) for 1 h and washed three times for 5 min each with TTBS, followed by ECL detection.
Quantitative Real-Time RT-PCR
RCC4 cells, 2 × 106, were seeded in 10-cm dishes 1 d before experiments. The following day medium was changed and cells were treated as indicated. Total RNA was isolated using the peqGOLD RNAPure kit (Peqlab, Erlangen, Germany). The reverse transcription was completed with a Clontech Advantage RT-for-PCR kit using hexamer random primers. The following primer pairs were selected for quantitative real-time PCR: human HIF-1α forward: 5′-CTCAAAGTCGGACAGCCTCA-3′; human HIF-1α backward: 5′-CCCTGCAGTAGGTTTCTGCT-3′; human actin forward: 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′; and human actin backward: 5′-CTAGAAGCATTTGCGGTCGACGATGGAGGG-3′.
The quantitative real-time PCR was performed by MyiQ (Bio-Rad). Reaction mixtures containing SYBR Green were composed according to the manufacturer's protocol. The cycling program was as follows: 50°C, 2 min; 95°C, 15 min; followed by 35 cycles at 95°C, 15 s; 55°C, 30 s; 72°C, 30 s. Values of HIF-1α were then normalized to the relative amounts of actin.
Cell Transfection and Reporter Assay
RCC4 cells, 2 × 105, were seeded in 6-cm dishes 1 d before transfection. At a rate of 60% confluence, cells were transfected with reporter plasmids or pHA-HIF-1α, using the Polyfect transfection reagent (Qiagen, Hilden, Germany) according to instructions of the manufacturer. Sixteen hours later medium was changed and incubations continued for another 8-h period. For reporter assays the medium was replaced and cells were treated as indicated for an additional 16-h period. Cells were lysed and luciferase activity was measured. Relative values of treated cells were calculated by normalizing to values of control cells (set as 100). For HA-HIF-1α expression, cells were treated for 4 h as indicated.
Peptide Array Assay
The peptide array containing HIF-1α peptide spots was generated after SPOT synthesis. Briefly, 272 overlapping peptide fragments of 15 amino acids in length with an offset of 3 amino acid residues were generated such that the complete HIF-1α protein sequence was covered. These HIF-1α peptides were chemically synthesized as an array of spots on an aminopegylated cellulose membrane as described previously (Frank and Overwin, 1996). All peptides are N-terminally acetylated and remain covalently attached to the membrane via their carboxy-termini. Binding studies were performed according to an adapted protocol from Frank and Overwin (1996). In brief, unspecific binding sites were blocked with buffer B (50 mM Tris/HCl, 150 mM NaCl, 5 mM EGTA, 5% glycerol, protease inhibitor cocktail, 1% BSA, pH 7.0). Afterward the membrane was incubated with 1 μg/ml calpain-1 in buffer B overnight at 4°C. For detection of binding an anti-calpain-1 antibody (1:1000 in buffer B) was added and incubated for 2 h at room temperature. Thereafter, the membrane was washed three times for 5 min each with phosphate-buffered saline (PBS; pH 7.0). For visualization, the membrane was incubated with a HRP-labeled goat anti-mouse secondary antibody (1:2000 in buffer B) for 1 h at room temperature and washed two times for 5 min each with TTBS and 5 min with PBS, followed by ECL detection. After signal documentation, the HIF-1α peptide array was stripped as described by Frank and Overwin (1996). Incubations were then repeated with the secondary antibody alone or with the combination of the anti-calpain-1 antibody and the secondary antibody in order to exclude unspecific signals originating from antibody bindings only.
HIF-1α–Calpain Coimmunoprecipitation
RCC4 cells, 2 × 106, were seeded in 10-cm dishes 1 d before experiments. After treatments, cells were scraped off from the dishes and collected. To each cell pellet 300 μl buffer C (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 0.5% NP-40, 5% glycerol, protease inhibitor cocktail, pH 7.5) was added, followed by immediate vortexing (3 × 15 s) and centrifugation (15000 × g for 30 min). Supernatants (1 mg protein) were transferred into fresh tubes, supplied with 1 μg anti-HIF-1α antibody, and incubated at 4°C for 1 h. Thereafter, 20 μl anti-mouse-DynaBeads were added and incubations continued at 4°C overnight. Beads were collected, washed three times with 100 μl buffer C, and finally supplemented with 50 μl 2× SDS-PAGE sample buffer and boiled at 95°C for 10 min. Beads were removed by centrifugation and supernatants were loaded on 7.5% SDS-PAGE. Western blot analysis was performed using calpain-1 or anti-HIF-1α antibodies.
Densitometric Quantification
Densitometric quantification (expressed as quantum levels) of HIF-1α and IκBα signals was performed with the Aida Image Software (Raytest Isotopenmessgeraete GmbH, Straubenhardt, Germany). Relative intensities were calculated from the Quantum levels ratios of HIF/IκBα by setting controls as 100.
Statistical Analysis
Each experiment was performed at least three times and representative data are shown. Data in bar graphs are given as mean values ± SEM. Means were checked for statistical differences by using the Student's t test with error probabilities of p < 0.05 (*).
RESULTS
Impact of Hypoxia/NO on HIF-1α Accumulation
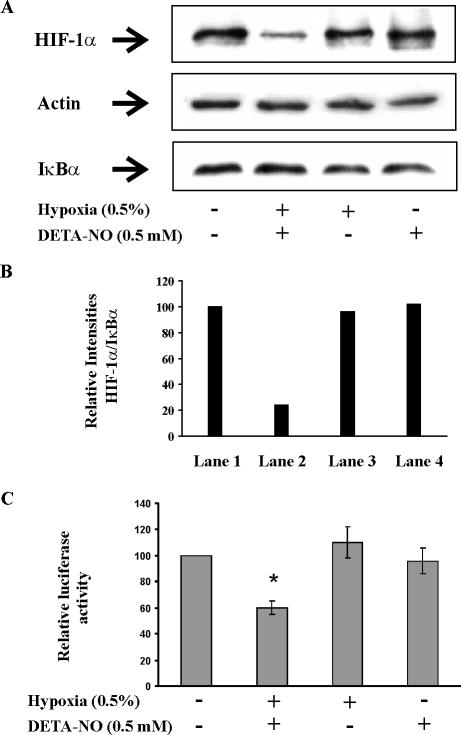
In RCC4 cells HIF-1α is constitutively expressed because a functional pVHL is lacking. Incubations of RCC4 cells for 4 h with 0.5 mM DETA-NO under hypoxia (0.5% O2) decreased HIF-1α protein level (Figure 1A). As controls, the expression of actin and IκBα remained constant under all treatments. Similar amounts of the housekeeping protein actin indicated equal loading of total protein to the gel. Constant expression of the rapidly turned-over IκBα implied that destruction of HIF-1α was not shared by other proteins that are degraded by the proteasome. Densitometric analysis indicated that only 24% of the HIF-1α signal remained under hypoxia/NO cotreatment compare to controls or hypoxia and/or DETA-NO added alone (Figure 1B). Destruction of HIF-1α was not seen when hypoxia or DETA-NO were supplied individually.
Figure 1.
Hypoxia and NO attenuate HIF-1 in RCC4 cells. (A) For protein analysis, RCC4 cells were exposed to hypoxia (0.5%), treated with 0.5 mM DETA-NO, a combination of hypoxia/DETA-NO for 4 h, or remained as controls. Western blotting was used to follow expression of HIF-1α, actin, and IκBα. Results are representative for three individual experiments. (B) For quantification of signals from A, densitrometric quantum levels of lanes 1–4 (from left to right) were measured, and relative intensities were calculated from ratios of HIF/IκBα by setting controls as 100. (C) For transactivation analysis, 2 × 105 RCC4 cells were transfected with 0.5 μg of the plasmid pGLEPOHRE and exposed to hypoxia (0.5%), 0.5 mM DETA-NO, combinations thereof for 16 h or remained as controls. After cell lysis, luciferase activity was measured and normalized compared with controls. Data are the mean ± SEM (n = 3). Significant alterations are expressed relative to controls.
Looking for the transactivation potential of HIF-1α we transfected RCC4 cells with a pGLEPOHRE reporter construct and followed expression of luciferase as a marker for HIF-1 activity (Figure 1C). As expected, we measured a basal activity that was neither increased by incubating cells under hypoxic conditions nor when treating cells with the NO-liberating compound DETA-NO. However, exposing cells to the combination hypoxia/DETA-NO lowered reporter activity by roughly 40%. Control experiments, by looking for trypan blue exclusion, excluded that cell viability was affected by any treatment.
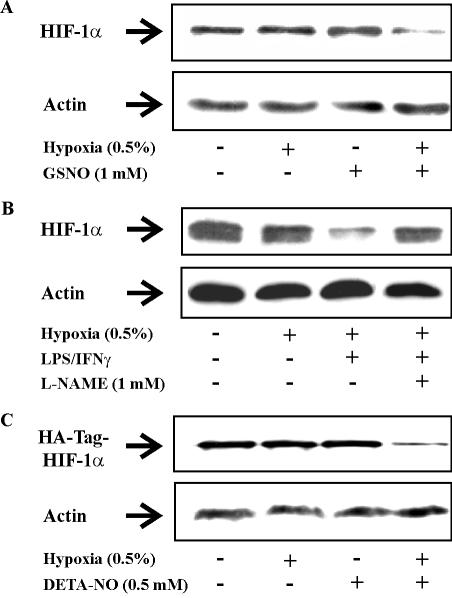
Further experiments with GSNO, an alternative NO donor, at a concentration of 1 mM, reproduced results seen with DETA-NO (Figure 2A). GSNO only decreased the amount of HIF-1α in combination with hypoxia.
Figure 2.
Hypoxia/NO attenuates endogenous and exogenous HIF-1α expression in RCC4 cells. (A) Cells were exposed for 4 h to hypoxia (0.5%), treated with 1 mM GSNO, a combination of hypoxia/GSNO, or remained as controls. (B) Cells were exposed to hypoxia (0.5%) for 16 h, a combination of hypoxia/LPS (1 μg/ml)/IFNγ (100 U/ml) for 16 h, a combination of hypoxia/LPS/IFNγ in the presence of 1 mM l-NAME for 16 h, or remained as controls. (C) Cells were transfected with 3 μg/dish pHA-HIF-1α plasmid. Twenty-four hours later, cells were exposed for 4 h to hypoxia (0.5%), treated with 0.5 mM DETA-NO, a combination of hypoxia/DETA-NO, or remained as controls. Western blotting was used to follow expression of HIF-1α by using anti-HIF-1α or anti-HA-Tag monoclonal antibodies relative to actin. Results are representative for three individual experiments.
To answer the question whether endogenously produced NO would suffice in lowering HIF-1α under hypoxia, we stimulated RCC4 cells with LPS/IFNγ, which is known to express inducible NO-synthase in these cells. The combination of hypoxia/LPS/IFNγ lowered the protein amount of HIF-1α (Figure 2B). Interestingly, blocking NO production with the NO-synthase inhibitor l-NAME restored HIF-1α expression, thus pointing to the essential role of NO in lowering protein expression of HIF-1α under hypoxia. Under normoxia the addition of LPS/IFNγ left HIF-1 expression unaltered (data not shown). To confirm destruction of exogenous HIF-1α under hypoxia/NO, we overexpressed HA-tagged HIF-1α in RCC4 cells by transfecting the pHA-HIF-1α plasmid (Figure 2C). The combination of hypoxia/NO lowered the amount of exogenous HIF-1α protein comparable to endogenous HIF-1α, whereas hypoxia or NO alone were ineffective.
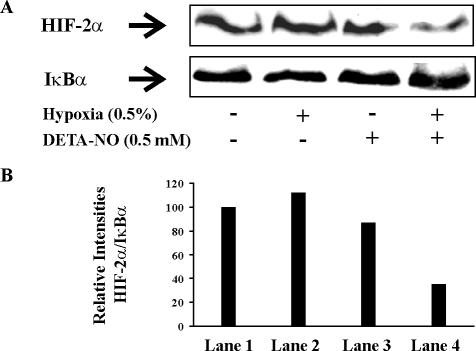
Similarly, expression of HIF-2α was reduced to 35% compared with controls under the influence of hypoxia/DETA-NO (Figure 3).
Figure 3.
Hypoxia and NO attenuate expression of HIF-2α in RCC4 cells. (A) Cells were exposed to hypoxia (0.5%), treated with 0.5 mM DETA-NO, a combination of hypoxia/NO donor for 4 h, or remained as controls. Western blotting was used to follow expression of HIF-2α relative to IκBα. Results are representative for three individual experiments. (B) Densitrometric quantum levels of lanes 1–4 (from left to right) from A were measured, and relative intensities were calculated from the ratios of HIF/IκBα by setting controls as 100.
Considering that expression regulation of HIF-2α and HIF-1α are very similar (Cockman et al., 2000), we concentrated on HIF-1α only. We conclude that in combination with hypoxia exogenously supplied or endogenously generated NO reduced HIF-1α accumulation and HIF-1 activity in RCC4 cells and thus in a pVHL-independent manner.
Hypoxia/NO Promotes pVHL-independent Degradation of HIF-1α
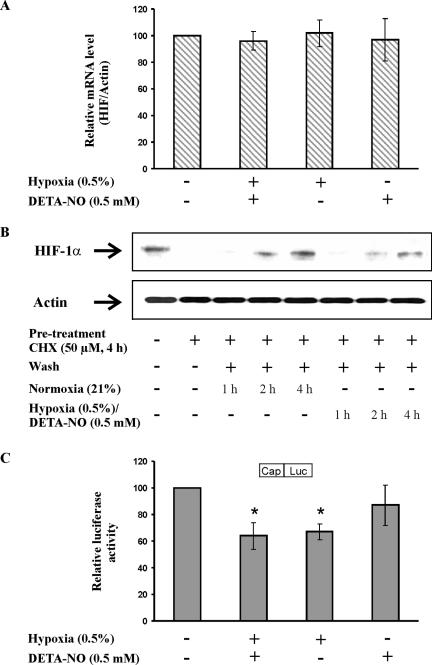
To rule out that variations in HIF-1α mRNA account for changes in HIF-1α protein under hypoxia/NO costimulation, we used quantitative real-time RT-PCR to follow HIF-1α mRNA (Figure 4A). HIF-1α mRNA remained constant and was neither affected by hypoxia or by DETA-NO given alone nor by coincubating hypoxia/DETA-NO.
Figure 4.
Hypoxia/NO and the transcriptional versus translational control of HIF-1α. (A) RCC4 cells were exposed to hypoxia (0.5%), 0.5 mM DETA-NO, combinations thereof for 4 h, or remained as controls. HIF-1α as well as actin mRNA was determined by quantitative real-time PCR. The ratio of HIF-1α/actin mRNA under control conditions was set to 100 and the mRNA ratios of samples are expressed relative to the control. Data are the mean ± SEM (n = 3). (B). RCC4 cells were pretreated with 50 μM cycloheximide (CHX) for 4 h or remained as a control. Afterward, cells were washed once with medium (wash) and incubations continued for 1, 2, or 4 h either under normoxia or under conditions of hypoxia/DETA-NO cotreatment. Western analysis was used to follow the expression of HIF-1α and actin. Results are representative for three individual experiments. (C) RCC4 cells, 2 × 105, were transfected with 0.5 μg of the plasmid cap-Luc and exposed to hypoxia (0.5%), 0.5 mM DETA-NO, combinations thereof for 16 h or left as controls. After cell lysis, luciferase activity was measured and normalized compared with controls. Data are the mean ± SEM (n = 3). Significant alterations are expressed relative to controls.
With the further experiment it was our intention to check a possible interference with translational control mechanisms of HIF-1α. To this end we exposed RCC4 cells for 4 h to 50 μM cycloheximide to block protein translation (Figure 4B). Thereafter, the medium was changed to wash out cycloheximide and incubations continued for 1–4 h under normoxia. This allowed recovering expression of HIF-1α. We then repeated the same experimental protocol in the presence of hypoxia/DETA-NO. After cycloheximide treatment HIF-1α disappeared but its expression recovered during the next 4 h even under conditions of hypoxia/DETA-NO. However, the reappearance of HIF-1α was weaker compared with the normoxic situation. Although indirect, the experiment might suggest that translation of HIF-1α was operating even under hypoxia/DETA-NO. To gain further support, we performed reporter assays to test cap-dependent translation in general (Figure 4C). As established, we reconfirmed that translation slowed down under hypoxia and noticed that there was no further alteration under hypoxia/DETA-NO cotreatments. Despite these results not completely eliminate the possibility of HIF-1α translational regulation, they showed that the combination of hypoxia/DETA-NO did not selectively impaired translation. Taking into account that the amount of exogenous HIF-1α is lowered by hypoxia/NO as well (Figure 2C) points to protein destruction as the underlying principle. Therefore, we focused on alternative pathways to explain HIF-1α destruction.
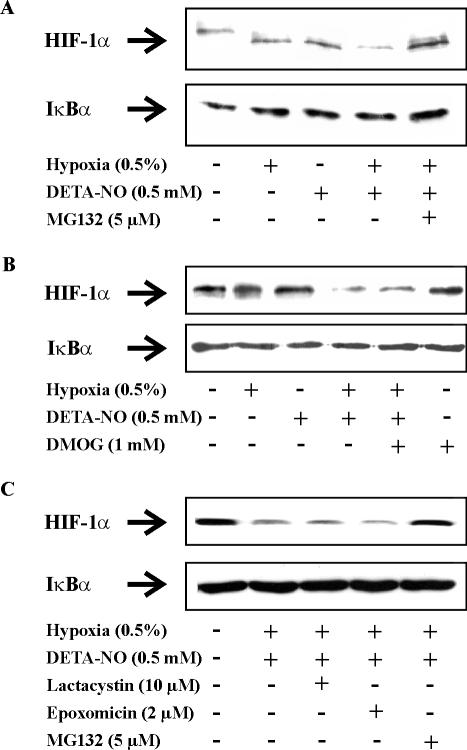
To address the importance of HIF-1α destruction, we asked whether inhibition of protein degradation with MG132 might affect HIF-1α disappearance under conditions when hypoxia and DETA-NO were supplied simultaneously. As seen in Figure 5A, attenuated accumulation of HIF-1α as a result of hypoxia/DETA-NO treatment was fully restored in the presence of the putative proteasome inhibitor MG132. This pointed to protein degradation in lowering HIF-1α protein level in the presence of hypoxia/DETA-NO.
Figure 5.
Hydroxylation- and 26S proteasome-independent degradation of HIF-1α. RCC4 cells were exposed for 4 h to hypoxia (0.5%), 0.5 mM DETA-NO, the combination of hypoxia/DETA-NO, the combination of hypoxia/DETA-NO with the addition of (A and C) the 26S proteasome inhibitor 5 μM MG132, (B) the prolyl hydroxylase inhibitor 1 mM DMOG, (C) the specific 26S proteasome inhibitor 10 μM lactacystin or 2 μM epoxomicin, or remained as controls. Expression of HIF-1α and IκBα were determined by Western blotting. Results are representative for three individual experiments.
Considering MG132 as a putative proteasomal inhibitor, results might point to an alternative pVHL-like E3 ligase and hydroxylation/ubiqutination/proteasome degradation system to be involved. Therefore, we assumed that the PHD inhibitor DMOG should be equally effective compared with MG132 because DMOG is supposed to block hydroxylation of HIF-1α and thus is expected to increase stabilization of HIF-1α. However, in contrast to MG132 the addition of DMOG under conditions of hypoxia/DETA-NO did not restore HIF-1α expression (Figure 5B). These observations are consistent with the notion that RCC4 cells do not contain the usual HIF-1α degradation system because pVHL is missing. More important, these results question the specificity of MG132 as a selective proteasomal inhibitor. We then applied more specific proteasome inhibitors such as lactacystin and epoxomicin. Interestingly, none of them reversed HIF-1α destruction under hypoxia/DETA-NO (Figure 5C). As these results ruled the 26S proteasome in degrading HIF-1α out, we searched for alternative destruction pathways that might explain the disappearance of HIF-1α in RCC4 cells when exposed to hypoxia and DETA-NO.
Calcium/Calpain-mediated HIF-1α Degradation
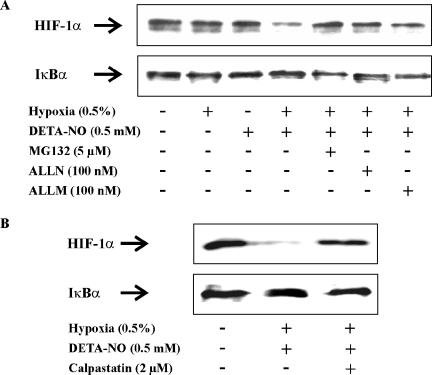
Considering that MG132, besides blocking proteasomal activity, also inhibits calpains and some lysosomal cysteine proteases (Tawa et al., 1997; Lee and Goldberg, 1998), we went on to test for the involvement of calpain in HIF-1α degradation. In a first approach we used two calpain inhibitors such as calpain inhibitor I (ALLN) and calpain inhibitor II (ALLM) and noticed their ability to fully restore HIF-1α stabilization that was compromised under the cotreatment of hypoxia/DETA-NO (Figure 6A). ALLN and ALLM, both at 100 nM, were equally effective compared with MG132 in blocking HIF-1α degradation.
Figure 6.
Calpain mediated HIF-1α destruction. (A) RCC4 cells were exposed for 4 h to hypoxia (0.5%), 0.5 mM DETA-NO, hypoxia/DETA-NO, the combination of hypoxia/DETA-NO with the addition MG132, ALLN, ALLM, or remained as a control. (B) RCC4 cells were exposed for 4 h to the combination of hypoxia/DETA-NO, the combination of hypoxia/DETA-NO with the addition of the calpain inhibitor calpastatin peptide (2 μM), or remained as a control. Expression of HIF-1α and IκBα were followed by Western analysis. Results are representative for three individual experiments.
To confirm a direct role of calpain in HIF-1α degradation, we applied the specific calpain inhibitor calpastatin, a 27-residue peptide inhibitor. Calpastatin peptide indeed blocked HIF-1α destruction under hypoxia/DETA-NO (Figure 6B). From these inhibitor studies we conclude that calpain is involved in HIF-1α degradation.
The Role of Calcium in Provoking Calpain-mediated HIF-1α Degradation
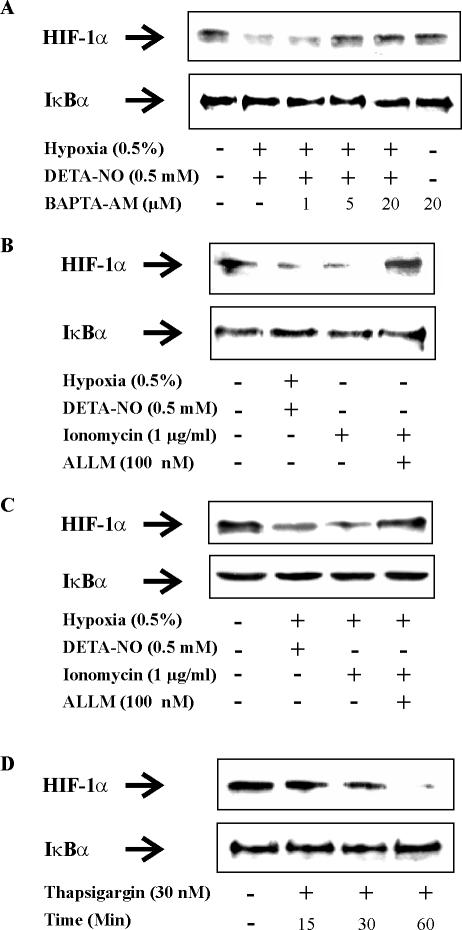
To establish a modulator role of calcium in affecting HIF-1α expression, we sought to inhibit the process by chelating intracellular calcium. Incubating RCC4 cells with 1, 5, or 20 μM BAPTA-AM dose-dependently reversed the disappearance of HIF-1α that was initiated by hypoxia/DETA-NO (Figure 7A). In the presence of 20 μM BAPTA-AM the effect of hypoxia/DETA-NO was nullified, whereas the intracellular calcium chelator alone did not affect HIF-1α accumulation in RCC4 cells.
Figure 7.
Ca2+ regulates the expression of HIF-1α in RCC4 cells. (A) RCC4 cells were exposed to combinations of hypoxia (0.5%)/DETA-NO (0.5 mM) either alone or in the presence of increasing concentrations of the Ca2+ chelator BAPTA-AM for 4 h. Alternatively, cells were treated with 20 μM BAPTA-AM under normoxia, or remained as a control. (B and C) RCC4 cells were exposed for 4 h to combinations of hypoxia (0.5%)/DETA-NO (0.5 mM), or treated with 1 μg/ml ionomycin with or without the addition of the calpain inhibitor ALLM (100 nM) under (B) normoxia or (C) hypoxia, or remained as controls. (D) RCC4 cells were exposed to 30 nM thapsigargin for 15, 30, or 60 min under normoxia or left untreated. Expression of HIF-1α and IκBα were followed by Western analysis. Results are representative for three individual experiments.
To proceed, we aimed at increasing intracellular calcium with the intention to destruct HIF-1α under normoxia. Exposing RCC4 cells for 4 h to 1 μg/ml the calcium ionophore ionomycin lowered expression of HIF-1α (Figure 7B). This effect was completely attenuated in the presence of the calpain inhibitor ALLM. Interestingly, degradation that was initiated by raising intracellular calcium with ionomycin was equally effective to the combination of hypoxia/DETA-NO in lowering the HIF-1α amount but left expression of IκB unaltered. As expected, under hypoxia ionomycin was equally effective in lowering the amount of HIF-1α (Figure 7C). In an additional experiment we have chosen the endoplasmic reticulum calcium (ER) pump inhibitor thapsigargin, known to promote a cytoplasmic calcium increase (Figure 7D). Incubating RCC4 cells under normoxia with 30 nM thapsigargin time-dependently lowered the amount HIF-1α, with the protein almost disappearing after 60 min. These results support the notion that calpain participates in regulating degradation of HIF-1α, in a pVHL-independent manner.
Calcium/Calpain-mediated HIF-1α Degradation in pVHL-containing Cells
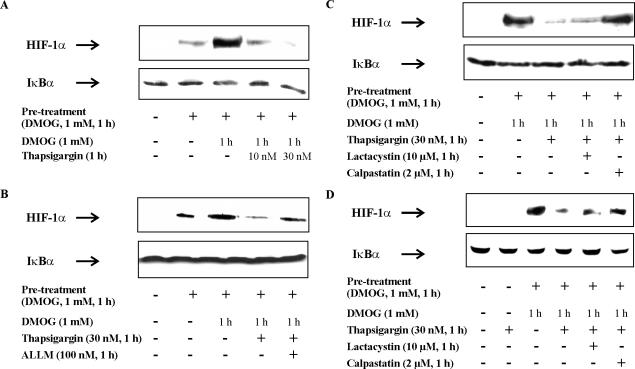
To answer the question whether calpain-mediated degradation of HIF-1α is restricted to RCC4 cells or can be observed in other cells as well, we performed additional experiments in pVHL-containing HEK293 cells. HEK293 cells were first incubated with 1 mM DMOG for 1 h to block PHD activity and to allow HIF-1α accumulation (Figure 8A). Afterward, incubations continued for 1 h either in the presence of DMOG alone or with the further addition of 10 versus 30 nM thapsigargin. As shown in Figure 8A, DMOG-induced HIF-1α accumulation was dose-dependently reduced by thapsigargin. Thapsigargin alone neither time- nor concentration-dependently accumulated HIF-1α (data not shown).
Figure 8.
Ca2+/calpain functions in pVHL-containing HEK293 and RCC4/VHL cells. HEK293 cells were pretreated with 1 mM PHD hydroxylase inhibitor DMOG for 1 h or remained as a control. Afterward, incubations continued for 1 h in the presence of DMOG alone or in the presence of DMOG with the addition of (A–C) 10 nM or 30 nM thapsigargin, (B) 100 nM calpain inhibitor ALLM, (C) 10 μM of the 26S proteasome inhibitor lactacystin, or 2 μM calpain inhibitor calpastatin peptide. Western analysis was used to follow the expression of HIF-1α and IκBα. (D) RCC4/VHL cells were pretreated for 1 h with 1 mM DMOG or 30 nM thapsigargin, or remained as a control. Pretreated cells were further exposed to DMOG alone, to DMOG with the addition of 30 nM thapsigargin, to DMOG with the addition of 30 nM thapsigargin and 10 μM lactacystin or 2 μM calpastatin peptide. Western analysis was used to follow the expression of HIF-1α and IκBα. Results are representative for three individual experiments.
Considering that PHD activity was blocked by DMOG suggests that besides the pVHL-dependent pathway an alternative, the calcium-regulated degradation system operates in HEK293 cells as well. Next we repeated the experimental protocol described in Figure 8A. However, applying ALLM and calpastatin peptide in combination with thapsigargin attenuated degradation of HIF-1α and restored expression of the protein close to a value seen with DMOG alone (Figure 8, B and C). As expected, the specific proteasome inhibitor lactacystin was unable to reverse HIF-1α destruction (Figure 8C). To match the RCC4 cell background, RCC4/VHL cells, which contain a stably reintroduced pVHL expression plasmid, were applied as a pVHL-containing cell to repeat experiments. DMOG accumulated HIF-1α in RCC4/VHL cells (Figure 8D). HIF-1α disappeared when cells were treated with thapsigargin and reappeared with the presence of calpastatin but not lactacystin. These results strengthen a role of calpain-mediated HIF-1α degradation.
Interaction between HIF-1α and Calpain-1
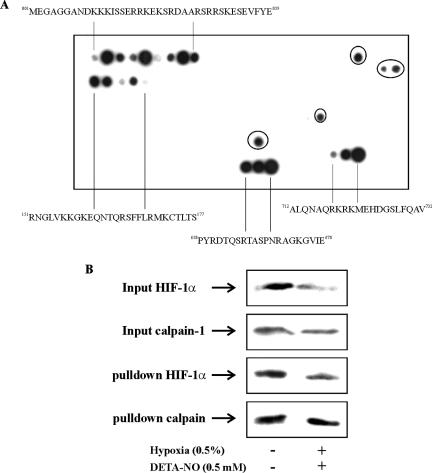
To provide complementary evidence to show an interaction between HIF-1α and calpain, we used a HIF-1α peptide spot array. The membrane was incubated with human calpain-1, followed by anti-calpain-1 antibody detection (Figure 9A).
Figure 9.
Human calpain-1 associates with HIF-1α. (A) Human calpain-1 protein was incubated with HIF-1α peptide fragments (each 15 amino acids long) spotted on a membrane. Incubations of human calpain-1 with the HIF-1α peptide array was performed as described in Material and Methods. Sequences indicating binding of calpain-1 were highlighted. Circled spots refer to unspecific binding of primary and/or secondary antibodies. (B) RCC4 cells were incubated under normoxia or the combination of hypoxia (0.5% O2) and DETA-NO (0.5 mM) for 4 h. Cells were then collected followed by immunoprecipitation as described under Materials and Methods. Input controls show protein expression in cell lysates. Immunoprecipitates were probed for HIF-1α and calpain-1. Western analysis was performed using anti-HIF-1α or anti-calpain-1 antibodies. The experiment was performed at least three times and representative data are shown.
This approach allowed us to identify four binding regions highlighted on the peptide array. We mapped four potential binding sites (deduced from the spot location) of human calpain-1 to HIF-1α with the sequences: 001MEGAGGANDKKKISSERRKEKSRDAARSRRSKESEVFYE039, 151RNGLVKKGKEQNTQRSFFLRMKCTLTS177, 658PYRDTQSRTASPNRAGKGVIE678, and 712ALQNAQRKRKMEHDGSLFQAV732. One should keep in mind that these potential calpain-1 binding sites not necessarily overlap with cleavage sites, especially as the binding assay has been performed with an inactive calpain enzyme, i.e., with the presence of EGTA. To further investigate the interaction between HIF-1α and calpain under cellular conditions, we used cell lysate of RCC4 cells either kept under normoxia or hypoxia/DETA-NO to perform coimmunoprecipitations using an anti-HIF-1α antibody for precipitation, followed by detection of calpain-1 (Figure 9B). Evidently, calpain bound to HIF-1α under normoxic and hypoxia/DETA-NO conditions. Control experiments leaving out the HIF-1α antibody used for immunoprecipitation revealed no unspecific binding of calpain-1 to the beads (data not shown). It should be mentioned that for these experiments calpain-1 activity again was blocked by using EGTA. Based on coimmunoprecipitation experiments one should be cautious to predict quantitative ratios of protein associations and therefore, further experiments need to determine whether calpain activation alters the amount of calpain bound to HIF-1α and whether this type of protein association is under the control of regulatory signaling pathways.
DISCUSSION
Degradation of HIF-1α under normoxia and its stabilization in hypoxia constitutes a major regulatory component in understanding cellular responses during the transition from ambient to low oxygen tension. We provide new evidence that calpain participates in destruction of HIF-1α, supporting the notion for a role of calcium in affecting expression of hypoxia-inducible genes. Studies in pVHL-deficient RCC4 cells and pVHL-containing RCC4/VHL or HEK293 cells imply that calpain-evoked destruction of HIF-1α must be considered an alternative system besides the established proteasomal degradation pathway to affect protein amount of HIF-1α. Moreover, our data provide an explanation to understand the HIF-1α-lowering capabilities of NO under hypoxia by showing destruction of HIF-1α independent of oxygen requirements.
Several independent lines of research have established the ability of RNI to stabilize HIF-1α protein and to exert genomic effects attributed to HIF-1 signaling (for references see Brüne and Zhou, 2003). Mechanistically, this is explained by direct inhibition of PHD activity (Metzen et al., 2003) and/or by enhancing phosphatidylinositol 3 kinase (PI3K)-dependent transcription/translation of HIF-1α (Kasuno et al., 2004). It can be assumed that both mechanisms may operate at different concentrations of RNI. One expects direct PHD inhibition at higher and activation of PI3K by lower doses of RNI although, as suggested for hypoxia, PI3K activation may not be ubiquitously operating (Alvarez-Tejado et al., 2002; Arsham et al., 2002). In contrast, seminal observations reported that NO attenuated hypoxia-evoked HIF-1α protein accumulation and blocked HIF-1-transactivating capabilities (Huang et al., 1999). Supporting studies noticed attenuated binding of HIF-1α to the DNA under hypoxic or CoCl2 treatments in combination with NO (Liu et al., 1998; Sogawa et al., 1998; Brüne and Zhou, 2003). Considering the importance of PHDs in hydroxylating HIF-1α set the ground to understand fundamentals of oxygen sensing (Epstein et al., 2001; Ivan et al., 2001; Jaakkola et al., 2001). This provoked the hypothesis to explain RNI actions in reverting HIF-1α accumulation by reactivating PHD activity under hypoxia. Indeed, Hagen et al. (2003) showed restored PHD activity by NO under hypoxic conditions. It is argued that NO blocks mitochondrial respiration, thereby leaving more oxygen available for PHDs to reactivate them. Alternatively, NO is supposed to directly activate PHD in vitro (Wang et al., 2002). Irrespective of underlying molecular mechanism the abovementioned suggestions center on reactivating PHD activity by NO under hypoxia to promote subsequent proteasomal degradation of HIF-1α. Results in RCC4 cells suggest an alternative, hydroxylation/ubiquitination/proteasome-independent degradation pathway that is calpain-mediated. Observations are based on the use of calpain inhibitors and manipulations of intracellular calcium to stimulate and/or inhibit calpain. These observations raise the interesting possibility that destruction of HIF-1α by calpain may represent an alternative proteolytic system that participates in adjusting the amount of HIF-1α to its demands and opens the possibility that two types of regulation, i.e., proteasomal versus calpain degradation may not be mutually exclusive. Of course further experiments need to determine the temporally and spatially contribution of calpain-mediated destruction of HIF-1α relative to the proteasome system.
Modulation of intracellular calcium is known to affect the expression of hypoxia-inducible genes and more recently, two reports provided evidence that lowering intracellular calcium by BAPTA-AM activates HIF-1 by attenuating hydroxylation of HIF-1α (Berchner-Pfannschmidt et al., 2004; Liu et al., 2004). Although the proposed mechanism will not be applicable to pVHL-deficient RCC4 cells, it supports the concept that calcium is involved in degradation of HIF-1α. Lowering calcium may not only block PHD activity but also attenuate calpain and both actions may synergize in stabilizing HIF-1α. However, on the basis of current information one would not assume a simple role of calcium as several reports noticed the involvement of calcium and/or calmodulin in signal transduction pathways, e.g., ERK activation (Mottet et al., 2003; Liu et al., 2004), leading to enhanced HIF-1 transcriptional activity (Yuan et al., 2005). The dual role of calcium is reflected by showing that chelating calcium provoked a transient HIF-1α increase by attenuating protein degradation, whereas the calcium ionophore A23187 induced HIF-1α mRNA expression at the same time (Liu et al., 2004). In other systems calcium failed to affect HIF-1α protein and/or HIF-1–dependent transcription (Salnikow et al., 2002). Therefore, the role of calcium in supporting or antagonizing hypoxic responses is not yet fully defined and variables such as the calcium concentrations, calcium compartments, and cell type–specific effects need to be sorted out.
As shown in RCC4 cells, the combination of hypoxia/DETA-NO, as well as an increase in calcium as a result of ionomycin or thapsigargin treatment, lowered accumulation of HIF-1α. The ability of BAPTA-AM to reverse these effects argues for a direct role of calcium. This suggests that hypoxia/DETA-NO raises calcium, which in turn activates calpain because HIF-1α digestion is suppressed by calpain inhibitors. Along that line a previous report indeed suggested a calcium increase and calpain activation by RNI (Sanvicens et al., 2004). In addition, experiments in HEK293 and RCC4/VHL cells that contain a functional pVHL-dependent proteolytic machinery showed that the calpain pathway can be activated when PHD activity is blocked. These observations may be relevant to understand HIF-1α disappearance under periods of long lasting and/or severe hypoxia in close association with calcium fluctuations. Alternatively, destruction of HIF-1α by NO via the calpain system might help to understand how NO mediates chemosensitivity in tumor cells, considering that hypoxia, i.e., expression of HIF-1α in tumors is associated with increased resistance to radiotherapy (Mitchell et al., 1998; Matthews et al., 2001). In addition, the biological relevance of this process can be envisioned when inducible NO-synthase is expressed as a HIF-1 target gene (Jung et al., 2000) and the production of NO may initiate a feedback regulatory loop to limit continuation of hypoxic responses.
Our studies suggest that activation of calpain participates in regulating the stability of HIF-1α, a pathway with importance to understand pVHL-independent degradation of the protein. Degradation of HIF-1α by calpain may help to realize how hypoxia and NO work together in attenuating HIF-1α expression as well as HIF-1 signaling.
Acknowledgments
The technical assistance of Sandra Christmann is highly appreciated. The work was supported by grants from Deutsche Forschungsgemeinschaft (BR999), Sander Foundation (2002.088.2), and Deutsche Krebshilfe.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-08-0770) on January 18, 2006.
References
- Agani, F. H., Puchowicz, M., Chavez, J. C., Pichiule, P., and LaManna, J. (2002). Role of nitric oxide in the regulation of HIF-1alpha expression during hypoxia. Am. J. Physiol. 283, C178–C186. [DOI] [PubMed] [Google Scholar]
- Alvarez-Tejado, M., Alfranca, A., Aragones, J., Vara, A., Landazuri, M. O., and del Peso, L. (2002). Lack of evidence for the involvement of the phosphoinositide 3-kinase/Akt pathway in the activation of hypoxia-inducible factors by low oxygen tension. J. Biol. Chem. 277, 13508–13517. [DOI] [PubMed] [Google Scholar]
- Arsham, A. M., Plas, D. R., Thompson, C. B., and Simon, M. C. (2002). Phosphatidylinositol 3-kinase/Akt signaling is neither required for hypoxic stabilization of HIF-1alpha nor sufficient for HIF-1-dependent target gene transcription. J. Biol. Chem. 277, 15162–15170. [DOI] [PubMed] [Google Scholar]
- Berchner-Pfannschmidt, U., Petrat, F., Doege, K., Trinidad, B., Freitag, P., Metzen, E., de Groot, H., and Fandrey, J. (2004). Chelation of cellular calcium modulates hypoxia-inducible gene expression through activation of hypoxia-inducible factor-1alpha. J. Biol. Chem. 279, 44976–44986. [DOI] [PubMed] [Google Scholar]
- Bruick, R. K. (2003). Oxygen sensing in the hypoxic response pathway: regulation of the hypoxia-inducible transcription factor. Genes Dev. 17, 2614–2623. [DOI] [PubMed] [Google Scholar]
- Bruick, R. K., and McKnight, S. L. (2001). A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340. [DOI] [PubMed] [Google Scholar]
- Brüne, B., and Zhou, J. (2003). The role of nitric oxide (NO) in stability regulation of hypoxia inducible factor-1alpha (HIF-1alpha). Curr. Med. Chem. 10, 845–855. [DOI] [PubMed] [Google Scholar]
- Cockman, M. E., Masson, N., Mole, D. R., Jaakkola, P., Chang, G. W., Clifford, S. C., Maher, E. R., Pugh, C. W., Ratcliffe, P. J., and Maxwell, P. H. (2000). Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275, 25733–25741. [DOI] [PubMed] [Google Scholar]
- Epstein, A. C. et al. (2001). C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54. [DOI] [PubMed] [Google Scholar]
- Frank, R., and Overwin, H. (1996). SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. In: Methods in Molecular Biology, Vol. 66, Totowa, NJ: Humana Press, 149–169. [DOI] [PubMed] [Google Scholar]
- Hagen, T., Taylor, C. T., Lam, F., and Moncada, S. (2003). Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science 302, 1975–1978. [DOI] [PubMed] [Google Scholar]
- Hart, T. W. (1997). Some observation concerning the S-nitroso and S-phenyl-sulfonyl derivatives of L-cysteine and glutathione. Tetrahedron Lett. 26, 2013–2016. [Google Scholar]
- Hewitson, K. S. et al. (2002). Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277, 26351–26355. [DOI] [PubMed] [Google Scholar]
- Huang, L. E., and Bunn, H. F. (2003). Hypoxia-inducible factor and its biomedical relevance. J. Biol. Chem. 278, 19575–19578. [DOI] [PubMed] [Google Scholar]
- Huang, L. E., Willmore, W. G., Gu, J., Goldberg, M. A., and Bunn, H. F. (1999). Inhibition of hypoxia-inducible factor 1 activation by carbon monoxide and nitric oxide. J. Biol. Chem. 274, 9038–9044. [DOI] [PubMed] [Google Scholar]
- Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S., and Kaelin, W. G., Jr. (2001). HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- Jaakkola, P. et al. (2001). Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472. [DOI] [PubMed] [Google Scholar]
- Jung, F., Palmer, L. A., Zhou, N., and Johns, R. A. (2000). Hypoxic regulation of inducible nitric oxide synthase via hypoxia inducible factor-1 in cardiac myocytes. Circ. Res. 86, 319–325. [DOI] [PubMed] [Google Scholar]
- Kaelin, W. G., Jr. (2002). How oxygen makes its presence felt. Genes Dev. 16, 1441–1445. [DOI] [PubMed] [Google Scholar]
- Kasuno, K., Takabuchi, S., Fukuda, K., Kizaka-Kondoh, S., Yodoi, J., Adachi, T., Semenza, G. L., and Hirota, K. (2004). Nitric oxide induces hypoxia-inducible factor 1 activation that is dependent on MAPK and phosphatidylinositol 3-kinase signaling. J. Biol. Chem. 279, 2550–2558. [DOI] [PubMed] [Google Scholar]
- Kimura, H., Weisz, A., Kurashima, Y., Hashimoto, K., Ogura, T., D'Acquisto, F., Addeo, R., Makuuchi, M., and Esumi, H. (2000). Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood 95, 189–197. [PubMed] [Google Scholar]
- Lando, D., Peet, D. J., Whelan, D. A., Gorman, J. J., and Whitelaw, M. L. (2002). Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science 295, 858–861. [DOI] [PubMed] [Google Scholar]
- Lee, D. H., and Goldberg, A. L. (1998). Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8, 397–403. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Moller, U., Flugel, D., and Kietzmann, T. (2004). Induction of plasminogen activator inhibitor I gene expression by intracellular calcium via hypoxia-inducible factor-1. Blood 104, 3993–4001. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Christou, H., Morita, T., Laughner, E., Semenza, G. L., and Kourembanas, S. (1998). Carbon monoxide and nitric oxide suppress the hypoxic induction of vascular endothelial growth factor gene via the 5′ enhancer. J. Biol. Chem. 273, 15257–15262. [DOI] [PubMed] [Google Scholar]
- Mahon, P. C., Hirota, K., and Semenza, G. L. (2001). FIH-1, a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15, 2675–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, N., Willam, C., Maxwell, P. H., Pugh, C. W., and Ratcliffe, P. J. (2001). Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20, 5197–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, N. E., Adams, M. A., Maxwell, L. R., Gofton, T. E., and Graham, C. H. (2001). Nitric oxide-mediated regulation of chemosensitivity in cancer cells. J. Natl. Cancer Inst. 93, 1879–1885. [DOI] [PubMed] [Google Scholar]
- Metzen, E., Zhou, J., Jelkmann, W., Fandrey, J., and Brüne, B. (2003). Nitric oxide impairs normoxic degradation of HIF-1alpha by inhibition of prolyl hydroxylases. Mol. Biol. Cell 14, 3470–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J. B., DeGraff, W., Kim, S., Cook, J. A., Gamson, J., Christodoulou, D., Feelisch, M., and Wink, D. A. (1998). Redox generation of nitric oxide to radiosensitize hypoxic cells. Int. J. Radiat. Oncol. Biol. Phys. 42, 795–798. [DOI] [PubMed] [Google Scholar]
- Mottet, D., Michel, G., Renard, P., Ninane, N., Raes, M., and Michiels, C. (2003). Role of ERK and calcium in the hypoxia-induced activation of HIF-1. J. Cell. Physiol. 194, 30–44. [DOI] [PubMed] [Google Scholar]
- Oehme, F., Ellinghaus, P., Kolkhof, P., Smith, T. J., Ramakrishnan, S., Hutter, J., Schramm, M., and Flamme, I. (2002). Overexpression of PH-4, a novel putative proline 4-hydroxylase, modulates activity of hypoxia-inducible transcription factors. Biochem. Biophys. Res. Commun. 296, 343–349. [DOI] [PubMed] [Google Scholar]
- Palmer, L. A., Gaston, B., and Johns, R. A. (2000). Normoxic stabilization of hypoxia-inducible factor-1 expression and activity: redox-dependent effect of nitrogen oxides. Mol. Pharm. 58, 1197–1203. [DOI] [PubMed] [Google Scholar]
- Pugh, C. W., and Ratcliffe, P. J. (2003). Regulation of angiogenesis by hypoxia: role of the HIF system. Nat. Med. 9, 677–684. [DOI] [PubMed] [Google Scholar]
- Salnikow, K., Kluz, T., Costa, M., Piquemal, D., Demidenko, Z. N., Xie, K., and Blagosklonny, M. V. (2002). The regulation of hypoxic genes by calcium involves c-Jun/AP-1, which cooperates with hypoxia-inducible factor 1 in response to hypoxia. Mol. Cell. Biol. 22, 1734–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandau, K. B., Zhou, J., Kietzmann, T., and Brüne, B. (2001). Regulation of the hypoxia-inducible factor 1alpha by the inflammatory mediators nitric oxide and tumor necrosis factor-alpha in contrast to desferroxamine and phenylarsine oxide. J. Biol. Chem. 276, 39805–39811. [DOI] [PubMed] [Google Scholar]
- Sanvicens, N., Gomez-Vicente, V., Masip, I., Messeguer, A., and Cotter, T. G. (2004). Oxidative stress-induced apoptosis in retinal photoreceptor cells is mediated by calpains and caspases and blocked by the oxygen radical scavenger CR-6. J. Biol. Chem. 279, 39268–39278. [DOI] [PubMed] [Google Scholar]
- Semenza, G. L. (2003). Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732. [DOI] [PubMed] [Google Scholar]
- Semenza, G. L., and Wang, G. L. (1992). A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12, 5447–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogawa, K., Numayama-Tsuruta, K., Ema, M., Abe, M., Abe, H., and Fujii-Kuriyama, Y. (1998). Inhibition of hypoxia-inducible factor 1 activity by nitric oxide donors in hypoxia. Proc. Natl. Acad. Sci. USA 95, 7368–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawa, N. E., Jr., Odessey, R., and Goldberg, A. L. (1997). Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J. Clin. Invest. 100, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D. D., Espey, M. G., Ridnour, L. A., Hofseth, L. J., Mancardi, D., Harris, C. C., and Wink, D. A. (2004). Hypoxic inducible factor 1alpha, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proc. Natl. Acad. Sci. USA 101, 8894–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Sekine, H., Kikuchi, Y., Takasaki, C., Miura, C., Heiwa, O., Shuin, T., Fujii-Kuriyama, Y., and Sogawa, K. (2002). HIF-1alpha-prolyl hydroxylase: molecular target of nitric oxide in the hypoxic signal transduction pathway. Biochem. Biophys. Res. Commun. 295, 657–662. [DOI] [PubMed] [Google Scholar]
- Wang, G. L., and Semenza, G. L. (1995). Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270, 1230–1237. [DOI] [PubMed] [Google Scholar]
- Wellman, T. L., Jenkins, J., Penar, P. L., Tranmer, B., Zahr, R., and Lounsbury, K. M. (2004). Nitric oxide and reactive oxygen species exert opposing effects on the stability of hypoxia inducible factor-1alpha (HIF-1alpha) in explants of human pial arteries. FASEB J. 18, 379–381. [DOI] [PubMed] [Google Scholar]
- Yuan, G., Nanduri, J., Bhasker, C. R., Semenza, G. L., and Prabhakar, N. R. (2005). Ca2+/calmodulin kinase-dependent activation of hypoxia inducible factor 1 transcriptional activity in cells subjected to intermittent hypoxia. J. Biol. Chem. 280, 4321–4328. [DOI] [PubMed] [Google Scholar]