Abstract
Two mutant strains of Desulfovibrio vulgaris Hildenborough lacking either the sod gene for periplasmic superoxide dismutase or the rbr gene for rubrerythrin, a cytoplasmic hydrogen peroxide (H2O2) reductase, were constructed. Their resistance to oxidative stress was compared to that of the wild-type and of a sor mutant lacking the gene for the cytoplasmic superoxide reductase. The sor mutant was more sensitive to exposure to air or to internally or externally generated superoxide than was the sod mutant, which was in turn more sensitive than the wild-type strain. No obvious oxidative stress phenotype was found for the rbr mutant, indicating that H2O2 resistance may also be conferred by two other rbr genes in the D. vulgaris genome. Inhibition of Sod activity by azide and H2O2, but not by cyanide, indicated it to be an iron-containing Sod. The positions of Fe-Sod and Sor were mapped by two-dimensional gel electrophoresis (2DE). A strong decrease of Sor in continuously aerated cells, indicated by 2DE, may be a critical factor in causing cell death of D. vulgaris. Thus, Sor plays a key role in oxygen defense of D. vulgaris under fully aerobic conditions, when superoxide is generated mostly in the cytoplasm. Fe-Sod may be more important under microaerophilic conditions, when the periplasm contains oxygen-sensitive, superoxide-producing targets.
Desulfovibrio spp. growing with sulfate or other oxidized sulfur anions as the electron acceptor have a certain tolerance to air exposure. The finding that Desulfovibrio spp. can reduce oxygen to water, with hydrogen or organic acids as electron donor, and can couple this process to the production of ATP led to the idea that these organisms also engage in aerobic respiration (5). Oxygen is reduced by a cytoplasmic pathway with rubredoxin:oxygen oxidoreductase (Roo) as the terminal oxidase (7, 30). Because this pathway does not involve a membrane-bound terminal oxidase, it is unclear how ATP synthesis is coupled to oxygen reduction. However, genes for a cytochrome c oxidase (Cox) have been found in Desulfovibrio vulgaris Miyazaki (14), and a cytochrome bd terminal oxygen reductase (Cbd) has been purified from D. gigas membranes (18). Genes for both Cox and Cbd are present in the genome sequence of D. vulgaris Hildenborough (http://www.tigr.org). An electron transport chain involving membrane-bound Cox or Cbd is thus a possibility in Desulfovibrio spp. and is more likely to couple proton-motive-force-dependent ATP synthesis to oxygen respiration.
Irrespective of whether oxygen is respired with or without coupled ATP synthesis, its presence and use will give rise to partially reduced, highly reactive oxygen species (ROS). D. vulgaris enzymes that detoxify ROS include superoxide reductase (Sor) (3, 13, 24, 34) and rubrerythrin (Rbr), which has NADH-dependent H2O2 reductase activity (4, 22, 28). Sor and Rbr reduce superoxide and hydrogen peroxide to water without regeneration of oxygen, a feature that is important for oxygen detoxification in anaerobes (13). These proteins are widely distributed in anaerobes and are sometimes found in a single operon (20). Another enzyme, superoxide dismutase (Sod), which is involved in the elimination of superoxide anions, has been identified in several species of Desulfovibrio (6, 11). Recently, the gene encoding Sod has been cloned and sequenced from D. vulgaris Hildenborough (20). This protein exhibits 50% sequence identity to Fe-Sod or Mn-Sod from E. coli. The N-terminal sequence contains a double-arginine motif characteristic of signal peptides of other bacterial redox proteins, suggesting this Sod to be periplasmic (20) in contrast to Sor, which is cytoplasmic. D. vulgaris also contains a cytoplasmic catalase that acts as a second system (in addition to Rbr) to eliminate hydrogen peroxide (20). Thus, D. vulgaris contains oxygen defense proteins that are typical of both the aerobic (Sod and Kat) and the anaerobic (Sor and Rbr) microbial world. D. vulgaris Hildenborough is one of the few microorganisms that contain both systems, with other examples being D. gigas and Methanothermobacterium thermoautotrophicum (6, 34, 36). In order to delineate the importance of these defense proteins, we constructed sod and rbr mutants of D. vulgaris Hildenborough. Their properties are compared here with those of the wild-type strain and of an sor mutant obtained earlier (42).
MATERIALS AND METHODS
Materials.
Restriction and DNA modification enzymes and bacteriophage λ DNA were obtained from Pharmacia. [α-32P]dCTP (10 mCi/ml; 3,000 Ci/mmol) was from ICN. Mixed gas (85% [vol/vol] N2, 10% [vol/vol] CO2, and 5% [vol/vol] H2) was from Praxair Products, Inc. Anti-mouse and anti-rabbit immunoglobulin G alkaline phosphatase (AP)-linked antibodies were from New England Biolabs. Other immunoblotting reagents, including nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate), as well as sequencing-grade modified trypsin, were from Promega. Reagent-grade chemicals were from either BDH, Fisher, or Sigma. Deoxyoligonucleotide primers were obtained from University Core DNA Services of the University of Calgary.
Bacterial strains, plasmids, and growth conditions.
Strains, plasmids, and primers used or constructed in the present study are listed in Table 1. Escherichia coli and D. vulgaris strains were grown as described elsewhere (8, 26, 41).
TABLE 1.
Bacterial strains, primers, vectors, and plasmids used
| Strain, plasmid, or primer | Genotype, sequence (position), and/or comment(s)a | Source or reference |
|---|---|---|
| Strains | ||
| D. vulgaris subsp. vulgaris Hildenborough | NCIMB 8303; isolated from clay soil near Hildenborough, United Kingdom | 27 |
| D. vulgaris SOD12 | Plasmid p2ΔSOD integrated into the chromosome; Sucs Cmr | This study |
| D. vulgaris SV2-6 | Plasmid pSV2 integrated into the chromosome; Sucs Cmr | 21 |
| D. vulgaris SOD100 | Δsod; Sucr Cmr | This study |
| D. vulgaris RBR100 | Δrbr; Sucr Cmr | This study |
| D. vulgaris L2 | Δsor; Sucr Cmr | 42 |
| E. coli S17-1 | thi pro hsdR hsdM+ recA RP4-2 (Tc::Mu Km::Tn7) | 35 |
| E. coli M15[pREP4] | Host strain for overexpression of pQE-encoded genes; Kmr | Qiagen, Inc. |
| Plasmids | ||
| pUC19Cm | pUC19 containing the cat gene | 8 |
| pNOT19 | Cloning vector pUC19; NdeI site replaced by a NotI site | 33 |
| pMOB2 | Contains oriT of plasmid RP4 and Bacillus subtilis sacBR genes on a 4.5-kb NotI fragment; Kmr Cmr | 33 |
| pLITMUS28 | Cloning vector; Apr | New England Biolabs, Inc. |
| pLITSOD, p2SODCat, p2NotSODCat, and p2ΔSOD | This study (see text) | |
| pSV2 | 21 | |
| pQE30 | Overexpression vector with an N-terminal His tag; Apr | Qiagen, Inc. |
| pQS10 | This study | |
| Primers | ||
| P154-f | 5′-GAGGATACCGTGTATGC (5-21)∗ | |
| P155-r | 5′-CAAAGGCCGGGAGTGTG (960-944)∗ | |
| P161-r | 5′-CATGCCTGCCCAGTAAAAG (479-461)∗ | |
| P162-f | 5′-CGGGAGGCGGAGGGAC (484-499)∗ | |
| P175-f | 5′-cctctagaCTCAACAAGGCCGTGGCGGG (336-355)∗ | |
| P176-r | 5′-ccgcatGCGTGCTCCCATACGTCGATGG (730-709)∗ | |
| P182-f | 5′-GCGGGCTTGACAGTCTCCTCC (665-685)† | |
| P183-r | 5′-GGGATGAGGTTGGAGGGATGC (1139-1119)† | |
| P184-f | 5′-GCATCCCTCCAACCTCATCCC (1119-1139)† | |
| P185-r | 5′-GAGTTCGAAGTGCGCCTTGGG (1714-1694)† | |
| P204-f | 5′-cgggatCCATGCCGCCCCTGCCCTATCCC (232-254)∗ | |
| P205-r | 5′-ggggtaccCTAGAGGCGTGTTGCCACC (824-806)∗ |
∗, Nucleotide positions in the 971-bp sequence containing the sod gene of D. vulgaris (accession number AF034841 [20]); †, nucleotide positions in the 3,270-bp sequence of the rbr operon of D. vulgaris (accession number M77011 [22]). Sucr, sucrose resistance; Cmr, CHL resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance. Lowercase letters in primer sequences indicate added restriction endonuclease cleavage sites
Construction of sod and rbr deletion mutants.
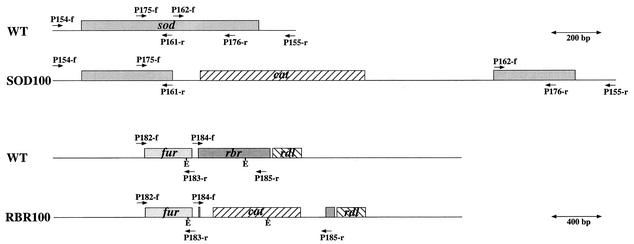
A 956-bp fragment containing the sod gene of D. vulgaris (Fig. 1) was amplified by PCR from genomic DNA by using the primers P154-f and P155-r and then cloned in the EcoRV site (GAT↓ATC) of pLITMUS28 to create plasmid pLITSOD. Plasmid pLITSOD was PCR amplified with primers P161-r and P162-f. This deleted bp 480 to 483 of the sequence with accession number AF034841. The PCR product was ligated with the 1.4-kb BamHI fragment from pUC19Cm to yield plasmid p2SODCat. The 2.4-kb insert of this plasmid was transferred to pNOT19 after digestion of both with KpnI and PstI to give plasmid p2NotSODCat. The suicide vector p2ΔSOD (9.6 kb) was generated by inserting the 4.5-kb NotI fragment from pMOB2 into the NotI site of plasmid p2NotSODCat. This plasmid was transferred to D. vulgaris by conjugation with E. coli S17-1(p2ΔSOD). A single crossover integrant, D. vulgaris SOD12, was selected. Growth of D. vulgaris SOD12 in the presence of chloramphenicol (CHL) and sucrose (8) yielded D. vulgaris SOD100 (Fig. 1). For generation of an rbr deletion mutant, suicide plasmid pSV2, containing the cat gene flanked by regions upstream and downstream from the rbr gene and constructed as described elsewhere (21), was transferred to D. vulgaris by conjugation, and a single crossover integrant D. vulgaris SV2-6 was selected. Treatment of D. vulgaris SV2-6 with CHL and sucrose gave replacement mutant D. vulgaris RBR100 (Fig. 1).
FIG. 1.
Maps for the rbr and sod regions. A scale of 200 bp is indicated for maps for D. vulgaris wild type (WT) and the marker replacement mutant D. vulgaris SOD100, as well as the hybridization positions of primers P154-f, P155-r, P161-r, P162-f, P175-f, and P176-r and the locations of the sod and cat genes. A scale of 400 bp is indicated for D. vulgaris wild type and the marker replacement mutant D. vulgaris RBR100, as well as the hybridization positions of primers P182-f, P183-r, P184-f, and P185-r; the locations of fur, rbr, rdl, and cat genes; and the positions of restriction sites for EcoRI (E).
Southern blot and PCR analysis for genotypic verification.
Replacement mutagenesis of the sod and rbr genes was verified by PCR with a Perkin-Elmer Gene Amp 2400 PCR system with TaqI DNA polymerase and primer pairs indicated in Fig. 1 and Table 1. Replacement mutagenesis of the rbr gene was also verified by Southern blotting (43) by using a probe generated by PCR with primers P182-f and P183-r and labeled with [α-32P]dCTP by the random hexamer procedure. Radioactive images of the blots were displayed with a Fuji BAS1000 Bioimaging Analyzer.
Immunoblotting.
Expression of sod and rbr genes was demonstrated by immunoblotting. A rabbit polyclonal antibody against Rbr, purified from D. vulgaris, was obtained from Don Kurtz, Jr., Department of Chemistry, University of Georgia. His-tagged Fe-Sod from D. vulgaris was overproduced in E. coli M15[pREP4](pQS10) with the QIAexpress system (Qiagen). Plasmid pQS10 was constructed by PCR amplification of the sod gene from plasmid pLITSOD with primers p204-f and p205-r and inserting the PCR product into plasmid pQE30 (Qiagen) after cleavage with BamHI and KpnI. Immunoreactive sera of suitable titers were obtained by injecting two mice intraperitoneally on days 1 and 26 with 400 μg of purified His-tagged Fe-Sod. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were carried out as described elsewhere (15, 26, 38).
Sod activity staining.
Early-stationary-phase cultures in 100 ml of medium C (27) were harvested by centrifugation (20 min, 4,000 × g) and washed once with 100 mM Tris-HCl (pH 8.0). The cell pellet was then resuspended in 0.8 ml of 100 mM Tris-HCl-100 mM EDTA (pH 9.0) (TE) and incubated at 37°C for 30 min (25, 39). After centrifugation (15,000 × g, 15 min at 6°C), the red supernatant (the TE-extract) was transferred into a clean microfuge tube. The TE pellet was resuspended in 3 ml of TE and lysed by two passes through a French press minicell at 1,800 lb/in2. Proteins were quantified by using the DC protein assay (Bio-Rad). Appropriate amounts of proteins were frozen in liquid nitrogen, dried in a speed vacuum, and then dissolved in 10 μl of H2O plus 10 μl of native protein sample buffer (100 mM Tris-Cl [pH 6.8], 20% glycerol, 0.2% bromophenol blue). Samples were loaded onto 12% (wt/vol) nondenaturing minigels and run at 4°C. Sod activity bands were located by staining as described elsewhere (2). Inhibition of the Sod activity in native polyacrylamide gels was tested by adding 10 mM azide (NaN3), cyanide (KCN), or hydrogen peroxide (H2O2) to solutions used for Sod activity staining. Sod activities were quantified spectrophotometrically by using the xanthine oxidase-cytochrome c system described by McCord and Fridovich (23).
Exposure to air.
Cultures (5 ml) of wild-type and mutant strains were grown anaerobically in medium C with kanamycin (25 μg/ml). At time zero, 0.5 ml of these cultures was diluted into 50 ml of aerobic medium C without yeast extract and then stirred with a magnetic stirrer continuously in air at room temperature. Also, 10 μl of anaerobic cultures were serially diluted in medium C. Aliquots (100 μl) of the 105- and 106-fold-diluted cultures were spread on medium E plates (27) to obtain the zero time count of CFU/ml. Aliquots (0.5 ml) of the air-exposed cultures were periodically transferred to the anaerobic hood, serially diluted, and plated. Colonies were counted after 1 week of anaerobic incubation at 32°C as the number of surviving CFU/ml. Data collected over a 72-h incubation period were fitted to the equation ln(Nt/N0) = −kt, where Nt and N0 are the recorded CFU/milliliter at times zero and t, respectively, and k is the first-order inactivation rate constant (per hour). Only k values derived from experiments for which the data fitted a straight line with a correlation coefficient r > 0.85 were used. Alternatively, colonies present on medium E plates after 5 days of anaerobic incubation at 32°C were exposed to air for 30, 50, or 66 h. At each of these times the plates were briefly returned to the hood. Part of a colony was streaked on a new medium E plate, whereas the other part was inoculated into 5 ml of anaerobic liquid medium C. Growth on the plates and in the liquid medium was scored after 7 and 5 days, respectively.
Exposure to increased concentrations of internally generated superoxide.
Experimental procedures were the same as described for exposure to air, except that 0.1 mM paraquat was included in the 50 ml of aerated medium into which 0.5 ml of the anaerobically grown cells was diluted.
Exposure to externally generated superoxide.
Aliquots (100 μl) of anaerobic cultures in medium C with kanamycin were pipetted into 1.5-ml sterile microfuge tubes in the anaerobic hood. The tubes were transferred to air and, after 30 min, xanthine and xanthine oxidase were added to final concentrations of 0.22 mM and 0.45 U/ml, respectively. After they were left in air for an additional 30 to 60 min, the tubes were returned to the anaerobic hood and serially diluted, and 100-μl portions of appropriate dilutions were spread on medium E plates. The fraction (f) of surviving cells was calculated by dividing the surviving CFU/milliliter by the count for unexposed aliquots. A time course for superoxide inactivation was done by transferring 30-μl aliquots of anaerobic cultures into 3 ml of aerobic medium C without lactate but containing xanthine (0.1 mM), xanthine oxidase (0.037 U/ml), and catalase (1 U/ml) and exposing the aliquots to air with shaking. Aliquots (20 μl) were removed after 5, 10, 15, and 30 min; transferred to the anaerobic chamber; and immediately plated onto medium E plates in duplicate. After incubation of the plates for 5 days at 32°C, colonies were counted, and the number of surviving CFU/milliliter was determined. This experiment was done in triplicate.
Exposure to H2O2.
Anaerobic cultures (100 μl, 108 cells/ml) were spread onto medium E plates with a glass spreader. Hydrogen peroxide (20 μl, 0.5 mM) was spotted onto a circular Whatman filter paper disk, which was placed on the cell lawn (20). The plates were incubated anaerobically for 4 days, and the diameter of the growth inhibition zone surrounding the disks was measured with a ruler.
Two-dimensional gel electrophoresis (2DE).
Strains were grown in 50 ml of a defined lactate-sulfate medium (41, 44). TE-extracts prepared from log-phase cells were stored frozen at −80°C. For isoelectric focusing, 5 to 10 μl of TE-extract (20 to 40 μg of protein) diluted in 345 to 340 μl of urea-containing rehydration buffer (9) was focused with 18-cm Immobiline Drystrip gels (pH 3-10NL; Amersham Pharmacia Biotech). For the second dimension, proteins were separated on 12.5% (wt/vol) polyacrylamide gels containing 0.1% (wt/vol) SDS (29). After overnight electrophoresis at 20°C, the gels were fixed and then stained either with silver or Coomassie brilliant blue (CBB). Data acquisition of stained gels was performed by using the IMAGE Scanner (Amersham Pharmacia Biotech).
MALDI-TOF/MS.
Selected protein spots were excised manually from either silver- or CBB-stained gels and placed in microfuge tubes. The gel pieces were washed, dried, and digested with trypsin solution (31, 45). Trypsin-digested samples were analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF/MS) on a Voyager DE-STR (Applied Biosystems, Foster City, Calif.). Peptide mass fingerprints were analyzed by using ProteinProspector software at http://prospector.ucsf.edu or were searched against a preliminary, annotated database of D. vulgaris provided by John Heidelberg from The Institute of Genomic Research by using the Knexus/Profound software package from Genomic Solutions.
RESULTS
Generation of marker replacement mutants.
Gene replacement was achieved by treating cultures of the single crossover integrants, D. vulgaris SOD12 and D. vulgaris SV2-6, with sucrose in the presence of CHL. For D. vulgaris SOD12 the procedures outlined by Fu and Voordouw (8), with rich media, were followed. Only a single colony, representing replacement mutant D. vulgaris SOD100, was eventually obtained. For D. vulgaris SV2-6, use of these same procedures was unsuccessful, even after extensive screening. However, plating on defined hydrogen-sulfate plates containing sucrose and CHL (26) yielded five colonies, three of which had the desired genotype.
Verification of genotype of marker replacement mutants.
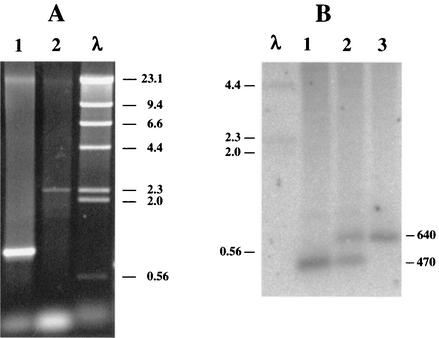
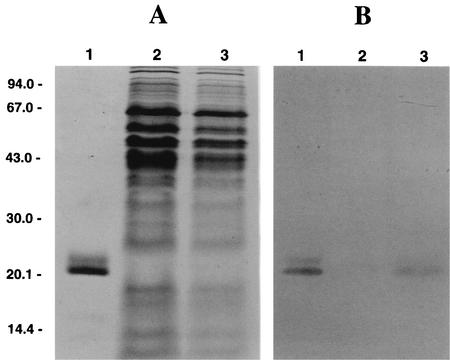
Amplification of DNA from the SOD100 strain with primers P154-f and P155-r gave a 2.3-kb PCR product, whereas a 0.95-kb product was obtained with DNA from the wild-type strain (Fig. 2A), indicating insertion of the CHL resistance (Cmr) marker. PCR of genomic DNA of the single crossover integrant yielded both products (not shown). Use of primers P175-f and P176-r (not shown) also confirmed the maps shown in Fig. 1. The genotype of the RBR100 strain was confirmed by Southern blotting (Fig. 2B). Hybridization of an EcoRI digest of wild-type DNA with the P182/P183 probe gave a 470-bp band (Fig. 2B, lane 1), whereas 470- and 640-bp bands were seen for DNA from the single crossover integrant, D. vulgaris SV2-6 (Fig. 2B, lane 2). The gene replacement mutant D. vulgaris RBR100 displayed only the 640-bp band (Fig. 2B, lane 3), in agreement with the maps shown in Fig. 1. Use of the P184-f and P185-r primer pair gave a 0.6-kb fragment for the wild-type, 0.6- and 1.1-kb fragments for the SV2-6, and a 1.1-kb fragment only for the RBR100 strain, respectively (data not shown). Immunoblotting confirmed the absence of the sod gene product in D. vulgaris SOD100 (data not shown) and of the rbr gene product in D. vulgaris RBR100 and their presence in the wild-type strain (Fig. 3).
FIG. 2.
(A) Agarose gel of PCR products of DNA from D. vulgaris wild-type (lane 1) and SOD100 (lane 2) strains with primers P154-f and P155-r. (B) Southern blot of EcoRI-digested chromosomal DNAs of D. vulgaris wild type (lane 1), strain SV2-6 (lane 2), and strain RBR100 (lane 3). The fragment sizes of a HindIII digest of bacteriophage λ DNA are indicated (lanes λ).
FIG. 3.
Immunoblot analysis of Rbr content. (A) CBB-stained gel after SDS-PAGE. Lane 1, purified Rbr, 10 μg; lane 2, RBR100 strain, 50 μg of protein; lane 3, wild-type strain, 30 μg of protein. (B) Immunoblot of gel as in panel A, except that only 1 μg of purified Rbr was loaded in lane 1.
Sod activity staining.
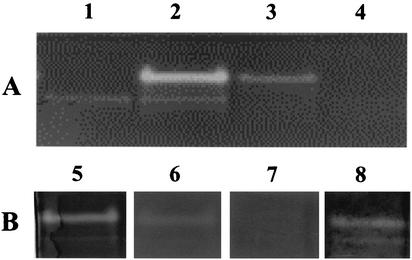
Sod activity staining of nondenaturing gels showed a major band, near the top of the gel in the wild-type TE-extract (Fig. 4A, lane 2), which was absent in the SOD100 TE-extract (Fig. 4A, lane 1), indicating that this band represents Sod. A second, less-intense band was detected in the TE-extracts of both strains. It is not yet clear which protein this represents. Experiments with purified D. vulgaris Sor indicated it to run as a diffuse band lower in the gel (not shown). When TE-pellet samples were loaded, no activity bands were detected for the sod mutant (Fig. 4A, lane 4), whereas a small amount of Sod (probably due to incomplete separation from TE-extract) was found for the wild type (Fig. 4A, lane 3). These results do not prove a periplasmic localization of Sod because TE-extracts also contain cytoplasmic proteins (Table 4). Thus, evidence for a periplasmic localization is primarily provided by the presence of an N-terminal, twin-arginine signal peptide (20). When more TE-extract protein was loaded, a weakly staining band was observed in the lower part of the gel for wild-type and sor and sod mutant strains but not for the Rbr100 strain (data not shown). This could thus represent Rbr. Lehman et al. (17) also reported that Rbr can be identified on native gels by Sod activity staining. Rbr can remove superoxide under the conditions used for the staining (2), even though the protein is neither a Sod nor a Sor.
FIG. 4.
Sod fractionation and sensitivity of Sod activity staining to inhibitors. Samples (75 μg of protein in all lanes 1 to 8) were run on a native 12% (wt/vol) polyacrylamide gel. (A) Fractionation into TE-extracts from SOD100 (lane 1) and wild-type (lane 2) strains and TE-pellets from wild-type (lane 3) and SOD100 (lane 4) strains. (B) Sensitivity to inhibitors as shown by activity staining without inhibitors (lane 5) or in the presence of 10 mM NaN3 (lane 6), 10 mM H2O2 (lane 7), or 10 mM KCN (lane 8).
TABLE 4.
MS identification of proteins from D. vulgaris separated by 2DE
| na | IDb | Sequence coveragec (%) | Significanced | pIe | Mrf | Locationg | Name, possible functionh |
|---|---|---|---|---|---|---|---|
| 30 | ORF03941 | 9 | 3.1 × 10−4 | 5.3 | 29.8 | C | Hypothetical protein |
| 31 | ORF03581 | 28 | 0.054 | 5.1 | 12.0 | C | Sulfite reductase, dissimilatory-type gamma subunit |
| 32 | ORFA00027 | 9 | 8.4 × 10−6 | 5.8 | 32.0 | M | SrpA, putative |
| 33 | ORF02650 | 38 | 2.2 × 10−4 | 5.4 | 15.0 | C | RNA-binding protein |
| 34 | ORF01017 | 49 | 6.4 × 10−5 | 5.4 | 10.2 | C | RNA-binding protein |
| 35 | ORF02955 | 17 | 3.9 × 10−4 | 4.2 | 16.1 | C | Hypothetical protein |
| 36 | ORF02453 | 28 | 3.5 × 10−13 | 5.7 | 12.6 | C | Uncharacterized ACR, COG1433 family |
| 37 | ORF03883 | 52 | 5.9 × 10−9 | 4.9 | 17.0 | C | Nitrogen assimilation regulatory protein |
| 38 | ORF00336 | 35 | 2.2 × 10−3 | 4.9 | 19.1 | C | Adenylyl sulfate reductase, beta subunit |
| 39 | ORF04271 | 50 | 0.046 | 6.0 | 14.3 | C | Sor, superoxide reductase |
| 40 | AF034841 | 36 | 1.437e0.04 | 6.6 | 22.1 | P | Sod, superoxide dismutase |
n, spot number in 2DE gels shown in Fig. 6.
ID, identification by MS in a database of coding genes for the D. vulgaris genome (http://www.tigr.org) provided by J. Heidelberg.
That is, the extent to which the combined peptide masses cover the indicated sequence.
That is, the significance of the match; probability that the match between the identified peptides and the indicated sequence is due to random chance.
pI is the average theoretical value calculated with programs found at http://www.embl-heidelberg.de/cgi/pi-wrapper.pl and at http://www.biologie.uni-freiburg.
Mr is the theoretical molecular mass derived from the genome sequence.
Most likely location—cytoplasmic (C), periplasmic (P), or in the membrane (M)—based on evaluation of the amino acid sequence.
Annotated function.
Search was performed by using MS-Fit software (http://prospector.ucsf.edu) against the NCBInr.6.6.2002 database. The indicated score is the MOWSE score.
Sod activity, as monitored by staining of native polyacrylamide gels, was observed in the presence of cyanide (Fig. 4B, lane 8) but not in the presence of H2O2 (Fig. 4B, lane 7), whereas only a slight stain was observed in the presence of azide (Fig. 4B, lane 6), indicating significant inhibition. These data suggest that D. vulgaris Sod is an iron-containing enzyme, similar to that of D. gigas (6). Fe-Sod is sensitive to H2O2 and azide but not to cyanide, whereas Mn-Sod is only inhibited by azide and CuZn-Sod is sensitive to cyanide and H2O2.
Most of the Sod activity, measured as the amount of enzyme required to inhibit the reduction of cytochrome c by 50%, was found in the wild-type TE-extract (33.6 ± 1.0 U/mg), whereas less activity was detected in the TE-pellet (9.1 ± 0.6 U/mg). No activity was detected in the TE-extract of the sod mutant, whereas an activity similar to that observed for the wild type was detected in the TE-pellet (10.7 ± 1.0 U/mg). Again, it is not clear which protein is represented by this activity, since the genome sequence does not indicate the presence of a second Sod.
Resistance to oxidative stress.
Because no statistically valid differences were observed for anaerobically grown log-phase or early-stationary-phase cells, all data were averaged. Average rate constants k for inactivation of D. vulgaris after dilution into fully aerated medium were very similar for the wild-type, the sod mutant, and the rbr mutant strains. Only the k value for the sor mutant was significantly higher (Table 2, 0.083 and 0.272 h−1, respectively). Surviving fractions can be calculated from these k values as follows: f = 14% for 24 h of air exposure of the wild-type, the sod mutant, and the rbr mutant strains and f = 0.15% for 24 h of air exposure of the sor mutant strain. Air exposure of colonies on plates had a more discriminating inactivating effect. The sor mutant was again the most sensitive and failed to grow even after 30 h of air exposure. However, both the sor and the sod mutants failed to grow after 66 h of air exposure, whereas growth of the wild-type and rbr mutant strains was observed in three of four and in two of four experiments, respectively (Table 3).
TABLE 2.
Effect of various oxidative stresses on D. vulgaris wild-type and mutant strains
| Stressa | Strain | Parameterb | nc | Mean ± avg deviationd |
|---|---|---|---|---|
| Air | Wild type | k | 10 | 0.083 ± 0.025 h−1 |
| Sod100 (sod) | k | 8 | 0.092 ± 0.025 h−1 | |
| L2 (sor) | k | 4 | 0.272 ± 0.050 h−1 | |
| Rbr100 (rbr) | k | 4 | 0.073 ± 0.032 h−1 | |
| Air+PQ | Wild type | k | 4 | 0.083 ± 0.037 h−1 |
| Sod100 (sod) | k | 3 | 0.124 ± 0.046 h−1 | |
| L2 (sor) | k | 6 | 0.304 ± 0.074 h−1 | |
| Rbr100 (rbr) | k | 4 | 0.110 ± 0.060 h−1 | |
| Air+X+XO | Wild type | f | 10 | 39% ± 13% |
| Sod100 (sod) | f | 10 | 28% ± 11% | |
| L2 (sor) | f | 10 | 15% ± 3% | |
| Rbr100 (rbr) | f | 10 | 35% ± 14% | |
| H2O2 | Wild type | d | 11 | 14.7 ± 0.8 mm |
| Sod100 (sod) | d | 11 | 15.3 ± 0.9 mm | |
| L2 (sor) | d | 11 | 14.3 ± 1.0 mm | |
| Rbr100 (rbr) | d | 11 | 14.7 ± 1.0 mm |
Air is a 100-fold dilution into air-saturated medium; Air+PQ is a 100-fold dilution into air-saturated medium containing 0.1 mM paraquat; Air+X+XO is exposure to air and xanthine plus xanthine oxidase for 30 to 60 min; H2O2 is exposure to 10 μmol of H2O2 applied to a disk on a lawn of bacteria.
k is the first-order inactivation constant; f is the fraction of surviving bacteria determined for a 0.5- to 1-h exposure in Air+X+XO; d is the diameter of the zone without growth surrounding the disk on which H2O2 was placed.
n is the number of experiments.
Mean is the mean value for n experiments. Deviation is the average deviation from the mean for n experiments.
TABLE 3.
Growth of single colonies of wild-type and mutant strains after exposure to aira
| Growth condition and time of exposure to air (h) | Growth of:
|
|||
|---|---|---|---|---|
| WT | Sod100 (sod) | L2 (sor) | Rbr100 (rbr) | |
| Growth on plates | ||||
| 30 | ++++ | ++++ | −−−± | ++++ |
| 50 | ++−+ | ++−+ | −−−− | ++−+ |
| 66 | ++−+ | −−−− | −−−− | −+−+ |
| Growth in liquid medium | ||||
| 30 | ++++ | ++−+ | +−−+ | ++++ |
| 50 | ++−+ | ++−+ | −−−− | ++−+ |
| 66 | ++−+ | −−−− | −−−− | −+−+ |
The results of four experiments are summarized. WT, wild type. Growth was scored as good (+), poor (±), or absent (−), after 7 days for plates and after 5 days for liquid medium.
In aerobic bacteria the oxidative stress level can also be increased by addition of paraquat (methylviologen), which increases the rate of cytoplasmic superoxide production in E. coli (10). As indicated in Table 2, the addition of 0.1 mM paraquat left the inactivation rate constant k of the wild type unchanged, whereas it increased somewhat for the mutant strains, corresponding to 24-h survivals of 14, 5, 0.07, and 7% for the wild-type and sod, sor, and rbr mutant strains, respectively. The insensitivity of wild-type D. vulgaris to paraquat agrees with an earlier study (20), in which a larger effect of paraquat on the inactivation rate of the sor mutant was found. The increased sensitivity of the SOD100 strain was unexpected, because paraquat is generally thought to only increase the production of cytoplasmic superoxide, which does not serve as a substrate for a periplasmic Sod since superoxide is membrane impermeable.
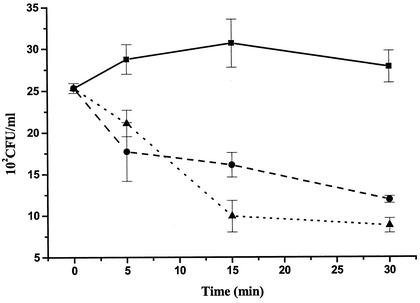
Exposure of D. vulgaris cells to limited amounts of external superoxide (0.10 mM xanthine, 0.037 U of xanthine oxidase/ml) did not result in a loss of viability of the wild type, whereas both sod and sor mutants were affected even after short exposure times (Fig. 5). After 30 min, only 50% of the sod mutant cells survived. However, the surviving fraction of sor mutant cells was even lower (33%). When D. vulgaris cells were exposed to high concentrations of externally produced superoxide (0.22 mM xanthine, 0.45 U of xanthine oxidase/ml), all strains lost viability. Relative to the wild-type strain (Table 2, 39% survival), the sor mutant was again affected the most (15% survival). The sod mutant was affected less strongly (28% survival), whereas the rbr mutation had little effect (35% survival). Thus, periplasmic Fe-Sod provided protection to externally produced superoxide. The inactivation of the sor mutant is in part caused by air exposure, i.e., for k = 0.272 h−1 a surviving fraction of f = 76% can be calculated for 1 h of exposure to air only. However, even after we corrected for that, cytoplasmically located Sor appears to be important for combating external superoxide stress.
FIG. 5.
Time course of loss of viability of the wild-type, SOD100, and L2 strains after exposure to limited amounts of extracellularly generated superoxide. Symbols: ▪, wild type; •, SOD100; ▴, L2.
Exposure to H2O2 did not present a lethal form of oxidative stress under the chosen assay conditions (Table 2). The presence of several rbr homologs in the D. vulgaris genome (http://www.tigr.org) may contribute to the lack of H2O2 sensitivity.
2DE.
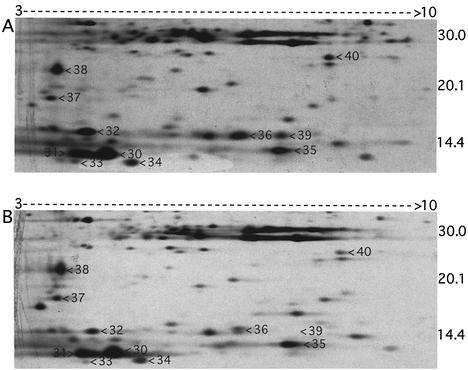
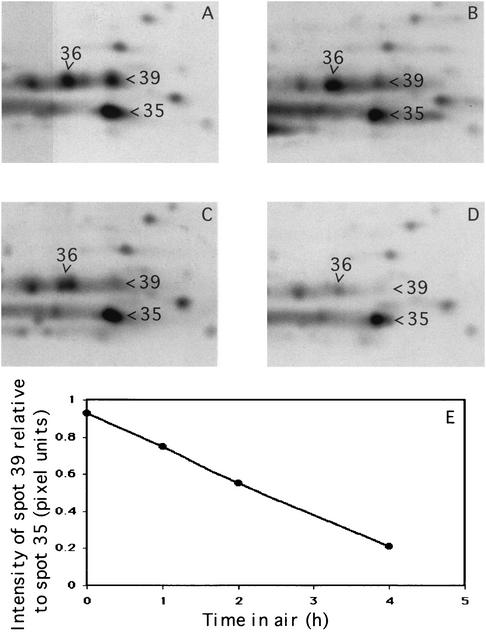
Comparison of silver-stained gels for the wild-type and SOD100 strains indicated that spot 40 in the wild-type TE-extract (Fig. 6A) was absent from the TE-extract of the SOD100 strain (not shown). MALDI-TOF/MS analysis of the protein digest, combined with a search of the NCBInr database with MS-Fit software, indicated spot 40 to be D. vulgaris Fe-Sod (Table 4). A 14-kDa protein in the wild-type TE-extract (Fig. 6A, spot 39) was missing from the sor mutant proteome (Fig. 6B). Analysis of the trypsin-digested samples by MALDI-TOF/MS and comparison of the determined peptide masses with a database derived from the D. vulgaris genome by using the program ProFound (Genomic Solutions) indicated ORF04271 Sor as the top score. Several other proteins were cut out from the CBB-stained gel and successfully identified by MALDI (Table 4). The position of Rbr (21 kDa) could not be identified by comparing protein patterns in silver-stained 2DE gels obtained for the rbr mutant and wild-type strains. In view of the central role of Sor in resistance to oxygen stress of D. vulgaris under fully aerated conditions (Table 2), we determined the dependence of the 2DE pattern on the length of air exposure. Prolonged exposure to air (1 to 4 h) led to a decrease in intensity of Sor (spot 39) relative to neighboring proteins in the 2DE patterns (Fig. 7).
FIG. 6.
2DE gels of TE-extracts of D. vulgaris Hildenborough wild-type (A) and sor mutant (B) strains. Only a part of the silver-stained gels is shown. Proteins were separated on 18-cm IPG strips (pH 3-10NL; from left to right) and by 2DE according to the molecular mass (indicated in kilodaltons). The numbered protein spots were cut out from CBB-stained gels. Their MALDI identifications are presented in Table 4.
FIG. 7.
Effect of air exposure on the D. vulgaris proteome. D. vulgaris wild type grown anaerobically in defined lactate-sulfate medium was exposed to air for 0 (A), 1 (B), 2 (C), and 4 (D) h. Part of the silver-stained 2DE gels of TE-extracts is shown. (E) Intensity of spot 39 (Sor) relative to that of spot 35 as a function of time.
DISCUSSION
Reduction of oxygen entering the cytoplasm of D. vulgaris Hildenborough by soluble Roo or membrane-bound Cox or Cbd results in generation of the ROS superoxide and H2O2. Sor and Rbr have been proposed to remove these cytoplasmically generated ROS by reduction (20). Because superoxide is not membrane permeable, periplasmic SOD has been proposed to protect D. vulgaris from superoxide generated in the periplasm, when oxygen reacts with reduced periplasmic redox proteins such as hydrogenase or cytochrome c3 (20). Interestingly, the D. vulgaris roo gene is located immediately downstream from the sor-rub operon (http://www.tigr.org). Thus, Roo and Sor may collaborate in reduction and detoxification of oxygen entering the cytoplasm through use of rubredoxin (Rub) as a common intermediary electron donor.
The lack of a strong oxygen stress phenotype for the rbr mutant (Table 2) is not surprising in view of the presence of genes for two Rbr homologs, Rbr2 and nigerythrin (Ngr), in the genome (http://www.tigr.org). D. vulgaris also has a kat gene encoding catalase, but this is located on a nif gene-containing plasmid that is lost when cells are cultured in ammonium chloride-containing media as were used here (G. Voordouw, unpublished data). Thus, the wild-type level of H2O2 resistance of the rbr mutant (Table 2) was caused by Rbr2 and Ngr and not catalase.
A somewhat unexpected finding was that resistance to internally and externally generated superoxide was not cleanly separated in the sor and sod mutants. In the case of internally generated superoxide, it has been shown that the stable paraquat monocation radical (PQ+.) formed from oxidized paraquat (PQ2+) in the cytoplasm can diffuse across the membrane, where it reacts with available oxygen to form superoxide (10). Hence, a role of Sod in remediating the additional oxidative stress induced by paraquat can be understood. The mechanism by which periplasmic superoxide induces additional oxidative stress in the cytoplasm is currently unknown but may be similar, i.e., through generation of membrane-transportable, lipophilic radicals.
Comparison of the sensitivity of sor, sod, and rbr mutants to oxidative stress indicates that under fully aerated conditions Sor is the key oxygen defense factor (Table 2). Apparently, superoxide is generated primarily in the cytoplasm under these conditions. The periplasmic redox protein pool may quickly drain electrons during exposure to fully aerated conditions as were used here, leaving no targets for periplasmic superoxide production. However, when superoxide is artificially generated outside the D. vulgaris cell, the presence of Sod appears to be advantageous, since the Sod mutant was found to be considerably more sensitive to such external superoxide stress than was the wild type (Table 2 and Fig. 5). Because D. vulgaris is not a pathogen, it is not exposed to large amounts of superoxide produced externally by other cells (e.g., macrophages). Sod is therefore more likely to function in the removal of periplasmic superoxide produced under microaerophilic conditions, when the periplasmic redox protein pool is at least partially reduced. Abdollahi and Wimpenny (1) have shown, in agreement with this conclusion, that in continuous cultures of D. desulfuricans Sod activity is maximal at 1% (vol/vol) oxygen. The activity was lower both in anaerobic cultures and in fully aerated cultures.
Periplasmic Sods in other microorganisms are either of the CuZn type (12, 32, 37) or the Mn type (16, 19). Although periplasmic CuZn-Sods have typical Sec system-dependent signal sequences, a twin-arginine leader sequence has been found in periplasmic Mn-Sod (19). To date, periplasmic Fe-Sods have only been found in Desulfovibrio spp. D. vulgaris Fe-Sod is exported through the Tat system (20), which only handles redox proteins with bound cofactors (40). The difference indicates that, prior to export, Desulfovibrio spp. assemble Fe-Sod in the cytoplasm, whereas metal acquisition by the enzymes exported via the Sec system is a periplasmic process. Evolution of cytoplasmically assembled, periplasmic Fe-Sod in Desulfovibrio spp. may have been necessitated by the fact that high sulfide concentrations produced by these organisms keep external concentrations of copper and zinc ions at extremely low levels. Fe-Sod or Mn-Sod in aerobic or facultative microorganisms, such as SodA and SodB in E. coli, are cytoplasmic enzymes. Because of their critical role in combating respiration-mediated superoxide stress, these are generally constitutive, whereas periplasmic CuZn-Sod is induced in stationary phase (12, 32, 37). A decreasing oxygen tension in stationary phase may induce oxygen-sensitive, periplasmic redox proteins (hydrogenases, nitrate reductases) associated with alternative respiratory chains, leading to increased periplasmic superoxide production. Hence, as in D. vulgaris, the need for periplasmic Sod in these other microorganisms may be greatest under conditions of low oxygen tension, when the periplasm has oxygen-sensitive targets.
In conclusion, it appears that under fully aerobic conditions Sor is the single most important oxygen defense factor. The decrease in Sor concentration recorded in wild-type cells during prolonged aeration (Fig. 7) may be caused by its inactivation upon continuous processing of highly reactive superoxide under conditions in which protein synthesis is compromised. This decrease may be a critical factor in causing the eventual cell death of D. vulgaris upon continued air exposure.
Acknowledgments
This work was supported by a grant from the Natural Science and Engineering Research Council of Canada (NSERC) to G.V. A database of the D. vulgaris Hildenborough genome was searched at the web site of The Institute for Genomic Research at http://www.tigr.org. Sequencing of this genome is financially supported by the U.S. Department of Energy.
M.F. and Y.Z. contributed equally to this study.
We thank Don Kurtz, Jr., and Heather Lumppio, Department of Chemistry, University of Georgia, for generating D. vulgaris SV2-6 and for polyclonal antibodies recognizing Rbr and D. Moinier from the IBSM Proteomics Facility for performing the Sod MALDI-TOF analysis. We thank John Heidelberg for making a preliminary annotation of the D. vulgaris genome available to us.
REFERENCES
- 1.Abdollahi, H., and J. W. T. Wimpenny. 1990. Effects of oxygen on the growth of Desulfovibrio desulfuricans. J. Gen. Microbiol. 136:1025-1030. [Google Scholar]
- 2.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 3.Brumlik, M. J., and G. Voordouw. 1989. Analysis of the transcriptional unit encoding the genes for rubredoxin (rub) and a putative rubredoxin oxidoreductase (rbo) in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 171:4996-5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulter, E. D., N. V. Shenvi, Z. M. Beharry, J. F. Smith, B. C. Prickril, and D. M. Kurtz, Jr. 2000. Rubrerythrin-catalyzed substrate oxidation by dioxygen and hydrogen peroxide. Inorg. Chim. Acta 297:231-241. [Google Scholar]
- 5.Cypionka, H. 2000. Oxygen respiration by Desulfovibrio species. Annu. Rev. Microbiol. 54:827-848. [DOI] [PubMed] [Google Scholar]
- 6.Dos Santos, W. G., I. Pacheco, M.-Y. Liu, M. Texeira, A. V. Xavier, and J. LeGall. 2000. Purification and characterization of an iron superoxide dismutase and a catalase from the sulfate-reducing bacterium Desulfovibrio gigas. J. Bacteriol. 182:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frazao, C., G. Silva, C. M. Gomes, P. Matias, R. Coelho, L. Sieker, S. Macedo, M. Y. Liu, S. Oliveira, M. Texeira, A. V. Xavier, C. Rodrigues-Pousada, M. A. Carrondo, and J. LeGall. 2000. Structure of a dioxygen reduction enzyme from Desulfovibrio gigas. Nat. Struct. Biol. 7:1041-1045. [DOI] [PubMed] [Google Scholar]
- 8.Fu, R., and G. Voordouw. 1997. Targeted gene replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143:1815-1826. [DOI] [PubMed] [Google Scholar]
- 9.Görg, A., C. Obermaier, G. Boguth, A. Harder, B. Scheibe, R. Wildgruber, and W. Weiss. 2000. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21:1053-1057. [DOI] [PubMed] [Google Scholar]
- 10.Hassan, H. M., and I. Fridovich. 1979. Paraquat and Escherichia coli: mechanism of production of extracellular superoxide radical. J. Biol. Chem. 254:10846-10852. [PubMed] [Google Scholar]
- 11.Hatchikian, E. C., and Y. A. Henry. 1977. An iron-containing superoxide dismutase from the strict anaerobe Desulfovibrio desulfuricans (Norway 4). Biochimie 59:153-161. [DOI] [PubMed] [Google Scholar]
- 12.Imlay, K. R. C., and J. A. Imlay. 1996. Cloning and analysis of sodC, encoding copper-zinc superoxide dismutase of Escherichia coli. J. Bacteriol. 178:2564-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenney, F. E., M. F. J. M. Verhagen, X. Cui, and M. W. W. Adams. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306-309. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura, M., K. Mizugai, M. Taniguchi, H. Akutsu, I. Kumagai, and T. Nakaya. 1995. A gene encoding a cytochrome c oxidase-like protein is located closely to the cytochrome c-553 gene in the anaerobic bacterium, Desulfovibrio vulgaris (Miyazaki F). Microbiol. Immunol. 39:75-80. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Leciere, V., A. Chotteau-Lelievre, F. Gancel, M. Imbert, and R. Blondeau. 2001. Occurrence of two superoxide dismutases in Aeromonas hydrophila: molecular cloning and differential expression of sodA and sodB genes. Microbiology 147:3105-3111. [DOI] [PubMed] [Google Scholar]
- 17.Lehman, Y., L. Meile, and M. Teuber. 1996. Rubrerythrin from Clostridium perfringens: cloning of the gene, purification of the protein, and characterization of its superoxide dismutase function. J. Bacteriol. 178:7152-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemos, R. S., C. M. Gomes, M. Santana, J. LeGall, A. V. Xavier, and M. Texeira. 2001. The “strict” anaerobe Desulfovibrio gigas contains a membrane-bound oxygen-reducing respiratory chain. FEBS Lett. 496:40-43. [DOI] [PubMed] [Google Scholar]
- 19.Li, T., X. Huang, R. Zhou, Y. Liu, B. Li, C. Nomura, and J. Zhao. 2002. Differential expression and localization of Mn and Fe superoxide dismutases in the heterocystous cyanobacterium Anabaena sp. J. Bacteriol. 184:5096-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lumppio, H. L., N. V. Shevni, R. P. Garg, A. O. Summers, G. Voordouw, and D. M. Kurtz, Jr. 2001. Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 183:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lumppio, H. L. 2000. Characterization of oxidative stress proteins from the anaerobe Desulfovibrio vulgaris. Ph.D. thesis. University of Georgia, Athens.
- 22.Lumppio, H. L., N. V. Shenvi, R. P. Garg, A. O. Summers, and D. M. Kurtz, Jr. 1997. A rubrerythrin operon and nigerythrin gene in Desulfovibrio vulgaris (Hildenborough). J. Bacteriol. 179:4607-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCord, J. M., and I. Fridovich. 1969. Superoxide dismutase: an enzymatic function for erythrocuprein (hemocuprein). J. Biol. Chem. 244:6049-6055. [PubMed] [Google Scholar]
- 24.Moura, I., P. Tavares, J. J. G. Moura, N. Ravi, B. H. Huynh, M.-Y. Liu, and J. LeGall. 1990. Purification and characterization of desulfoferrodoxin: a novel protein from Desulfovibrio desulfuricans (ATCC 27774) and from Desulfovibrio vulgaris (strain Hildenborough) that contains a distorted rubredoxin center and a mononuclear ferrous center. J. Biol. Chem. 265:21596-21602. [PubMed] [Google Scholar]
- 25.Pierik, A. J., R. B. G. Wolbert, G. L. Portier, M. F. J. M. Verhagen, and W. R. Hagen. 1993. Nigerythrin and rubrerythrin from Desulfovibrio vulgaris each contain two mononuclear iron centers and two dinuclear iron clusters. Eur. J. Biochem. 212:237-245. [DOI] [PubMed] [Google Scholar]
- 26.Pohorelic, B. K. J., J. K. Voordouw, E. Lojou, A. Dolla, J. Harder, and G. Voordouw. 2002. Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism. J. Bacteriol. 184:679-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Postgate, J. R. 1984. The sulfate-reducing bacteria, 2nd ed. Cambridge University Press, Cambridge, England.
- 28.Prickrill, B. C., D. M. Kurtz, Jr., J. LeGall, and G. Voordouw. 1991. Cloning and sequencing of the gene for rubrerythrin from Desulfovibrio vulgaris (Hildenborough). Biochemistry 30:11118-11123. [DOI] [PubMed] [Google Scholar]
- 29.Rabus, R., D. Gade, R. Helbig, M. Bauer, F. O. Glöckner, M. Kube, H. Schlesner, R. Reinhardt, and R. Amann. 2002. Analysis of N-acetylglucosamine metabolism in the marine bacterium Pirellula sp. strain 1 by a proteomic approach. Proteomics 2:649-655. [DOI] [PubMed] [Google Scholar]
- 30.Santos, H., P. Fareleira, A. V. Xavier, L. Chen, M.-Y. Liu, and J. LeGall. 1993. Aerobic metabolism of carbon reserves by the “obligate anaerobe” Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 195:551-557. [DOI] [PubMed] [Google Scholar]
- 31.Schevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 32.Schnell, S., and H. M. Steinman. 1995. Function and stationary-phase induction of periplasmic copper-zinc superoxide-dismutase and catalase/peroxidase in Caulobacter crescentus. J. Bacteriol. 177:5924-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 34.Silva, G., J. LeGall, A. V. Xavier, M. Texeira, and C. Rodrigues-Pousada. 2001. Molecular characterization of Desulfovibrio gigas neelaredoxin, a protein involved in oxygen detoxification in anaerobes. J. Bacteriol. 183:4413-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon, R., U. Priefer, and A. Pühler. 1983. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 36.Smith, D. R., L. A. Doucette-Stamm, C. Deloughery, H. Lee, J. Dubois, T. Aldredge, R. Bashirzadeh, D. Blakely, R. Cook, K. Gilbert, D. Harrison, L. Hoang, P. Keagle, W. Lumm, B. Pothier, D. Qiu, R. Spadafora, R. Vicare, Y. Wang, J. Wierzbowski, R. Gibson, N. Jiwani, A. Caruso, D. Bush, H. Safer, D. Patwell, S. Prabhakar, S. McDougall, G. Shimer, A. Goyal, S. Pietrovski, G. M. Church, C. J. Daniels, J.-L. Mao, P. Rice, J. Nölling, and J. N. Reeve. 1997. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J. Bacteriol. 179:7135-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.St. John, G., and H. M. Steinman. 1996. Periplasmic copper-zinc superoxide dismutase of Legionella pneumophila: role in stationary-phase survival. J. Bacteriol. 178:1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van den Westen, H., S. G. Mayhew, and C. Veeger. 1978. Separation of hydrogenase from intact cells of Desulfovibrio vulgaris. FEBS Lett. 86:122-126. [DOI] [PubMed] [Google Scholar]
- 40.Voordouw, G. 2000. A universal system for the transport of redox proteins: early roots and latest developments. Biophys. Chem. 86:131-140. [DOI] [PubMed] [Google Scholar]
- 41.Voordouw, G. 2002. Carbon monoxide cycling by Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 184:5903-5911. [DOI] [PMC free article] [PubMed]
- 42.Voordouw, J. K., and G. Voordouw. 1998. Deletion of the rbo gene increases the oxygen sensitivity of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 64:2882-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voordouw, G., J. D. Strang, and F. R. Wilson. 1989. Organization of the genes encoding [Fe] hydrogenase in Desulfovibrio vulgaris subsp. oxamicus Monticello. J. Bacteriol. 171:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 4. Springer-Verlag, New York, N.Y.
- 45.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]