Abstract
Bacillus subtilis PhoP is a member of the OmpR/PhoB family of response regulators that is directly required for transcriptional activation or repression of Pho regulon genes in conditions under which Pi is growth limiting. Characterization of the PhoP protein has established that phosphorylation of the protein is not essential for PhoP dimerization or DNA binding but is essential for transcriptional regulation of Pho regulon genes. DNA footprinting studies of PhoP-regulated promoters showed that there was cooperative binding between PhoP dimers at PhoP-activated promoters and/or extensive PhoP oligomerization 3′ of PhoP-binding consensus repeats in PhoP-repressed promoters. The crystal structure of PhoPN described in the accompanying paper revealed that the dimer interface between two PhoP monomers involves nonidentical surfaces such that each monomer in a dimer retains a second surface that is available for further oligomerization. A salt bridge between R113 on one monomer and D60 on another monomer was judged to be of major importance in the protein-protein interaction. We describe the consequences of mutation of the PhoP R113 codon to a glutamate or alanine codon and mutation of the PhoP D60 codon to a lysine codon. In vivo expression of either PhoPR113E, PhoPR113A, or PhoPD60K resulted in a Pho-negative phenotype. In vitro analysis showed that PhoPR113E was phosphorylated by PhoR (the cognate histidine kinase) but was unable to dimerize. Monomeric PhoPR113E∼P was deficient in DNA binding, contributing to the PhoPR113E in vivo Pho-negative phenotype. While previous studies emphasized that phosphorylation was essential for PhoP function, data reported here indicate that phosphorylation is not sufficient as PhoP dimerization or oligomerization is also essential. Our data support the physiological relevance of the residues of the asymmetric dimer interface in PhoP dimerization and function.
The natural environment of Bacillus subtilis is the soil, an environment in which inorganic phosphate is the critical limiting nutrient and the levels of inorganic phosphate are often 2 to 3 orders of magnitude lower than the levels of other required ions (38). The B. subtilis inorganic phosphate deficiency response is controlled by at least two global regulatory systems, the alternative transcription factor which is activated by environmental or energy stress, sigma B (1), and the two-component signal transduction regulator, PhoP-PhoR (43, 44). The PhoP and PhoR proteins, whose primary role is a role in the phosphate deficiency response, are part of a signal transduction network that also includes ResD-ResE, which also has a role in respiratory regulation, and the Spo0A phosphorelay required for initiation of the stationary phase and sporulation (5, 17, 19, 47).
PhoP is a member of the winged helix-turn-helix family of response regulators based on similarity to the output domain of OmpR (35). This is the largest subfamily of response regulators, with 14 paralogues in B. subtilis alone. The 240-amino-acid PhoP protein is composed of a regulatory domain (119 amino acids), a long linker region connecting the regulatory domain to the output domain (19 amino acids), and a 102-amino-acid OmpR conserved DNA binding domain. PhoP is the proposed B. subtilis orthologue of PhoB of Escherichia coli as PhoP is activated by PhoR in response to phosphate starvation and activates a number of the same genes, such as an alkaline phosphatase(s) and a high-affinity Pi transporter. Among the differences between the two systems recently reviewed (17), PhoP is a transcription factor capable of activation or repression depending on the gene of interest, while PhoB is only known to activate genes of the E. coli Pho regulon.
Certain B. subtilis genes that are induced upon phosphate limitation (∼0.1 mM Pi) were shown to directly require the PhoP-PhoR two-component signal transduction regulator for this induction (11, 29-31). At the same level of Pi a second set of genes is repressed in a PhoP-PhoR-dependent manner (5, 28). In vitro transcription studies showed that the phosphorylated response regulator, PhoP∼P, was essential for the induction or repression of all Pho regulon genes tested (39, 40). Additional genes whose expression was dependent on phoPR in vivo may be directly regulated by PhoP-PhoR (1, 24, 42)
DNA binding studies indicated that either PhoP or PhoP∼P could bind PhoP-activated promoters, although PhoP∼P usually required lower protein concentrations for binding. Analysis of PhoP DNase I protection studies at activated promoters (10, 29-31) revealed traits common to all of the promoters. PhoP or PhoP∼P protected all promoters between approximately −20 and −60 bp upstream of a sigma A transcriptional start site. Direct repeats of a 6-bp consensus sequence (10), TT(A/C/T)A(T/C)A, separated by four to six nonconserved base pairs were located at approximately positions −25, −35, −45, and −55 in each promoter, a region referred to as the PhoP core binding region. Mutagenesis of the PhoD promoter showed that the specific bases of the consensus repeat are essential for promoter activation as well as PhoP binding and that binding of PhoP dimers at the core binding region is highly cooperative (10). Differences in PhoP-activated promoters included additional pairs of consensus repeats found directly 5′ (30) or more than 100 bp 5′ (10) of the core binding region or in two promoters (31) located in the coding region >75 bp downstream of the +1 transcriptional site. The secondary binding sites were necessary for maximal promoter activity; they were required for 45 to 95% of the full promoter activity depending on the promoter (10, 31).
Other OmpR family response regulators have been reported to bind to tandemly repeated consensus sequences. Nuclear magnetic resonance studies (37) and the crystal structure of the PhoB DNA binding domain-phoA promoter complex (6) have shown that PhoB (E. coli) binds to TG of the TGTCA conserved sequence and makes direct contact within the last four bases of an 11-bp sequence repeated in each Pho box. Two PhoB molecules bind in a tandem array on the Pho box. While the consensus sequences for OmpR binding exhibit less similarity, tandem repeats of 10-bp half-sites were proposed, suggesting that there is a tandem arrangement of OmpR monomers (12, 13, 16, 33), which was confirmed by a DNA affinity cleavage analysis that also established the head-to-tail orientation of the OmpR monomers on the DNA (14). The positions of the PhoB and PhoP consensus repeats are similar (starting at about position −20 relative to the transcriptional start site), while OmpR consensus repeats start at approximately position −40 (32-34).
PhoP-repressed promoters have PhoP binding patterns that are similar to each other (5, 28) and differ from binding patterns at PhoP-activated promoters. In each repressed promoter, PhoP binds to regions that overlap the transcription initiation site, and protection extends into the coding regions of the genes, as far as 168 bp (31). Repressed promoters which contain only two PhoP consensus repeats require phosphorylation of PhoP for binding, while PhoP or PhoP∼P binds at the repressed promoter with four consensus repeats. Phosphorylation is nevertheless required for apparent oligomerization of PhoP∼P, which results in protection of DNA far into the coding regions of repressed genes.
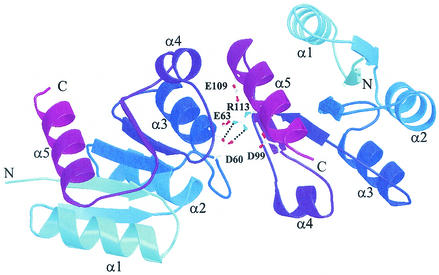
The cumulative data for characterization of the PhoP protein (29) and characterization of PhoP binding at both activated and repressed promoters indicated that PhoP phosphorylation was not essential for PhoP dimerization or DNA binding. The highly cooperative binding of PhoP or PhoP∼P to the repeated dimer binding sites in the core binding regions of Pho-activated promoters (10) and the extensive PhoP∼P protection downstream of a single or repeated dimer binding site(s) in the PhoP-repressed promoters suggested that there is protein-protein interaction between PhoP dimers and/or PhoP oligomerization along the DNA. The crystal structure of the PhoP receiver domain indeed showed that the interface between two PhoP monomers involves nonidentical surfaces of each molecule (Fig. 1) that make oligomerization of the protein possible.
FIG. 1.
Salt bridge interaction (dotted lines) between D60 and R113 in the PhoPN dimer interface. Each protomer is represented by ribbons, and the color varies from light blue (N terminus) to purple (C terminus). α helixes are labeled. Red atoms, oxygen; blue atoms, nitrogen.
The studies described here were designed to disrupt by mutagenesis the PhoP dimerization-oligomerization interface observed in the PhoPN structure (3) and to ask how the changes affect PhoP function in vivo and in vitro. For these studies, mutations at R113 or D60 of PhoP were investigated as they were judged to be of major importance in PhoP-PhoP associations due to formation of a salt bridge between two PhoP monomers (Fig. 1). The in vivo phenotype of strains with a mutation in the R113 codon or D60 was Pho negative. In vitro data presented here provide evidence for an essential role for R113 in Pho dimerization but not for association with the cognate kinase or for phosphorylation. PhoPR113E∼P was DNA binding deficient.
MATERIALS AND METHODS
Strains and plasmids.
Table 1 shows the strains and plasmids used in this study. E. coli DH5α was used as the host for plasmid construction. E. coli BL21(DE3)/pLysS (Novagen) served as the host for overexpressing the PhoP proteins. B. subtilis JH642 and MH5913 (a phoPR deletion derivative of JH642) were used for in vivo Pho induction experiments.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | Lab stock | |
| BL21(DE3)/pLysS | Novagen | |
| B. subtilis strains | ||
| JH642 | pheA1 trpC2 | J. A. Hoch |
| MH5913 | pheA1 trpC2 ΔphoPR::Tetr | This study |
| MH6101 | MH5913 ΩpCH24 | This study |
| MH6102 | MH5913 ΩpCH25 | This study |
| MH6103 | MH5913 ΩpCH26 | This study |
| MH6104 | MH5913 ΩpCH28 | This study |
| Plasmids | ||
| pHT4phoPR | pHT315::Sau3AI fragment of B. subtilis chromosome containing phoPR operona | T. Masdek |
| pHT315 | oriEc ori1030 Ampr Ermr | 2 |
| pJM103 | oriEc Ampr | J. A. Hoch |
| pES35 | pJM103 disrupted EcoRI | This study |
| pES135 | pES35::3′-mdh (259 bp) phoPR 5′-polA (750 bp) | This study |
| pES136 | pES135 phoPRΔEcoRI | This study |
| pXH35 | pES136 phoPRΔEcoRI::Tetr | This study |
| pKL1036 | pJM103::phoP | This study |
| pBluescript II KS(+) | oriEc lac Ampr | Stratagene |
| pKL20 | pBluescript II KS(+)::phoP | This study |
| pDH88 | oriEc Pspac Cmr | 15 |
| pWL29 | pDH88::phoPWT | This study |
| pCH22 | pDH88::phoPR113E | This study |
| pCH23 | pDH88::phoPR113A | This study |
| pCH24 | pDH88::phoPWTR 5′-polA (750 bp) | This study |
| pCH25 | pDH88::phoPR113ER 5′-polA (750 bp) | This study |
| pCH26 | pDH88::phoPR113AR 5′-polA (750 bp) | This study |
| pCH27 | pDH88::phoPD60K | This study |
| pCH28 | pDH88::phoPD60KR 5′-polA (750 bp) | This study |
| pCR2.1 | oriEc lac Ampr | Invitrogen |
| pWL31 | pCR2.1::phoP | 29 |
| pTZ18U | oriEc lac Ampr | Bio-Rad |
| pET16b | oriEc T7 lac Ampr | Novagen |
| pCH01 | pTZ18U::phoP | This study |
| pCH05 | pTZ18U::phoPR113E | This study |
| pCH13 | pET16b::phoPR113E | This study |
| pYQ22 | pCR2.1::pstS | 41 |
See text.
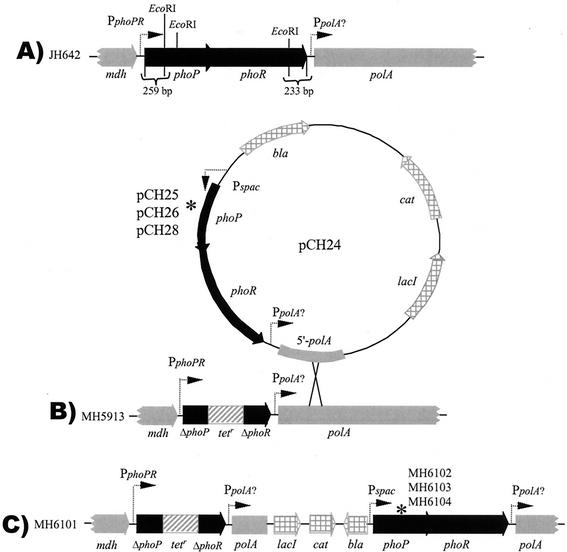
Several plasmids were used to construct MH5913. Plasmid pHT4phoPR was a kind gift from Tarek Masdek (Department des Biotechnologies, Unite de Biochimie Microbienne, Institut Pasteur, Paris, France). A 3,828-bp fragment from a partial Sau3AI digest of B. subtilis 168 chromosomal DNA was cloned into the BamHI site of pHT315 (2), yielding pHT4phoPR. The insert in pHT4phoPR contains (5′ to 3′) 259 bp of the 3′ coding region of the mdh gene, the phoPR operon, and the first 750 bp of the polA gene coding region. The genomic insert in pHT4phoPR was released by digestion with KpnI and PstI and cloned into the same sites of pES35 to construct pES135. (pES35 is a derivative of pJM103 in which the single EcoRI site was destroyed by digestion with EcoRI, followed by treatment with Klenow DNA polymerase and religation.) The two EcoRI fragments within the phoPR operon (Fig. 2A) in plasmid pES135 were deleted by EcoRI digestion and religation of pES135 to form pES136. A tetracycline resistance cassette (Tetr) released from pUC19 by EcoRI digestion (21) was cloned into the EcoRI site in pES136, yielding pXH35. pXH35 linearized by digestion with PstI was used to transform to JH642 to generate PhoPR deletion strain MH5913. Transformants were selected by Tetr and screened for Cms. The chromosomal DNA of MH5913 has a Tetr cassette inserted between bp 259 in the phoP open reading frame and bp 1504 in the phoR open reading frame.
FIG. 2.
Chromosomal structure of the phoPR locus in various B. subtilis strains: phoPR loci in wild-type strain JH642 (A), phoPR-negative strain MH5913 (B), and IPTG-inducible phoPR wild-type and phoPR113 mutant strains (C). phoPR or portions of phoPR genes are indicated by thick solid arrows. Other chromosomal genes (mdh and polA) are indicated by thick gray arrows. pDH88 vector genes are indicated by thick gray arrows with windowpanes. The Tetr insertion into the phoPR EcoRI deletion is indicated by a cross-hatched box. pCH24 used for Campbell insertion-duplication in MH5913 is also shown. The dotted arrows indicate promoters. An asterisk indicates the site of amino acid codon replacement in PhoP (R113E, R113A, and D60K in MH6102, MH6103, and MH6104, respectively).
For site-directed mutagenesis of phoP, the phoP gene was amplified by PCR by using primers FMH102 (5′-CTC CTA GGC ACG GTA TTT ATT-3′) and FMH103 (5′-ACG TCG ACA ATA CTG GAG GCA CAG-3′) with B. subtilis JH642 chromosomal DNA as the template. The PCR product was cloned into the SmaI site of pJM103 to make pKL1036. The orientation of the phoP gene was in the direction from HindIII to EcoRI. The HincII-KpnI fragment of pJM1036 containing the phoP gene was subcloned into the HincII-KpnI site of pBluescript II KS(+) (Stratagene) to construct pKL20. pKL20 was digested with KpnI, the cohesive ends were made blunt with the Klenow fragment, and the phoP gene was released by digestion with XbaI. The phoP gene was cloned into the blunted ClaI site and the XbaI site of pDH88 to make pWL29. The R113 or D60 amino acid codon of phoP in pWL29 was mutated by using a QuickChange site-directed mutagenesis kit (Stratagene) with primers FMH451 (5′-GGA AGT AAA TGC GGA AGT CAA AGC G-3′) and FMH462 (5′-CGC TTT GAC TTCCGC ATT TAC TTC C-3′), primers FMH450 (5′-GGA AGT AAA TGC GGC AGT CAA AGC G-3′) and FMH461 (5′-CGC TTT GAC TGC CGC ATT TAC TTC C-3′), and primers FMH669 (5′-GTG ATG CTT CCA AAA TTG AAA GGA ATC GAA GTA TGC AAG C-3′) and FMH 670 (5′-GCT TGC ATA CTT CGA TTC CTT TCA ATT TTG GAA GCA TCA C-3′) according to the instruction manual to obtain pCH22, pCH23, and pCH27, respectively (the substituted codons are underlined). The site-directed mutations were confirmed by DNA sequencing. To construct a complete phoPR operon on each plasmid, pCH22, pCH23, and pCH27 were digested with BpuI102I and SphI and were ligated with the 2.9-kbp (2,875-bp) BpuI102I/SphI fragment obtained from pHT4phoPR, yielding pCH25, pCH26, and pCH28, respectively. pCH25, pCH26, and pCH28 contained the Pspac promoter, a ribosome binding site, a mutated phoPR operon (PR113ER, PR113AR, and PD60K, respectively), and a partial 5′ polA gene. pCH24 was constructed as described above without mutagenesis. Plasmids pCH24, pCH25, pCH26, and pCH28 were transformed into MH5913, and representative transformants were designated MH6101, MH6102, MH6103, and MH6104, respectively. Construction of the mutant strains was confirmed by PCR and DNA sequencing.
To construct a plasmid for overexpressing mutant PhoP protein, the phoP gene was released from pWL31 (29) by digestion with HindIII and XbaI and cloned into pTZ18U (Bio-Rad) at the same sites, and the resulting plasmid was designated pCH01. pCH05 was obtained from pCH01, which was mutated by using primers FMH451 and FMH462 and the method described above. The mutated phoP gene was then released from pCH05 by NdeI and BamHI digestion and cloned into the NdeI and BamHI sites of pET16b (Novagen), yielding pCH13. The mutation was confirmed by DNA sequencing. pCH13 contains a T7 lac promoter, the codons for 10 histidine residues, and an engineered Xa factor cleavable site upstream of the phoP gene; the mutant PhoP produced by this plasmid overexpressed in E. coli contained a 10-His tag and another 11 amino acids at the N terminus (His10-PhoPR113E).
Growth conditions and APase activity assay.
Alkaline phosphatase (APase) activity was measured in cells that had been grown in low-phosphate defined medium (LPDM) as described previously (20) either with isopropyl-β-d-thiogalactoside (IPTG) present at a final concentration of 1 mM throughout growth or without IPTG. Culture density and APase activity were determined every hour by using cells grown under culture conditions described previously (9).
Western immunoblotting.
B. subtilis cells were grown in LPDM as described above. The cells were collected by centrifugation at 5,000 × g for 10 min at 4°C. The cells were washed with sonication buffer (0.3 M NaCl, 5 mM MgCl2, 10 mM dithiothreitol [DTT], 50 mM Tris-HCl [pH 7.8]). The cells were suspended in sonication buffer containing lysozyme (1.6 mg/ml) and incubated at 32°C for 10 min. After 1 mM phenylmethylsulfonyl fluoride was added, the cells were immediately disrupted by sonication and centrifuged at 80,000 × g for 1 h at 4°C. Each supernatant fraction was mixed with 0.25 volume of 6× sodium dodecyl sulfate (SDS) loading buffer. The proteins were separated on an SDS-12.5% polyacrylamide gel electrophoresis (PAGE) gel and transferred to a polyvinylidene fluoride membrane (Millipore) by using a Mini Trans-Blot transfer cell (Bio-Rad) according to the manufacturer's instructions. Rabbit anti-PhoPC polyclonal antiserum (Biologic Resources Laboratory, University of Illinois at Chicago, Chicago, Ill.) was used as the primary antibody, and goat anti-rabbit immunoglobulin G conjugated with APase (Bio-Rad) was used as the secondary antibody. Immunodetection was performed by the method described previously (45).
Overexpression and purification of His10-PhoPR113E.
E. coli BL21(DE3)/pLysS harboring pCH13 was incubated overnight at 37°C in Luria-Bertani medium containing ampicillin (100 μg/ml) and was then inoculated into 6 liters of the same medium at a ratio of 1:100. The cells were grown at 20°C until the optical density at 600 nm of the culture reached about 0.4. Then 1 mM IPTG was added to the culture, and the preparation was incubated for another 12 h. The cells were harvested by centrifugation at 4°C and washed with buffer A (1 M NaCl, 5 mM MgCl2, 10 mM DTT, 50 mM Tris-HCl [pH 7.8]). Cell pellets (wet weight, 20 g) were then suspended on ice in 250 ml of buffer A containing 1 mM phenylmethylsulfonyl fluoride and were immediately subjected to sonication. Lysis of the cells was confirmed by microscopy. After centrifugation at 40,000 × g for 1 h at 4°C, the supernatant fraction was filtered through a 0.45-μm-pore-size membrane and applied to a 2-ml Ni-nitrilotriacetic acid (NTA) (Qiagen) affinity column, which was attached to a Waters 650E fast protein liquid chromatography system. The column was washed with buffer A until the optical density at 280 nm of the elute was less than 0.08. The column was further washed with 30 ml of buffer A containing 20 mM imidazole. The protein bound to the column was eluted by using a linear gradient of 20 to 300 mM imidazole in buffer A. The protein peak fractions containing His10-PhoPR113E were pooled (5 ml) and dialyzed stepwise at 4°C against buffer A with decreasing concentrations of NaCl (1, 0.8, 0.6, 0.4, 0.2, and 0.1 M). The protein was concentrated to a volume of 1 ml with an Amicon concentrator by low-speed centrifugation and loaded onto an HR10/30 Superdex 200 column (Pharmacia) that was preequilibrated with buffer B (0.1 M KCl, 5 mM MgCl2, 10 mM DTT, 50 mM Tris-HCl [pH 7.5]). The protein was eluted at a flow rate of 0.5 ml/min and collected in 0.5-ml fractions. His10-PhoPR113E purified by gel filtration was used throughout the experiments described below.
Phosphorylation of PhoP by PhoR.
Glutathione S-transferase (GST)-*PhoR (45), prepared in phosphorylation buffer (50 mM KCl, 5 mM MgCl2, 50 mM HEPES [pH 8.0]), was used to phosphorylate His10-PhoPR113E. Boiled glutathione beads (400 mg) were washed with phosphorylation buffer and incubated with 780 μg of GST-*PhoR on a rocker at room temperature for 10 min. The unbound component was washed off the beads with 20 volumes of phosphorylation buffer, and the extra buffer was removed by microcentrifugation for 10 s. Then 20 μl of [γ-32P]ATP (10 mCi/ml) was added to the beads, and autophosphorylation of GST-*PhoR was performed at room temperature for 20 min. The beads were thoroughly washed with phosphorylation buffer until the flowthrough was free of ATP. His10-PhoPR113E (800 μg) containing 10 mM DTT and 50 mM KCI was added to the beads. After incubation at room temperature for 20 min, the His10-PhoPR113E was collected by centrifugation and passed through a syringe filter (Osmonics Inc.) to remove any remaining beads before it was used in size exclusion chromatography experiments.
For experiments in which phosphotransfer to PhoPWT or PhoPR113E was analyzed, PhoP proteins were phosphorylated with *PhoR∼P as described previously (45).
Determination of protein concentration.
Protein concentration was determined by the Bradford method (8) by using a Bio-Rad protein assay kit as instructed by the manufacturer.
SDS-PAGE and native PAGE.
SDS-PAGE was performed as described by Laemmli (25). A 12.5% polyacrylamide separating gel was used for detection of His10-PhoPR113E. An 8% native PAGE gel was prepared as an SDS-PAGE separating gel without any SDS, and the percentage of cross-linker in the gel was set at 2.6% (acrylamide/bisacrylamide ratio, 37.5:1). After the gel was electrophoresed at 100 V for 1 h at 4°C, the samples were loaded and the gel was electrophoresed for another 4 h.
Quantitation of radioactivity.
The radioactivity after gel filtration was measured with a Beckman LS 6500 scintillation system (Beckman Coulter, Inc., Fullerton, Calif.). The radioactivity of proteins on SDS-PAGE and native PAGE gels was detected either with Fuji medical X-ray film [Fuji Photo Film (Europe) GmbH, Dusseldorf, Germany] or with a PhosphorImager (Molecular Dynamics, Inc., Sunnyvale, Calif.) and was quantified by using the ImageQuant software (version 1.0).
Determination of solution status of His10-PhoPR113E and His10-PhoPR113E ∼P by gel filtration.
His10-PhoPR113E (0.4 ml of a 2-mg/ml solution) or freshly prepared His10-PhoPR113E∼P (see above) was loaded onto an HR10/30 Superdex 200 column (Pharmacia) that had been preequilibrated with buffer B, and then the protein was eluted at a flow rate of 0.5 ml/min and collected in 0.5-ml fractions. Protein molecular weight standards (MW-GF-200 kit; Sigma) were applied to the column under the same conditions. The elution volume and molecular weight of each standard were used to generate a standard curve to determine the molecular weight of the PhoP protein.
Molecular mass determination by light scattering.
High-performance liquid size exclusion chromatography was performed with a TSK G3000SW column by using a solution containing 100 mM KCl, 5 mM MgCl2, 200 mM imidazole, and 50 mM Tris-HCl (pH 7.5). The elution profiles were monitored by multiangle static light scattering at 690 nm and differential refractometry (DAWN-EOS and Optilab instruments, respectively; Wyatt Technology Corp., Santa Barbara, Calif.). The molecular mass was determined by using a specific refractive index increment of 0.182 and the Debye plotting formalism of the Astra software supplied by Wyatt Technology Corp.
Gel shift assays.
Gel shift assays were performed as described previously (29). The probe used was a 364-bp fragment containing the pstS promoter released from pYQ22 (41) by digestion with XbaI and SpeI and end labeled with the Klenow enzyme in the presence of 4 μl of [α-32P]dATP (10 mCi/ml). The His tag was released from His10-PhoP or His10-PhoPR113E by treatment with factor Xa proteinase to generate PhoP and PhoPR113E. The amounts of the PhoP, PhoPR113E, and GST-*PhoR proteins used for gel shift assays are indicated below. For PhoP and PhoPR113E phosphorylation, ATP was included in the reaction mixtures at a final concentration of 4 mM.
RESULTS
Construction of B. subtilis strains that contain a single copy of the wild-type phoPR operon or an operon containing a mutation affecting R113 or D60 of PhoP under control of an IPTG-inducible promoter.
According to the crystal structure of PhoPN (3), residues D60 and R113 are located central to the asymmetric dimer interface and were judged to be critical for electrostatic balance (Fig. 1). To determine the importance of the interaction in vivo, an inducible PhoPR production system was constructed in a phoPR deletion strain. The wild-type phoPR operon and phoPR operons containing site-mutated phoPR113E, phoPR113A, and phoPD60K were cloned into plasmid pDH88 downstream of the IPTG-inducible Pspac promoter to construct pCH24 (Pspac phoPR), pCH25 (Pspac phoPR113ER), pCH26 (Pspac phoPR113AR), and pCH28 (Pspac phoPD60KR), respectively. Plasmids containing the required phoPR operon and 750 bp of the downstream polA gene were each transformed into B. subtilis strain MH5913 with selection for Cmr transformants. The parent strain, MH5913 (ΔphoPR::Tetr), has a deletion-insertion (Tetr) at the phoPR locus (Fig. 2B). Analysis of the transformants showed that each plasmid was integrated downstream of the Tetr gene in MH5913 (data not shown) via a Campbell-like insertion duplication (Fig. 2B) to produce B. subtilis MH6101 (Pspac phoPR), MH6102 (Pspac phoPR113ER), MH6103 (Pspac phoPR113AR), and MH6104 (Pspac phoPD60KR). The chromosomal structures of these strains are shown in Fig. 2C.
Strains with mutations producing amino acid substitutions at arginine 113 or aspartic acid 60 of PhoP have a Pho-negative phenotype.
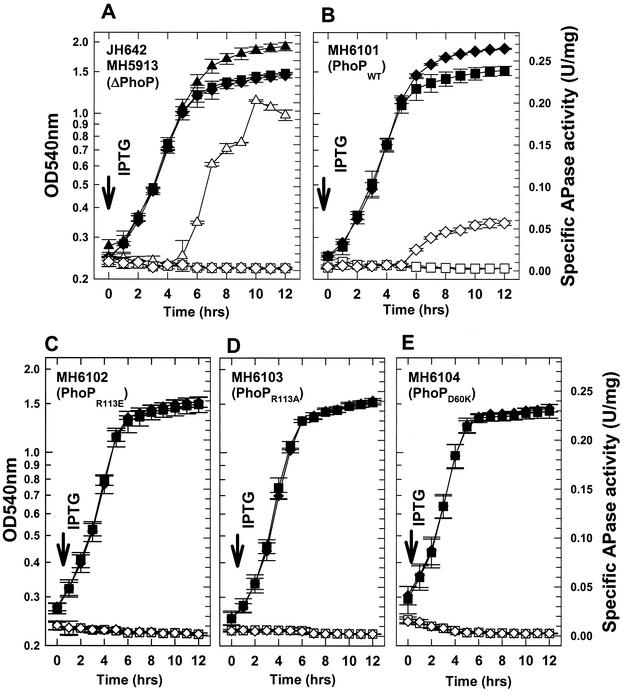
Total APase specific activity was measured as a reporter of Pho induction. PhoA, PhoB, and PhoD contribute approximately 65% (22), 30% (7), and <5% (18) of the total APase specific activity, respectively. Strains were grown for 12 h in LPDM, in which parental strain B. subtilis JH642 induced APase production at approximately h 5 (Fig. 3A), while MH5913 (phoPR::Tetr) failed to induce APase production with or without IPTG (Fig. 3A). The phoPR deletion strain showed a decreased growth phenotype (h 5 to 12) compared to parental strain JH642, which is typical of other Pho− strains upon phosphate limitation (45). The MH5913 derivative that contains the wild-type phoPR operon controlled by the Pspac promoter, MH6101 (phoPWT) (Fig. 3B), showed the same growth and APase phenotype as MH5913 when it was grown without IPTG, but when IPTG (1 mM) was added to the culture at time zero, it exhibited a growth phenotype similar to that of JH642 and induced APase at h 5, albeit with fourfold-reduced specific activity compared to the specific activity of JH642 (Fig. 3A). No strain containing a mutated phoPR operon under Pspac control, including MH6102 (PhoPR113E) (Fig. 3C), MH6103 (PhoPR113A) (Fig. 3D), and MH6104 (PhoPD60K) (Fig. 3E), showed any change in growth or APase production in the presence of IPTG compared to the growth and APase production without IPTG; all strains maintained the growth and APase phenotypes of ΔphoPR::Tetr parental strain MH5913 (Fig. 3A). phoA-lacZ promoter fusion expression was assayed in the PhoPR113E and PhoPR113A strains with and without IPTG. The results (data not shown) were consistent with the total APase expression in these strains (Fig. 3C and D).
FIG. 3.
Growth of and APase production by B. subtilis strains MH5913, MH6101, MH6102, MH6103 and MH6104 cultured for 12 h in LPDM. The strains used are indicated in the panels. Symbols: ▴, cell growth of JH642 (wild-type control); ▵, APase specific activity of JH642; ⧫, cell growth with 1 mM IPTG; ▪, cell growth without IPTG; ◊, APase production with 1 mM IPTG; □, APase production without IPTG. The error bars indicate standard deviations for duplicates in at least three independent growth or APase experiments. OD540nm, optical density at 540 nm.
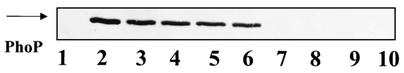
Strains with the phoPR operon under control of the Pspac promoter contained similar concentrations of PhoP that were less than the PhoP concentration in the wild-type strain.
It seemed possible that different levels of PhoP protein in the various strains might contribute to the different Pho phenotypes shown in Fig. 3. The relative strengths of the wild-type phoPR promoter and the Pspac-induced promoter were unknown, and the in vivo turnover rates for wild-type and mutated proteins might differ. To investigate the role of PhoP protein concentration in these strains, Western immunoblotting was performed by using polyclonal PhoPC antibody (Fig. 4). The wild-type (JH642), phoPR deletion (MH5913), and PhoP mutant strains were grown in LPDM with and without 1 mM IPTG until the postexponential stage, when the cells were collected for soluble protein sample preparation. Under IPTG induction conditions, the PhoP proteins of strains MH6101, MH6102, MH6103, and MH6104 were expressed (Fig. 4, lanes 3 to 6, respectively), while no PhoP proteins were detected in these strains cultured without IPTG (Fig. 4, lanes 7 to 10, respectively) or in the phoPR deletion strain (Fig. 4, lane 1). The levels of PhoP protein were similar in strains with the phoPR operon under control of the Pspac IPTG-induced promoter (Fig. 4, lanes 3 to 6), but the levels were less than the PhoP protein level in the phoPR wild-type parent strain, JH642 (Fig. 4, lane 2). These data indicate that PhoP protein concentrations produced from the phoPR operon with its native promoter in JH642 were somewhat higher than the concentration produced from the phoPR operon under control of the Pspac promoter in MH6101 (Fig. 4, compare lanes 2 and 3; also data not shown). These protein levels are consistent with the decreased IPTG induction of total APase in MH6101 compared to the induction of APase in JH642 (Fig. 3A and B). The finding that the soluble PhoP protein (native PhoP or mutant PhoP) concentrations were similar in all four IPTG-induced Pspac phoPR strains (Fig. 4, lanes 3 to 6) indicates that the in vivo turn-over rate of PhoPR113E, PhoPR113A, or PhoPD60K was similar to that of native PhoP and that the PhoP protein concentration or solubility was not a factor in the Pho-negative phenotype of strains producing PhoPR113E, PhoPR113A, or PhoPD60K protein (Fig. 3C, D, and E). Taken together, these results show that when the arginine 113 residue was changed to a negatively charged side chain amino acid, glutamic acid, or to a noncharged side chain amino acid, alanine, the mutant PhoP proteins lost in vivo response regulator function, as did the D60K mutant PhoP protein.
FIG. 4.
Western immunoblot detection of PhoP protein from B. subtilis strains. The cells were grown for 11 h and collected by centrifugation, and soluble proteins were extracted as described in Materials and Methods. The same amount (26 μg) of protein for each sample was separated by SDS-PAGE, transferred onto a polyvinylidene fluoride membrane, and immunodetected by using anti-PhoPC polyclonal sera. The migration position of purified PhoPWT (50 ng) used as a control is indicated by an arrow. Lane 1, MH5913; lane 2, JH642; lanes 3 and 7, MH6101; lanes 4 and 8, MH6102; lanes 5 and 9, MH6103; lanes 6 and 10, MH6104; lanes 3, 4, 5, and 6, with induction; lanes 7, 8, 9, and 10, without IPTG.
Overexpression and purification His10-PhoPR113E.
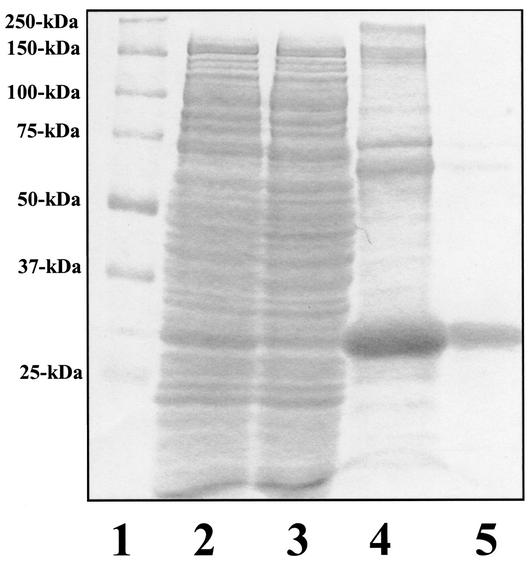
To better characterize the mutated PhoPR113E protein with respect to response regulator function, the mutated PhoP protein was overexpressed in E. coli and purified by multiple chromatography steps. The mutated protein, His10-PhoPR113E, was produced at the same level as His10-PhoPWT (29), but only 10% of the total His10-PhoPR113E protein was in the soluble fraction after centrifugation at 40,000 × g for 1 h, in contrast to 80% of the protein for His10-PhoPWT (29; data not shown). Decreasing the growth temperature to 20°C only slightly improved the solubility of His10-PhoPR113E, which was purified by Ni-NTA affinity chromatography (Fig. 5, lanes 2 to 4), followed by gel filtration (Fig. 5, lane 5). During size exclusion chromatography, approximately one-half of the Ni-NTA affinity-purified His10-PhoPR113E eluted immediately after the void volume (data not shown), corresponding to a highly aggregated form of the His10-PhoPR113E protein. This His10-PhoPR113E single peak was judged to be >95% pure by SDS-PAGE (Fig. 5, lane 5). A minor protein band migrated at ∼70 kDa, which is approximately the molecular mass of a His10-PhoPR113E dimer. Upon treatment with a higher concentration of reducing agent (0.5 M DTT), the dimeric form of His10-PhoPR113E migrated at 30,000 Da on SDS-PAGE gels (data not shown), suggesting that the ∼70-kDa species was a disulfide bond-mediated dimer. Such dimers were previously reported during purification of PhoPWT and could not be phosphorylated by PhoR∼P (27).
FIG. 5.
Purification of overexpressed His10-PhoPR113E from E. coli. Each purification step was analyzed by SDS-PAGE, and the gel was stained with Biosafe Coomassie blue (Bio-Rad). Lane 1, prestained protein standards; lane 2, supernatant fraction (20 μg); lane 3, nonbinding fractions from Ni-NTA chromatography; lane 4, eluate of His10-PhoPR113E from 30 to 300 mM imidazole gradient (10 μg); lane 5, gel filtration-purified His10-PhoPR113E (8 μg). The molecular masses of protein markers (lane 1) are indicated on the left.
Phosphotransfer rates from PhoR∼P to PhoPR113E and to PhoPWT were identical.
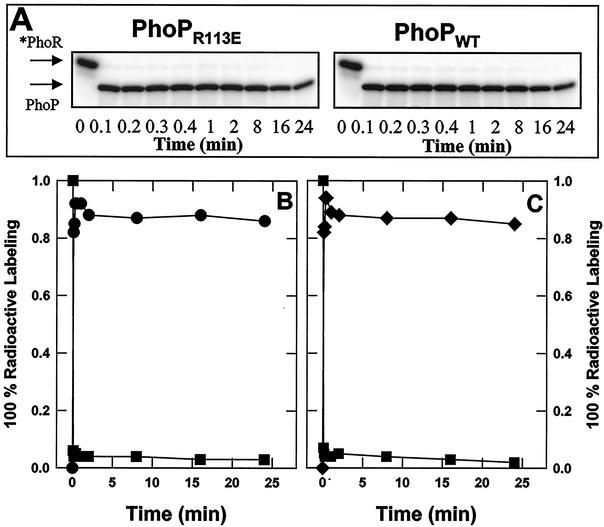
The native PhoP protein is capable of dimerization and DNA binding in the unphosphorylated state (29) but requires phosphorylation by PhoR to activate transcription of Pho promoters (29, 31, 39, 40). To explore whether the Pho induction-negative phenotypes of PhoPR113E mutants are due to defects in the phosphorylation ability of the protein, an in vitro phosphotransfer reaction from PhoR∼P to the response regulator was performed. Phosphorylated *PhoR was incubated with each PhoP protein (PhoPR113E or PhoPWT). The progress of the reaction over time was monitored and analyzed by SDS-PAGE, followed by autoradiography. When PhoPR113E or PhoPWT was added to autophosphorylated *PhoR, the phosphotransfer was nearly complete within 6 s (Fig. 6A). Quantitation of the phosphotransfer reaction showed that the phosphate transfer rates between phosphorylated PhoR and PhoPR113E and between phosphorylated PhoR and PhoPWT were similar, with the maximal level of PhoP∼P observed after less than 24 s (Fig. 6B and C). Both PhoPR113E∼P and PhoPWT∼P showed only a slight loss of the total radioactive label over the time monitored, 24 min (Fig. 6B and C). Thus, the efficiency of phosphotransfer from PhoR∼P to PhoPR113E was unchanged compared to the efficiency of phosphotransfer from PhoR∼P to PhoPWT. These results suggest that the Pho-negative phenotype of the strain producing the PhoPR113E protein (MH6102) is not the result of a phosphorylation-defective protein as PhoPR113E is efficiently phosphorylated in vitro.
FIG. 6.
Phosphotransfer rates from *PhoR∼P to PhoPR113E and PhoPWT are similar. GST-*PhoR was phosphorylated by incubating it with [γ-32P]ATP and was bound to glutathione-agarose. After excess [γ-32P]ATP was removed, 10 U of thrombin was added to the beads and mixed at room temperature for 20 min. The PhoR∼P was separated from the beads and mixed with an equimolar amount of PhoPR113E or PhoPWT. Samples having the same volume were removed from each reaction mixture at different times, as indicated, and the reaction was stopped with SDS loading buffer. Samples were then subjected to SDS-PAGE. The gels were dried, and the radioactivity was quantified with a PhosphorImager. Phosphorylated protein contents were expressed in arbitrary units. (A) Radioactivity in SDS-PAGE profile. The migration positions of *PhoR∼P and PhoP∼P are indicated by arrows. (B) *PhoR∼P incubated with PhoPR113E. (C) *PhoR∼P incubated with PhoPWT. Symbols: ▪, *PhoR∼P; •, PhoPR113E∼P; ⧫, PhoPWT∼P.
PhoPR113E is a monomeric species in its unphosphorylated and phosphorylated forms.
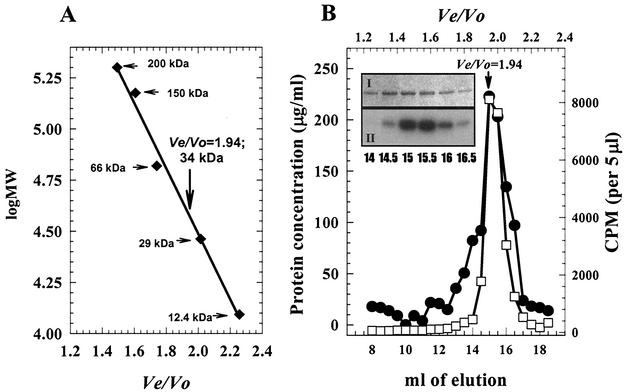
In addition to phosphorylation, dimerization of the PhoP protein likely plays a key role in controlling the activity of PhoP as a response regulator. Previous analyses have shown that native PhoP exists as a noncovalent dimer and that phosphorylation of this dimer does not change the PhoP oligomeric state in solution (27, 29). Purified His10-PhoPR113E was likely unphosphorylated as overexpression of the protein in E. coli under high-phosphate growth conditions would not favor the formation of His10-PhoPR113E∼P. His10-PhoPR113E and His10-PhoPR113E phosphorylated by GST-*PhoR were individually applied to a Superdex 200 gel filtration column. Unphosphorylated His10-PhoPR113E was eluted as a 34-kDa globular protein (Fig. 7A), whose molecular mass was similar to that of His10-PhoPR113E deduced from the DNA sequence (30,076 Da). This result strongly suggested that His10-PhoPR113E was a monomer. When phosphorylated His10-PhoPR113E was loaded onto the same column, the elution pattern showed that the single radioactivity peak was coincident with the protein peak of unphosphorylated His10-PhoPR113E (Fig. 7). The peak fractions were identified as His10-PhoPR113E∼P by SDS-PAGE followed by protein staining (Fig. 7B, panel I) and autoradiography (Fig. 7B, panel II). These data indicate that PhoPR113E is a monomer in solution and that phosphorylation does not change the solution state of the protein. The leading PhoP protein shoulder (elution volume, 13 to 14.5 ml) contained disulfide PhoP dimer which could not be phosphorylated (note the PhoP protein concentration [Fig. 7B, panel I] relative to the radioactivity [panel II]).
FIG. 7.
Determination of the molecular masses of His10-PhoPR113E and His10-PhoPR113E∼P by gel filtration. (A) Protein molecular standard curve, on which the elution position of His10-PhoPR113E is indicated by a vertical arrow. The horizontal arrows indicate the positions of molecular mass standards. (B) Elution profile for His10-PhoP R113E∼P on a Superdex 200 column. Symbols: •, protein concentration; □, radioactive counts. (Inset) SDS-PAGE profile of protein peak fraction. Panel I, Biosafe Coomassie blue stain; panel II, radioactivity. Ten microliters of each fraction indicated was used for SDS-PAGE.
The difference in the migration patterns of His10-PhoPR113E and His10-PhoPWT on native PAGE gels was not changed by phosphorylation of the proteins.
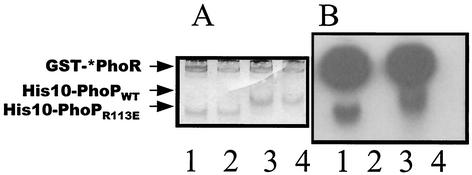
Unphosphorylated and phosphorylated His10-PhoPR113E were compared with His10-PhoPWT by using native PAGE. Previous results (27, 29) showed that native PhoP forms a dimer in solution independent of the phosphorylation state; results described above suggest that the R113E PhoP mutant protein has lost the ability to dimerize but could be phosphorylated by PhoR∼P. The PhoP proteins were incubated with GST-*PhoR with or without [γ-32P]ATP and were applied to a native PAGE gel (Fig. 8). As the migration positions of proteins in a native PAGE gel are dependent on the ratio of the molecular size to the net charge and since the net charge of the PhoP proteins at pH 8.0 is changed by only 10% by the R113E mutation, the difference in the migration positions of His10-PhoPR113E and His10-PhoPWT should reflect the molecular size difference between these two proteins. His10-PhoPR113E migrated faster than His10-PhoPWT (Fig. 8, lanes 2 and 4), which is consistent with the solution state of His10-PhoPR113E as a monomer and the solution state of His10-PhoPWT as a dimer (27, 29). Autoradiography showed that phosphorylation of these proteins did not change the migration patterns (Fig. 8, lanes 1 and 3), indicating that the oligomeric state of His10-PhoPR113E∼P or His10-PhoPWT∼P was not changed upon phosphorylation.
FIG. 8.
Biosafe Coomassie blue staining (A) and radioactivity (B) of native PAGE gel. The positions of GST-*PhoR, His10-PhoPWT, and His10-PhoPR113E are indicated on the left. GST-*PhoR and His10-PhoPR113E or His10-PhoPWT (1 μg each) were mixed with or without 10 μCi of [γ-32P]ATP in phosphorylation buffer. After incubation at room temperature for 20 min, the mixture was loaded onto the native gel. Lanes 1, GST-*PhoR with His10-PhoPR113E in the presence of [γ-32P]ATP; lanes 2, GST-*PhoR with His10-PhoPR113E; lanes 3, GST-*PhoR with His10-PhoPWT in the presence of [γ-32P]ATP; lanes 4, GST-*PhoR with His10-PhoPWT.
Molecular mass of PhoPR113E is consistent with the monomer state.
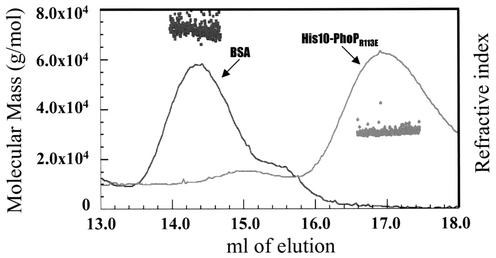
His10-PhoPR113E eluted as a 34-kDa protein when size exclusion chromatography was performed. To further establish the solution state of His10-PhoPR113E, the protein was analyzed by using an on-line multiangle static light scatter detector that allows direct determination of molecular mass as the light scatter intensity is proportional to the product of the molecular mass and the concentration of the particle under study. The pattern of elution of His10-PhoPR113E from a TSK G3000SW column is shown in Fig. 9, in which the protein concentration is represented by the change in refractive index. His10-PhoPR113E eluted as a major peak centered at an elution volume of 17 ml. Light-scattered intensity data were collected at 0.25-s intervals across the PhoP protein peak with 11 angle detectors. The calculated molecular mass of His10-PhoPR113E at each interval is plotted in Fig. 9. The molecular mass across the peak, 30,450 Da, is very close to the expected molecular mass of the monomeric form of His10-PhoPR113E (30,076 Da). The minor peak in the same elution profile (elution volume, 15 ml) had a calculated molecular mass of approximately 60,000 Da, a value consistent with the molecular mass of the nonphosphorylatable disulfide bond-mediated dimer of His10-PhoPR113E (data not shown). The first peak at ∼14.3 ml in Fig. 9 represents elution of the bovine serum albumin (BSA) (molecular mass, 66,000 Da) used to demonstrate the accuracy of this method.
FIG. 9.
Molecular mass of His10-PhoPR113E. His10-PhoPR113E or BSA was injected into a TSK G3000SW column. The refractive index signals (PhoPR113E or BSA) are displayed along with the molecular mass (•, PhoP; ▪, BSA) calculated for each successive 0.25 s along the peak.
PhoPR113E is DNA binding deficient.
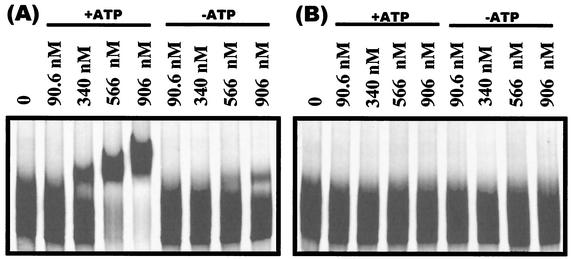
The data described above suggest that PhoPR113E retains the ability to interact with the cognate histidine kinase in order to be phosphorylated but that neither the phosphorylated protein nor the unphosphorylated protein dimerizes. Gel shift assays were preformed to compare the DNA binding efficiencies of PhoP and PhoPR113E in both their phosphorylated and unphosphorylated states. The pstS promoter was chosen for these studies as it is one of the strongest Pho regulon promoters and PhoP∼P activation of this promoter has been well documented (31, 39). Gel shift assays showed that PhoP∼P binds DNA at much lower concentrations than unphosphorylated PhoP binds DNA (Fig. 10A) but that neither PhoPR113E nor PhoPR113E∼P produces a shift (Fig. 10B) at protein concentrations up to 0.906 μM. These data show that PhoP∼PR113E is highly defective in DNA binding compared to the native PhoP∼P, a fact consistent the Pho-negative phenotype of strains expressing the PhoPR113E∼P protein (Fig. 3C).
FIG. 10.
Gelshift assays of the pstS promoter bound by PhoP∼P, PhoP, PhoPR113E∼P, or PhoPR113E. The 364-bp pstS promoter was incubated with PhoP (A) or PhoPR113E (B) and GST-*PhoR in the presence or absence of ATP. The concentrations of PhoP and PhoPR113E used in the reactions are indicated above the lanes. Each reaction mixture contained 1 μM GST-*PhoR.
DISCUSSION
Dimerization of unphosphorylated PhoP or PhoPN in solution (29) and of PhoPN in the crystal structure has been observed (Fig. 1) (3). The crystal structure revealed that the interface between two monomers involves nonidentical surfaces on the two molecules such that each monomer in a dimer has a remaining surface free for association with an additional PhoPN dimer. Thus, the PhoP protein-protein interactions observed in the crystal structure are consistent with cooperative binding of PhoP dimers at Pho-regulated (Pho-activated or Pho-repressed) promoters which contain two tandem PhoP dimer binding sites between positions −20 and −60 relative to the transcriptional start site. Phosphorylation of PhoP extends the footprint at these promoters 3′ and/or 5′ of the tandem binding sites at some promoters (pstS [31] or tuaA [30]) but not at other promoters (phoB [29] or phoD [11]). PhoP-repressed promoters that have only one dimer binding site (5, 28) require PhoP phosphorylation for binding as well as for oligomerization of PhoP∼P along the DNA >100 bp into the coding region 3′ of the dimer binding site.
Our studies revealed that PhoP proteins resulting from mutations in the codons for R113 and D60, residues that form a salt bridge buried in the hydrophilic dimer interface observed in the PhoPN crystal (3), caused a Pho-negative phenotype in vivo. The fact that the PhoPR113A negative in vivo phenotype was as strong as the negative phenotype of PhoPR113E emphasizes the importance of the positively charged arginine for charge balance in the asymmetric interface. In vitro characterization of the PhoPR113E protein showed that phosphorylation of this protein by PhoR∼P was not affected compared to phosphorylation of PhoPWT, suggesting that the R113E mutation did not alter the overall folding of the protein. The suggestion that the R113E mutation did not grossly alter the overall conformation of the protein indicates that the effect of this mutation on dimerization and consequently DNA binding is specific, as the structural model predicts.
Previous studies have shown that the unphosphorylated dimeric form of PhoP could bind regulated promoters but could not activate transcription until it was phosphorylated (31, 40), while the present findings indicate that the monomeric PhoPR113E protein can be phosphorylated but is defective in DNA binding in vitro and transcription activation in vivo. Together, these data support the hypothesis that both PhoP phosphorylation and dimerization are essential for PhoP transcriptional regulation of Pho regulon promoters.
It is interesting to compare the properties of PhoPR113E with the properties of two ompR mutations that each caused an OmpF− OmpC− phenotype irrespective of the medium osmolarity (36). The ompR mutations mapped to codons affecting R115 and E96, which align with R113 and E94 in PhoP, both of which are residues that are involved in the asymmetric interface between PhoP monomers in the PhoPN crystal structure (3). For both OmpRR115S and OmpRE96A phosphorylation by EnvZ was normal, but in vitro DNA binding was severely affected. Further studies suggested that both OmpRR115S and OmpRE96A were defective in dimerization or oligomerization. Does this imply that there are similar dimerization interfaces between monomers of different members of the OmpR family? The PhoP-OmpR data are consistent with such an idea. In contrast, PhoBN structural studies have predicted a very different monomer interface for PhoB dimers involving a symmetrical hydrophobic surface that includes α helix 1, loop β5α5, and the N terminus of α5 (46).
Our findings support the importance for physiological function of the protein-protein interface between PhoPN monomers, which is nearly 10% of the solvent-exposed surface in each monomer. However, as the PhoP in the PhoPN crystal is unphosphorylated, questions remain concerning how phosphorylation may affect the structure, as activated FixJ and NtrC have exhibited appreciable activation-induced changes (4, 23). Still, activation of CheY was judged not to cause large structural changes (26). Because the footprint protection patterns of PhoP and PhoP∼P are similar and even identical at certain promoters (phoB [29] or phoD [11]), it has been hypothesized that transcriptional activation by PhoP∼P may result from conformational changes that affect interactions with RNA polymerase and/or affect the affinity of PhoP for DNA binding. The differences in the PhoP∼P concentrations required for full DNase I promoter protection compared to the concentrations of PhoP range from 4-fold reduction (11) to as little as 1.5-fold reduction (29), depending on the promoter. At promoters containing only one dimer binding site, PhoP∼P is essential for DNA binding, oligomerization of PhoP∼P 3′ along DNA into the coding region (5, 28), and transcriptional repression (40). While the current structure accommodates PhoP dimerization, cooperative binding of PhoP dimers, and PhoP oligomerization very nicely, information concerning how the dimerization interfaces defined by the PhoPN structure are influenced by PhoP phosphorylation awaits PhoPN∼P structural analysis.
Acknowledgments
We thank L. Randall for performing the molecular mass determination and Wei Liu and T. Masdek for providing plasmids.
This work was supported by National Institutes of Health grant GM-33471 to F.M.H. and by grants from CNRS and from Le Programme de Recherche en Microbiologie Fondamentale of the French Ministry of Research to J.-P.S.
REFERENCES
- 1.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics approach and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Birck, C., Y. Chen, F. M. Hulett, and J.-P. Samama. 2003. The crystal structure of the regulatory domain in PhoP reveals a functional tandem association mediated by an asymmetric interface. J. Bacteriol. 185:254-261. [DOI] [PMC free article] [PubMed]
- 4.Birck, C., L. Mourey, P. Gouet, B. Fabry, J. Schumacher, P. Rousseau, D. Kahn, and J. P. Samama. 1999. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure Fold Des. 7:1505-1515. [DOI] [PubMed] [Google Scholar]
- 5.Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 6.Blanco, A. G., M. Sola, F. X. Gomis-Ruth, and M. Coll. 2002. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10:701-713. [DOI] [PubMed] [Google Scholar]
- 7.Bookstein, C., C. W. Edwards, N. V. Kapp, and F. M. Hulett. 1990. The Bacillus subtilis 168 alkaline phosphatase III gene: impact of a phoAIII mutation on total alkaline phosphatase synthesis. J. Bacteriol. 172:3730-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Chesnut, R. S., C. Bookstein, and F. M. Hulett. 1991. Separate promoters direct expression of phoAIII, a member of the Bacillus subtilis alkaline phosphatase multigene family, during phosphate starvation and sporulation. Mol. Microbiol. 5:2181-2190. [DOI] [PubMed] [Google Scholar]
- 10.Eder, S., W. Liu, and F. M. Hulett. 1999. Mutational analysis of the phoD promoter in Bacillus subtilis: implications for PhoP binding and promoter activation of Pho regulon promoters. J. Bacteriol. 181:2017-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eder, S. C. 1998. The mechanism of PhoP transcriptional activation of Bacillus subtilis phoD and other PHO regulon genes. M.S. thesis. University of Illinois at Chicago, Chicago.
- 12.Forst, S., I. Kalve, and W. Durski. 1995. Molecular analysis of OmpR binding sequences involved in the regulation of ompF in Escherichia coli. FEMS Microbiol. Lett. 131:147-151. [DOI] [PubMed] [Google Scholar]
- 13.Harlocker, S. L., L. Bergstrom, and M. Inouye. 1995. Tandem binding of six OmpR proteins to the ompF upstream regulatory sequence of Escherichia coli. J. Biol. Chem. 270:26849-26856. [DOI] [PubMed] [Google Scholar]
- 14.Harrison-McMonagle, P., N. Denissova, E. Martinez-Hackert, R. H. Ebright, and A. M. Stock. 1999. Orientation of OmpR monomers within an OmpR:DNA complex determined by DNA affinity cleaving. J. Mol. Biol. 285:555-566. [DOI] [PubMed] [Google Scholar]
- 15.Henner, D. J. 1990. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 185:223-228. [DOI] [PubMed] [Google Scholar]
- 16.Huang, K. J., and M. M. Igo. 1996. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J. Mol. Biol. 262:615-628. [DOI] [PubMed] [Google Scholar]
- 17.Hulett, F. M. 2002. The Pho regulon, p. 193-203. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 18.Hulett, F. M. 1993. Regulation of phosphorus metabolism, p. 229-235. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 19.Hulett, F. M. 1996. The signal-transduction network for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:933-939. [DOI] [PubMed] [Google Scholar]
- 20.Hulett, F. M., C. Bookstein, and K. Jensen. 1990. Evidence for two structural genes for alkaline phosphatase in Bacillus subtilis. J. Bacteriol. 172:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hulett, F. M., J. Lee, L. Shi, G. Sun, R. Chesnut, E. Sharkova, M. F. Duggan, and N. Kapp. 1994. Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis. J. Bacteriol. 176:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapp, N. V., C. W. Edwards, R. S. Chesnut, and F. M. Hulett. 1990. The Bacillus subtilis phoAIV gene: effects of in vitro inactivation on total alkaline phosphatase production. Gene 96:95-100. [DOI] [PubMed] [Google Scholar]
- 23.Kern, D., B. Volkman, P. Luginbuhl, M. Nohalle, S. Kustu, and D. Wemmer. 1999. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature 402:849-899. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, K., M. Ogura, H. Yamaguchi, K. Yoshida, N. Ogasawara, T. Tanaka, and Y. Fujita. 2001. Comprehensive DNA microarray analysis of Bacillus subtilis two-component regulatory systems. J. Bacteriol. 183:7365-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. Y., H. S. Cho, J. G. Pelton, D. Yan, E. A. Berry, and D. E. Wemmer. 2001. Crystal structure of activated CheY. Comparison with other activated receiver domains. J. Biol. Chem. 276:16425-16431. [DOI] [PubMed] [Google Scholar]
- 27.Liu, W. 1997. Biochemical and genetic analyses establish a dual role for PhoP in Bacillus subtilis PHO regulation. Ph.D. thesis. University of Illinois at Chicago, Chicago.
- 28.Liu, W., S. Eder, and F. M. Hulett. 1998. Analysis of Bacillus subtilis tagAB and tagDEF expression during phosphate starvation identifies a repressor role for PhoP-P. J. Bacteriol. 180:753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, W., and F. M. Hulett. 1998. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology 144:1443-1450. [DOI] [PubMed] [Google Scholar]
- 31.Liu, W., Y. Qi, and F. M. Hulett. 1998. Sites internal to the coding regions of phoA and pstS bind PhoP and are required for full promoter activity. Mol. Microbiol. 28:119-130. [DOI] [PubMed] [Google Scholar]
- 32.Maeda, S., and T. Mizuno. 1988. Activation of the ompC gene by the OmpR protein in Escherichia coli. The cis-acting upstream sequence can function in both orientations with respect to the canonical promoter. J. Biol. Chem. 263:14629-14633. [PubMed] [Google Scholar]
- 33.Maeda, S., and T. Mizuno. 1990. Evidence for multiple OmpR-binding sites in the upstream activation sequence of the ompC promoter in Escherichia coli: a single OmpR-binding site is capable of activating the promoter. J. Bacteriol. 172:501-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda, S., K. Takayanagi, Y. Nishimura, T. Maruyama, K. Sato, and T. Mizuno. 1991. Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: a study involving synthetic OmpR-binding sequences. J. Biochem (Tokyo) 110:324-327. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109-124. [DOI] [PubMed] [Google Scholar]
- 36.Nakashima, K., K. Kanamaru, H. Aiba, and T. Mizuno. 1991. Signal transduction and osmoregulation in Escherichia coli. A novel type of mutation in the phosphorylation domain of the activator protein, OmpR, results in a defect in its phosphorylation-dependent DNA binding. J. Biol. Chem. 266:10775-10780. [PubMed] [Google Scholar]
- 37.Okamura, H., S. Hanaoka, A. Nagadoi, K. Makino, and Y. Nishimura. 2000. Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box. J. Mol. Biol. 295:1225-1236. [DOI] [PubMed] [Google Scholar]
- 38.Ozanne, P. G. 1980. Phosphate nutrition of plants—a general treatise, p. 559-585. In E. Khasswenh (ed.), The role of phosphorus in agriculture. American Society of Agronomy, Madison, Wis.
- 39.Qi, Y., and F. M. Hulett. 1998. PhoP-P and RNA polymerase sigmaA holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP-P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187-1197. [DOI] [PubMed] [Google Scholar]
- 40.Qi, Y., and F. M. Hulett. 1998. Role of PhoP∼P in transcriptional regulation of genes involved in cell wall anionic polymer biosynthesis in Bacillus subtilis. J. Bacteriol. 180:4007-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi, Y., Y. Kobayashi, and F. M. Hulett. 1997. The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the Pho regulon. J. Bacteriol. 179:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robichon, D., M. Arnaud, R. Gardan, Z. Pragai, M. O'Reilly, G. Rapoport, and M. Debarbouille. 2000. Expression of a new operon from Bacillus subtilis, ykzB-ykoL, under the control of the TnrA and PhoP-PhoR global regulators. J. Bacteriol. 182:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seki, T., H. Yoshikawa, H. Takahashi, and H. Saito. 1987. Cloning and nucleotide sequence of phoP, the regulatory gene for alkaline phosphatase and phosphodiesterase in Bacillus subtilis. J. Bacteriol. 169:2913-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seki, T., H. Yoshikawa, H. Takahashi, and H. Saito. 1988. Nucleotide sequence of the Bacillus subtilis phoR gene. J. Bacteriol. 170:5935-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, L., and F. M. Hulett. 1999. The cytoplasmic kinase domain of PhoR is sufficient for the low phosphate-inducible expression of Pho regulon genes in Bacillus subtilis. Mol. Microbiol. 31:211-222. [DOI] [PubMed] [Google Scholar]
- 46.Sola, M., F. X. Gomis-Ruth, L. Serrano, A. Gonzalez, and M. Coll. 1999. Three-dimensional crystal structure of the transcription factor PhoB receiver domain. J. Mol. Biol. 285:675-687. [DOI] [PubMed] [Google Scholar]
- 47.Sun, G., S. M. Birkey, and F. M. Hulett. 1996. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19:941-948. [DOI] [PubMed] [Google Scholar]