Abstract
In Bacillus subtilis, the alternative sigma factor σB is activated in response to environmental stress or energy depletion. The general stress regulon under the control of σB provides the cell with multiple stress resistance. Experiments were designed to determine how activated σB replaces σA as a constituent of the RNA polymerase holoenzyme. Studies of the transcription of the σA-dependent stress gene clpE under σB-inducing conditions showed that expression was higher in a sigB mutant background than in the wild type. The relative affinities of σA and σB for binding to the core RNA polymerase (E) were determined by means of indirect surface plasmon resonance. The results showed that the affinity of σB for E was 60-fold lower than that of σA. Western blot analyses with antibodies against σA, σB, and E showed that, after exposure to ethanol stress, the concentration of σB was only twofold higher than those of σA and E. Thus, the concentration of σB after stress is not high enough to compensate for its relatively low affinity for E, and it seems that additional mechanisms must be invoked to account for the binding of σB to E after stress.
Bacillus subtilis has 17 different σ factors, which are synthesized and activated at various times during development or after changes in environmental conditions. The active σ factors bind to core RNA polymerase (E) to recognize specific promoter sequences and thus to catalyze gene expression that is appropriate to the conditions.
If several σ factors are active at the same time, what mechanisms determine which of them binds to the core RNA polymerase? In particular, do they compete with one another for binding, or is core RNA polymerase present in excess, with the result that they can all be accommodated? By investigating the composition of the holoenzyme during sporulation in Bacillus subtilis, Fujita concluded that core RNA polymerase is indeed in excess in the cell, so that successive σ factors do not need to displace each other from the holoenzyme (14). However, this conclusion, which was based on the finding that there is twofold more E than σA in the cell, is open to question, as two-thirds of the molecules of E are known to be involved in transcription elongation (9) and are therefore not in a state in which they can bind any σ factor. Furthermore, other measurements of the intracellular concentration of E and σA have suggested that the two proteins are present at approximately the same molar concentration in sporulating cells (26, 36). In addition, expression studies have suggested that σA and σH in B. subtilis compete for binding to the core RNA polymerase, as do σ70 and σS in Escherichia coli, since in both systems overexpression of one σ factor leads to a decrease in the gene expression that is dependent on the other σ factor (13, 18).
In this study, we wanted to understand how the general stress σ factor of B. subtilis, σB, replaces σA in the holoenzyme. EσB transcribes genes whose products provide the cell with nonspecific, general, and multiple stress resistance. It is known to be activated after energy depletion or after a variety of environmental stresses such as heat, ethanol, acid, and osmotic and oxidative stress through cascades of PP2C phosphatases. σB is held inactive by its anti-σ factor RsbW as long as the anti-anti-σ factor RsbV is phosphorylated. After environmental stress or energy depletion, RsbV is dephosphorylated by the PP2C phosphatases RsbU and RsbP, and the resulting RsbV binds to RsbW, which thereupon liberates σB (for recent reviews, see references 16 and 33).
Although this mechanism of activating σB is relatively well understood, what is not known is how the activated σB competes successfully with σA for binding to E. In the present study, we examined whether the genetic loss of σB affects the expression of σA-dependent general stress genes under conditions that would normally induce σB, determined the relative affinities of the two σ factors for E, and measured the intracellular concentrations of E and of the two σ factors before and during a period of ethanol stress.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used were B. subtilis 168 (trpC2) (2) and ML6 (trpC2 sigB::ΔHindIII-EcoRV::cat) (20). B. subtilis strains were grown at 37°C to an optical density at 500 nm (OD500) of 0.4 in synthetic medium as described previously (3) or to an OD600 of 0.4 in Luria-Bertani (LB) medium and exposed to heat shock (50°C) or to 4% (vol/vol) ethanol for 10, 20, or 30 min. The strains used for purification of σA and σB were E. coli BL21(DE3)/pLys/pRSETA[sigA] and BL21(DE3)/pLys/pRSETA[sigB] (see below). E. coli strains were grown at 37°C in LB medium.
DNA techniques.
Plasmid isolation, restriction enzyme analysis, transformation of E. coli, and ligation of DNA fragments were performed by standard methods (35). Chromosomal DNA from B. subtilis was isolated as described by Meade et al. (30). Transformation of naturally competent B. subtilis cells was carried out as described by Hoch (19). The sigA and sigB genes of B. subtilis were amplified from chromosomal DNA of B. subtilis 168 with the primers sigAfor (5′-GGAGGATCCATGGCTGATAAACAAACCCA-3′), sigArev (5′-CGGGGTACCTTATTCAAGGAAATCTTTCA-3′), sigBfor (5′-GGAGGATCCTTGATCATGACACAACCATC-3′), and sigBrev (5′-CGGGGTACCTTACATTAACTCCATCGAGG-3′), containing cleavage sites for BamHI (forward primer) and KpnI (reverse primer). The amplified fragments were cleaved with KpnI and BamHI and cloned into plasmid pRSETA (Invitrogen), resulting in pRSETA[sigA] and pRSETA[sigB], respectively.
Purification of σA and σB.
The plasmids pRSETA[sigA] and pRSETA[sigB] were transformed into E. coli BL21(DE3)/pLysS, generating E. coli BL21(DE3)/pLys/pRSETA[sigA] and BL21(DE3)/pLys/pRSETA[sigB], respectively. Freshly transformed cells were grown at 37°C in 2YT medium containing 1% (wt/vol) glucose. At an OD600 of 0.5, 1 mM isopropylthiogalactopyranoside (IPTG) was added for 2 h, and the cells were harvested, resuspended in native lysis buffer (0.05 M Na2HPO4/NaH2PO4 [pH 8.0], 0.3 M NaCl, 0.01 M imidazole) and disrupted by sonication. In the first step, His6-σA and His6-σB were purified by Ni-nitrilotriacetic acid (NTA) affinity chromatography under native conditions (wash buffer: 0.05 M Na2HPO4/NaH2PO4 [pH 8.0], 0.3 M NaCl, 0.02 M imidazole; elution buffer: 0.05 M Na2HPO4/NaH2PO4 [pH 8.0], 0.3 M NaCl). His6-σA was further purified by gel filtration on Superdex-75 with buffer G (50 mM Tris-Cl [pH 7.5], 0.2 M NaCl, 0.5 M EDTA, 1 mM dithiothreitol [DTT]).
His6-σB-containing fractions were loaded onto a DEAE-Sepharose column, and a linear 0 to 600 mM gradient of NaCl in buffer A (20 mM Tris-Cl [pH 8.0], 1 mM DTT) was applied to the column. The fractions containing His6-σB were then loaded onto Superdex-75. Pure fractions of His6-σA and His6-σB were pooled and dialyzed against storage buffer (50 mM Tris-Cl [pH 7.5], 50% [vol/vol] glycerol, 1 mM EDTA, 1 mM DTT, 50 mM NaCl). The purity of His6-σA and His6-σB was >98%, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of E.
Core RNA polymerase (E) was purified from B. subtilis 168 by modifications of previously published protocols (5, 10, 15, 26). Cell pellets from a 4-liter culture were resuspended in 80 ml of lysis buffer (50 mM Tris-Cl [pH 8.0], 10 mM MgCl2, 2 mM EDTA, 0.1 mM DTT, 1 mM β-mercaptoethanol, 233 mM NaCl, 10% [vol/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride) and sonicated. After Polymin P fractionation and ammonium sulfate precipitation (65% saturation), the precipitate was resuspended and dialyzed against TED (10 mM Tris-Cl [pH 8.0], 0.1 mM EDTA, 0.1 mM DTT) containing 0.05 M NaCl and then subjected to DNA-agarose affinity chromatography. Elution of E was accomplished with 0.4 M NaCl in TED.
E-containing fractions were pooled and precipitated again with ammonium sulfate (65% saturation). The precipitate was resuspended in TGED (10 mM Tris-Cl [pH 8.0], 10% [vol/vol] glycerol, 0.1 mM EDTA, 0.1 mM DTT) containing 0.5 M NaCl and loaded onto a Superdex-200 column. E-containing fractions were dialyzed against TGED with 0.24 M NaCl and applied to a MonoQ HR 5/5 column. E was eluted with a linear gradient of 0.24 to 0.56 M NaCl in TGED and dialyzed into storage buffer (see above). Protein purity was greater than 95%, as judged by SDS-PAGE, and the level of residual σA was <0.5%. No σB was detectable.
Immobilization of σA to sensor chip surface.
Purified His6-σA at a concentration of 0.3 mg/ml was dialyzed extensively against phosphate-buffered saline (pH 7.4) containing 1 mM DTT and immobilized on the dextran surface of one flow cell of sensor chip CM5 by the amine coupling method as described previously (26).
Measurement of free E after incubation with His6-σA or His6-σB.
Immobilized His6-σA was used as a sensor to determine the concentration of free E after incubation of 100 or 50 nM E with different amounts of His6-σA and His6-σB. Surface plasmon resonance (SPR) measurements with the Biacore were performed as described previously (26).
Competition experiments.
E at a concentration of 0.1 μM was incubated with either 0.1 μM His6-σA or 4 μM His6-σB for 10 min at room temperature, and His6-σB and His6-σA were added to final concentrations ranging from 0 to 48 μM and 0.03 to 1.2 μM, respectively. After incubation for 10 min, the mixtures were separated on 8% native polyacrylamide gels, the proteins were transferred to a nitrocellulose membrane, and His6-σA was detected by Western blotting with anti-σA polyclonal antibody and peroxidase-conjugated anti-rabbit immunoglobulin antibodies (Sigma). The detection of peroxidase was carried out with the ECL Western blotting detection reagent (Amersham Pharmacia).
Transcription analysis.
Total RNA of B. subtilis was isolated from cells before and after exposure to stress by the acid phenol method of Majumdar et al. (28). Northern blot analyses were performed as described previously (38). Briefly, an internal fragment of the clpE gene was amplified with the clpE-specific primers CLPEfor (5′-TTCCGTTCATAAACAGATGG-3′) and CLPErev (5′-CTAATACGACTCACTATAGGGAGAATAGCCTGTTCAATTGAAGG-3′). (Note that the 3′ primer for the amplified gene contains a T7 promoter sequence.) This amplified fragment was used for the T7 RNA polymerase-directed synthesis of the digoxigenin-labeled clpE-specific RNA probe. Slot blot analyses were performed with decreasing amounts of total RNA as described by Maul et al. (29). The luminograms were quantified with the Lumi-Imager (Boehringer Mannheim), with the level of the control transcript set to 1.
Western blot experiments.
B. subtilis cells grown in LB medium to an OD600 of 0.4 were harvested by centrifugation before or after 10, 20, or 30 min of exposure to 4% (vol/vol) ethanol. The resulting cell pellets were washed twice in TE buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride), resuspended in TE buffer, and disrupted by sonication. Cell debris was removed by centrifugation, and the protein concentration of the soluble cell fraction was determined by the method of Bradford (8). Dilutions of purified standard proteins (His6-σA, His6-σB, and E) and of cell extracts were separated on an SDS-10% to 12% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. σA, σB, and E were visualized with specific polyclonal antibodies and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin antibodies (Sigma). Alkaline phosphatase was detected with the substrate CDP-Star (Tropix) and the Lumi-Imager system (Boehringer Mannheim). The quantities of σA, σB, and E in the different protein extracts were determined with the help of the LumiAnalyst 3.0 software (Boehringer Mannheim). For E, the combined densities of the β and β′ subunits were used for the quantitation. Results are expressed as femtomoles of sigma or E per microgram of whole cell protein.
RESULTS
clpE induction by heat stress and ethanol stress in wild-type B. subtilis and sigB mutant.
In glucose-starved or stressed cells of B. subtilis, a significant portion of the RNA polymerase is probably engaged in σB-dependent transcription, because more than 150 σB-dependent genes are strongly induced (31, 34). Transcriptional studies of stress- and starvation-inducible genes in the wild type and a sigB mutant have suggested that in these circumstances, σB competes with other sigma factors (mainly σA) (4, 32). In a sigB mutant, one would expect the proportion of core enzyme engaged in the transcription of σB-dependent genes to decrease, leading to overexpression of σB-independent stress genes. We have now investigated this possibility for clpE, which encodes a heat stress-inducible chaperone/ATPase controlled by the global repressor CtsR at a σA-dependent promoter (11).
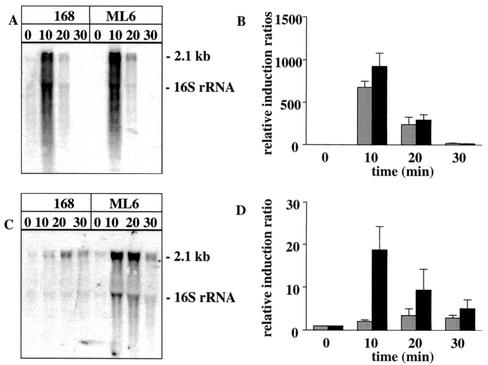
RNA preparations from heat-stressed and ethanol-stressed cells were used to analyze the transcription of clpE in wild-type cells and in a sigB mutant. Northern blot and slot blot results showed that treatment of the cells for 10 min at 50°C greatly increased the transcription of clpE. Transcription was slightly greater in the sigB mutant than in the wild type (Fig. 1A and B). Ethanol stress led to a weaker induction than heat stress, but clpE induction in the presence of ethanol was about 10-fold greater in the sigB mutant than in the wild type (Fig. 1C and D). (We note that Petersohn et al. [31] have shown that ethanol stress is much more effective than heat stress in σB-dependent genes.) These results, which suggest (but do not prove) that there may be competition between σA and σB for the core enzyme (at least under ethanol stress), encouraged us to analyze the affinity of both sigma factors for the core enzyme.
FIG. 1.
Transcript analysis of clpE-specific mRNA in wild-type B. subtilis (168) and the sigB mutant (ML6) after heat shock (A and B) and ethanol stress (C and D). Total RNA was isolated from cells grown in a synthetic medium before (0 min) and at different times (10, 20, and 30 min) after the exposure to stress. For the Northern blotting experiments, samples of 2 μg (A) or 10 μg (C) of total RNA were applied. The relative induction ratios of clpE-specific mRNA in the wild type (light grey bars) and sigB mutant (dark grey bars) were determined in slot blot experiments, where the level of the clpE mRNA in the control was set to 1 (B and D). The error bars indicate the standard error of the mean.
Comparison of relative affinities of σA and σB for core RNA polymerase.
We measured the affinities of σA and σB for core RNA polymerase (E) by SPR by the backtitration method described by Lord et al. (26). This method determines the concentration of each sigma factor that is needed to diminish a standard concentration of free E by 50%. We used σA immobilized on the sensor chip to report the concentration of free E in a mixture of E and a sigma factor by noting the response in terms of resonance units (RU) when each mixture was passed over the chip.
We first made a standard curve that related the RU to the concentration of E (not shown). We then prepared a series of mixtures, each containing a known concentration of E but with different concentrations of σA or σB. In each mixture, the sigma factor interacts with E to form holoenzyme, but a certain concentration of E remains free; we were able to measure this concentration of free E by passing the mixture over the immobilized σA and comparing the resultant reading with the standard curve.
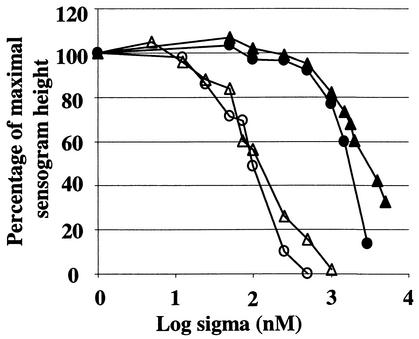
When 100 nM E was incubated with increasing concentrations of σA, the sensor chip reported (as expected) that the concentration of free E in the mixture gradually diminished. The concentration of σA needed to reduce the concentration of free E to 50 nM was 70 nM (mean of three experiments, with a range of 58 to 89 nM; open circles in Fig. 2). When this experiment was repeated with a starting concentration of 50 nM E, the concentration of σA needed to reduce this to 25 nM was found to be 50 nM (open triangles in Fig. 2).
FIG. 2.
Sequestration of free E by His6-σA and His6-σB. Immobilized His6-σA was used as a sensor in SPR experiments to determine the concentration of free E after incubation of a constant amount of E with different amounts of His6-σA or His6-σB. The percentage of maximal sensorgram height was plotted against the log10 σ factor concentration. Symbols indicate binding of σA to 100 nM E (○) or 50 nM E (▵) and binding of σB to 100 nM E (•) and 50 nM E (▴).
Much higher concentrations of σB were needed to reduce the concentration of free E by 50%. With a starting concentration of 100 nM E, 1,330 nM σB was required to reduce the free E concentration to 50 nM (mean of three experiments, with a range of 1,202 to 1,413 nM; solid circles in Fig. 2), and with a starting concentration of 50 nM E, 1,320 nM σB was required to reduce the free E concentration to 25 nM (solid triangles in Fig. 2).
We can use these results to calculate the relative affinity of E for the two sigma factors. For σA, the concentration of free sigma factor in solution when E was half-saturated was 22.5 nM (the mean of 20 and 25 nM). This figure is identical to that reported by Lord et al. (26). For σB, the equivalent figure was 1,287.5 nM (the mean of 1,280 and 1,295 nM). Thus, we conclude that the affinity of E for σA is approximately 60 times greater than that for σB.
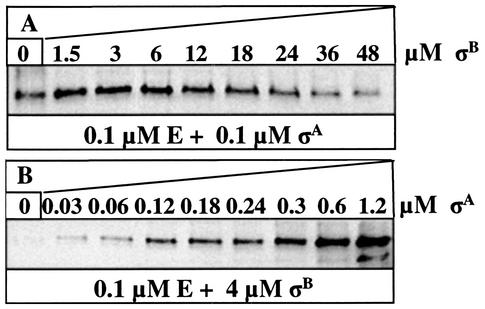
We confirmed this conclusion by performing a series of experiments in which we attempted to displace σA from a σA-containing holoenzyme by incubating it with σB and vice versa. In the first experiment, we incubated E and σA (0.1 μM each) together for 10 min at room temperature and then added σB at final concentrations ranging from 0 to 48 μM. After continuing the incubation for 10 min, we separated the mixtures on native polyacrylamide gels, transferred the proteins to a nitrocellulose membrane, and detected σA by Western blotting with specific anti-σA polyclonal antibody. Figure 3A shows that the quantity of σA in the holoenzyme was not reduced until the concentration of σB exceeded 6 μM. In the reciprocal experiment, when 0.1 μM E was incubated with 4 μM σB, a concentration of σA as low as 0.06 μM was sufficient to yield a detectable quantity of E-σA holoenzyme (Fig. 3B). We were unable to confirm whether all of the σB was active in binding to core RNA polymerase, but to exclude the possibility that some of the σB was degraded, we incubated it with its specific anti-sigma factor RsbW and found that all of it was able to form a complex with RsbW (results not shown).
FIG. 3.
Amount of σA bound to E after competition with σB and vice versa. Mixtures of 0.1 μM His6-σA and 0.1 μM E (A) or 4 μM His6-σB and 0.1 μM E (B) were incubated before the addition of increasing amounts of His6-σB or His6-σA, respectively. Proteins were analyzed by native PAGE, Western blotting, and immunodetection with His6-σA-specific polyclonal antibodies. The amount of σA in the holoenzyme is shown.
Taking all these experiments together, we conclude that the affinity of E for σB is about 60-fold lower than that for σA.
Intracellular concentrations of σA, σB, and E.
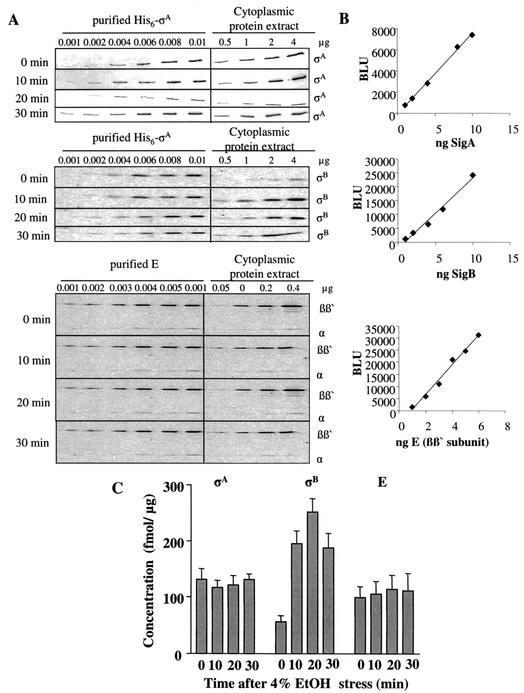
The results just described have shown that the replacement of σA by σB under conditions of environmental stress cannot be explained on the basis that the latter sigma factor has a higher affinity for E. Another possible explanation is that environmental stress leads to an increase in the concentration of σB, with the result that mass action drives some of the core enzyme to form σB-containing holoenzyme. We therefore subjected cultures of wild-type cells to stress by growing them in LB medium and exposing them to 4% (vol/vol) ethanol. Samples were taken at intervals both before and after the addition of the ethanol, and the intracellular concentrations of E, σA, and σB were measured by Western blotting.
The results (Fig. 4) show that the intracellular concentrations of E and σA remained roughly constant throughout the experiments, whereas the concentration of σB increased fivefold within 20 min of exposure to ethanol and then fell rapidly. Figure 4 also shows that the concentrations of E and σA were roughly equal (around 100 to 120 fmol/μg) both before and during the period of stress. The concentration of σB, by contrast, was only about 56 fmol/μg before ethanol was added to the culture, but rose to a maximum of about 250 fmol/μg after 20 min of exposure to ethanol, so that the concentration of σB at its maximum was twofold higher than that of σA. The results were not significantly different when this experiment was repeated with cells grown in minimal medium (results not shown).
FIG. 4.
Determination of intracellular concentrations of σA, σB, and E in wild-type B. subtilis during ethanol stress. Extracts were prepared from cells before and after exposure to ethanol (EtOH) stress. Dilutions of purified standard proteins (His6-σA, His6-σB, and E) and of cell extracts were used for Western blotting and immunodetection with σA-, σB-, and E-specific antibodies (A). Quantification of intracellular σA, σB, and E was performed as described in Materials and Methods. Standard curves were generated by using dilutions of purified standard proteins (B). The intracellular concentrations of σA, σB, and E were then determined (C). The error bars indicate the standard error of the mean. BLU, Boehringer light units.
DISCUSSION
The majority of bacteria apparently contain more than one sigma factor, and in B. subtilis, 18 potential σ factor genes have been predicted from the genome sequence, encoding 17 different σ factors (24). The mechanisms that determine which of these sigma factors interact with the core enzyme at any given moment are still largely unknown, but it is possible that such mechanisms constitute a significant step in the regulation of gene expression.
There are by now several lines of evidence in favor of the view that, in some circumstances, different σ factors compete for a limiting pool of the core enzyme, both in E. coli and in B. subtilis (13, 18, 26, 36). The experiments with a sigB mutant reported here support this view, since they show that σA-dependent transcription is much more strongly induced by stress in the mutant cells than in the wild type (Fig. 1). Petersohn et al. likewise found higher induction in a sigB mutant than in the wild type for many stress-inducible genes (31). Similarly, Antelmann et al. and Pragai and Harwood reported data suggesting that σB and σA compete for core polymerase in phosphate-starved cells (4, 32). The ability of σB to compete successfully with σA for the core enzyme would also explain the fact that stress leads to an extremely rapid expression of general stress genes (17, 31, 34), with the result that up to 20% of the translational capacity quickly becomes devoted to the synthesis of σB-dependent general stress proteins (7).
One obvious possible explanation for the ability of σB to compete with σA would be that the affinity of the former for the core RNA polymerase is higher than that of the latter. However, this possibility has been excluded by our experimental results (Fig. 2 and 3), which showed that the affinity of σB for the core enzyme was 60-fold lower than that of σA. In a similar way, σF was found to have a 25-fold-lower affinity for core RNA polymerase than σA (26), even though σF must be able to compete effectively with σA, given that substantial σF-dependent transcription is induced early in sporulation. Similarly, in competitive transcription assays in vitro, σH and σE (though not σK) bind to E with lower affinity than σA (14). In E. coli, too, the housekeeping σ factor σ70 has a higher affinity than other σ factors for E (27). The low affinity of σB for the core may be one mechanism for ensuring that σB-dependent transcription is negligible in the absence of stress—a mechanism that is reinforced by the presence of the anti-sigma factor RsbW, which binds to σB and prevents it from interacting with the core enzyme (6).
A second possible explanation for the ability of σB to compete with σA would be that, during stress, the concentration of the former in the cells becomes higher than that of the latter. Our results (Fig. 4) suggest that the concentration of σB does indeed increase fivefold during stress and that, at its maximum, its concentration becomes at least twofold higher than that of σA. It is also known that, during stress, RsbW switches partners, binding to RsbV rather than to σB, so that one can expect the bulk of the newly synthesized σB to be free to form σB-containing holoenzyme (1, 37, 39). Nonetheless, given the 60-fold-lower affinity of σB than of σA for core RNA polymerase, one would expect that in these circumstances only some 3% of transcriptional activity would be σB dependent, whereas in fact the true figure is believed to be 10 to 20% (7). This argument suggests that there may be some additional factor(s) controlling the competition between σB and σA.
Two possibilities are differential stabilization of the promoter-holoenzyme complex and a specific anti-σA factor similar to the anti-σ70 factor Rsd reported in E. coli (21, 22). A further possibility is that the δ factor of B. subtilis may play a role in σ factor switching, as suggested by Lopez de Saro et al. (25). Yet another mechanism could involve the stringent response, given that recent experiments in E. coli suggest that the stringent response, mediated through ppGpp, may alter the relative competitiveness of σ factors in accordance with cellular demands during physiological stress (23). There is some evidence that ppGpp acts in a similar way in B. subtilis. After norvaline treatment, which induces amino acid starvation, the induction of strong σB-dependent genes in a spo0H spo0A double mutant requires both the stringent response and RsbP, the metabolic inducer of the general stress response (12). One or more of these mechanisms (or possibly another mechanism still to be discovered) is apparently responsible for altering the ability of σB to compete with σA in the cell.
Acknowledgments
We thank Jörg Mostertz for strain BL21(DE3)/pLys/pRSETA[sigB] and Renate Gloger for technical assistance with the transcription experiments.
C.R., J.K., H.A., and M.H. were supported by grants from the Deutsche Forschungsgemeinschaft (DFG), the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BMFT), European Union grant QLK3-CT-1999-00413, the Fonds der Chemischen Industrie to M.H., and Genencor International (Palo Alto, Calif.). Experiments in M.D.Y.'s laboratory were supported by the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., S. Engelmann, R. Schmid, A. Sorokin, A. Lapidus, and M. Hecker. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor σB in Bacillus subtilis. J. Bacteriol. 179:7251-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antelmann, H., C. Scharf, and M. Hecker. 2000. Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol. 182:4478-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anthony, L. C., I. Artsimovitch, V. Svetlov, R. Landick, and R. R. Burgess. 2000. Rapid purification of His(6)-tagged Bacillus subtilis core RNA polymerase. Protein Expr. Purif. 19:350-354. [DOI] [PubMed] [Google Scholar]
- 6.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernhardt, J., U. Volker, A. Volker, H. Antelmann, R. Schmid, H. Mach, and M. Hecker. 1997. Specific and general stress proteins in Bacillus subtilis — a two-dimensional protein electrophoresis study. Microbiology 143:999-1017. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Bremer, H., and P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell growth rate, p. 1553-1569. In F. Neidhardt, R. Curtiss, J. Ingraham, E. Lin, K. Low, B. Magasanik, W. Reznikoff, M. Riley, M. Schaechter, and H. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 10.Burgess, R. R., and J. J. Jendrisak. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634-4638. [DOI] [PubMed] [Google Scholar]
- 11.Derre, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 12.Eymann, C., and M. Hecker. 2001. Induction of σB-dependent general stress genes by amino acid starvation in a spo0H mutant of Bacillus subtilis. FEMS Microbiol. Lett. 199:221-227. [DOI] [PubMed] [Google Scholar]
- 13.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, M. 2000. Temporal and selective association of multiple sigma factors with RNA polymerase during sporulation in Bacillus subtilis. Genes Cells 5:79-88. [DOI] [PubMed] [Google Scholar]
- 15.Hager, D. A., D. J. Jin, and R. R. Burgess. 1990. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry 29:7890-7894. [DOI] [PubMed] [Google Scholar]
- 16.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 17.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks, K. A., and A. D. Grossman. 1996. Altering the level and regulation of the major sigma subunit of RNA polymerase affects gene expression and development in Bacillus subtilis. Mol. Microbiol. 20:201-212. [DOI] [PubMed] [Google Scholar]
- 19.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204:305-320. [DOI] [PubMed] [Google Scholar]
- 20.Igo, M., M. Lampe, C. Ray, W. Schafer, C. P. Moran, Jr., and R. Losick. 1987. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J. Bacteriol. 169:3464-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jishage, M., and A. Ishihama. 1998. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc. Natl. Acad. Sci. USA 95:4953-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jishage, M., and A. Ishihama. 1999. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J. Bacteriol. 181:3768-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 25.Lopez de Saro, F. J., N. Yoshikawa, and J. D. Helmann. 1999. Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis. J. Biol. Chem. 274:15953-15958. [DOI] [PubMed] [Google Scholar]
- 26.Lord, M., D. Barillà, and M. D. Yudkin. 1999. Replacement of vegetative σA by sporulation-specific σF as a component of the RNA polymerase holoenzyme in sporulating Bacillus subtilis. J. Bacteriol. 181:2346-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majumdar, D., Y. J. Avissar, and J. H. Wyche. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. BioTechniques 11:94-101. [PubMed] [Google Scholar]
- 29.Maul, B., U. Volker, S. Riethdorf, S. Engelmann, and M. Hecker. 1995. Sigma B-dependent regulation of gsiB in response to multiple stimuli in Bacillus subtilis. Mol. Gen. Genet. 248:114-120. [DOI] [PubMed] [Google Scholar]
- 30.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pragai, Z., and C. R. Harwood. 2002. Regulatory interactions between the Pho and sigma(B)-dependent general stress regulons of Bacillus subtilis. Microbiology 148:1593-1602. [DOI] [PubMed] [Google Scholar]
- 33.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 34.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Tjian, R., and R. Losick. 1974. An immunological assay for the sigma subunit of RNA polymerase in extracts of vegetative and sporulating Bacillus subtilis. Proc. Natl. Acad. Sci. USA 71:2872-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 38.Wetzstein, M., U. Volker, J. Dedio, S. Lobau, U. Zuber, M. Schiesswohl, C. Herget, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bacteriol. 174:3300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]