Abstract
The first step in the pentachlorophenol (PCP) degradation pathway in Sphingobium chlorophenolicum has been believed for more than a decade to be conversion of PCP to tetrachlorohydroquinone. We show here that PCP is actually converted to tetrachlorobenzoquinone, which is subsequently reduced to tetrachlorohydroquinone by PcpD, a protein that had previously been suggested to be a PCP hydroxylase reductase. pcpD is immediately downstream of pcpB, the gene encoding PCP hydroxylase (PCP monooxygenase). Expression of PcpD is induced in the presence of PCP. A mutant strain lacking functional PcpD has an impaired ability to remove PCP from the medium. In contrast, the mutant strain removes tetrachlorophenol from the medium at the same rate as does the wild-type strain. These data suggest that PcpD catalyzes a step necessary for degradation of PCP, but not for degradation of tetrachlorophenol. Based upon the known mechanisms of flavin monooxygenases such as PCP hydroxylase, hydroxylation of PCP should produce tetrachlorobenzoquinone, while hydroxylation of tetrachlorophenol should produce tetrachlorohydroquinone. Thus, we proposed and verified experimentally that PcpD is a tetrachlorobenzoquinone reductase that catalyzes the NADPH-dependent reduction of tetrachlorobenzoquinone to tetrachlorohydroquinone.
Pentachlorophenol (PCP) is a widely used and highly toxic wood preservative. It was first introduced as a pesticide in 1936 (7) and is not known to be a natural product. Despite its recent introduction into the environment and its high toxicity, several strains of Sphingobium chlorophenolicum (previously Sphingomonas chlorophenolica) (24) that can mineralize PCP have been identified. The best studied of these are strains ATCC 39723 (19), RA-2 (23), and UG30 (6). It appears that S. chlorophenolicum has assembled a new metabolic pathway capable of converting this anthropogenic compound into a recognizable metabolite. Our previous studies suggest that this pathway has been assembled by patching together enzymes from at least two different metabolic pathways (8). PCP hydroxylase (PCP monooxygenase; EC 1.14.13.50) and 2,6-dichlorohydroquinone dioxygenase may have originated from enzymes that hydroxylated a naturally occurring chlorinated phenol and then cleaved the resulting hydroquinone. Tetrachlorohydroquinone (TCHQ) dehalogenase appears to have originated from a glutathione-dependent double bond isomerase such as maleylacetoacetate isomerase or maleylpyruvate isomerase (which are involved in degradation of tyrosine and benzoate, respectively) (2). If this pathway has evolved recently in response to the introduction of PCP into the environment, then it would not be expected to perform at the high level characteristic of pathways that have evolved over periods of millions or billions of years. Indeed, the PCP degradation pathway shows signs of immaturity in several respects. First, PCP hydroxylase, the first enzyme in the pathway, is very inefficient in vitro (P. M. Kiefer and S. D. Copley, unpublished data), and appears to severely limit the flux of PCP through the pathway in vivo (17). Second, TCHQ dehalogenase is profoundly inhibited by its aromatic substrate (K. Anandarajah, P. M. Kiefer, and S. D. Copley, unpublished data). Third, TCHQ dehalogenase expression is not regulated in tandem with the other known enzymes in the pathway but is apparently constitutive (21). All of these findings are consistent with the idea that the PCP degradation pathway has been patched together rather recently and has not been fine-tuned to perform as effectively as do most bacterial metabolic pathways.
The gene encoding PCP hydroxylase (pcpB) is immediately upstream of two additional genes. pcpR encodes a regulatory protein that responds to PCP (5). pcpD, which is immediately downstream of pcpB, resembles genes for the reductase components of two-component oxygenases, some of which hydroxylate aromatic compounds. Based upon this resemblance, it has been proposed that PcpD is a reductase that facilitates the hydroxylation of PCP by PCP hydroxylase (19), and the annotation of PcpD in GenBank states that it is PCP 4-monooxygenase reductase. We suspected that this assignment was incorrect because PCP hydroxylase is a flavin monooxygenase, and such enzymes do not generally require reductases. Consequently, we undertook studies to determine whether PcpD is required for degradation of PCP. We find that transcription of pcpD is induced by PCP, as previously reported for pcpA (29) and pcpB (20). A mutant strain in which PcpD has been knocked out is able to remove PCP from the medium when it is present at low concentrations, but not when it is present at high concentrations. In contrast, the knockout strain can remove tetrachlorophenol (TCP) from the medium as well as the wild-type strain, even at high concentrations. These results suggest that PcpD may catalyze a step that is critical for degradation of PCP but not TCP and therefore must involve the chlorine at the 4 position of PCP. Based upon the expected mechanism of the hydroxylase reaction, the sequence of PcpD, and our experimental results, we propose that PcpD is a tetrachlorobenzoquinone (TCBQ) reductase required for degradation of PCP but not TCP.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. chlorophenolicum strain RA-2 was obtained from S. Schmidt (University of Colorado at Boulder). S. chlorophenolicum strain ATCC 39723 and Pseudomonas aeruginosa PAO1-Lac were obtained from the American Type Culture Collection. Escherichia coli XL1-Blue and pBluescript were obtained from Stratagene. E. coli M15[pReP4] and pQE30 were obtained from Qiagen. The E. coli-Pseudomonas shuttle vector pUCP-Nde (10) was a generous gift from C. N. Cronin (Veterans Affairs Medical Center, San Francisco, Calif.). Growth medium for S. chlorophenolicum contained (per liter) 2.9 g of Na2HPO4, 0.3 g of KH2PO4, 0.1 g of NH4NO3, 0.02 g of MgSO4 · 7H2O, 10 mg of FeSO4 · 7H2O, and either 4 g of glucose or 4 g of sodium glutamate. PCP was added to various final concentrations as indicated in the text. Cells were grown at 25°C with shaking. E. coli cells were grown on either Luria-Bertani (LB) or 2YT medium supplemented, when required for maintenance of plasmids, with ampicillin (100 μg/ml) and/or kanamycin (25 μg/ml), at 37°C with shaking.
Sequence analysis.
BLAST searches (1) were carried out at the NCBI website. Multiple sequence alignments were performed using ClustalW (25), and uncorrected distances were obtained using the Distances algorithm in the Genetics Computer Group package. Motif analyses were performed using the MEME algorithm (3) at the San Diego Supercomputing Center.
RT-PCR to detect transcription of pcpD.
S. chlorophenolicum strain ATCC 39723 was grown on minimal medium with 0.4% (wt/vol) sodium glutamate in the presence or absence of 100 μM PCP. After growth for 4 days, RNA was isolated using a Qiagen RNeasy kit. After isolation, the RNA was treated with RNase-free DNase (2 μg/ml) for 1 h at 37°C. Reverse transcription (RT) was then performed using a Qiagen Omniscript reverse transcriptase kit according to the manufacturer's instructions. A forward primer for pcpD (5′-GGGGTACCATGACAAACCCCGTTTCGACAAT-3′) was used as a template for cDNA synthesis in the reverse transcriptase reaction. The resulting cDNA was amplified by PCR using the same forward primer and a reverse primer to the 3′-end of pcpD (5′-GCTCTAGATTCGAGCGGTCGATTCAGATGT-3′). The PCR program consisted of a denaturation step of 5 min at 95°C followed by 28 cycles of denaturation at 95°C for 50 s, annealing at 52°C for 50 s, and extension at 72°C for 50 s, with a final extension step at 72°C for 10 min. PCR products were subjected to agarose gel electrophoresis and visualized by staining with ethidium bromide.
Construction of pSCDK1 for allelic exchange of pcpD.
A kanamycin resistance gene from plasmid pRep4 (Qiagen) was inserted into pcpD using an overlap-extension PCR method (13). A 562-bp fragment (D1) from the 5′ region of pcpD was amplified using primers pDKpnI (5′-GGGGTACCACAAACCCCGTTTCGACA-3′) (which includes a KpnI site, designated by the underlined letters) and pD1 (5′-GTCTTCCATCCTCCAGAAATTTGCCGTCGGCCTCTTCA-3′). The remaining 544-bp fragment (D2) at the 3′ end of the gene was amplified using primers pD2 (5′-AAACTGCCAACTCCGTCATGAAGAGGCCGACGGCAAAT-3′) and pDHindIII (5′-TTCGAATTCGAGCGGTCGATTCAG-3′) (which contains a HindIII site, designated by the underlined letters). PCRs were performed in 100-μl reaction mixture volumes containing 8 μl of 2.5 mM deoxynucleoside triphosphates, 200 ng of template DNA, 100 pmol of each set of primers, 2.5 U of Taq polymerase, and reaction buffer supplied by the manufacturer (Promega). Amplification was carried out for 30 cycles. Each cycle consisted of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s. The kanamycin resistance gene from plasmid pRep4 (Qiagen) was amplified using primers p3 (5′-TTCTGGAGGATGGAAGACATGGCGATAGCTAGACT-3′) and p4 (5′-TGACGGAGTTGGCAGTTTGGACGGTCATTTCGAACC-3′). Amplification was carried out for 28 cycles. Each cycle consisted of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 60 s. The primers for amplification of the three fragments (D1, the kanamycin resistance gene, and D2) were designed so that pD1 and p3 contained complementary regions to allow splicing of the D1 fragment to the 5′ end of the kanamycin resistance gene, and pD2 and p4 contained complementary regions to allow splicing of the D2 fragment to the 3′ end of the kanamycin resistance gene. The three amplified fragments were purified from 1% agarose gels and then used in an overlap-extension reaction with pDKpnI and pDHindIII primers. Amplification was carried out for five cycles. Each cycle consisted of denaturation at 95°C for 30 s, annealing at 37°C for 30 s, and extension at 72°C for 60 s. Annealing at low temperatures was necessary to allow efficient pairing of the complementary ends of the fragments. Subsequently, an additional 25 cycles were carried out to amplify the full-length product, with each cycle consisting of denaturation at 95°C for 60 s, annealing at 55°C for 60 s, and extension at 72°C for 150 s. The full-length PCR products were digested with KpnI and HindIII and inserted into the multiple cloning site of pBluescript which had been digested with KpnI and HindIII. The ligation product was transformed into E. coli XL1-Blue and transformants were selected on LB plates containing ampicillin (100 μg/ml) and kanamycin (50 μg/ml). This plasmid was designed pSCDK1.
Construction of pcpD knockout strains of S. chlorophenolicum strains ATCC 39723 and RA-2.
S. chlorophenolicum cells were made competent for transformation by washing four times with sterile 10% (vol/vol) glycerol and then were resuspended in 10% (vol/vol) glycerol. pSCDK1 was introduced into competent cells by electroporation using a 5.9-ms pulse (2.5 kV) on a Bio-Rad Micropulser in 0.2-cm gapped electroporation cuvettes. Following electroporation, one-quarter-strength tryptic soy broth was added and the cells were allowed to recover with shaking at room temperature for 12 h. The cells were then plated on one-quarter-strength tryptic soy broth agar medium containing kanamycin (25 μg/ml). pSCDK1 would not be expected to replicate in S. chlorophenolicum, and indeed, loss of pSCDK1 from colonies that grew on the plate was confirmed by PCR using primers for the T3 and T7 promoter regions of the parent plasmid, pBluescript. Thus, cells can only survive if the kanamycin resistance gene is transferred to the genomic DNA by homologous recombination of the disrupted pcpD into the intact genomic pcpD. Homologous recombination of the disrupted pcpD into genomic DNA was confirmed by PCR. A band of the expected size was obtained using a forward primer specific for pcpB (the gene immediately upstream of pcpD) and a reverse primer specific for the kanamycin resistance gene.
Degradation of PCP and TCP by wild-type and mutant strains of S. chlorophenolicum.
Wild-type and mutant strains of S. chlorophenolicum were grown on minimal medium containing sodium glutamate and either PCP or TCP for 20 h at 25°C. PCP and TCP concentrations were determined at intervals using high-performance liquid chromatography (HPLC). Samples were centrifuged for 3 min at 10,000 × g, and aliquots of the supernatant were injected onto a Rainin C18 column (4.6 by 50 mm). The column was eluted at 1 ml/min with 45% acetonitrile-55% 0.1% acetic acid in water. The detector was set at 210 nm.
Cloning and expression of PCP hydroxylase and PcpD.
Genomic DNA was isolated from S. chlorophenolicum strain ATCC 39723 using a DNeasy tissue kit (Qiagen). The gene encoding PCP hydroxylase (pcpB) was amplified by PCR using the forward primer 5′-CGGAATTCTCGGGCAAGACCACC-3′ and the reverse primer 5′-TAGCAAGCTTGTTCTTGACCACCTT-3′ The forward primer was designed to start directly after the ribosome binding site and ATG initiation codon. EcoRI and HindIII restriction sites that were incorporated in the forward and reverse primers, respectively, are underlined in the primer sequences. For amplification of pcpD, the forward primer was 5′-AATTCCATATGACAAACCCCGTTTCGA-3′ and the reverse primer was 5′-TTGGTACCTTATCAGTGGTGGTGGTGGTGGTGGTGGTGGTGGTGGTGGTGGATGTCCAGCACCAGCCGC-3′. The forward primer was designed to start directly after the ribosome binding site and ATG initiation codon. The reverse primer encodes a His12 tag at the C terminus of the protein. NdeI and KpnI sites that were incorporated in the forward and reverse primers, respectively, are underlined in the primer sequences. PCR amplification in both cases was performed in a total volume of 100 μl containing 200 ng of genomic DNA, 2.5 U of Taq polymerase, 2.0 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, and 100 pmol of primers. Amplification was carried out for 28 cycles. Each cycle consisted of denaturation at 95°C for 60 s, annealing at 60°C for 60 s, and extension at 72°C for 120 s. The amplified fragments were sequenced to confirm that no mutations had been introduced during PCR amplification.
The amplified pcpB and the pQE30 vector were digested with EcoRI and HindIII. The digestion products were then ligated according to standard procedures and transformed into competent E. coli XL1-Blue cells. Transformants were selected on LB agar with ampicillin (100 μg/ml). Plasmid DNA was prepared from a culture grown from a single colony, and the presence of the desired insert was verified by agarose gel electrophoresis after digestion with EcoRI and HindIII. This plasmid was designated pMD-PcpB. For protein expression, pMD-PcpB was transformed into E. coli M15[pReP4], and transformants were selected on LB medium containing ampicillin (100 μg/ml) and kanamycin (25 μg/ml).
The amplified pcpD and the pUCP-Nde vector were digested with NdeI and KpnI, and the digestion products were then ligated according to standard procedures and transformed into competent E. coli XL1-Blue cells. White colonies were selected on LB agar with ampicillin (100 μg/ml) in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (800 μg/plate) and IPTG (isopropyl-β-d-thiogalactopyranoside) (800 μg/plate) Plasmid DNA was prepared from a culture grown from a single colony, and the presence of the desired insert was verified by agarose gel electrophoresis after digestion with KpnI and XhoI. This plasmid was designated pMD-PcpD. For protein expression, pMD-PcpD was transformed into P. aeruginosa strain PAO1-Lac. Transformants were selected on LB agar containing carbenicillin (200 μg/ml) and tetracycline (15 μg/ml).
Purification of PCP hydroxylase and PcpD.
A 50-ml culture of E. coli M15[pReP4] containing pMD-PcpB was grown overnight in 2YT medium and then used to inoculate 1 liter of 2YT medium supplemented with ampicillin (100 μg/ml) and kanamycin (25 μg/ml) in a 4-liter flask. The cells were grown with vigorous shaking at 37°C until an optical density at 600 nm (OD600) of 0.5 to 0.7 was reached. Expression of PcpB was induced by addition of IPTG (0.5 mM). Cells were harvested after a further 3 h of growth by centrifugation at 4,000 × g for 10 min at 4°C. Cultures of P. aeruginosa containing pMD-PcpD were grown similarly in 2YT containing carbenicillin (200 μg/ml) and tetracycline (15 μg/ml). The cell pellets were resuspended in 100 ml of buffer A [50 mM sodium phosphate, pH 8.0, containing 300 mM NaCl, 100 mM imidazole, and 0.1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF)] and lysed by four passes through a French press at a pressure of 12,000 lb/in2. The lysates were subjected to centrifugation at 12,000 × g for 30 min at 4°C, and the His6-tagged PCP hydroxylase and His12-tagged PcpD were purified on Ni2+-nitrilotriacetic acid (NTA) resin according to the protocol in the QIAexpress kit manual (Qiagen). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis indicated that PcpB had been purified to homogeneity, and PcpD had been purified to >95% homogeneity. Protein concentrations were determined using a Bio-Rad protein assay kit.
Characterization of products formed in enzymatic reactions.
For gas chromatography-mass spectrometry (GC/MS) analysis of reaction products formed by PCP hydroxylase, PCP hydroxylase (10 μM) was incubated with 250 μM PCP or 100 μM TCHQ and 1 mM NADPH in 100 mM potassium phosphate, pH 8.0, at 37°C for 2 min. Samples were centrifuged for 3 min at 10,000 × g to remove particulates, and aliquots of the supernatant were injected onto a Rainin C18 column (4.6 by 50 mm). The column was eluted with 25% acetonitrile-75% 0.1% acetic acid in water at a flow rate of 1 ml/min. The detector was set at 210 nm. The product peak was collected and extracted three times with an equal volume of ethyl acetate. The ethyl acetate was removed by rotary evaporation, and the residue was redissolved in a small volume of ethyl acetate prior to analysis by GC/MS with a Hewlett-Packard 5988A gas chromatograph-mass spectrometer equipped with a Hewlett-Packard fused-silica capillary column (length, 25 m; inner diameter, 0.2 mm; film thickness, 0.33 μm) containing HP-5 cross-linked 5% phenyl methyl silicone. GC/MS analysis of the product formed by PcpD was analyzed similarly using reaction mixtures containing His12-tagged PcpD (74 μg/ml), 50 μM TCBQ, and 1 mM NADPH in 100 mM potassium phosphate, pH 8.0.
For quantification of TCBQ formed by PCP hydroxylase, His6-tagged PCP hydroxylase (100 μM) was incubated with 100 μM PCP and 1 mM NADPH in 100 mM potassium phosphate, pH 8.0, at 37°C for 1 min. Glutathione was added to a final concentration of 2 mM to convert TCBQ to 2,3,5-trichloro-6-S-glutathionylbenzoquinone (GS-TriCBQ) (26, 27). The His6-tagged PCP hydroxylase was removed by treatment of the reaction mixture with 5 μl of Ni2+-NTA resin (Qiagen). Samples were centrifuged for 2 min at 10,000 × g to remove the resin, and aliquots of the supernatant were injected onto a Rainin C18 HPLC column (4.6 by 50 mm). For quantification of GS-TriCBQ, the column was eluted at 1 ml/min with a gradient of 0 to 40% acetonitrile in 0.1% trifluoroacetic acid (vol/vol) in water over a period of 10 min. The detector was set at 290 nm. A standard curve was generated by incubating TCBQ (12.5 to 100 μM) with 2 mM glutathione and then injecting aliquots onto the reverse-phase HPLC column. Under these conditions, GS-TriCBQ eluted at 5.0 min. For quantification of PCP, the column was eluted at 1 ml/min with 45% acetonitrile-55% 0.1% trifluoroacetic acid (vol/vol) in water. The detector was set at 210 nm. Under these conditions, PCP eluted at 3.8 min.
Enzyme assays.
PCP hydroxylase activity was measured by monitoring the disappearance of PCP in a reaction mixture containing PCP hydroxylase, 100 μM PCP, and 200 μM NADPH in 50 mM potassium phosphate, pH 8.0. Reaction mixtures were incubated at 25°C, and enzyme activity was quenched by heating the tubes in a 140°C sand bath for 1 min. (PCP was determined to be stable under these conditions.) After centrifugation for 3 min to remove particulates, aliquots of the supernatant were injected onto a Rainin C18 column (4.6 by 50 mm). The column was eluted as described above.
Reduction of TCBQ by NADPH was assayed spectrophotometrically by measuring the decrease in OD340 in a 0.1-cm-path-length quartz cuvette. Reaction mixtures contained 50 μM TCBQ, 1 mM NADPH, and variable amounts of PcpD in 100 mM potassium phosphate, pH 8.0.
Incubation of PCP hydroxylase with TCBQ.
PCP hydroxylase (10 μM) was incubated with either TCBQ or TCHQ (0 to 80 μM) in 50 mM potassium phosphate, pH 7.5, in a glove box filled with N2 for 30 min at room temperature. Samples were then removed, and PCP hydroxylase activity was assayed as described above.
RESULTS AND DISCUSSION
Sequence analysis predicts that PcpD is a reductase containing a flavin and a 2Fe2S cluster.
As previously reported (19), a BLAST search (1) using PcpD as a query sequence reveals that this protein is most closely related to a number of proteins that provide the reductase component of class IA two-component oxygenase enzymes (4, 15, 18, 22). The functions of several of the highest scoring relatives are listed in Table 1. The oxygenase components of these enzymes typically contain a nonheme iron and a Rieske-type 2Fe2S cluster (5). Oxygen is activated at the nonheme iron site for attack upon an aromatic substrate, either upon the ring itself or upon an alkyl substituent. These enzymes require the input of electrons from NADH to the active site iron atom for catalytic turnover. However, electrons cannot be transferred directly from NADH to nonheme iron. NADH is a two-electron reductant, providing reducing equivalents in the form of hydride. Nonheme iron can only accept reducing equivalents in the form of single electrons. Thus, the reductase component of two-component oxygenases acts as a transducer, accepting hydride from NADH to reduce a bound flavin. The flavin then transfers electrons singly through iron-sulfur clusters to the oxygenase active site.
TABLE 1.
Relatives of PcpD with known functions identified in the nonredundant NCBI database
| gi | Protein function | % Identity |
|---|---|---|
| 628500 | Phenoxybenzoate dioxygenase beta subunit | 44 |
| 15600098 | Vanillate O-demethylase oxidoreductase | 39 |
| 1790868 | Toluenesulfonate methyl monooxygenase reductase component | 39 |
| 3914349 | Phthalate dioxygenase reductase | 34 |
| 9820076 | 3-Chlorobenzoate 3,4-dioxygenase reductase | 34 |
The sequence of PcpD leaves little doubt that it is a reductase. It contains the three domains typical of the reductase components of two-component oxygenases: a flavin binding domain (pFam 00970; E = 2 × 10−6), an NAD-binding domain (pFam 00175; E = 0.004), and a ferrodoxin domain (pFam 00111; E = 6 × 10−10). Furthermore, the sequences of motifs involved in binding the flavin and NAD, as well as the ligands for the iron-sulfur cluster, are conserved in PcpD. Figure 1 shows a multiple sequence alignment of a divergent set of reductases related to PcpD. (A divergent set in which none of the members has no more than 45% pairwise identity to any other member helps to pinpoint regions of the sequence that are conserved for reasons of structure and/or function.) Highly conserved motifs identified by the MEME algorithm, as well as amino acid residues that are involved in direct contacts with the flavin, NAD, or iron atoms in the structurally characterized phthalate dioxygenase reductase (9), are highlighted in the figure. PcpD has all of these motifs except for one of the three motifs involved in NAD binding. This motif is the most variable of the motifs found and is also missing in another member of the set.
FIG. 1.
Multiple sequence alignment of PcpD and related reductases. Secondary structural elements for the flavin (F), NAD (N), and iron-sulfur (I) domains of the phthalate dioxygenase structure (PDB 2PIA) are designated under each row. Conserved motifs involved in binding the flavin and NAD are shown in magenta and blue, respectively, and the motif containing the ligands for the iron sulfur cluster is shown in green. Residues highlighted in yellow are involved in specific interactions with the various cofactors. Abbreviations: Phbendiox.Pp, phenoxybenzoate dioxygenase, Pseudomonas pseudoalcaligenes (gi628500); vandemeth.Pa, vanillate O-demethylase oxidoreductase, P. aeruginosa (gi15600098); putdioxsubunit.Ec, putative dioxygenase beta subunit, E. coli O157:H7 (gi15831766); putoxidored.Sm, putative oxidoreductase/oxygenase, Sinorhizobium meliloti (gi14523944); oxidoredsubunit.Sc, iron-sulfur oxidoreductase beta subunit, Streptomyces coelicolor (gi7480270); tolsulf-methylmonoox.Ct, toluenesulfonate methyl-monooxygenase reductase component, Comomonas testosteroni (gi1790868); Phthdioxred.Bc, phthalate dioxygenase reductase, Burkholderia cepacia (gi349931, PDB 2PIA); 3Cbdioxred.Alsp, 3-chlorobenzoate-3,4-dioxygenase reductase, Alcaligenes sp. Tn5271 (gi9820076); PcpD.Sc, S. chlorophenolicum (gi881298).
Although PcpD appears to have the motifs necessary to function as a reductase, the proposal that PcpD is a reductase for PCP hydroxylase seems unlikely based upon our understanding of the mechanism of the enzyme. Although the reaction reported to be catalyzed by PCP hydroxylase (Fig. 2) does resemble those catalyzed by some of the two-component oxygenases, PCP hydroxylase is a flavin monooxygenase whose mechanism is entirely different. Flavin monooxygenases catalyze the NAD(P)H-dependent hydroxylation of phenolic substrates by reaction of the phenol with a C4a-hydroperoxyflavin species generated by reaction of reduced flavin with O2 (11, 12, 16). In these enzymes, NAD(P)H directly reduces the flavin at the active site, and there is no need for an intermediary reductase. Thus, it seemed unlikely that PcpD is a reductase for PCP monooxygenase, but this remained to be established experimentally. Furthermore, it is possible that PcpD might play an as-yet-uncharacterized role in the pathway. We therefore undertook experiments to explore the role of PcpD in PCP degradation.
FIG. 2.
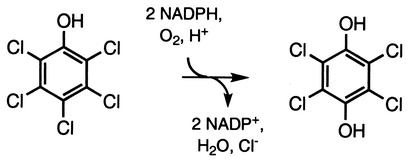
Reaction reported by Xun and Orser (29, 30) to be catalyzed by PCP hydroxylase.
PcpD expression is induced by PCP.
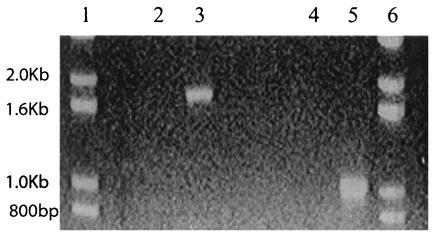
RT-PCR was used to determine whether PcpD was expressed in response to PCP. Cultures of S. chlorophenolicum strain ATCC 39723 were grown on minimal medium plus sodium glutamate (0.4% [wt/vol]) in the absence and presence of 100 μM PCP. The cells were harvested, and total mRNA was isolated. RT-PCR was carried out with primers specific for pcpD and for pcpB, whose transcription is known to be regulated by PCP. Figure 3 shows that bands corresponding to the expected sizes are seen only with samples prepared from cells grown in the presence of PCP. This result demonstrates that transcription of pcpD is induced by PCP. Additional experiments (data not shown) in which RT-PCR was carried out with a forward primer specific for pcpB and a reverse primer specific for pcpD showed that a band of the size expected for both genes (2.6 kb) was amplified, demonstrating that a single transcript was produced from pcpB and pcpD.
FIG. 3.
RT-PCR amplification of pcpB (lanes 2 and 3) and pcpD (lanes 4 and 5) fragments from RNA prepared from S. chlorophenolicum strain ATCC 39723 cells grown in the absence (lanes 2 and 4) or presence (lanes 3 and 5) of 100 μM PCP. Lanes 1 and 6 contain size markers.
Creation of a mutant strain lacking PcpD.
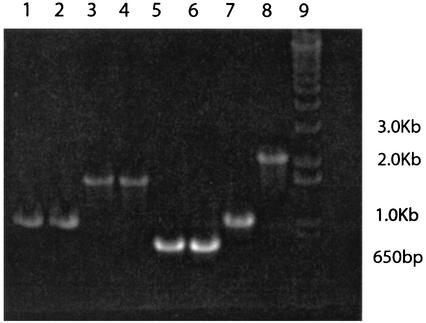
In order to determine whether PcpD is required for degradation of PCP, we created mutant strains of S. chlorophenolicum ATCC 39723 and RA-2 in which pcpD was interrupted with a kanamycin resistance gene. A DNA fragment in which a kanamycin resistance gene had been inserted into the middle of pcpD was created using overlap-extension PCR. This fragment was ligated into pBluescript, and the resulting plasmid (pSCDK1) was transformed into E. coli XL1-Blue. pSCDK1 was then isolated and used to transform S. chlorophenolicum strains ATCC 39723 and RA-2. Homologous recombination of the interrupted gene into the genomes of several transformants was confirmed by PCR. PCR amplification using primers specific for pcpD gave 2.2-kb bands, consistent with the size of the interrupted pcpD (Fig. 4, lane 8). Additional experiments using one primer specific for pcpB and another for the kanamycin resistance gene gave a band consistent with the size expected for the sum of pcpB, the initial fragment of pcpD, and the kanamycin resistance gene (data not shown). PCR amplification using primers specific for the T3 and T7 regions of pSCDK1 gave a band of the expected size for control experiments with pSCDK1 DNA but no band when DNA from the mutant strains was used (data not shown), thus confirming the loss of pSCDK1. Figure 4 summarizes PCR results obtained with genomic DNA from the mutant RA-2 strain using primers for pcpA, pcpB, pcpC, and pcpD. The mutant strain shows bands equivalent to those seen for the wild-type strain for amplification of pcpA, pcpB, and pcpC from genomic DNA. However, the band obtained with primers for pcpD is larger in the case of the mutant strain. Similar results were obtained for the mutant strain obtained from S. chlorophenolicum strain ATCC 39723. Functional PcpD cannot be produced in the mutant strain because there is a stop codon at the end of the kanamycin resistance gene. If a fusion protein consisting of the N-terminal region of PcpD and the kanamycin resistance protein is produced, it will lack part of the NAD-binding domain and the entire iron-sulfur cluster domain and therefore be unable to function as a reductase.
FIG. 4.
PCR amplification of pcpD and the three genes known to be required for degradation of PCP in the wild-type strain S. chlorophenolicum strain RA-2 (lanes 1, 3, 5, and 7) and the pcpD knockout (lanes 2, 4, 6, and 8). Lanes 1 and 2, pcpA; lanes 3 and 4, pcpB; lanes 5 and 6, pcpC; lanes 7 and 8, pcpD. Lane 9 contains size markers.
Lack of functional PcpD affects the ability of S. chlorophenolicum to degrade PCP at high concentrations, but not at low concentrations.
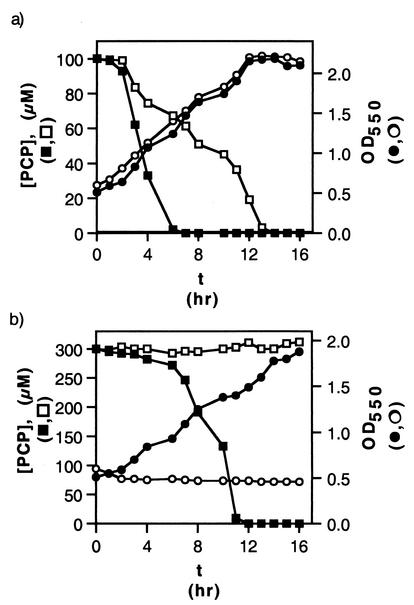
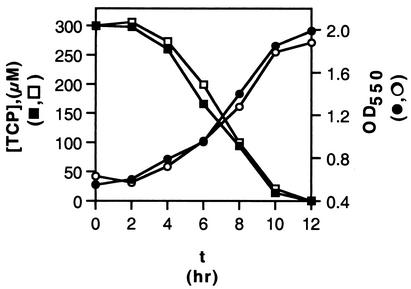
We followed the time course of cell growth and PCP degradation for wild-type and mutant versions of S. chlorophenolicum strain ATCC 39723. As shown in Fig. 5a, PCP disappeared from the medium in a culture of the knockout strain with 100 μM PCP, although at a reduced rate compared to that seen for the wild-type strain. However, in medium containing 300 μM PCP, the knockout strain was unable to either remove PCP from the medium or to continue growing (Fig. 5b). Similar results were obtained with wild-type and mutant versions of S. chlorophenolicum strain RA-2 (data not shown). If PcpD is a PCP hydroxylase reductase, it should be required for removal of PCP at all levels. Thus, these results are inconsistent with the hypothesis that PcpD is a PCP hydroxylase, unless an alternative reductant has been recruited to replace the function of the missing PcpD.
FIG. 5.
PCP disappearance and cell growth in cultures of wild-type and pcpD-knockout versions of S. chlorophenolicum strain ATCC 39723. (a) Initial concentration of PCP, 100 μM. [PCP] in wild-type culture (▪) and pcpD-knockout culture (□); OD550 in wild-type culture (•) and pcpD-knockout culture (○). (b) Initial concentration of PCP, 300 μM. [PCP] in wild-type culture (▪) and pcpD-knockout culture (□); OD550 in wild-type culture (•) and pcpD-knockout culture (○).
Three possible explanations could be advanced to explain these results. First, PcpD may be involved in a step downstream of the initial hydroxylation of PCP, and lack of functional PcpD may prevent carbon from flowing through the degradation pathway. Furthermore, absence of PcpD may result in accumulation of a toxic metabolite that overcomes the cells at high levels of PCP, resulting in decreased growth and PCP removal at high PCP concentrations. Second, PcpD might be involved in the bacterial response to the toxicity of PCP or one of its metabolites and may be essential for viability at high levels of PCP. Finally, disruption of pcpD might have altered levels of PcpR, the regulatory protein that controls expression of Pcp hydroxylase (4).
Lack of functional PcpD does not affect the ability of S. chlorophenolicum strain ATCC 39723 to remove TCP from the medium.
Important insights into the role of PcpD were obtained from experiments in which the abilities of wild-type and pcpD-knockout versions of strain ATCC 39723 to remove TCP from the medium were examined. Figure 6 shows that the rates of disappearance of TCP from the medium were identical for both strains. This result rules out the possibility that disruption of pcpD has altered levels of PcpR, since PCP hydroxylase must be present at comparable levels in the wild-type and knockout strains. Furthermore, since the toxic effects of PCP and TCP should be similar, these results suggest that PcpD is not involved in protection against toxicity. Rather, these results suggest that PcpD is required for a step that occurs during degradation of PCP, but not of TCP.
FIG. 6.
TCP disappearance and cell growth in cultures of wild-type and pcpD-knockout versions of S. chlorophenolicum strain ATCC 39723. Initial concentration of TCP, 300 μM. [TCP] in wild-type culture (▪) and pcpD-knockout culture (□); OD550 in wild-type culture (•) and pcpD-knockout culture (○).
Suggestion for the role of PcpD.
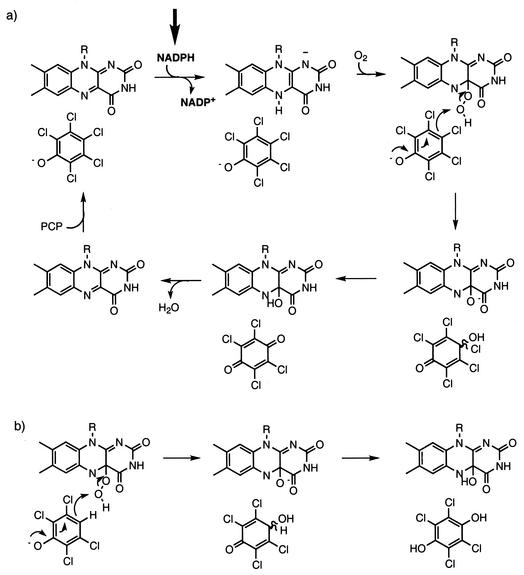
Since PCP and TCP differ only in the nature of the substituent at the 4 position, our observations suggest that PcpD is required when the hydroxylation reaction catalyzed by PCP hydroxylase involves replacement of a chlorine substituent with a hydroxyl group. Consideration of the reaction catalyzed by PCP hydroxylase provides a novel suggestion for the role of PcpD. Detailed mechanistic studies for phenol and p-hydroxybenzoate hydroxylases (11, 12, 16) have been carried out. A model for the reaction catalyzed by PCP hydroxylase based upon this extensive body of literature is shown in Fig. 7a. The product of this reaction should be TCBQ, which is formed by gem elimination of HCl from the initial hydroxylation product. TCBQ must undergo two-electron reduction to generate TCHQ, which is the substrate for TCHQ dehalogenase. We propose that PcpD is a TCBQ reductase that catalyzes reduction of TCBQ to TCHQ. This proposal is consistent with the observation that PcpD is not required for degradation of TCP. Based upon the known chemistry of phenol hydroxylases, PCP hydroxylase would be expected to convert TCP directly to TCHQ (Fig. 7b). Thus, a quinone reductase would not be required for degradation of TCP. If, however, PcpD were indeed a PCP hydroxylase reductase as previously suggested, the knockout strain should be unable to degrade either PCP or TCP, as the initial hydroxylation step is required in both cases.
FIG. 7.
(a) Predicted mechanism for hydroxylation of PCP by PCP hydroxylase based upon those of phenol hydroxylase and p-hydroxybenzoate hydroxylase. The arrow indicates the step in which NADPH is expected to directly reduce flavin at the active site of the enzyme. (b) Part of an analogous mechanism for hydroxylation of TCP showing that the product is TCHQ.
The product formed by PCP hydroxylase is TCBQ, not TCHQ.
The mechanism shown in Fig. 7a suggests that the product formed from PCP by PCP hydroxylase should be TCBQ. Contrary to this expectation, PCP hydroxylase has been reported to form TCHQ from PCP (30, 31). We reinvestigated the nature of the product formed from PCP by PCP hydroxylase. We used GC/MS to identify the product formed from PCP after reaction with PCP hydroxylase and NADPH for 5 min. The reaction mixture was injected onto a reverse-phase HPLC column, and the product peak was collected, extracted into ethyl acetate, and analyzed by GC/MS. (This procedure was necessary because we found that TCBQ could not be extracted directly from the reaction mixture, probably because it associates tightly with the denatured protein.) The mass spectrum of the peak eluting from the GC at 14.2 min (the retention time for TCBQ) was identical to that of authentic TCBQ (data not shown). A small amount of TCHQ was also detectable in the sample. These results could be explained in two ways. First, if TCBQ is the initial product, some TCHQ could be formed during the incubation period by nonenzymatic reduction by NADPH. Alternatively, if TCHQ is the initial product, some TCBQ might be formed during the incubation period by oxidation. To address these possibilities, we incubated TCBQ and TCHQ under comparable reaction conditions for 5 min and analyzed the products in the same way. GC/MS analysis indicated that some nonenzymatic reduction of TCBQ occurred during the incubation period. However, no oxidation of TCHQ occurred during the incubation period. These results support the conclusion that TCBQ is the initial product formed from PCP by PCP hydroxylase and the conclusion that TCHQ detected in the reaction mixture is formed by nonenzymatic reduction of TCBQ by NADPH.
In order to further confirm that TCBQ was the product formed from PCP by PCP hydroxylase, we quantified the amount of TCBQ formed from PCP using a large amount of enzyme so that all of the substrate could be converted in a short period of time, thereby minimizing the extent of nonenzymatic reactions. Since TCBQ and TCHQ coeluted under a variety of HPLC conditions, we devised a procedure in which TCBQ was converted to an adduct that eluted from the HPLC column at a retention time different from that of TCHQ. Treatment of TCBQ with glutathione results in rapid conversion to GS-TriCBQ, which can be quantified by HPLC. Thus, we incubated His6-tagged PCP hydroxylase with 100 μM PCP and 1 mM NADPH for 1 min at 37°C and then added glutathione to trap TCBQ as GS-TriCBQ. This analysis showed that all of the PCP had been converted to product and 95 μM GS-TriCBQ was formed (data not shown). No TCHQ was detected after the short 1-min reaction time. These data provide conclusive evidence that PCP is converted directly to TCBQ by PCP hydroxylase.
The evidence previously presented that PCP hydroxylase produces TCHQ from PCP must now be reexamined. In the initial report of the purification of PCP hydroxylase (30), the product identification was based upon the coelution of the product and authentic TCHQ on HPLC. We have found that TCHQ and TCBQ are very difficult to separate by HPLC. Furthermore, retention time is never sufficient to unambiguously assign structure. In later experiments (31), GC/MS was performed to determine whether the oxygen atom added to PCP derived from H2O or O2. In these experiments, reaction mixtures were incubated for 30 min and then extracted with ether prior to GC/MS analysis. While this is generally an adequate procedure for recovering aromatic compounds from aqueous solutions, we have found that TCBQ cannot be recovered under these conditions. We suspect that the TCHQ recovered in these experiments resulted from some nonenzymatic reduction of TCBQ over the 30-min incubation time.
PcpD catalyzes the formation of TCHQ from TCBQ.
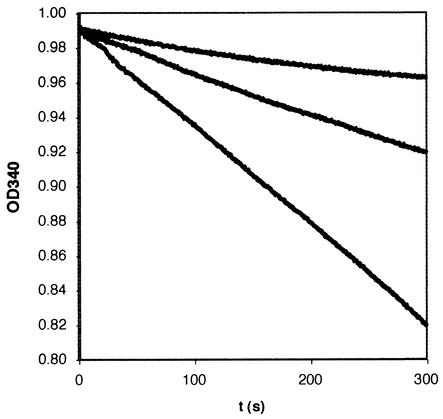
In order to assess the ability of PcpD to catalyze the reduction of TCBQ to TCHQ, we cloned and expressed the protein in P. aeruginosa PAO1-Lac. Only low levels of protein were obtained, because most of the protein was found in inclusion bodies. In order to produce enough protein for initial characterization, we created a construct bearing a His12-affinity tag at the C terminus of the protein. The tagged protein was purified to greater than 95% homogeneity by Ni2+-NTA affinity chromatography. Figure 8 shows that addition of PcpD to a reaction mixture containing 50 μM TCBQ and 1 mM NADPH substantially accelerates the reduction reaction in a concentration-dependent manner. GC/MS analysis confirmed that the product of the reaction was indeed TCHQ (data not shown). The specific activity of the protein under these reaction conditions was 1.2 μmol/min/mg, corresponding to a turnover of 44 substrate molecules per enzyme per min. (This number does not necessarily correspond to kcat since the concentrations of TCBQ and NADPH may not be saturating.)
FIG. 8.
UV-visible assays of the reduction of 50 μM TCBQ by 1 mM NADPH at 25°C in 50 mM potassium phosphate, pH 8.0, in the absence and presence of PcpD monitored by observing the change in absorbance at 340 nm. The upper trace shows the rate of the nonenzymatic reaction; the middle and lower traces show the rate in the presence of His12-tagged PcpD at 37 and 74 μg/ml.
TCBQ inactivates PCP hydroxylase.
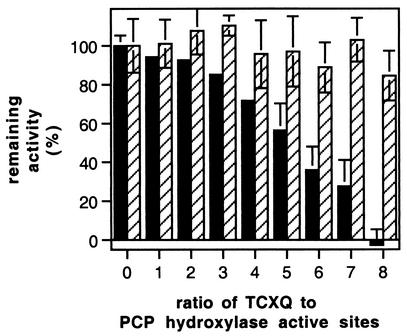
Since PcpD is involved in a step downstream of PCP hydroxylase, it remains for us to explain the observation that the pcpD-knockout strain can remove PCP from the medium when it is present at low levels but not when it is present at high levels. TCBQ is a highly reactive electrophile and can react with a variety of cellular nucleophiles (14, 26, 28). We have previously reported that TCBQ is severely toxic to E. coli spheroplasts at concentrations lower than 1 μM (17). We incubated TCBQ with PCP hydroxylase for 30 min and found that TCBQ inactivates the enzyme (Fig. 9), presumably by reacting with nucleophilic groups in the vicinity of the active site. Incubation with TCHQ (under anoxic conditions to prevent oxidation of TCHQ to TCBQ) resulted in no loss of activity. The inactivation of PCP hydroxylase by TCBQ is likely to explain the observation that the pcpD-knockout strain of ATCC 39723 can remove PCP from the medium and grow at 100 μM PCP, but not at 300 μM PCP. At higher PCP concentrations, the accumulation of toxic TCBQ would be expected to decrease the ability of cells to remove PCP from the medium because of the inactivation of PCP hydroxylase and, furthermore, would compromise cell viability.
FIG. 9.
Inactivation of PCP hydroxylase after incubation with TCBQ but not TCHQ for 30 min under anoxic conditions. Each bar indicates the average of three independent determinations of activity. Solid bars, TCBQ; hatched bars, TCHQ; error bars, standard deviation.
Conclusions.
PcpD is expressed in S. chlorophenolicum in response to PCP, and the mRNA is cotranscribed with that for PcpB. We have shown that PcpD is involved in degradation of PCP, but not in degradation of TCP. Based upon this finding, an understanding of the mechanism of the PCP monooxygenase reaction, and the sequence of PcpD, we suggested and verified experimentally that PcpD is a TCBQ reductase. Thus, we have identified a previously unrecognized step in the PCP degradation pathway and have corrected the misassignment of the function of PcpD. Efforts to express PcpD at levels sufficient for further biochemical and kinetic characterization are in progress.
Acknowledgments
This work was supported by the U.S. Army Research Office under grant DAAD19-99-1-0301.
Protein sequence analyses were performed at the Pittsburgh Supercomputing Center.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anandarajah, K., P. M. Kiefer, and S. D. Copley. 2000. Recruitment of a double bond isomerase to serve as a reductive dehalogenase during biodegradation of pentachlorophenol. Biochemistry 39:5303-5311. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 4.Batie, C., D. P. Ballou, and C. C. Corell. 1991. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases, p. 543-556. In F. Müller (ed.), Chemistry and biochemistry of flavoenzymes, vol. 3. CRC Press, Boca Raton, Fla.
- 5.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassidy, M. B., H. Lee, J. T. Trevors, and R. B. Zablotowicz. 1999. Chlorophenol and nitrophenol metabolism by Sphingomonas sp. UG30. J. Ind. Microbiol. Biotechnol. 23:232-241. [DOI] [PubMed] [Google Scholar]
- 7.Cline, R. E., R. H. Hill, D. L. Phillips, and L. L. Needham. 1989. Pentachlorophenol measurements in body fluids of people in log homes and workplaces. Arch. Environ. Contam. Toxicol. 18:475-481. [DOI] [PubMed] [Google Scholar]
- 8.Copley, S. D. 2000. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem. Sci. 25:261-265. [DOI] [PubMed] [Google Scholar]
- 9.Correll, C. C., C. J. Batie, D. P. Ballou, and M. L. Ludwig. 1992. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleoptides to [2Fe-2S]. Science 258:1604-1610. [DOI] [PubMed] [Google Scholar]
- 10.Cronin, C. N., and W. S. McIntire. 1999. pUCP-Nco and pUCP-Nde: Escherichia-Pseudomonas shuttle vectors for recombinant protein expression in Pseudomonas. Anal. Biochem. 272:112-115. [DOI] [PubMed] [Google Scholar]
- 11.Detmer, K., and V. Massey. 1984. Effect of monovalent anions on the mechanism of phenol hydroxylase. J. Biol. Chem. 259:11265-11272. [PubMed] [Google Scholar]
- 12.Entsch, B., and W. J. H. Van Berkel. 1995. Structure and mechanism of para-hydroxybenzoate hydroxylase. FASEB J. 9:476-483. [DOI] [PubMed] [Google Scholar]
- 13.Horton, R. M. 1993. In vitro recombination and mutagenesis of DNA. In B. A. White (ed.), PCR protocols: current methods and applications. Humana Press, Totowa, N.J.
- 14.Lin, P.-H., J. Nakamura, S. Yamaguchi, P. B. Upton, D. K. La, and J. A. Swenberg. 2001. Oxidative damage and direct adducts in calf thymus DNA induced by the pentachlorophenol metabolites, tetrachlorohydroquinone and tetrachloro-1,4-benzoquinone. Carcinogenesis 22:627-634. [DOI] [PubMed] [Google Scholar]
- 15.Locher, H. H., T. Leisinger, and A. M. Cook. 1991. 4-Toluene sulfonate methyl-monooxygenase from Comamonas testosteroni T-2: purification and some properties of the oxygenase component. J. Bacteriol. 173:3741-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda-Yorita, K., and V. Massey. 1993. On the reaction mechanism of phenol hydroxylase: new information obtained by correlation of fluorescence and absorbance stopped flow studies. J. Biol. Chem. 268:4134-4144. [PubMed] [Google Scholar]
- 17.McCarthy, D. L., A. Claude, and S. D. Copley. 1997. In vivo levels of chlorinated hydroquinones in a pentachlorophenol-degrading bacterium. Appl. Environ. Microbiol. 63:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakatsu, C. H., N. A. Straus, and R. C. Wyndham. 1995. The nucleotide sequence of the TN5271 3-chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology 141:485-495. [DOI] [PubMed] [Google Scholar]
- 19.Orser, C. S., and C. C. Lange. 1994. Molecular analysis of pentachlorophenol degradation. Biodegradation 5:277-288. [DOI] [PubMed] [Google Scholar]
- 20.Orser, C. S., C. C. Lange, L. Xun, T. C. Zahrt, and B. J. Schneider. 1993. Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orser, C. S., J. Dutton, C. Lange, P. Jablonski, L. Xun, and M. Hargis. 1993. Characterization of a Flavobacterium glutathione-S-transferase gene involved in reductive dehalogenation. J. Bacteriol. 175:2640-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priefert, H., J. Rabenhorst, and A. Steinbüchel. 1997. Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate. J. Bacteriol. 179:2595-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radehaus, P. M., and S. K. Schmidt. 1992. Characterization of a novel Pseudomonas sp. that mineralizes high concentrations of pentachlorophenol. Appl. Environ. Microbiol. 58:2879-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi, M., K. Hamana, and H. Akira. 2001. Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analyses. Int. J. Syst. E vol. Microbiol. 51:1405-1417. [DOI] [PubMed] [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Ommen, B., A. E. P. Adang, L. Brader, M. A. Posthumus, F. Mueller, and P. J. van Bladeren. 1986. The microsomal metabolism of hexachlorobenzene. Biochem. Pharmacol. 35:3233-3238. [DOI] [PubMed] [Google Scholar]
- 27.van Ommen, B., C. Den Besten, A. L. M. Rutten, J. H. T. M. Ploemen, R. M. E. Vos, F. Müller, and P. J. Van Bladeren. 1988. Active site-directed irreversible inhibition of glutathione-S-transferases by the glutathione conjugate of tetrachloro-1,4-benzoquinone. J. Biol. Chem. 263:12939-12942. [PubMed] [Google Scholar]
- 28.van Ommen, B., J. W. Voncken, F. Muller, and P. J. van Bladeren. 1988. The oxidation of tetrachloro-1,4-hydroquinone by microsomes and purified cytochrome P-450b. Implications for covalent binding to protein and involvement of reactive oxygen species. Chem.-Biol. Interact. 65:247-259. [DOI] [PubMed] [Google Scholar]
- 29.Xun, L., and C. S. Orser. 1991. Purification of a Flavobacterium pentachlorophenol-induced periplasmic protein (PcpA) and nucleotide sequence of the corresponding gene (pcpA). J. Bacteriol. 173:2920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xun, L., and C. S. Orser. 1991. Purification and properties of pentachlorophenol hydroxylase, a flavoprotein from Flavobacterium sp. strain ATCC 39723. J. Bacteriol. 173:4447-4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xun, L., E. Topp, and C. S. Orser. 1992. Confirmation of oxidative dehalogenation of pentachlorophenol by a Flavobacterium pentachlorophenol hydroxylase. J. Bacteriol. 174:5745-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]