Abstract
The effects of sublethal concentrations of four different classes of translation inhibitors (puromycin, tetracycline, chloramphenicol, and erythromycin) on global transcription patterns of Streptococcus pneumoniae R6 were determined by microarray analyses. Consistent with the general mode of action of these inhibitors, relative transcript levels of genes that encode ribosomal proteins and translation factors or that mediate tRNA charging and amino acid biosynthesis increased or decreased, respectively. Transcription of the heat shock regulon was induced only by puromycin or streptomycin treatment, which lead to truncation or mistranslation, respectively, but not by other antibiotics that block translation, transcription, or amino acid charging of tRNA. In contrast, relative transcript amounts of certain genes involved in transport, cellular processes, energy metabolism, and purine nucleotide (pur) biosynthesis were changed by different translation inhibitors. In particular, transcript amounts from a pur gene cluster and from purine uptake and salvage genes were significantly elevated by several translation inhibitors, but not by antibiotics that target other cellular processes. Northern blotting confirmed increased transcript amounts from part of the pur gene cluster in cells challenged by translation inhibitors and revealed the presence of a 10-kb transcript. Purine metabolism genes were negatively regulated by a homologue of the PurR regulatory protein, and full derepression in a ΔpurR mutant depended on optimal translation. Unexpectedly, hierarchical clustering of the microarray data distinguished among the global transcription patterns caused by antibiotics that inhibit different steps in the translation cycle. Together, these results show that there is extensive control of transcript amounts by translation in S. pneumoniae, especially for de novo purine nucleotide biosynthesis. In addition, these global transcription patterns form a signature that can be used to classify the mode of action and potential mechanism of new translation inhibitors.
Regulation at the transcription level plays a major role in controlling gene expression in prokaryotes. Because transcription and translation are coupled in bacteria, there are a number of mechanisms by which the translation process participates in regulating the amount of mRNA transcribed. One of the best examples of such a mechanism is attenuation, in which transcription termination is regulated by translation of a leader peptide (reviewed in reference 25). This regulatory mechanism was first characterized in depth for the trp operon of Escherichia coli and was later found to be common for various amino acid biosynthetic genes in gram-negative bacteria (25). In this case, transcription of the trp operon structural genes is prevented by factor-independent transcription termination that occurs when ribosomes fully translate a leader peptide containing tandem tryptophan residues. If ribosomes stall during translation at the tryptophan codons, then an alternate antiterminator structure forms in the leader transcript, allowing read-through transcription into the trp structural genes. Other variations of the attenuation mechanism that involve coupled translation exist, such as for the pyrBI operon of E. coli. In this case, the factor-independent terminator overlaps the coding region of the leader peptide. If the cellular amount of UTP is low, RNA polymerase pauses at a run of uridine residues inside the region encoding the leader peptide, allowing the translating ribosome to catch up and prevent formation of the terminator structure (37, 38).
In addition, there are forms of attenuation that do not involve direct translation coupling, such as the TRAP (tryptophan RNA-binding attenuation protein), S-box, and T-box mechanisms of gram-positive bacteria (1, 13, 18, 20). The TRAP and S-box mechanisms do not utilize components of the translation machinery, whereas the T-box mechanism utilizes charged or uncharged tRNA molecules to regulate attenuation of genes encoding amino acid biosynthetic and aminoacyl-tRNA synthetases (AARSs)(14, 16, 20). Individual components of the translation machinery can also function as transcription regulators. For example, ribosomal protein L4 stimulates transcription termination by a NusA-RNA polymerase complex paused in the upstream region of the nascent transcript of the E. coli S10 operon (51).
Two previous studies reported global gene expression patterns of E. coli (46) and Haemophilus influenzae (11) cells exposed to translation-inhibiting antibiotics. In both of these studies, bacterial cells were treated with a sublethal amount of translation inhibitors and protein synthesis patterns were determined by using two-dimensional electrophoresis. VanBogelen and Neidhardt found that certain antibiotics, such as puromycin and aminoglycosides (kanamycin and streptomycin), elicited a heat shock response in E. coli, whereas treatment with other antibiotics, such as chloramphenicol, erythromycin, and tetracycline, mimicked a cold shock response (46). However, the identities of many of the affected peptides were not determined. Evers and coworkers reported that the rates of synthesis of several ribosomal proteins and RNA polymerase subunits were induced by treatment with chloramphenicol, erythromycin, fusidate, puromycin, and tetracycline in H. influenzae (11). Although only a subset (≈600) of peptide spots were resolved and identified, these studies provided insight into the regulatory role of translation on global gene expression in H. influenzae (11).
Here we report analogous studies using microarray analyses to determine the changes in global transcription patterns in a nonvirulent model of the important human pathogen Streptococcus pneumoniae following treatment with sublethal amounts of four different classes of translation inhibitors (puromycin, tetracycline, chloramphenicol, and erythromycin). Our results reveal for the first time patterns of translation control of transcript amounts in this important bacterial pathogen. In particular, we found that the transcript levels of genes related to translation, heat shock, and purine nucleotide biosynthesis changed in response to decreased translation capacity. Hierarchical clustering showed a surprising ability to distinguish transcription regulation patterns for antibiotics that act at different stages of the translation cycle. This clustering serves as a signature to classify new potential classes of translation-inhibiting antibiotics.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. pneumoniae R6 was routinely cultured in chemically defined medium without methionine supplement (CDM [45]) at 37°C without shaking in the presence of 5% CO2. Mupiracin was obtained from Pliva Chemical Company (Zagreb, Croatia). Translation inhibitors and other antibacterial agents were purchased from Sigma (St. Louis, Mo.). Each compound was added from a 100× stock dissolved in dimethyl sulfoxide (DMSO). Hence, the final concentration of solvent in each treatment was 1% (vol/vol) DMSO. A culture containing 1% (vol/vol) DMSO lacking antibiotics was used as the control. Overnight bacterial cultures were diluted in fresh CDM to an optical density at 620 nm (OD620) of ≈0.02 (≈107 CFU per ml), and the diluted cultures were grown to mid-exponential phase (OD620 ≈ 0.2; ≈108 CFU per ml) before addition of antibiotics. Total RNA was extracted after 10 min of treatment and purified as described previously (36).
A mutant in which ≈90% of the purR open reading frame (ORF) was replaced by aad9 (spectinomycin resistance) was constructed in S. pneumoniae R6 as previously described (36). Briefly, primers KK0021 (CGAACGAAAATCGATGACAACCATCCGATCACTTCTTC) and KK0022 (ACCGTCGTACCATAGCTATCGAGCG) were used to PCR amplify the 5′ flanking region of purR, while primers KK0017 (CTTCAGACATATCGTTACCTTCCTTGAAAACG) and KK0018 (GAAATATTCATTCTAACGATGTTGAGGTTGGCAATATC) were used to amplify the 3′ flanking region of purR from purified R6 genomic DNA. A spectinomycin resistance cassette was PCR amplified using primers KK0019 (CCAACCTCAACATCGTTAGAATGAATATTTCC) and KK0020 (GATCGGATGGTTGTCATCGATTTTCGTTCGTGAAT). These three amplicons were joined together by PCR and transformed into S. pneumoniae R6. Spectinomycin-resistant transformants were screened by PCR for the presence of the ΔpurR mutation. A spectinomycin-resistant clone carrying the ΔpurR mutation was designated EL1232.
Microarray analysis.
The S. pneumoniae R6 microarray was designed and manufactured by Affymetrix based on the published S. pneumoniae R6 genome sequence, as previously described (36). Fragmentation, labeling of total RNA, and hybridization of labeled RNA to the microarray were performed as previously described (36). By using this method, we routinely detect expression of ≈60% of total ORFs of S. pneumoniae R6, which is comparable to previously published results from other bacterial species (26, 39).
Northern blot analysis.
Northern blot analyses were performed as described previously (36), except that radioactively labeled probe was used. Radioactive labeling of probe was performed by using the Prime-a-Gene labeling system (Promega, Madison, Wis.) following the manufacturer's instructions. A PCR product amplified from purified S. pneumoniae R6 genomic DNA using primers WN0038 (TATTGAAAGTCTGGTTTGCTGAG) and WN0040 (CGGGATCCCTGCTCAGAGAAAATGTGC) was used to prepare the probe for the purCL region. Primers WN0023 (AGCAAGTCTCCTGACCCTCGC) and WN0028 (AAGGAAATCGCTGAAACTAC) were used to amplify clpL and its flanking regions. The resulting PCR amplicon was digested with EcoRI, and a 980-bp fragment corresponding to the internal coding region of clpL was used to prepare the clpL-specific probe.
Microarray data analysis and hierarchical clustering.
Microarray data were analyzed using Affymetrix Microarray suite 5.0. A detailed description of the analysis algorithms can be obtained from the Affymetrix website. Each experiment was performed twice. Data obtained from cultures treated with 1% (vol/vol) DMSO were used for baseline comparisons. Average relative fold changes were calculated from the average of the signal log ratio (SLR) from two separate experiments by using the following equations: for an SLR of ≥0, average relative fold change = 2SLR; for an SLR of <0, average relative fold change = −1 × 2(−1 × SLR). Relative changes of ≥2-fold in independent experiments were considered indicative of differences in transcript amounts, on the basis of previous comparisons in which transcript amounts were also determined by Northern blotting or reverse transcription-PCR (e.g., see reference 36). Hierarchical clustering of microarray data was performed by an implementation of the published algorithm in reference 10.
RESULTS
Overview of global transcript quantitation profiles of translation-inhibited cells.
We used microarray analysis to determine the patterns of relative transcript amounts in cells exposed to sublethal concentrations of four different translation inhibitors. These four inhibitors were chosen because they inhibit different steps in the translation cycle. Puromycin is an aminoacyl-tRNA analogue that is incorporated into the nascent peptide chain, thereby causing premature termination and release (30). Tetracycline prevents the binding of charged tRNA to the A site of the ribosome (3, 33). Chloramphenicol inhibits the peptidyl transferase reaction of the large subunit of the ribosome (30, 42). Erythromycin and other macrolides block the ribosome exit tunnel, thereby preventing movement and release of the nascent peptide (42).
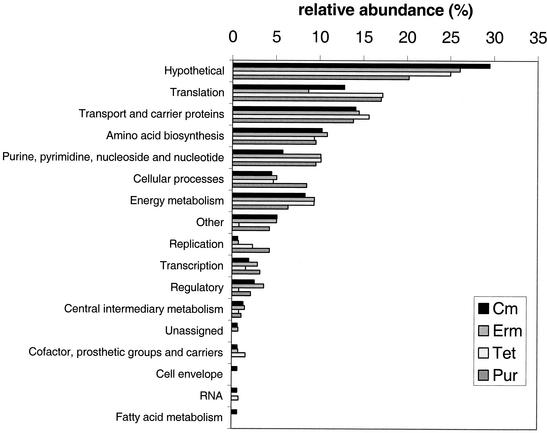
The amount of translation inhibitor used in each experiment was determined empirically by titrating exponentially growing cultures (OD620 ≈ 0.2; ≈108 CFU per ml) with different concentrations of inhibitors. We chose the concentrations at which the doubling time of the culture increased from 70 to 80 min for the DMSO control to 120 to 180 min following addition of compounds in DMSO (see Materials and Methods) (Table 1). Total RNA was extracted directly from each culture 10 min after addition of each translation inhibitor or DMSO (control). This relatively short interval was chosen for these initial studies to minimize possible secondary effects that might arise during prolonged treatment with the drugs. Hybridization intensity for every gene tiled onto the microarray was compared for the antibiotic-treated and DMSO control samples. Genes whose relative transcript levels reproducibly changed by ≥2-fold in two independent experiments were evaluated further (see Materials and Methods) (Table 1). Transcript amounts changed for ≈100 genes in response to treatment with each translation inhibitor compared to the control (Table 1). The complete list of genes displaying altered transcript levels is listed at http://www.streppneumoniae.com/microarray_data.htm. We routinely detected transcription from about 60% (≈1,200) of the total number of ORFs encoded by S. pneumoniae R6 (21). Therefore, only a small percentage (≈10%) of the genes whose transcripts were detected in these analyses changed in the presence of translation-inhibiting antibiotics. Of the genes showing altered transcript amounts in response to each antibiotic treatment (Fig. 1), ≈20 to 30% encoded hypothetical proteins without known functions, ≈10 to 15% encoded proteins involved in translation or transport, which are plentiful in the S. pneumoniae genome (21), and about 10% encoded proteins involved in amino acid biosynthesis or in purine and pyrimidine metabolism. Genes related to other cellular processes or energy metabolism comprised a smaller subset (<10%) of the total whose transcription was affected. Additional descriptions and experiments for some of these classes of genes are presented in the next sections.
TABLE 1.
Summary of global transcription patterns caused by treatment of S. pneumoniae R6 with sublethal concentrations of translation inhibitors in CDMa
| Translation inhibitor | Concn (μg/ml)b | Doubling time (min)c | Genes with change in transcript amt
|
||
|---|---|---|---|---|---|
| Total no. | No. with decrease | No. with increase | |||
| Puromycin | 16 | 123 | 94 | 52 | 42 |
| Tetracycline | 0.25 | 166 | 128 | 87 | 41 |
| Chloramphenicol | 4 | 154 | 156 | 103 | 53 |
| Erythromycin | 0.12 | 181 | 138 | 91 | 47 |
Growth conditions and microarray analyses are described in Material and Methods.
Drug concentrations were chosen to give 50% growth inhibition compared to control (DMSO) cultures (see Materials and Methods).
The reported doubling times are averages from two to three independent experiments. The doubling time of S. pneumoniae R6 control cultures treated with the solvent (1% [vol/vol] DMSO) used to dissolve the antibiotics was ≈70 to 80 min.
The number of genes whose transcript amounts showed relative changes ≥2-fold in at least two independent experiments are indicated (see Materials and Methods). Of the total number, genes whose transcript amounts increased or decreased are tabulated.
FIG. 1.
Distribution of functions of genes whose relative transcript amounts were affected by treatment of cells with translation inhibitors. The relative abundance is expressed as a percentage of the total number of genes whose relative transcript amount was affected by each inhibitor (Table 1) and includes increased and decreased transcript amounts.
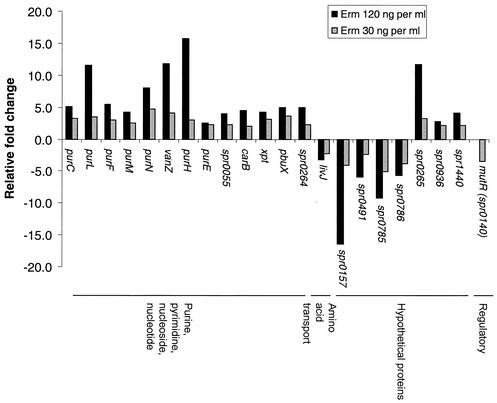
We tested whether there was a dose dependence for treatment with erythromycin. Instead of the standard concentration of 120 ng per ml used in all other experiments (Table 1), we added 30 ng of erythromycin per ml, which only slightly increased the doubling time by ≈20%. At the lower concentration of erythromycin, changes in transcript amounts from genes involved in purine and pyrimidine metabolism were still detected, but they were significantly reduced compared to those detected at the higher concentration of erythromycin (Fig. 2). The transcript amounts of other classes of genes, including those involved in translation or transport and most of those involved in amino acid biosynthesis, were not changed by the treatment with the lower concentration of erythromycin. It is unlikely that this dose-dependent pattern of transcript amounts for erythromycin was due to changes in growth rate per se, because completely different transcription patterns were detected for various concentrations of antibiotics that act by mechanisms other than translation inhibition, such as triclosan (a possible fatty acid biosynthesis inhibitor) or novobiocin (a DNA supercoiling inhibitor) (data not shown; see cluster analysis results, below). The dose dependence for other translation inhibitors was not determined.
FIG. 2.
Effect of translation inhibitor dosage on global transcription regulation. In two independent experiments, the relative transcript amount of 22 genes changed ≥2-fold in cells treated with 30 ng of erythromycin per ml for 10 min. The corresponding relative fold changes of these genes in cells treated with 120 ng of erythromycin per ml are included for comparison.
Altered transcript amounts from genes encoding the translation apparatus and amino acid biosynthetic enzymes.
Previous reports demonstrated that treating the gram-negative bacteria E. coli and H. influenzae with translation inhibitors increased the relative synthesis rate of a number of ribosomal proteins and translation factors (11, 46). Therefore, we tabulated the changes in relative transcript amounts of the translation apparatus of S. pneumoniae in the presence of translation inhibitors. We found that the relative transcript levels of 20 out of the total 55 genes encoding ribosomal proteins in the R6 genome (21) were increased by ≥2-fold (Table 2). Most of the affected genes are members of the S10 gene cluster that shares a similar organization to that of other bacterial species (27). Except for the genes encoding RF-3 (prfC) and IF-3 (infC), the relative transcript amounts from genes encoding accessory translation factors remained unchanged. The transcript levels from prfC and infC were increased ≈2-fold by chloramphenicol treatment. In addition, transcript amounts from infC were increased ≈2-fold by tetracycline treatment (Table 2).
TABLE 2.
Relative fold changes of transcript amounts of genes that encode ribosomal proteins, AARSs, and amino acid biosynthetic enzymes in response to sublethal concentrations of translation inhibitors
| Functional category and gene | Protein | Entry no. in R6 genome database | Avg relative fold changea
|
|||
|---|---|---|---|---|---|---|
| Ermb | Cmb | Purb | Tetb | |||
| Ribosomal proteins | ||||||
| rpsD | 30S ribosomal protein S4 | spr0078 | 1.9 | 1.1 | 1.1 | 2.8 |
| rpsJ | 30S ribosomal protein S10 | spr0187 | 1.4 | 1.6 | 2.1 | 1.8 |
| rplC | 50S ribosomal protein L3 | spr0188 | 1.5 | 1.5 | 2.0 | 1.8 |
| rplD | 50S ribosomal protein L4 | spr0189 | 1.6 | 1.8 | 2.0 | 2.0 |
| rplW | 50S ribosomal protein L23 | spr0190 | 1.6 | 1.7 | 2.1 | 2.0 |
| rplB | 50S ribosomal protein L2 | spr0191 | 1.6 | 1.7 | 2.0 | 1.9 |
| rpsS | 30S ribosomal protein S19 | spr0192 | 1.7 | 1.8 | 2.1 | 2.1 |
| rplV | 50S ribosomal protein L22 | spr0194 | 1.8 | 2.4 | 2.1 | 1.9 |
| rplP | 50S ribosomal protein L16 | spr0196 | 1.7 | 2.0 | 2.0 | 1.8 |
| rpmC | 50S ribosomal protein L29 | spr0197 | 1.7 | 2.0 | 2.0 | 1.7 |
| rplX | 50S ribosomal protein L24 | spr0200 | 1.9 | 2.4 | 2.0 | 2.2 |
| rplE | 50S ribosomal protein subunit L5 | spr0201 | 1.7 | 2.1 | 2.0 | 2.0 |
| rplF | 50S ribosomal protein subunit L6 | spr0204 | 1.7 | 2.0 | 2.0 | 2.1 |
| rplR | 50S ribosomal protein L18 | spr0205 | 1.6 | 1.9 | 2.0 | 1.9 |
| rplK | 50S ribosomal protein L11 | spr0555 | 1.3 | 2.2 | 1.8 | 1.6 |
| rpsP | 30S ribosomal protein S16 | spr0682 | 1.4 | 1.7 | −1.1 | 2.1 |
| rpmI | 50S ribosomal protein L35 | spr0862 | 1.3 | 2.3 | 1.7 | 2.3 |
| rplT | 50S ribosomal protein L20 | spr0863 | 1.6 | 2.9 | 1.9 | 3.2 |
| rplJ | 50S ribosomal protein L10 | spr1212 | 1.0 | 1.7 | 1.6 | 1.9 |
| rpsR | 30S ribosomal protein S18 | spr1394 | 2.3 | 2.4 | 1.9 | 2.9 |
| AARSs | ||||||
| serS | Seryl-tRNA synthetase | spr0372 | −1.9 | −2.2 | −1.8 | −2.0 |
| valS | Valyl-tRNA synthetase | spr0492 | −3.0 | −2.1 | −1.5 | −2.5 |
| pheS | Phenylalanyl-tRNA synthetase alpha chain | spr0507 | −13.2 | −4.7 | −2.3 | −13.5 |
| pheT | Phenylalanyl-tRNA synthetase beta chain | spr0509 | −2.7 | −2.4 | −2.1 | −4.0 |
| glyS | Glycyl-tRNA synthetase beta chain | spr1328 | −2.2 | −1.9 | −1.4 | −2.3 |
| glyQ | Glycyl-tRNA synthetase alpha chain | spr1329 | −2.5 | −2.8 | −1.8 | −2.7 |
| thrS | Threonyl-tRNA synthetase 1 | spr1472 | −2.3 | −1.9 | −1.2 | −2.1 |
| ileS | Isoleucyl-tRNA synthetase | spr1502 | −3.6 | −2.4 | −1.5 | −3.3 |
| tyrS | Tyrosyl-tRNA synthetase 1 | spr1910 | −2.2 | −2.0 | −1.4 | −1.5 |
| hisS | Histidyl-tRNA synthetase | spr1931 | −2.2 | −1.5 | −1.4 | −1.7 |
| Translation factors | ||||||
| prfC | Peptide chain release factor 3 | spr0396 | 1.2 | 2.0 | 1.2 | −1.1 |
| infC | Translation initiation factor 3 | spr0861 | 1.4 | 2.1 | 1.8 | 2.4 |
| Amino acid biosynthesis | ||||||
| aspC | Aspartate aminotransferase | spr0035 | −1.8 | −2.1 | −1.4 | −1.4 |
| ilvN | Acetolactate synthase small subunit | spr0402 | −3.2 | −3.3 | −3.2 | −5.1 |
| ilvC | Ketol acid reductoisomerase | spr0403 | −2.2 | −2.9 | −3.4 | −3.2 |
| metE | Tetrahydropteroyltriglutamate methyltransferase | spr0514 | −6.5 | −8.1 | −4.2 | −5.4 |
| metF | 5,10-Methylenetetrahydrofolate reductase | spr0515 | −4.5 | −3.8 | −3.5 | −3.6 |
| ilvE | Branched-chain-amino-acid transaminase | spr0758 | −3.1 | −2.5 | −2.3 | −3.9 |
| asd | Aspartate beta-semialdehyde dehydrogenase | spr0918 | −3.0 | −3.1 | −2.5 | −3.8 |
| dapA | Dihydrodipicolinate synthase | spr0919 | −2.1 | −2.2 | −2.9 | −2.2 |
| glyA | Serine hydroxymethyltransferase | spr0928 | −2.0 | −1.7 | −1.4 | −1.2 |
| metY-truncation | O-Acetylhomoserine sulfhydrylase, truncation | spr1095 | −3.5 | −5.5 | −2.9 | −2.8 |
| leuD-truncation | 3-Isopropylmalate dehydratase small subunit, truncation | spr1133 | 2.0 | 1.2 | 1.5 | 1.2 |
| gdhA | NADP-specific glutamate dehydrogenase | spr1181 | −3.1 | −3.5 | −1.7 | −2.0 |
| metB | Cystathionine gamma-synthase | spr1377 | −2.9 | −3.2 | −2.1 | −1.8 |
| metA | Homoserine O-succinyltransferase | spr1434 | −2.1 | −1.4 | −1.1 | −1.8 |
| trpA | Tryptophan synthase alpha chain | spr1631 | −2.0 | −2.0 | −1.3 | −2.0 |
| trpC | Indole-3-glycerol-phosphate synthase | spr1634 | −2.2 | −2.1 | −1.3 | −2.0 |
| trpD | Anthranilate phosphoribosyltransferase | spr1635 | −2.4 | −2.4 | −1.4 | −2.3 |
| trpG | Anthranilate synthase component II (glutamine amido-transferase) | spr1636 | −5.1 | −6.1 | −6.0 | −12.0 |
| ilvD | Dihydroxyacid dehydratase | spr1935 | −2.5 | −2.2 | −1.8 | −1.8 |
Microarray analyses were performed as described in Materials and Methods. Only genes whose transcript amounts showed a relative change of ≥2-fold in two independent experiments in response to at least one of the translation inhibitors are listed (see Materials and Methods). Negative values indicate a decrease in average relative transcript amount.
Erm, erythromycin; Cm, chloramphenicol; Pur, puromycin; Tet, tetracycline, at concentrations shown in Table 1.
In addition, 10 of the 21 genes encoding AARSs showed relative decreases in transcript amounts following treatment with translation inhibitors (Table 2). The affected genes encode eight different AARSs, including those that charge glycine, histidine, isoleucine, phenylalanine, serine, threonine, tyrosine, and valine onto their respective tRNAs. Of this subset, relative transcript amounts from the linked pheS and pheT genes showed large decreases in response to all four antibiotics (Table 2).
As a group, genes encoding enzymes involved in amino acid biosynthesis also showed decreased relative transcription in the presence of translation inhibitors (Table 2). Transcript amounts from a number of genes in this category, including asd, dapA, ilvC, ilvE, ilvN, metE, metF, and metY, decreased in all four antibiotic treatments. We were not able to identify possible changes in the relative amounts of different tRNAs, because the corresponding oligonucleotides were not included on the microarray chips.
Heat shock response induced by puromycin and streptomycin treatment.
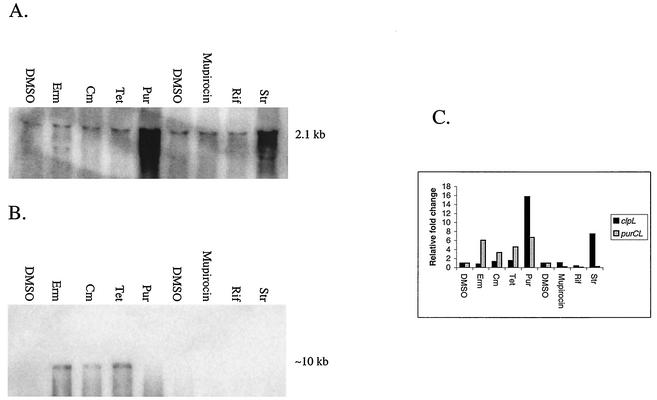
The relative transcription of a subset of the S. pneumoniae heat shock regulon, including clpE, clpL, hrcA-grpE-dnaK-dnaJ, and groEL-groES, was highly induced by puromycin, but not by addition of the other three translation inhibitors (see http://www.streppneumoniae.com/microarray_data.htm). However, we did not observe increased transcript amounts from the clpP and clpC genes, which showed weaker induction upon heat shock compared to the other regulon genes in a previous microarray analysis (36). To confirm these microarray data and to investigate whether other antibacterial agents induced the transcription of the heat shock regulon, we performed Northern blot analyses. A radioactively labeled probe internal to the clpL ORF was hybridized to total RNA extracted from cells treated with puromycin, tetracycline, chloramphenicol, erythromycin, mupirocin (an isoleucyl-AARS inhibitor [34, 50]), rifampin (an RNA polymerase inhibitor [4, 35]), or streptomycin (an aminoglycoside causing mistranslation [5]) at concentrations that increased the doubling time of the culture by ≈2-fold. Hybridization signals were detected and quantified by using a phosphorimager (Fig. 3). A hybridization band corresponding to the size of a full-length clpL monocistronic transcript (2.1 kb) was detected in all samples (Fig. 3). Of all the compounds tested, only puromycin or streptomycin, which lead to peptide-chain truncation or mistranslation, respectively, caused a significant change (15- or 7-fold increase, respectively; Fig. 3) in the signal intensities of the clpL transcript.
FIG. 3.
Northern blot analysis of clpL and purCLFMN-vanZ-purH transcripts from S. pneumoniae R6 cells treated with various antibiotics. Growth of cultures, treatment with compounds, and Northern blotting were performed as described in Materials and Methods. (A) Expression of the 2.1-kb monocistronic clpL transcript hybridized to a probe internal to the clpL ORF. (B) Expression of a ≈10-kb purCLFMN-vanZ-purH operon transcript hybridized to a probe that extends from the end of purC to the beginning of purL. (C) Hybridization signals from both blots were analyzed using ImageQuant software (Molecular Dynamics). The relative fold changes in transcript amounts for each treatment were compared to that of the control (DMSO). DMSO, solvent control; Erm, erythromycin; Cm, chloramphenicol; Tet, tetracycline; Pur, puromycin; Rif, rifampin; Str, streptomycin.
Increased transcript amounts of purine biosynthetic, salvage, and transport genes in translation-inhibited cells.
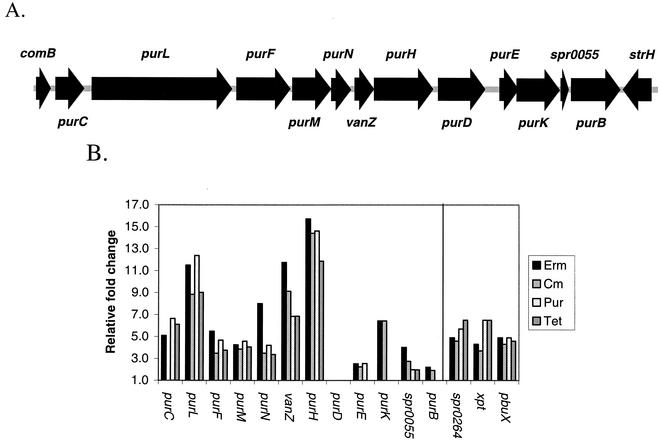
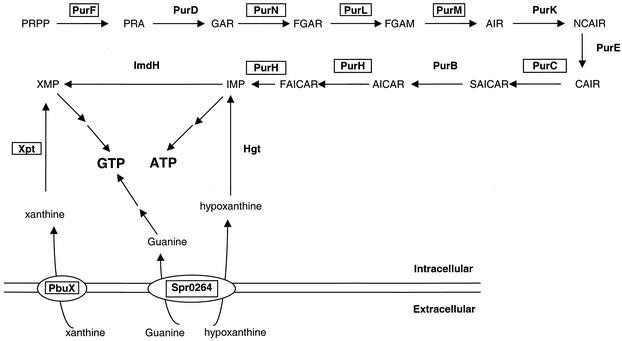
The relative transcript levels of several genes that encode enzymes of the de novo purine nucleotide biosynthetic pathway were strongly induced 4- to 15-fold in cells treated separately with the four classes of translation inhibitors (Fig. 3B and 4). The genes required for the conversion of PRPP to IMP are located in a single cluster flanked by comB and strH in the S. pneumoniae R6 genome (Fig. 4 and 5). Using this method of microarray analysis, relative transcript levels from the first seven members of this gene cluster (purCLFMN-vanZ-purH) were increased by treatment with all of the translation inhibitors tested except for purC, whose transcript level did not seem to be induced by chloramphenicol (Fig. 4B). The relative amount of purD transcript did not appear to change in response to any of the translation inhibitors, whereas the relative transcript levels of the downstream genes in the pur cluster increased ≥2-fold in response to at least one treatment (Fig. 4B).
FIG. 4.
Organization of the S. pneumoniae R6 pur gene cluster and relative transcript amounts from its genes in cells subjected to translation inhibition. (A) Organization of the pur gene cluster that encodes the biosynthetic enzymes that convert PRPP to IMP (see Fig. 5). Each arrow represents an ORF. Flanking ORFs comB and strH are included for reference. (B) The relative fold change in transcript amounts of each gene in the pur cluster determined by independent microarray analyses of RNA from cells treated with sublethal concentrations of translation inhibitors (see Materials and Methods and Results; Table 1). Also shown are the relative fold changes in transcript amounts from the spr0264, xpt, and pbuX genes that mediate purine salvage and uptake but that are not genetically linked to the pur gene cluster (see Results and Discussion). Erm, erythromycin; Cm, chloramphenicol; Pur, puromycin; Tet, tetracycline.
FIG. 5.
Purine nucleotide biosynthesis and transport in S. pneumoniae. Proposed pathways for purine transport and the de novo biosynthetic pathway for purine nucleotides are shown. The function of each gene product was assigned based on sequence homology (21). PbuX and Spr0264 are proposed to be transporters. A homologue of gde that mediates the conversion of xanthine to guanine was not identified in the S. pneumoniae R6 genome (21). Enzymes shown in boxes are those whose relative transcript amounts increased significantly in response to puromycin, tetracycline, chloramphenicol, or erythromycin treatment (Fig. 4).
To confirm these microarray data and to better understand the organization of the pur cluster, we performed Northern analyses by hybridizing a radioactively labeled probe that extends from the 3′ end of purC to the 5′ end of purL (Fig. 4A) to total RNA extracted from cells treated with sublethal concentrations of the translation inhibitors, an AARS inhibitor, and a transcription inhibitor (Fig. 3B). Under the hybridization conditions used, a signal was detected only for samples treated with puromycin, tetracycline, chloramphenicol, or erythromycin, but not for the control (1% [vol/vol] DMSO) or for samples treated with mupirocin, rifampin, or streptomycin (Fig. 3). In the samples treated with erythromycin, chloramphenicol, and tetracycline, we also detected a discrete ≈10-kb band (Fig. 3B), which is the predicted size of a polycistronic transcript extending from purC through purH. Thus, both the microarray and Northern data suggest the existence of a purCLFMN-vanZ-purH multigene operon (Fig. 4). We presently do not know why we detect increased purC transcript as part of the operon by Northern blotting but not by microarray analysis of chloramphenicol-treated cells.
The relative transcript levels of some purine salvage and uptake genes also increased significantly in cells treated with translation inhibitors (Fig. 4B and 5). Relative transcript levels from the xpt, pbuX, and spr0264 genes were increased ≈5-fold by treatment with each of the four translation inhibitors (Fig. 4B). xpt encodes the enzyme that converts xanthine into XMP at the expense of PRPP (Fig. 5), and pbuX and spr0264 likely encode transporters of xanthine and guanine/hypoxanthine, respectively (Fig. 5) (40). Transcript levels from most of the genes that convert hypoxanthine to IMP (hgt), IMP to GTP (imdH, guaA, gmk, and ndk), and IMP to ADP (purA, purB, and adk) (Fig. 5) (44) did not respond to translation inhibition, except for imdH, purB, and adk. The relative imdH transcript level dropped by 2.5-fold in response to tetracycline, whereas the relative transcript levels of purB or adk increased >2.2-fold in response to erythromycin (see http://www.streppneumoniae.com/microarray_data.htm). Moreover, the adk transcript amount also increased about threefold in response to puromycin.
The genome of S. pneumoniae R6 contains a putative PurR regulator (ORF spr1793). To assess the role of PurR on purine regulation and the effects of translation inhibitors on the pur cluster transcription, we constructed a ΔpurR::aad9 (spectinomycin resistance) insertion-deletion mutant (see Materials and Methods). Microarray analyses demonstrated that the PurR regulon consists of genes that are in the pur cluster (Fig. 4) and that mediate purine transport and salvage (Table 3). In addition, reproducible increases in transcript levels from genes involved in folate metabolism were detected. The decrease in lysA transcript amounts may be a polar effect, since lysA is located downstream from purR. Transcript amounts of all of the genes in the pur gene cluster were markedly increased in the ΔpurR mutant (Table 3). This pattern contrasts with that observed for treatment with translation inhibitors, which led mainly to increased transcript amounts from the purC to purH genes in the cluster (Fig. 4).
TABLE 3.
Genes with altered transcript amounts in a ΔpurR::aad9 mutant compared to its isogenic purR+ parent
| Functional group and R6 genome entry no. | Gene | Avg fold change in ΔpurR mutanta | Protein |
|---|---|---|---|
| Purine gene cluster | |||
| spr0045 | purC | 13.5 | Phosphoribosylaminoimidazole-succinocarboxamide synthetase |
| spr0046 | purL | 26.2 | Phosphoribosylformylglycinamide synthetase |
| spr0047 | purF | 12.5 | Amidophosphoribosyl transferase |
| spr0048 | purM | 10.6 | Phosphoribosylaminoimidazole synthetase |
| spr0049 | purN | 14.6 | 5′-Phosphoribosylglycinamide transformylase 1 |
| spr0050 | vanZ | 18.6 | Teicoplanin resistance protein |
| spr0051 | purH | 23.4 | Phosphoribosylaminoimidazolecarboxamide formyltransferase |
| spr0052 | purD | 8.7 | Phosphoribosylglycinamide synthetase |
| spr0053 | purE | 5.0 | Phosphoribosylaminoimidazole carboxylase, catalytic subunit |
| spr0054 | purK | 12.4 | Phosphoribosyl glucinamide formyltransferase |
| spr0055 | spr0055 | 4.7 | Hypothetical protein |
| spr0056 | purB | 3.9 | Adenylosuccinate lyase |
| Purine transport and salvage | |||
| spr0264 | spr0264 | 6.5 | Guanine/hypoxanthine transporter? |
| spr1128 | guaC | 5.5 | GMP reductase |
| spr1662 | xpt | 3.0 | Xanthine phosphoribosyltransferase |
| spr1663 | pbuX | 3.2 | Nucleobase:cation symporter for xanthine |
| Folate metabolism | |||
| spr0266 | sulA | 2.5 | Dihydropteroate synthase |
| spr0267 | sulB | 2.6 | Dihydrofolate synthetase |
| spr0268 | sulC | 2.5 | GTP cyclohydrolase |
| spr0269 | sulD | 2.1 | Aldolase-pyrophosphokinase |
| spr1109 | fhs | 2.2 | Formate-tetrahydrofolate ligase |
| Hypothetical and other | |||
| spr0105 | spr0105 | 2.6 | Hypothetical transporter-truncation |
| spr0265 | spr0265 | 6.5 | Conserved hypothetical protein |
| spr0639 | copY | 2.7 | COPAB ATPases metal-fist-type repressor |
| spr0641 | ctpA | 2.2 | P-type ATPase, probable copper transporter |
| spr1440 | spr1440 | 2.5 | Conserved hypothetical protein |
| spr1441 | oxlT | 3.6 | Major facilitator:oxalate:formate antiporter |
| spr1792 | lysA | −3.9 | Diaminopimelate decarboxylase |
| spr1793 | purR | −27.2 | Repressor of purine biosynthetic genes |
Microarray analyses were performed as described in Materials and Methods on bacteria grown in CDM to mid-exponential phase. Only genes whose transcript amounts showed a relative change of ≥2-fold in three independent experiments are listed. A negative value indicates a decrease in average relative transcript amount.
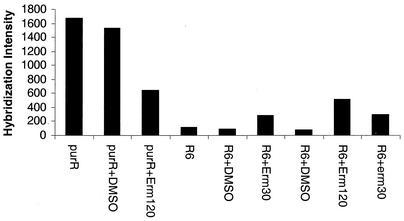
Finally, we performed microarray analyses of the ΔpurR::aad9 mutant treated with a sublethal concentration of erythromycin to determine if the effects of the purR mutation and translation inhibition were independent or somehow linked. We compared hybridization intensities of mRNA to each of the pur genes on the microarrays, which is operationally equivalent to a blotting experiment (Fig. 6). The comparison for purL in Fig. 6 is representative of hybridizations to other genes in the pur cluster. The untreated ΔpurR mutant contained the greatest amount of purL transcript. Instead of increasing this transcript amount further in an additive or multiplicative way, addition of erythromycin reduced the amount of purL transcript to approximately that observed in the purR+ R6 strain treated with erythromycin (Fig. 6). As a control, the hybridization intensities of genes that did not respond to translation inhibitors or the ΔpurR mutation were comparable for the different conditions (data not shown).
FIG. 6.
Hybridization intensities to the purL oligonucleotides on an Affymetrix chip of RNA isolated from bacteria subjected for 10 min to the indicated treatments. Growth and treatment of bacteria and microarray analyses are described in Materials and Methods. Hybridization intensities are in arbitrary fluorescent units. The hybridization intensities of genes not affected by treatment with translation inhibitors remained essentially constant for the different strains and treatments (data not shown). purR refers to the ΔpurR mutant and R6 refers to its isogenic purR+ parent (see Materials and Methods). Additions: no indication, nothing added; +DMSO, DMSO added lacking drug; +Erm30 and +Erm120, addition of 30 or 120 ng of erythromycin per ml.
Hierarchical clustering of translation inhibitors.
We used hierarchical clustering to classify the global transcription patterns caused by sublethal treatment with translation inhibitors that act via different mechanisms compared to that of the antibacterial agent triclosan, which may inhibit fatty acid biosynthesis (Fig. 7 and Table 4) (W.-L. Ng and M. E. Winkler, unpublished data). In this analysis, correlation values of +1 or 0 indicate identical or unrelated global transcription patterns, respectively (Table 4) (10). We found that the global transcription patterns observed for treatment with each translation inhibitor were highly similar to each other, but still distinct, with correlation coefficients that ranged from ≈0.4 to 0.6 (Table 4). In contrast, treatment with an amount of triclosan that increased the culture doubling time by about twofold exhibited a very low (<0.05) correlation coefficient compared to the translation inhibitors, except for puromycin (Table 4). The slight correlation (0.14) between puromycin and triclosan is probably due to induction of heat shock gene expression by both drugs (W.-L. Ng and M. E. Winkler, unpublished data). Finally, we determined the transcription pattern for roxithromycin, which is a semisynthetic macrolide antibiotic derivative of erythromycin (22). The transcription pattern for roxithromycin clustered with that of erythromycin, but not with the other translation inhibitors tested (Fig. 7).
FIG. 7.
Hierarchical clustering of global transcription patterns determined by microarray analyses. Hierarchical clustering was performed as described in Materials and Methods and the Discussion. The dendrogram shows the relationship between each translation inhibitor and the antibacterial agent triclosan. See Table 4 for the matrix of correlation coefficients used to generate the dendrogram.
TABLE 4.
Correlation coefficients calculated from microarray profiles
| Agent | Correlation coefficienta
|
|||||
|---|---|---|---|---|---|---|
| Triclosan | Chloramphenicol | Erythromycin | Puromycin | Roxithromycin | Tetracycline | |
| Triclosan | 1.000 | 0.045 | 0.035 | 0.138 | −0.023 | 0.009 |
| Chloramphenicol | 1.000 | 0.674 | 0.420 | 0.551 | 0.640 | |
| Erythromycin | 1.000 | 0.429 | 0.675 | 0.655 | ||
| Puromycin | 1.000 | 0.269 | 0.495 | |||
| Roxithromycin | 1.000 | 0.561 | ||||
| Tetracycline | 1.000 | |||||
A correlation value of +1 or 0 indicates identical or unrelated global transcription patterns, respectively.
DISCUSSION
We report here that there are extensive effects of translation inhibition on relative transcript amounts in the gram-positive human pathogen S. pneumoniae. Some patterns emerged that are consistent with those observed previously in other bacteria. For example, induction of heat shock genes by puromycin treatment was previously reported in both E. coli and Bacillus subtilis (12, 28). Generally, translation inhibition led to increased transcript levels from genes encoding ribosomal proteins (Table 2), consistent with mechanisms that balance ribosome amount and function. This effect was not observed at lower doses of erythromycin, but was not attributable to changes in growth rate per se (see Results; Fig. 2). In E. coli, ribosomal protein synthesis is controlled by a negative feedback mechanism that couples ribosomal protein and rRNA amounts (reviewed in reference 31). Considerably less is known about the control of ribosomal protein synthesis in B. subtilis and other gram-positive bacteria (19). Similar to B. subtilis, the rpsD gene encoding protein S4 appears to be monocistronic in S. pneumoniae (21). In B. subtilis, S4 regulates its own expression by translational repression (15, 17), which might also operate to allow increased rpsD transcript amounts in S. pneumoniae R6 when challenged with translation inhibitors (Table 2). S. pneumoniae contains an extended cluster of ribosomal protein genes containing rpsJ (S10 protein), and transcript amounts from many of the genes in this cluster increased in response to translation inhibitors (Table 2). However, not all of the genes in the cluster are listed in Table 2. Several genes within the cluster were excluded from Table 2 {e.g., spr0195 [rpsC (S3)], spr0198 [rpsQ (S17)], spr0199 [rplN (L14)], spr0202 [rpsN (S14)], and spr0203 [rpsH (S8)]}, because their relative transcript amounts did not increase by at least twofold in response to any of the translation inhibitors, although most showed increases slightly below twofold (data not shown). In S. pneumoniae, it is not clear that rplK and rplA form an L11-L1 operon as in other bacteria (19), because the two genes are separated by an intercistronic space of 210 bp (21). Within the limits of this microarray analysis, transcription of rplK (L11 protein), but not that of rplA (L1 protein), was observed to increase in translation-inhibited cells (Table 2). These patterns provide a starting point for further transcript analyses of ribosomal protein synthesis in S. pneumoniae.
Transcript amounts from genes that encode AARSs and amino acid biosynthetic genes generally decreased in S. pneumoniae R6 treated with translation inhibitors (Table 2). Repression of some of these genes was particularly strong (e.g., pheS, metE, metF, asd, metY, and trpG) (Table 2). As noted above, we may have missed some genes that respond to translation inhibition, but the overall pattern is distinctive. In gram-positive bacteria, genes that encode AARSs are generally regulated by the T-box mechanism (19, 20). Computer-based searches revealed putative T boxes upstream of the valS, pheS, pheT, glyQ, glyS, thrS, and ileS genes (data not shown). Except for pheS and ileS, these putative T boxes were separated from the AARS gene by additional ORFs, whose transcript levels usually decreased in response to one or more of the translation inhibitors (data not shown). T-box regulation can be tested in further experiments.
A surprising result of this study was the strong increase in transcript amounts from branches of the de novo purine biosynthetic, salvage, and uptake pathways (Fig. 3, 4, and 5). The organization of the pur gene cluster in S. pneumoniae (Fig. 4) is different from that in B. subtilis and Lactococcus lactis (7, 29, 32), which is metabolically related to S. pneumoniae. In B. subtilis, there is an inversion so that purEKB are upstream of purC, and two more genes, purSQ, are inserted between purC and purL (7). In L. lactis, the pur genes are separated into several unlinked clusters: purDEK, purCSQL, purMN, and purH (29, 32). The PurL enzyme (FGAR to FGAM; Fig. 5) of S. pneumoniae contains the separate domains encoded by purQ and purL in B. subtilis, and purS is only found in bacteria with separate purQ and purL genes (41). Our Northern blot and microarray analyses (Fig. 3 and 4) suggest that purCLFMN-vanZ-purH are cotranscribed as a single 10-kb mRNA. On the basis of the discontinuous pattern of transcript amounts observed in response to translation inhibitors (Fig. 4B), purD and purEK-spr0055-purB may be transcribed separately. However, at this stage, we cannot rule out additional events such as transcript processing. Transcript levels also increased from the xpt and pbuX genes in response to translation inhibitors (Fig. 4B). xpt and pbuX are adjacent and are probably cotranscribed (Fig. 4).
The amount of transcript from the pur cluster increased with erythromycin, chloramphenicol, tetracycline, and puromycin treatments, which block different steps in translation (Fig. 3B and 4). Interestingly, streptomycin did not lead to an increased amount of pur cluster mRNA (Fig. 4B). Streptomycin causes mistranslation, but it does not block the translation cycle like the other four translation inhibitors do. The increase in pur cluster transcript was also specific to these four translation inhibitors and was not observed following treatment of cells with mupirocin, rifampin, novobiocin-norfloxacin, or triclosan, which inhibit tRNA charging, transcription, DNA gyrase, or possibly fatty acid biosynthesis, respectively (Fig. 3B and C and data not shown).
The regulation of purine nucleotide biosynthesis has been studied in B. subtilis (reviewed in reference 44), but not in S. pneumoniae. In B. subtilis, the expression of the 12-member purEKBCSQLFMNHD operon, which encodes all of the enzymes required to convert PRPP to IMP (Fig. 5), as does the pur cluster in S. pneumoniae (Fig. 4), is regulated by two different mechanisms. Initiation of transcription is repressed by adenine-adenosine, mediated by the PurR repressor, which releases from its operator in response to increasing cellular levels of PRPP (Fig. 5) (48, 49). Recently, xpt-pbuX, pbuO, and pbuG were added to the PurR regulon of B. subtilis (40). By contrast, the PurR homologue in L. lactis seems to act as a positive regulator of purC and purD expression (23). The PurR homologue in S. pneumoniae seems to act as a repressor of the transcription of all of the genes in the pur cluster (Table 3). Besides purine biosynthetic, salvage, and transport genes, the transcription of genes involved in folate metabolism were up regulated in a ΔpurR mutant (Table 3). The products of the sulABCD genes convert GTP produced by the purine biosynthetic pathway to 7,8-dihydrofolate (24). fhs encoding formyl-tetrahydrofolate synthetase catalyzes the formation of 10-formyltetrahydrofolate from formate and tetrahydrofolate in an ATP-dependent fashion (6). Thus, there is a direct metabolic link between the purine and folate biosynthetic pathways that may require coordinate control.
We did not detect any change in the amount of purR transcript in response to any of the antibiotics tested herein. Also, the effect of translation inhibitors was discontinuous across the cluster (Fig. 4B), whereas transcript amounts from all the genes in the cluster increased in the ΔpurR mutant (Table 3). Addition of translation inhibitors to the ΔpurR mutant caused a decrease in the hybridization to each gene in the pur cluster compared to the untreated ΔpurR strain (Fig. 6 and data not shown). Thus, full derepression of pur cluster transcription appears to depend on optimal translation efficiency. Molecular genetic experiments are in progress to determine whether this apparent coupling is direct or indirect. In addition, experiments are in progress to measure the effects, if any, of translation inhibition or the purR mutation on nucleotide and magic spot [i.e., (p)ppGpp] pools in S. pneumoniae.
There may be another level of control by which translation inhibitors affect the transcription of the purCLFMN-vanZ-purH operon and other purine salvage and uptake genes in S. pneumoniae. In B. subtilis, addition of guanine/guanosine promotes transcription termination of the pur operon independently of PurR (8, 9). This induction is mediated by an attenuation mechanism that involves formation of mutually exclusive terminator or antiterminator structures in the pur operon leader transcript (7, 8). However, it is unknown whether this attenuation mechanism uses coupled translation of a leader peptide or a TRAP-like mechanism, such as occurs for the E. coli or B. subtilis trp operon, respectively (see the introduction) (1, 13, 25). Initial computer analyses indicate potential hairpin structures upstream of purC; however, operon punctuation and signals are presently not well characterized in S. pneumoniae. Alternatively, translation inhibitors may be acting at the level of transcript stability. Erythromycin and tetracycline stabilize certain transcripts in B. subtilis due to ribosome stalling in the leader peptide coding regions (2, 47). Further experiments are needed to distinguish whether there is control by attenuation or RNA stability modulation of the pur gene expression in S. pneumoniae.
Finally, hierarchical clustering of these transcription patterns revealed an unexpected result. Hierarchical clustering can identify genes that may be coregulated by a signal based on expression patterns in response to different treatments or conditions (10). In addition, this method can group samples with similar cellular phenotypes (e.g., breast tumors) on the basis of gene expression patterns (43). The transcription pattern caused by treatment with sublethal concentrations of triclosan (a possible fatty acid biosynthesis inhibitor) was distinct and distant from those caused by translation inhibitors (Fig. 7). In preliminary experiments, the transcription pattern from treatment with novobiocin (a DNA supercoiling inhibitor) also clustered away from those of triclosan and the translation inhibitors (data not shown). Unexpectedly, the clustering analysis showed that the transcription patterns were distinguishable for translation inhibitors that inhibit different steps in the translation cycle. Puromycin leads to premature peptide release (30), whereas tetracycline prevents entry of charged tRNA to the A site (3, 33). Chloramphenicol and erythromycin inhibit translation at later stages of the translation cycle by inhibiting the peptidyl transferase reaction and by blocking the ribosome exit tunnel, respectively (30, 42). The dendrogram in Fig. 7 suggests that global transcription patterns in S. pneumoniae are not just sensitive to the broad class of antibiotic (i.e., fatty acid versus translation inhibitors), but are also sensitive to the mechanism of inhibition within a subclass of antibiotics, such as the translation inhibitors. This correlation may reflect a fine-tuning mechanism that regulates transcript amounts in response to different kinds of inhibitors. In addition, this hierarchical clustering provides a highly useful signature for the classification of the mode of action and potential mechanism of new compounds that inhibit translation. To test this notion, we performed microarray analyses on cells treated with a sublethal concentration of the macrolide antibiotic roxithromycin, which is a semisynthetic derivative of erythromycin. The global transcription pattern for roxithromycin resembled that of erythromycin more closely than that of the other translation inhibitors (Table 4 and Fig. 7).
Acknowledgments
We thank Qingqin Li for help with clustering analyses, John Glass and Yong Yang for help with bioinformatics, Dalai Yan, John Richardson, and Jennifer Glass for critical discussions, and Thalia Nicas and Dan Mytelka for reviewing drafts of the manuscript.
This work was supported by resources provided by the Lilly Research Laboratories, and Krystyna M. Kazmierczak and Gregory T. Robertson were supported by Lilly Postdoctoral Fellowships.
REFERENCES
- 1.Babitzke, P. 1997. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol. Microbiol. 26:1-9. [DOI] [PubMed] [Google Scholar]
- 2.Bechhofer, D. H., and K. H. Zen. 1989. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171:5803-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 5.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 6.Crowley, P. J., J. A. Gutierrez, J. D. Hillman, and A. S. Bleiweis. 1997. Genetic and physiologic analysis of a formyl-tetrahydrofolate synthetase mutant of Streptococcus mutans. J. Bacteriol. 179:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebbole, D. J., and H. Zalkin. 1987. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J. Biol. Chem. 262:8274-8287. [PubMed] [Google Scholar]
- 8.Ebbole, D. J., and H. Zalkin. 1988. Detection of pur operon-attenuated mRNA and accumulated degradation intermediates in Bacillus subtilis. J. Biol. Chem. 263:10894-10902. [PubMed] [Google Scholar]
- 9.Ebbole, D. J., and H. Zalkin. 1989. Interaction of a putative repressor protein with an extended control region of the Bacillus subtilis pur operon. J. Biol. Chem. 264:3553-3561. [PubMed] [Google Scholar]
- 10.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evers, S., K. Di Padova, M. Meyer, H. Langen, M. Fountoulakis, W. Keck, and C. P. Gray. 2001. Mechanism-related changes in the gene transcription and protein synthesis patterns of Haemophilus influenzae after treatment with transcriptional and translational inhibitors. Proteomics 1:522-544. [DOI] [PubMed] [Google Scholar]
- 12.Goff, S. A., and A. L. Goldberg. 1985. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell 41:587-595. [DOI] [PubMed] [Google Scholar]
- 13.Gollnick, P. 1994. Regulation of the Bacillus subtilis trp operon by an RNA-binding protein. Mol. Microbiol. 11:991-997. [DOI] [PubMed] [Google Scholar]
- 14.Grundy, F. J., M. T. Haldeman, G. M. Hornblow, J. M. Ward, A. F. Chalker, and T. M. Henkin. 1997. The Staphylococcus aureus ileS gene, encoding isoleucyl-tRNA synthetase, is a member of the T-box family. J. Bacteriol. 179:3767-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundy, F. J., and T. M. Henkin. 1992. Characterization of the Bacillus subtilis rpsD regulatory target site. J. Bacteriol. 174:6763-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundy, F. J., and T. M. Henkin. 1994. Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in gram-positive bacteria. J. Mol. Biol. 235:798-804. [DOI] [PubMed] [Google Scholar]
- 17.Grundy, F. J., and T. M. Henkin. 1991. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J. Bacteriol. 173:4595-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 19.Henkin, T. M. 2002. Ribosomes, protein synthesis factors, and tRNA synthetases, p. 313-322. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 20.Henkin, T. M. 1994. tRNA-directed transcription antitermination. Mol. Microbiol. 13:381-387. [DOI] [PubMed] [Google Scholar]
- 21.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, R. N., A. L. Barry, and C. Thornsberry. 1983. In vitro evaluation of three new macrolide antimicrobial agents, RU28965, RU29065, and RU29702, and comparisons with other orally administered drugs. Antimicrob. Agents Chemother. 24:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilstrup, M., and J. Martinussen. 1998. A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis. J. Bacteriol. 180:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacks, S. A., B. Greenberg, and P. Lopez. 1995. A cluster of four genes encoding enzymes for five steps in the folate biosynthetic pathway of Streptococcus pneumoniae. J. Bacteriol. 177:66-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landick, R., C. L. Turnbough, Jr., and C. Yanofsky. 1996. Transcription attenuation, p. 1263-1286. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 26.Lee, J. M., S. Zhang, S. Saha, S. Santa Anna, C. Jiang, and J. Perkins. 2001. RNA expression analysis using an antisense Bacillus subtilis genome array. J. Bacteriol. 183:7371-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, X., L. Lindahl, Y. Sha, and J. M. Zengel. 1997. Analysis of the Bacillus subtilis S10 ribosomal protein gene cluster identifies two promoters that may be responsible for transcription of the entire 15-kilobase S10-spc-α cluster. J. Bacteriol. 179:7046-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mogk, A., A. Volker, S. Engelmann, M. Hecker, W. Schumann, and U. Volker. 1998. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J. Bacteriol. 180:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson, D., and M. Kilstrup. 1998. Cloning and expression of the Lactococcus lactis purDEK genes, required for growth in milk. Appl. Environ. Microbiol. 64:4321-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 31.Nomura, M. 1999. Regulation of ribosome biosynthesis in Escherichia coli and Saccharomyces cerevisiae: diversity and common principles. J. Bacteriol. 181:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peltonen, T., and P. Mantsala. 1999. Isolation and characterization of a purC(orf)QLF operon from Lactococcus [correction of Lactobacillus] lactis MG1614. Mol. Gen. Genet. 261:31-41. [DOI] [PubMed] [Google Scholar]
- 33.Pioletti, M., F. Schlunzen, J. Harms, R. Zarivach, M. Gluhmann, H. Avila, A. Bashan, H. Bartels, T. Auerbach, C. Jacobi, T. Hartsch, A. Yonath, and F. Franceschi. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20:1829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pope, A. J., K. J. Moore, M. McVey, L. Mensah, N. Benson, N. Osbourne, N. Broom, M. J. Brown, and P. O'Hanlon. 1998. Characterization of isoleucyl-tRNA synthetase from Staphylococcus aureus. II. Mechanism of inhibition by reaction intermediate and pseudomonic acid analogues studied using transient and steady-state kinetics. J. Biol. Chem. 273:31691-31701. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes, G., and M. J. Chamberlin. 1975. Kinetic analysis of ribonucleic acid chain initiation by Escherichia coli ribonucleic acid polymerase bound to DNA. J. Biol. Chem. 250:9112-9120. [PubMed] [Google Scholar]
- 36.Robertson, G. T., W. L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roland, K. L., C. G. Liu, and C. L. Turnbough, Jr. 1988. Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 85:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roland, K. L., F. E. Powell, and C. L. Turnbough, Jr. 1985. Role of translation and attenuation in the control of pyrBI operon expression in Escherichia coli K-12. J. Bacteriol. 163:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:E112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saxild, H. H., K. Brunstedt, K. I. Nielsen, H. Jarmer, and P. Nygaard. 2001. Definition of the Bacillus subtilis PurR operator using genetic and bioinformatic tools and expansion of the PurR regulon with glyA, guaC, pbuG, xpt-pbuX, yqhZ-folD, and pbuO. J. Bacteriol. 183:6175-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxild, H. H., and P. Nygaard. 2000. The yexA gene product is required for phosphoribosylformylglycinamidine synthetase activity in Bacillus subtilis. Microbiology 146:807-814. [DOI] [PubMed] [Google Scholar]
- 42.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 43.Sorlie, T., C. M. Perou, R. Tibshirani, T. Aas, S. Geisler, H. Johnsen, T. Hastie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, T. Thorsen, H. Quist, J. C. Matese, P. O. Brown, D. Botstein, P. Eystein Lonning, and A. L. Borresen-Dale. 2001. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 98:10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Switzer, R. L., H. Zalkin, and H. H. Saxild. 2002. Purine, pyrimidine, and pyridine nucleotide metabolism, p. 255-270. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 45.Talkington, D. F., D. C. Voellinger, L. S. McDaniel, and D. E. Briles. 1992. Analysis of pneumococcal PspA microheterogeneity in SDS polyacrylamide gels and the association of PspA with the cell membrane. Microb. Pathog. 13:343-355. [DOI] [PubMed] [Google Scholar]
- 46.VanBogelen, R. A., and F. C. Neidhardt. 1990. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, Y., and D. H. Bechhofer. 2002. Tetracycline induces stabilization of mRNA in Bacillus subtilis. J. Bacteriol. 184:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng, M., P. L. Nagy, and H. Zalkin. 1995. Identification of the Bacillus subtilis pur operon repressor. Proc. Natl. Acad. Sci. USA 92:7455-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weng, M., and H. Zalkin. 2000. Mutations in the Bacillus subtilis purine repressor that perturb PRPP effector function in vitro and in vivo. Curr. Microbiol. 41:56-59. [DOI] [PubMed] [Google Scholar]
- 50.Yanagisawa, T., J. T. Lee, H. C. Wu, and M. Kawakami. 1994. Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli: a proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase. J. Biol. Chem. 269:24304-24309. [PubMed] [Google Scholar]
- 51.Zengel, J. M., and L. Lindahl. 1996. A hairpin structure upstream of the terminator hairpin required for ribosomal protein L4-mediated attenuation control of the S10 operon of Escherichia coli. J. Bacteriol. 178:2383-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]