Abstract
The seven rRNA operons in Escherichia coli each contain two promoters, rrn P1 and rrn P2. Most previous studies have focused on the rrn P1 promoters. Here we report a systematic analysis of the activity and regulation of the rrnB P2 promoter in order to define the intrinsic properties of rrn P2 promoters and to understand better their contributions to rRNA synthesis when they are in their natural setting downstream of rrn P1 promoters. In contrast to the conclusions reached in some previous studies, we find that rrnB P2 is regulated: it displays clear responses to amino acid availability (stringent control), rRNA gene dose (feedback control), and changes in growth rate (growth rate-dependent control). Stringent control of rrnB P2 requires the alarmone ppGpp, but growth rate-dependent control of rrnB P2 does not require ppGpp. The rrnB P2 core promoter sequence (−37 to +7) is sufficient to serve as the target for growth rate-dependent regulation.
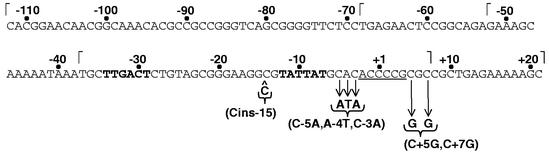
To meet the requirement for protein synthesis as nutritional conditions change, bacteria modulate ribosome synthesis primarily by controlling transcription initiation from rRNA promoters. Escherichia coli contains seven rRNA operons (rrnA, -B, -C, -D, -E, -G, and -H), each of which has two promoters, rrn P1 and rrn P2, separated by ∼120 bp (16). The rrn P1 and rrn P2 promoters have many sequence characteristics in common: near consensus −10 and −35 hexamers, separated by 16 bp, that bind the σ subunit of RNA polymerase (RNAP) (16); an A+T-rich region upstream of the −35 hexamer (UP element) that increases transcription by binding the C-terminal domains of the α subunits of RNAP (27, 28); and a G+C-rich region (the discriminator) (38) between the −10 hexamer and the transcription start site that is required for proper regulation (4, 8, 15, 25) (see Fig. 1 for the rrnB P2 sequence).
FIG. 1.
Sequence of the rrnB P2 promoter region from −112 to +21. The most frequent rrnB P2 transcription start site is designated +1. The −10 and −35 hexamers are indicated in bold, start sites are underlined, and the upstream (−112, −68, −53, and −37) or downstream (+7 and +21) endpoints used for construction of lacZ fusions are indicated by ⌈ or ⌉, respectively. The −15C insertion is indicated by the symbol ^, and substitutions are identified below the arrows.
Regulation of rRNA transcription initiation is often analyzed in terms of the responses of rRNA promoters to three experimental situations. First, rrn P1 activity changes in proportion to the steady-state growth rate (growth rate-dependent control [12, 22; reviewed in reference 13]), coordinating the number of ribosomes with the need for protein synthesis. Second, rrn P1 activity changes in inverse proportion to changes in rRNA gene dose (feedback regulation [14; reviewed in reference 13]), maintaining ribosome synthesis homeostatically. Third, relA-dependent inhibition of rrn P1 activity is observed following amino acid starvation (stringent control [reviewed in reference 6]), preventing an overinvestment of energy in ribosome synthesis when the substrates for protein synthesis are unavailable.
The literature is less clear about the regulation of the rrn P2 promoters. Early studies indicated that rrn P2 promoters are growth rate dependent (but less so than rrn P1 promoters) and not stringently controlled (31-33), but subsequent studies suggested the opposite, that rrn P2 promoters are not growth rate dependent (12, 40) and are stringently controlled (11, 15, 19). Studies employing rrnB P2-lacZ or rrnB P2-cat fusions as reporters (12, 40) suggested that rrn P2 promoters are more active than rrn P1 promoters, in contrast to conclusions based on direct measurements of RNAs transcribed from rrn P1 and rrn P2 promoters (7, 20, 31, 40). Furthermore, inhibition of rrn P2 promoters by transcription originating upstream from the rrn P1 promoters (promoter occlusion [11]) qualifies conclusions about the intrinsic activity and regulation of rrn P2 derived from examination of constructs containing both promoters (7, 19, 20, 31-33).
Here we systematically reinvestigated the activity and regulation of rrnB P2 by using newly constructed reporters of rrnB P2 promoter activity, as well as direct measurement of transcripts by primer extension. We found that rrnB P2 is less active during steady-state growth than we concluded previously and that it is regulated in response to growth rate, amino acid starvation, and rRNA gene dose. In addition, we show that stringent control of rrnB P2 requires ppGpp but that growth rate-dependent regulation of rrnB P2 does not require ppGpp. The rrnB P2 core promoter sequence is sufficient to serve as the regulatory target for growth rate-dependent control.
MATERIALS AND METHODS
Media and growth conditions.
Cells were grown at 30°C in Luria-Bertani (LB) medium, brain heart infusion medium (Difco), or either M9 (21) or morpholinepropanesulfonic acid (23) minimal medium containing 0.4% glucose or glycerol, with or without 0.8% Casamino Acids (Difco) plus tryptophan (40 μg/ml). When required, ampicillin (Sigma) was included at 100 μg/ml. To induce a stringent response, the serine analog serine hydroxamate (1 mg/ml; Sigma) was added to an exponentially growing culture supplemented with amino acids (37).
Strains.
The strains and plasmids used in this study are listed in Table 1. Antibiotic resistance cassettes replacing the relA and spoT genes were transduced into λ lysogens by using phage P1vir (10). All promoter derivatives were generated by PCR from plasmids containing a wild-type promoter(s). Primers for the PCR were designed to include an EcoRI site at the upstream junction of the promoter sequence and a HindIII site at the downstream junction. The PCR products were cleaved with EcoRI and HindIII, ligated into plasmid pRLG770 (29), sequenced, and then cloned into bacteriophage λ derivatives containing lacZ to form promoter-lacZ fusions as previously described (26). In all lacZ fusion constructs (referred to as system 1 [26]), the DNA sequence downstream from the indicated promoter fragment is identical and derives from trp-lac DNA sequences. Promoter constructs are named by the endpoints of the DNA fragments used for their construction, where +1 is the transcription start site. rrnB P2 utilizes multiple transcription start sites; +1 refers to the most frequently utilized site, a C residue 7 bp downstream from the −10 hexamer (Fig. 1; H.D.M. and R.L.G., data not shown).
TABLE 1.
Strains, plasmids, and promoter constructs used in this study
| Strain or plasmid | Genotype or description | Source |
|---|---|---|
| Strains | ||
| VH1000 | MG1655 pyrE lacl lacZ (RLG3499) | 9 |
| CF1693 | relA251::kan spoT207::cam (RLG857) | 10, 39 |
| RLG3848 | VH1000 λrrnB P1(−61 to +50)-lacZ | This work |
| RLG3851 | VH1000 λrrnB P2(−37 to +7)-lacZ | This work |
| RLG3863 | VH1000 λrrnB P2(−52 to +7)-lacZ | This work |
| RLG3866 | VH1000 λrrnB P2(−37 to +7)-lacZ relA spoT | This work |
| RLG3871 | VH1000 λrrnB P1P2(−152 of P1 to +7 of P2)-lacZ | This work |
| RLG3897 | VH1000 λrrnB P2(−112 to +7; C-5A, A-4T, C-3A)-lacZ | This work |
| RLG3898 | VH1000 λrrnB P2(−112 to +7; Cins−15)-lacZ | This work |
| RLG3914 | VH1000 λrrnB P2(−112 to +21)-lacZ | This work |
| RLG3915 | VH1000 λrrnB P2(−112 to +7; C+5G,C+7G)-lacZ | This work |
| RLG4296 | VH1000 λrrnB P2(−68 to +7; C+5G,C+7G)-lacZ | This work |
| RLG4757 | VH1000 λrrnB P1(−152 to +50)-lacZ | This work |
| RLG4993 | VH1000 λlacUV5 (−59 to +36)-lacZ | This work |
| RLG5014 | VH1000 λrrnB P2(−112 to +7)-lacZ | This work |
| RLG6982 | VH1000 λrrnB P2(−112 to +7)-lacZ relA | This work |
| Plasmids | ||
| pRLG770 | General transcription vector | 29 |
| pRLG3858 | pRLG770 rrnB P1 (−152 of P1 to +7 of P2) | This work |
| pRLG3859 | pRLG770 rrnB P2 (−52 to +7) | This work |
| pRLG3865 | pRLG770 rrnB P2 (−37 to +7) | This work |
| pRLG3890 | pRLG770 rrnB P2 (−112 to +7; C-5A, A-4T,C-3A) | This work |
| pRLG3893 | pRLG770 rrnB P2 (−112 to +7; Cins−15) | This work |
| pRLG3895 | pRLG770 rrnB P2 (−112 to +21) | This work |
| pRLG3896 | pRLG770 rrnB P2 (−112 to +7; C+5G,C+7G) | This work |
| pRLG4871 | pRLG770 rrnB P2 (−112 to +7) | This work |
| pNO1301 | pBR322 expressing an intact rrnB operon | 14 |
| pNO1302 | pNO1301 with 2.4-kb deletion within 16S and 23S genes | 14 |
The rrnB P2-lacZ fusion described previously (12) was constructed by cleavage of rrnB DNA with FnuDII, ligation with a HindIII linker, and then fusion of this fragment to a λ phage arm containing lacZ. This resulted in an rrnB P2 promoter with the downstream DNA sequence 5′-CCCGGGGAAGCTT…-3′, beginning with position +1. This sequence differs from the wild-type rrnB P2 sequence by C-to-G transversions at +5 and +7 (Fig. 1).
In vivo β-galactosidase assays.
Cultures of lysogens containing promoter-lacZ fusions were started (A600 of ∼0.02) from fresh colonies and shaken at 30°C for about 4 generations to an A600 of ∼0.35, and β-galactosidase activity was measured following sonication as previously described (12, 21).
In vivo primer extension.
Cultures of lysogens containing promoter-lacZ fusions were started from fresh colonies (starting A600 of ∼0.02) and shaken at 30°C for about 4 generations to an A600 of ∼0.35. RNA was extracted by a boiling lysis procedure, and reverse transcription was performed as previously described (15, 35). Note that the absolute activities of promoters making different RNAs are not comparable, since the efficiency of extension by reverse transcriptase varies on different templates. To ensure that any differences in RNA levels observed at different growth rates did not result from changes in RNA half-life, half-lives were measured directly after addition of rifampin. No differences in RNA half-life were detected (data not shown). Furthermore, control promoters whose activities did not change with growth rate when examined by using promoter-lacZ fusions but that had the same mRNA sequences as the growth rate-dependent promoters examined here did not change with growth rate when examined by primer extension (data not shown).
Feedback assays.
A multicopy plasmid containing an intact rrnB operon coding for functional 16S, 23S, and 5S rRNAs (pNO1301) or an rrnB operon coding for nonfunctional 16S and 23S rRNAs (pNO1302) was transformed into lysogens carrying promoter-lacZ fusions. Cultures were shaken at 30°C in M9 medium supplemented with glucose and ampicillin to an A600 of ∼0.35, and β-galactosidase activity was measured as described above. As described in the figure legends, the activity of a promoter-lacZ fusion in a strain containing pNO1301 and that of a promoter-lacZ fusion in a strain containing pNO1302 were compared and the ratio was then normalized to that obtained from a control strain containing a lacUV5 promoter-lacZ fusion transformed with the same plasmids (12).
RESULTS
Sequence determinants for rrnB P2 activity.
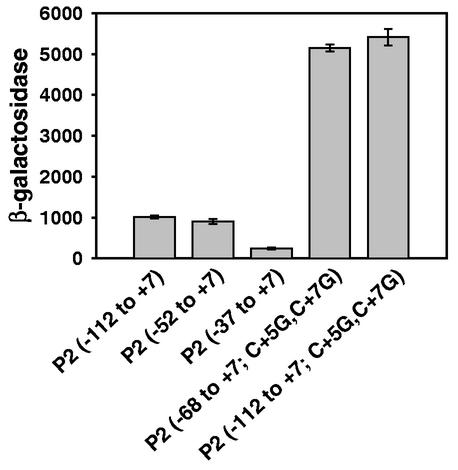
In order to investigate the behavior of rrnB P2 systematically, we created a series of rrnB P2-lacZ fusions containing promoter fragments with different upstream endpoints and the same downstream endpoint: −112 to +7, −52 to +7, and −37 to +7 (Fig. 1). We measured the activities of these fusions at a relatively low growth rate (i.e., when most of the rRNA transcription in the cell originates from the rrn P2 promoters [see below and reference 31]). rrnB P2 promoters with upstream endpoints of −112 and −52 had similar activities and were fourfold more active than the −37 to +7 construct (Fig. 2). The stimulatory effect of the −37 to −52 region is consistent with the previous finding that rrnB P2 contains an UP element; i.e., that the rrnB P2 sequence upstream of the −35 hexamer stimulates transcription both in vivo and in vitro (27, 28). These results also suggest that, unlike the rrn P1 promoters, which are activated by the transcription factor Fis (29), rrnB P2 does not require activation by a transcription factor that binds upstream of the UP element.
FIG. 2.
Relative activities of rrnB P2 promoter variants. β-Galactosidase activities from single-copy rrnB P2 promoter-lacZ fusions were measured in M9 minimal medium containing 0.4% glycerol as a carbon source. The rrnB P2(−68 to +7; C+5G,C+7G) fusion is the construct reported in reference 12. The average and standard deviation of at least three independent experiments are shown for each promoter.
The rrnB P2-lacZ fusion used in our previous studies (12, 15) was five- to sixfold more active than the P2(−112 to +7) or P2(−52 to +7) fusion described above (Fig. 2). This construct [now referred to as P2(−68 to +7; C+5G,C+7G)] contains a different upstream endpoint and C-to-G transversions at positions +5 and +7 (i.e., downstream of the transcription start site [Fig. 1 and Materials and Methods]). Since sequences upstream of −52 did not appear to affect transcription, we tested whether the difference in activity between the original and new constructs resulted from the substitutions at +5 and +7 by introducing these transversions into the P2(−112 to +7)-lacZ fusion. The P2(−112 to +7; C+5G,C+7G) construct was five- to sixfold more active than the wild-type P2(−112 to +7) promoter (Fig. 2).
In theory, the increase in β-galactosidase activity could have arisen from either an increase in mRNA half-life or a more favorable RNAP-promoter interaction. We found that the C+5G,C+7G mutations did not alter mRNA half-life in vivo, but the mutations did alter characteristics of the open complex formed with RNAP in vitro (data not shown; see Discussion). Therefore, we now consider our previously constructed rrnB P2 promoter-lacZ fusion, P2(−68 to +7; C+5G,C+7G), to be a mutant that does not accurately reflect wild-type promoter interactions with RNAP. As a result, we reevaluated our previous conclusions concerning the regulation of the rrnB P2 promoter (12, 15).
The rrnB P2 promoter is subject to growth rate-dependent control.
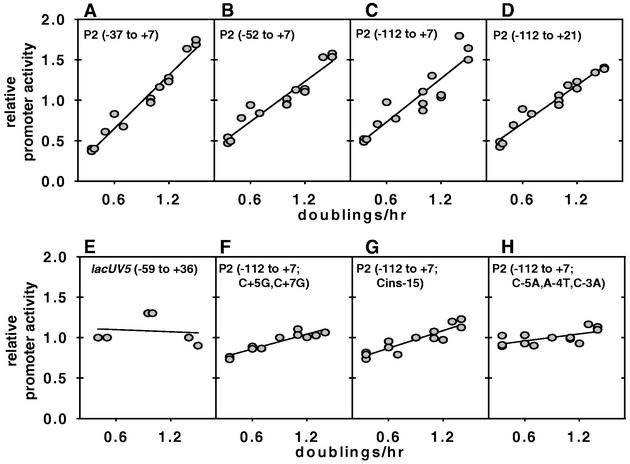
To evaluate growth rate dependence of transcription from the rrnB P2 promoter, cultures were grown in various media supporting different steady-state growth rates and promoter activities were determined. The activities of promoter-lacZ fusions containing the rrnB P2 −37 to +7, −52 to +7, or −112 to +7 sequence increased at least threefold over a fourfold range of growth rates (Fig. 3A to C). Promoter-lacZ fusions with an rrnB P2 downstream endpoint of +21 (with an upstream endpoint of either −112 [Fig. 3D] or −37 [data not shown]) exhibited regulation similar to that of rrnB P2 promoters with a downstream endpoint of +7, suggesting that these junctions represent wild-type promoter behavior. These regulation patterns are quite different from those exhibited by the control lacUV5 promoter (Fig. 3E) or by the mutant promoter construct P2(−112 to +7; C+5G,C+7G) (Fig. 3F and reference 12; see also reference 40). These results suggest that sequences upstream from −37 or downstream from +7 are not required for growth rate-dependent regulation of rrnB P2 and that the substitutions at +5 and/or +7 affect properties of the rrnB P2 promoter that are important for its regulation (see Discussion).
FIG. 3.
Promoter activities as a function of growth rate. β-Galactosidase activities were measured at different growth rates, obtained by growing cells in different media as described in Materials and Methods: M9 medium with 0.4% glycerol, M9 medium with 0.4% glucose, M9 medium with 0.4% glycerol plus 0.8% Casamino Acids plus tryptophan, M9 medium with 0.4% glucose plus 0.8% Casamino Acids plus tryptophan, and LB medium. Linear regressions were drawn by using SigmaPlot 5.0 (Jandel Scientific). The endpoints of the wild-type and mutant rrnB P2 promoter fragments used to construct the fusions are indicated in the panels. The lacUV5 promoter-lacZ fusion shown in panel E has been described previously (12). To enable visual comparison of the slopes, the activity of each promoter was normalized to a value of 1.0 at a growth rate of 0.9 doubling per hour (8). Strain designations and observed promoter activities (in Miller units) at a growth rate of 0.9 doubling per hour are as follows: A, RLG3851, 784 ± 34 U; B, RLG3863, 1,749 ± 72 U; C, RLG5014, 1,958 ± 236 U; D, RLG3914, 4,189 ± 252 U; E, RLG4993, 408 ± 66 U; F, RLG3915, 7,211 ± 300 U; G, RLG3898, 3,167 ± 281 U; H, RLG3897, 2,240 ± 104 U. Data from at least two independent experiments are shown for each construct.
Previous studies of rrnB P1 revealed a number of features in the core promoter important for regulation, including the length of the spacer (16 bp) between the −10 and −35 hexamers and the identity of the sequence in the discriminator region between the −10 hexamer and the transcription start site (4, 8). We constructed mutations in the rrnB P2 spacer or discriminator region to determine whether changing these features would alter regulation of this promoter. Either a 1-bp insertion in the rrnB P2 spacer, P2(−112 to +7; Cins−15), or a 3-bp substitution in the discriminator region, P2(−112 to +7; C−5A,A−4T,C−3A), significantly reduced growth rate regulation of the promoter (Fig. 3G and H), suggesting that similar sequence features in rrn P1 and rrn P2 might contribute to their regulation.
Growth rate-dependent regulation of the rrnB P2 promoter in tandem with rrnB P1.
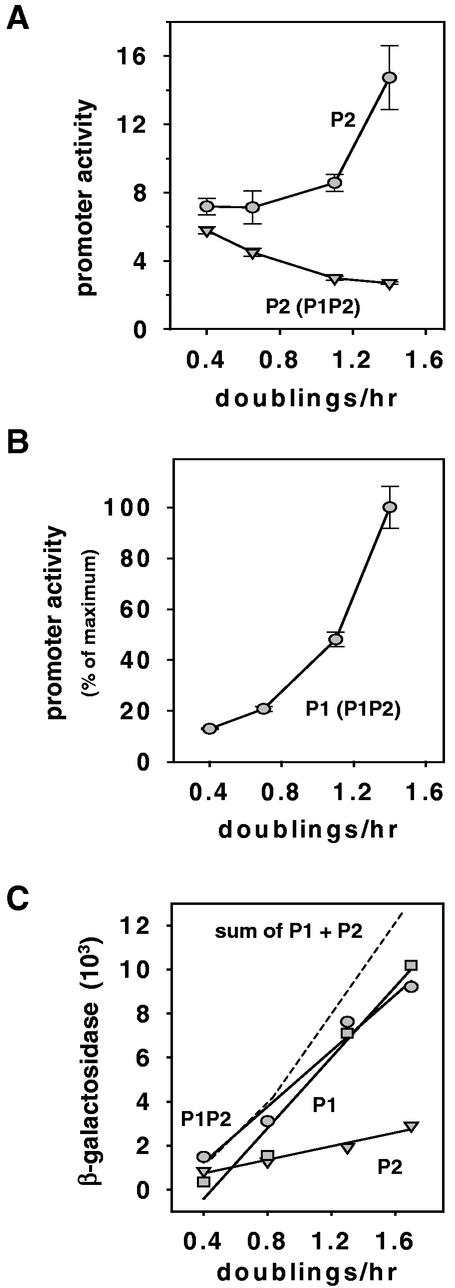
In their natural setting in rRNA operons, the rrn P2 promoters are located about 120 bp downstream from the rrn P1 promoters. To measure the activities of rrnB P2 when it is alone or in tandem with rrnB P1, we directly measured RNA synthesis by using quantitative primer extension. Transcription from the isolated rrnB P2 promoter increased as a function of growth rate (Fig. 4A), in agreement with the results obtained with lacZ fusions. However, in the presence of transcription from rrnB P1 (promoter activity illustrated in Fig. 4B), rrnB P2 promoter activity did not increase with growth rate and actually decreased slightly (Fig. 4A). These results are consistent with previous indications (11) that when the rrn P2 promoter is in its natural setting, its activity is occluded by transcription originating from rrn P1. Occlusion is most apparent when rrnB P1 activity is greatest, i.e., at high growth rates, and is sufficient to mask the intrinsic growth rate dependence of rrnB P2.
FIG. 4.
Transcription from rrnB P2 in the presence and absence of rrnB P1. (A) RNA transcribed from rrnB P2 promoter-lacZ fusions was measured directly by primer extension from lysogens grown in morpholinepropanesulfonic acid medium supplemented with 0.4% glycerol, 0.4% glucose, or 0.4% glucose plus 0.8% Casamino Acids plus tryptophan or in LB medium. Symbols: •, RLG5014, rrnB P2(−112 to +7); ▾, RLG3871, rrnB P1P2(−152 of P1 to +7 of P2). The average and standard deviation of at least three independent experiments are shown. Promoter activities are expressed in arbitrary units. (B) RNA transcribed from rrnB P1, in the context of the rrnB P1P2(−152 of P1 to +7 of P2) promoter-lacZ fusion, was measured directly by primer extension from lysogens grown in the same media used for the experiment whose results are shown in panel A. The average and standard deviation of at least three independent experiments are shown. Promoter activity is expressed as a percentage of maximum promoter activity. Absolute promoter activity should not be compared to that of rrnB P2 (see Materials and Methods). (C) β-Galactosidase activities from the same promoter-lacZ fusions described in panels A and B and from an rrnB P1-lacZ fusion. Symbols: •, RLG3871, rrnB P1P2(−152 of P1 to +7 of P2); ▪, RLG4757, rrnB P1(−152 to +50); ▾, RLG5014, rrnB P2(−112 to +7). The average of two independent experiments is shown for each promoter. The dashed line is a plot of the sum of the activities from the isolated rrnB P1-lacZ and rrnB P2-lacZ fusion constructs.
We next measured whether a lacZ fusion to a DNA fragment containing the intact rrnB P1P2 region would report the sum of the two isolated promoter activities (Fig. 4C). The isolated rrnB P1 and rrnB P2 promoters were each growth rate dependent (although to different extents [31]). As expected from the results shown in Fig. 4A, the activity of the tandem rrnB P1P2-lacZ fusion at low growth rates reflected the sum of the activities from the isolated promoters (Fig. 4C, dashed line), since occlusion of rrnB P2 is negligible when rrnB P1 is least active. In contrast, at high growth rates, the activity of the rrnB P1P2 fusion was substantially lower than that of the sum of the two isolated promoters (Fig. 4C, dashed line), consistent with the observed occlusion of rrnB P2 by rrnB P1 under these growth conditions (Fig. 4A). We conclude that the lacZ fusion to the tandem rRNA promoters accurately reports expression from the intact rrnB P1P2 promoter region, describing the sum of the activities from the rrnB P1 promoter and the occluded rrnB P2 promoter.
rrnB P2 is stringently controlled but does not require ppGpp for growth rate-dependent control.
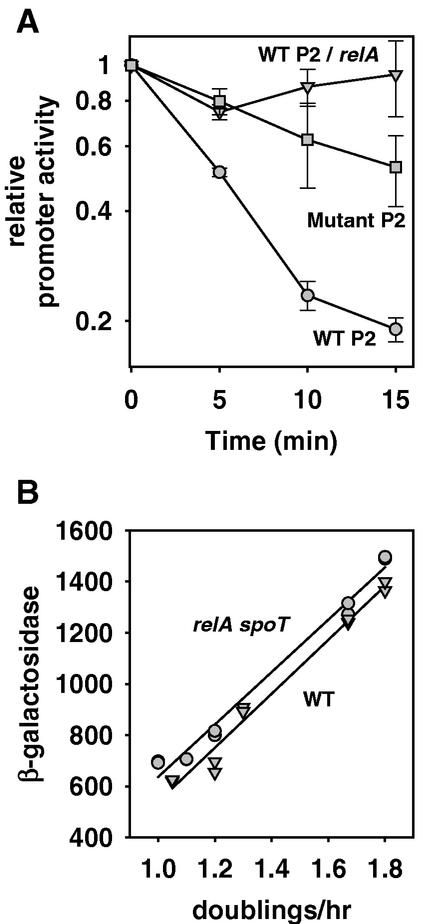
To determine whether the rrnB P2 promoter is stringently controlled, we used primer extension to measure its expression following amino acid starvation in a wild-type strain. Figure 5A shows that transcription from wild-type rrnB P2 was strongly inhibited, as reported previously for rrnB P1 (11, 15, 19). The rrnB P2 promoter with an altered discriminator sequence, P2(−112 to +7; C−5A,A−4T,C−3A), was inhibited only slightly (15, 38), and inhibition of wild-type rrnB P2 did not occur in a relA mutant (Fig. 5A). Thus, the results confirm previous conclusions (11, 15, 19, 35) that rrnB P2 is stringently controlled by the relA product, ppGpp.
FIG. 5.
rrnB P2 is stringently controlled but does not require ppGpp for growth rate-dependent regulation. (A) RNA transcribed from rrnB P2 promoter-lacZ fusions was measured directly by primer extension following amino acid starvation induced by serine hydroxamate addition to a culture growing exponentially in LB medium (A600 of ∼0.3). The identity of the rrnB P2 promoter and the strain background are indicated for each sample. Symbols: •, RLG5014, wild-type (WT) rrnB P2(−112 to +7); ▪, RLG3987, mutant rrnB P2(−112 to +7; C−5A,A−4T,C−3A); ▾, RLG6982, wild-type rrnB P2(−112 to +7) in a relA strain. The average and standard deviation of three independent experiments are shown. (B) β-Galactosidase activity from an rrnB P2 core promoter-lacZ fusion, P2(−37 to +7), as a function of growth rate in wild-type and relA spoT mutant strains. Symbols: ▾, RLG3851, wild-type strain; •, RLG3866, relA spoT mutant strain. Data from two independent experiments are shown for each strain.
ppGpp concentrations change inversely with growth rate (6, 30). To determine whether ppGpp is required for growth rate-dependent control of rrnB P2, we measured the activity of an rrnB P2-lacZ fusion at different growth rates in a wild-type strain and in a strain lacking the two genes capable of ppGpp synthesis, relA and spoT. Growth rate regulation of rrnB P2 was very similar in the mutant and wild-type strains (Fig. 5B), indicating that a regulator(s) other than ppGpp is sufficient to maintain normal levels of transcription from rrnB P2 in steady-state growth. However, we have not ruled out the possibility that ppGpp, when present, contributes to this regulation.
Increased rRNA gene dose reduces transcription from rrnB P2.
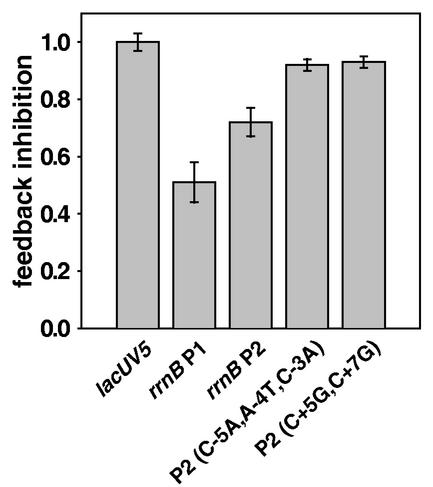
Total rRNA synthesis is gene dose independent, because transcription from individual rRNA operons is reduced in inverse proportion to the increase in rRNA gene number (14). It was shown previously that rrnB P1 promoter-lacZ fusions were inhibited by the presence of extra rRNA operons on multicopy plasmids, while the mutant rrnB P2 promoter-lacZ fusion P2(−68 to +7; C+5G,C+7G) was unaffected (12). To determine whether the wild-type rrnB P2 promoter is feedback regulated, we compared the activities of wild-type P2(−112 to +7) and two mutant promoter-lacZ fusions, P2(−112 to +7; C+5G,C+7G) and P2(−112 to +7; C−5A,A−4T,C−3A), in the presence of a multicopy plasmid harboring an intact rrnB operon or control plasmids (see the legend to Fig. 6 and reference 12). The wild-type rrnB P2 and rrnB P1 promoters were inhibited about 30 and 50%, respectively, while the mutant rrnB P2 promoters were inhibited less than 10% by the presence of extra rRNA operons (Fig. 6). Thus, rrnB P2 promoter activity is feedback regulated, although to a lesser extent than rrnB P1.
FIG. 6.
rrnB P2 promoter activity is inhibited by the presence of extra rRNA operons. Lysogens containing promoter-lacZ fusions were transformed with a multicopy plasmid containing either an intact rrnB operon (pNO1301) or an rrnB operon containing a large deletion in the 16S and 23S rRNA genes (pNO1302). Promoter activity is expressed as the ratio of the β-galactosidase activity from a promoter-lacZ fusion in a strain containing pNO1301 to that in a strain containing pNO1302 and then normalized to the value obtained from a control lacUV5 promoter as previously described (12). RLG4993, lacUV5 (−59 to +36); RLG3848, rrnB P1 (−61 to +50); RLG5014, rrnB P2(−112 to +7); RLG3987, rrnB P2(−112 to +7; C−5A,A−4T,C−3A); RLG3915, rrnB P2(−112 to +7, C+5G,C +7G). The average results and standard deviations of experiments with three independent transformants are shown.
DISCUSSION
rrn P2 promoters are regulated.
We have shown that the rrnB P2 promoter (in the absence of rrnB P1) has intrinsic regulatory characteristics similar to those of the rrn P1 promoters. rrnB P2 activity increases with growth rate, core promoter sequences are sufficient for this growth rate-dependent regulation, and ppGpp is not required for this control. Furthermore, rrnB P2 is feedback inhibited by the presence of extra rRNA operons and is stringently controlled (11, 15). Thus, our studies suggest that both the rrn P1 and rrn P2 promoters are coordinately regulated (although not to the same extent). As a consequence of being regulated to different degrees, the rrn P1 and rrn P2 promoters have different functions: the rrn P1 promoters are most responsible for the large amounts of rRNA required during rapid growth, while the rrn P2 promoters are responsible for most of the rRNA expression that occurs at low growth rates (Fig. 4B and reference 31).
In some previous studies (18, 41), changes in rrnB P2 promoter activity with growth rate and amino acid limitation were attributed to changes in the concentration of free RNAP. We suggest that this conclusion is not likely to be correct, since (i) UP elements reduce the concentration of RNAP required for transcription (26), yet growth rate-dependent regulation of rrnB P2 was not affected by deletion of its UP element (Fig. 3); (ii) rrnB P2 activity was unaffected by conditions that should reduce the effective RNAP concentration within the cell (i.e., by the presence of multicopy plasmid pNO1302, whose transcription titrates out RNAP [reference 2 and data not shown]); and (iii) rrnB P2 activity was affected by conditions that do not alter the cell's RNAP concentration (i.e., by the presence of multicopy plasmid pNO1301 [Fig. 6 and references 2, 11, 26, and 30]). Thus, the activity of rrnB P2 is unlikely to be a valid indicator of the free cellular RNAP concentration.
Mechanism of regulation of rrn P2 promoters.
The rrnB P2 promoter forms a relatively short-lived open complex with RNAP (H.D.M. and R.L.G., unpublished data), accounting for its sensitivity to inhibition by ppGpp (3). However, since rrnB P2 is growth rate dependent even in relA spoT mutant strains, mechanisms in addition to those involving ppGpp must contribute to control of the rrn P2 promoters. One mechanism contributing to regulation of the rrn P1 promoters involves sensing of changing initiating nucleoside triphosphate concentrations (4, 5, 9, 35). rrnB P2 responds to changing concentrations of its initiating nucleoside triphosphate, primarily CTP, in vitro and in vivo (H.D.M and R.L.G., unpublished data). We suggest, therefore, that when the rrn P2 promoters are responsible for most rRNA synthesis, ribosome production is determined, at least in part, by CTP levels.
The small DNA-binding proteins Fis and H-NS affect the activity of the rrn P1 promoters in vitro and in vivo (1, 29, 36). We tested rrnB P2 promoter activity in strains with fis or hns deleted in vivo and observed that rrnB P2 activity was increased in both mutants (data not shown). However, neither protein has been implicated in direct negative control of rrn P2 promoters (e.g., see reference 36). Since rrnB P2 promoter activity is regulated homeostatically (Fig. 6), it is likely that the increase in rrnB P2 promoter activity in the strain lacking fis results indirectly from the loss of activation of rrn P1 (feedback derepression [13]). In contrast, the activities of numerous promoters have been shown to increase in hns strains (34). Further work is required to determine whether the effect of hns on rrnB P2 promoter activity is specific and/or direct.
The loss of regulation of the rrnB P2 promoter that resulted from the transversions at +5 and +7 was somewhat unexpected, since sequences downstream of +1 that affect regulation of rrn P1 promoters have not been identified (8). The C+5G and C+7G mutations increase the half-life of the rrnB P2 open complex in vitro (H.D.M. and R.L.G. unpublished), consistent with the model in which regulation of rrnB P2 results from its intrinsic kinetic characteristics. Since RNAP clamps down on the template in the downstream region of the transcription elongation complex (17) and this region is protected by RNAP in footprints of transcription initiation complexes (24), it is possible that direct contacts of RNAP with DNA are affected by the mutations at +5 and/or +7. We will address the molecular mechanism of regulation of rrnB P2 in a subsequent report.
Acknowledgments
We thank Wilma Ross and other members of our laboratory for helpful comments and strain construction.
H.D.M. was supported by a National Institutes of Health Genetics Predoctoral Training Grant. Research in our laboratory is supported by grant RO1 GM37048 from the National Institutes of Health to R.L.G.
REFERENCES
- 1.Afflerbach, H., O. Schroder, and R. Wagner. 1998. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol. Microbiol. 28:641-653. [DOI] [PubMed] [Google Scholar]
- 2.Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689-702. [DOI] [PubMed] [Google Scholar]
- 3.Barker, M. M., T. Gaal, C. A. Josaitis, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673-688. [DOI] [PubMed] [Google Scholar]
- 4.Barker, M. M., and R. L. Gourse. 2001. Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J. Bacteriol. 183:6315-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett, M. S., T. Gaal, W. Ross, and R. L. Gourse. 1998. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J. Mol. Biol. 279:331-345. [DOI] [PubMed] [Google Scholar]
- 6.Cashel, M., D. R. Gentry, V. H. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and J. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C.
- 7.de Boer, H., and M. Nomura. 1979. In vivo transcription of rRNA operons in Escherichia coli initiates with purine nucleoside triphosphates at the first promoter and with CTP at the second promoter. J. Biol. Chem. 254:5609-5612. [PubMed] [Google Scholar]
- 8.Dickson, R. R., T. Gaal, H. A. deBoer, P. L. deHaseth, and R. L. Gourse. 1989. Identification of promoter mutants defective in growth-rate-dependent regulation of rRNA transcription in Escherichia coli. J. Bacteriol. 171:4862-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 10.Gaal, T., and R. L. Gourse. 1990. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:5533-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gafny, R. S., S. Cohen, N. Nachaliel, and G. Glaser. 1994. Isolated P2 rRNA promoters of E. coli are strong promoters that are subject to stringent control. J. Mol. Biol. 243:152-156. [DOI] [PubMed] [Google Scholar]
- 12.Gourse, R. L., H. A. de Boer, and M. Nomura. 1986. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell 44:197-205. [DOI] [PubMed] [Google Scholar]
- 13.Gourse, R. L., T. Gaal, M. S. Bartlett, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645-677. [DOI] [PubMed] [Google Scholar]
- 14.Jinks-Robertson, S., R. L. Gourse, and M. Nomura. 1983. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell 33:865-876. [DOI] [PubMed] [Google Scholar]
- 15.Josaitis, C. A., T. Gaal, and R. L. Gourse. 1995. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc. Natl. Acad. Sci. USA 92:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keener, J., and M. Nomura. 1996. Regulation of ribosome synthesis, p. 1417-1428. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and J. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, vol. 1. ASM Press, Washington, D.C.
- 17.Korzheva, N., A. Mustaev, M. Kozlov, A. Malhotra, V. Nikiforov, A. Goldfarb, and S. A. Darst. 2000. A structural model of transcription elongation. Science 289:619-625. [DOI] [PubMed] [Google Scholar]
- 18.Liang, S., M. Bipatnath, Y. Xu, S. Chen, P. Dennis, M. Ehrenberg, and H. Bremer. 1999. Activities of constitutive promoters in Escherichia coli. J. Mol. Biol. 292:19-37. [DOI] [PubMed] [Google Scholar]
- 19.Liebig, B., and R. Wagner. 1995. Effects of different growth conditions on the in vivo activity of the tandem Escherichia coli ribosomal RNA promoters P1 and P2. Mol. Gen. Genet. 249:328-335. [DOI] [PubMed] [Google Scholar]
- 20.Lund, E., and J. E. Dahlberg. 1979. Initiation of Escherichia coli ribosomal RNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 76:5480-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Miura, A., J. H. Krueger, S. Itoh, H. A. deBoer, and M. Nomura. 1981. Growth rate-dependent regulation of ribosome synthesis in E. coli: expression of the lacZ and galK genes fused to ribosomal promoters. Cell 25:773-782. [DOI] [PubMed] [Google Scholar]
- 23.Neidhardt, F. C., C. A. Bloch, and D. F. Smith. 1974. Culture media for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozoline, O. N., and M. A. Tsyganov. 1995. Structure of open promoter complexes with Escherichia coli RNA polymerase as revealed by the DNase I footprinting technique: compilation analysis. Nucleic Acids Res. 23:4533-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokholok, D. K., M. Redlak, C. L. Turnbough, Jr., S. Dylla, and W. M. Holmes. 1999. Multiple mechanisms are used for growth rate and stringent control of leuV transcriptional initiation in Escherichia coli. J. Bacteriol. 181:5771-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao, L., W. Ross, J. A. Appleman, T. Gaal, S. Leirmo, P. J. Schlax, M. T. Record, Jr., and R. L. Gourse. 1994. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J. Mol. Biol. 235:1421-1435. [DOI] [PubMed] [Google Scholar]
- 27.Ross, W., S. E. Aiyar, J. Salomon, and R. L. Gourse. 1998. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J. Bacteriol. 180:5375-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 29.Ross, W., J. F. Thompson, J. T. Newlands, and R. L. Gourse. 1990. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 9:3733-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryals, J., R. Little, and H. Bremer. 1982. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J. Bacteriol. 151:1261-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarmientos, P., and M. Cashel. 1983. Carbon starvation and growth rate-dependent regulation of the Escherichia coli ribosomal RNA promoters: differential control of dual promoters. Proc. Natl. Acad. Sci. USA 80:7010-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarmientos, P., S. Contente, G. Chinali, and M. Cashel. 1983. Ribosomal RNA operon promoters P1 and P2 show differential regulatory responses, p. 65-74. In D. H. Hamer and M. Rosenburg (ed.), Gene expression. Alan R. Liss, Inc., New York, N.Y.
- 33.Sarmientos, P., J. E. Sylvester, S. Contente, and M. Cashel. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32:1337-1346. [DOI] [PubMed] [Google Scholar]
- 34.Schmid, M. B. 1990. More than just “histone-like” proteins. Cell 63:451-453. [DOI] [PubMed] [Google Scholar]
- 35.Schneider, D. A., T. Gaal, and R. L. Gourse. 2002. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc. Natl. Acad. Sci. USA 99:8602-8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tippner, D., H. Afflerbach, C. Bradaczek, and R. Wagner. 1994. Evidence for a regulatory function of the histone-like Escherichia coli protein H-NS in ribosomal RNA synthesis. Mol. Microbiol. 11:589-604. [DOI] [PubMed] [Google Scholar]
- 37.Tosa, T., and L. I. Pizer. 1971. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J. Bacteriol. 106:972-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Travers, A. A. 1984. Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res. 12:2605-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 40.Zacharias, M., H. U. Goringer, and R. Wagner. 1989. Influence of the G\CGC discriminator motif introduced into the ribosomal RNA P2- and tac promoter on growth-rate control and stringent sensitivity. EMBO J. 8:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, X., and H. Bremer. 1995. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 270:11181-11189. [DOI] [PubMed] [Google Scholar]