Abstract
Allantoinase is a suspected dinuclear metalloenzyme that catalyzes the hydrolytic cleavage of the five-member ring of allantoin (5-ureidohydantoin) to form allantoic acid. Recombinant Escherichia coli allantoinase purified from overproducing cultures amended with 2.5 mM zinc, 1 mM cobalt, or 1 mM nickel ions was found to possess ∼1.4 Zn, 0.0 Co, 0.0 Ni, and 0.4 Fe; 0.1 Zn, 1.0 Co, 0.0 Ni, and 0.2 Fe; and 0.0 Zn, 0.0 Co, 0.6 Ni, and 0.1 Fe per subunit, respectively, whereas protein obtained from nonamended cultures contains near stoichiometric levels of iron. We conclude that allantoinase is incompletely activated in the recombinant cells, perhaps due to an insufficiency of a needed accessory protein. Enzyme isolated from nonsupplemented cultures possesses very low activity (kcat = 34.7 min−1) compared to the zinc-, cobalt-, and nickel-containing forms of allantoinase (kcat values of 5,000 and 28,200 min−1 and 200 min−1, respectively). These rates and corresponding Km values (17.0, 19.5, and 80 mM, respectively) are significantly greater than those that have been reported previously. Absorbance spectroscopy of the cobalt species reveals a band centered at 570 nm consistent with five-coordinate geometry. Dithiothreitol is a competitive inhibitor of the enzyme, with significant Ki differences for the zinc and cobalt species (237 and 795 μM, respectively). Circular dichroism spectroscopy revealed that the zinc enzyme utilizes only the S isomer of allantoin, whereas the cobalt allantoinase prefers the S isomer, but also hydrolyzes the R isomer at about 1/10 the rate. This is the first report for metal content of allantoinase from any source.
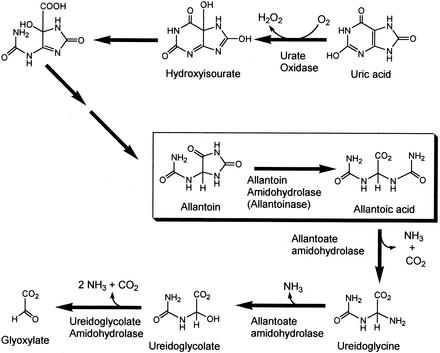
Allantoinase (EC 3.5.2.5), present in a wide variety of bacteria, fungi, and plants, as well as a few animals, such as fish and amphibians, catalyzes the conversion of allantoin (5-ureidohydantoin) to allantoic acid by hydrolytic cleavage of the five-member hydantoin ring (33, 52). Allantoin is a key intermediate in nitrogen metabolism of leguminous plants, such as soybean (Fig. 1), where ureides provide the major transport form of symbiotically fixed dinitrogen (29, 43, 48, 55). Until recently, it was thought that allantoin was formed directly from uric acid by the action of uric acid oxidase (39); however, it has now been established that the true product of soybean uric acid oxidase is 5-hydroxyisourate (22), which is then acted upon by the enzyme hydroxyisourate hydrolase to yield 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (42). This compound is then converted to allantoin, most likely by a yet unidentified enzyme. The product of allantoin hydrolysis, allantoate, is sequentially converted in soybean to ureidoglycine, ureidoglycolate, and glyoxylate with the ammonia released during these reactions utilized for plant cell growth. Variations of this pathway occur in other organisms, including Escherichia coli, which can grow on allantoin as the sole nitrogen source, but not as the sole carbon source. The E. coli allantoinase gene is part of a 12-gene regulon involved in allantoin degradation and nitrogen assimilation, and these genes are expressed only when the cells are grown under anaerobic conditions (10).
FIG. 1.
Allantoinase reaction in the context of soybean uric acid metabolism. The boxed reaction is catalyzed by allantoinase.
On the basis of limited sequence similarities, allantoinase was suggested to belong to a broad family of metal-dependent amidohydrolases (17). Members of this family contain either mononuclear metal sites, such as the zinc site found in adenosine deaminase (56) and the iron site in cytosine deaminase (57), or dinuclear sites, such as the di-nickel site documented to occur in urease (20) and the di-zinc sites of dihydroorotase (47) and hydantoinase (1, 2, 8). Some, but not all, dinuclear hydrolase family members have their two metals bridged by a carbamylated lysine ligand (1, 2, 4, 7, 8, 20, 47). Significantly, we find that allantoinase sequences align with the active site regions of crystallized hydantoinases (1, 2, 8), dihydroorotase (47), and urease (20), as well as with a subgroup of hydantoinases that have not been structurally characterized (Fig. 2). The ligands to the metal centers in these enzymes appear to be conserved in allantoinases, even including the lysine that is carbamylated in the crystallized enzymes. Thus, allantoinase is likely to possess a dinuclear metal site bridged by a carbamylated lysine side chain. Surprisingly, despite its purification from several sources (21, 24, 25, 33, 52, 55), the metal content of allantoinase has not been reported. Here we focus on recombinant allantoinase from E. coli and examine how supplementation of the medium with different metal ions affects the enzyme metal content, kinetic parameters, and substrate stereospecificity.
FIG. 2.
Sequence comparison of regions of selected amidohydrolases. Regions of sequence similarity are shown for allantoinases (All) of Escherichia coli, Deinococcus radiodurans (D.rad.), Bacillus subtilis (B.sub.), Bacillus halodurans (B.hal.), Streptomyces coelicolor (S.coe), and Saccharomyces cerevisiae (S.cer.); hydantoinases (Hyd) of Agrobacterium tumefaciens (A.tum.), Ralstonia pickettii (R.pic.), Agrobacterium sp. strain IP I-671 (A. sp.), Geobacillus stearothermophilus (formerly Bacillus stearothermophilus [G.ste.]), S. coelicolor, Brucella melitensis (B.mel.), Pseudomonas putida (P.put.), Sulfolobus tokodaii (S.tok.), a Thermus sp. (Ther.), and Arthrobacter aurescens (A.aur.); E. coli dihydroorotase (DHO), and Klebsiella aerogenes urease large subunit (K.aer. Ure). Crystal structures have been reported for the D-specific hydantoinases from Thermus (2) and G. stearothermophilus (8) (Protein DataBank accession no. 1GKP and 1K1D), the L-specific hydantoinase from A. aurescens (1) (accession no. 1GKR), E. coli dihydroorotase (47) (accession no. 1J79), and K. aerogenes urease (20) (accession no. 1FJW). The metallocenter ligands in these proteins are underlined with carbamylated lysines bridging the two metals.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli K-12 strain W3110 (16) was used as the source of genomic DNA for cloning the allantoinase gene. Production of recombinant allantoinase was performed with cells of E. coli strain C41(DE3) (31) as the host cells, which were grown in TB medium, consisting of 12 g of tryptone, 24 g of yeast extract (Difco), and 4 ml of glycerol per 900 ml (41) supplemented with 100 ml of filter-sterilized 0.5 M 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7.2), in place of the standard phosphate buffer. The MOPS buffer allowed addition of high concentrations of metal ions without precipitation of the phosphate salts. All other recombinant DNA manipulations were done with E. coli DH5α or XL1-Blue (Stratagene) and grown in Luria-Bertani (LB) medium according to standard protocols (3).
PCR cloning and overexpression of allantoinase gene.
Genomic DNA was isolated from E. coli by standard methods (3). The PCR primers used were 5′-AAGAAACGTAGACCAGCAG-3′ for the forward direction and 5′-GCAGAAGGTACCGTAGGAG-3′ for the reverse, where the underlined bases represent base changes relative to the genomic sequence that introduced a KpnI site before the 5′ end of the allantoinase gene and an AccI site after the 3′ end. The amplified DNA fragment encoding the allantoinase gene was isolated from a 1% agarose gel by using QIAEX II gel purification reagents (Qiagen) and digested with KpnI and AccI. Vector pGEM-4Z (Promega) was also digested with KpnI and AccI, and the reaction products were ligated to produce plasmid pEA5. The entire allantoinase gene was sequenced by automated sequencing methods at the Michigan State University DNA Sequencing Facility.
The allantoinase gene was subcloned from pEA5 into pET11a to achieve a high level of expression. Site-directed mutagenesis was first performed with PCR with the complementary primers 5′-CAATCTGTATTTTACAAGGAGTTTCATATGTCTTTTGATTTAATCATTAAAAACG-3′ (forward), and 5′-CGTTTTTAATGATTAAATCAAAAGACATATGAAACTCCTTGTAAAATACAGATTG-3′ (reverse). The underlined bases represent base changes to introduce an NdeI site at the start ATG codon. The amplified circular plasmid products were transformed into E. coli XL1-Blue and analyzed for the presence of a new NdeI restriction site (one new site at the ATG start codon and one existing site after the 3′ end of the gene in the pGEM-4Z vector). The allantoinase gene was resequenced to verify that no unintended mutations occurred, excised from pEA5 by using NdeI to produce a 1.1-kb fragment, isolated, and ligated into NdeI-cut pET11a (Novagen). The plasmid construct, designated pEA14, was checked by digestion with PstI to verify proper insert orientation with respect to the T7 promoter.
Allantoinase assays.
Enzyme assays were performed as previously described (40) by the o-phenylenediamine method. The standard assay mixture contained 50 mM allantoin (from Acros or Sigma) and 50 mM HEPES (pH 7.4) at 37°C. Allantoin solutions were prepared by adding the solid to hot water to aid in dissolution. When higher concentrations of allantoin were required (up to 125 mM), the solutions were held at 37°C and used within several min. For determination of Km values, substrate concentrations ranged from 1.5 to 119 mM. Data were fit to the Michaelis-Menten equation with SigmaPlot (Jandell Scientific). Units of activity are defined as micromoles of allantoic acid produced per minute per milligram of protein. Values of kcat were calculated with an Mr of 49,500 for the allantoinase monomer.
Effect of metal ions on levels of enzyme activity in cell cultures.
Because allantoinase was suspected to be a metalloenzyme (see the introduction), the activity of recombinant enzyme was examined in crude extracts prepared from cultures grown in the presence of various metal ions. Cultures of E. coli C41(DE3)(pEA14) were grown aerobically at 30°C with rapid shaking in 10 ml of TB medium containing 250 μg of ampicillin per ml or 100 μg of carbenicillin per ml and 1 mM concentrations of the following metal salts: FeSO4, Zn acetate, CoCl2, NiCl2, CdCl2, CuSO4, MgCl2, MnCl2, Na2MO4, and Na2VO4. Additional anaerobic cultures were grown either without added metal ions or with 2 mM ferrous ammonium sulfate in a sealed vial in which the headspace was replaced with nitrogen. When the cultures reached an A600 of 0.1 to 0.2, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 100 μM, and the cultures were moved to room temperature and grown overnight. Allantoinase activities were determined after centrifuging 1 ml of each culture, resuspending the pellets in 1 ml of 20 mM HEPES (pH 7.8), and disrupting them by sonication with a Branson Sonifier 250. Assays of the anaerobically grown cells were performed with an assay mix that was placed into centrifuge tubes and vials that were sealed and subjected to 10 cycles of evacuation and argon filling. The anaerobic cells were resuspended in degassed buffer containing 20 mM Tris (pH 8) with 0.1 mM EDTA, 0.1 mg of lysozyme per ml, and 1 mM sodium hydrosulfite. After 20 min at room temperature, the cells were frozen and thawed twice, and aliquots were removed and assayed under aerobic and anaerobic conditions.
Enzyme purification.
E. coli C41(DE3) cells were transformed with plasmid pEA14 and grown in TB medium supplemented with 250 μg of ampicillin per ml plus either 2.5 mM zinc acetate, 1 mM cobalt chloride, or 1 mM nickel chloride at 30°C with rapid shaking. When the cultures reached an A600 of ∼0.1, IPTG was added to 50 μM, the flasks were shifted to room temperature, and growth continued for an additional 16 to 18 h. Preliminary studies aimed at optimizing expression conditions utilized 10- to 50-ml cultures, whereas routine large-scale purification was performed with 250-ml cultures grown in 2.8-liter Fernbach flasks. Cells were chilled on ice, harvested by centrifugation, and frozen at −20°C. After thawing, cells were resuspended in 50 mM HEPES buffer (pH 8.1) and disrupted by sonication with ice cooling between pulses. The disrupted cell suspensions were adjusted to 0.5 mM in phenylmethylsulfonyl fluoride and 2% (wt/vol) in streptomycin sulfate, mixed thoroughly, and centrifuged at 50,000 × g for 25 min. The supernatant solutions were chromatographed at room temperature with DEAE Sepharose (2.5-cm diameter by 35 cm; Pharmacia). After being washed with 50 mM HEPES (pH 8.1), allantoinase was eluted with a 0 to 1 M KCl gradient in 50 mM HEPES (pH 8.1) in a total volume of 250 ml. Fractions were examined for the presence of allantoinase protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, and those containing enzyme of sufficient purity were pooled and dialyzed in 50 mM HEPES (pH 8.1) at 4°C overnight. The dialyzed preparation was divided into two parts, and each was chromatographed on a Mono-Q HR 10/10 column by using a Pharmacia fast protein liquid chromatography system. Enzyme was eluted with a 0 to 1 M KCl gradient in a buffer of 50 mM HEPES (pH 8.1) with a two-step gradient of 0 to 0.4 M KCl in 80 ml followed by 0.4 to 1 M in 20 ml. The fractions were examined by SDS-PAGE analysis, and those containing >95% pure allantoinase were pooled and stored on ice. Protein concentrations for samples from early stages in the purification were determined with the Bio-Rad protein assay reagent with bovine serum albumin as the standard. The concentrations of purified enzyme samples were measured by using a calculated extinction coefficient at 280 nm of 35,700 M−1 cm−1 (35).
In a similar manner, allantoinase was purified from cell cultures of E. coli C41(DE3)(pEA14) to which no metal ions were added. Cultures grown in this way exhibited allantoinase activities comparable to those of background controls, yet showed significant allantoinase protein production based on SDS-PAGE analysis. The nearly inactive enzyme was isolated to 90% purity with the same DEAE-Sepharose/Mono-Q protocol described above, except that, for some preparations, buffers contained 0.1 mM dipicolinic acid to chelate trace metals in the solutions. Final purification, yielding at least 95% homogeneous protein, was achieved by concentrating fractions from the Mono-Q column in a Centricon 30 microconcentrator and chromatographing the sample on a 2.5-cm by 47-cm Superose 6 column equilibrated in 50 mM HEPES buffer (pH 8.1) containing 50 μM EDTA and 50 μM dipicolinic acid. The fractions of highest purity as judged by SDS-PAGE analysis were pooled and stored on ice.
Effect of added metal ions on allantoinase activity.
Activation studies were performed to assess the effect of added metal ions on allantoinase activity. Buffers were treated with Chelex-100 resin to remove trace metal contaminants. Solutions of zinc sulfate and cobalt sulfate utilized Puratronic grade reagents (Johnson Mathey Chemicals, Ltd.). Typical experiments (100 μl) contained 2.4 nmol of allantoinase in 50 mM Tris buffer (pH 8) containing 100 μM zinc sulfate or 2 or 20 mM cobalt. In addition, some experiments included 1 to 3.6 M urea or 10 mM bicarbonate. After mixing allantoinase with the desired additives, samples were incubated at room temperature and assayed periodically over several days.
Characterization of allantoinase.
Metal contents of allantoinase preparations were measured by inductively coupled plasma emission-mass spectrometry in the Food Safety and Toxicology Center at Michigan State University or by inductively coupled plasma emission spectroscopy at the Chemical Analysis Laboratory of the University of Georgia. Prior to analyses, buffer components and excess metal ions were removed by several concentration/dilution steps in a Centricon-30 ultrafilter with metal-analysis-grade water. The resulting protein samples were made 5% in nitric acid (Fisher, TraceMetal grade) and centrifuged at 13,000 rpm for 10 min to remove the precipitated protein. The supernatants were used for metal analyses.
The pH dependence of allantoinase activity was measured in solutions of 50 mM allantoin containing 50 mM of the appropriate buffering agent. Because allantoin is a weak acid, the pH of each solution of allantoin was measured and adjusted by addition of NaOH as needed. Nonenzymatic degradation of allantoin, known to occur under acidic and alkaline conditions (51), was measured at pH values above 8.5. No significant nonenzymatic allantoin hydrolysis was observed under acidic conditions down to pH 5.4. The stability of allantoinase as a function of pH was measured by diluting enzyme samples into solutions containing 50 mM 2-(4-morpholino)ethanesulfonic acid (MES), HEPES, 2-[N-cyclohexylamino]ethanesulfonic acid (CHES), or 3-[cyclohexylamino]-1-propanesulfonic acid (CAPS) over a wide pH range. After exposure of the enzyme to the indicated pH conditions for 1 h at room temperature, the samples were tested for activity with the standard assay described above.
During preliminary experiments with allantoinase, it was found that thiols inhibited the enzyme. Allantoinase inhibition by dithiothreitol (DTT) was examined in detail for the cobalt and zinc forms by measuring enzyme activity over a range of DTT and allantoin concentrations.
CD spectroscopy.
For circular dichroism (CD) spectroscopy, the enantiomeric specificities of the zinc and cobalt forms of allantoinase were examined by monitoring the changes in optical activity of solutions containing 1 mM racemic allantoin in the presence of 20 mM sodium phosphate buffer (pH 7) in a volume of 800 μl. Samples were placed in a 1-cm quartz cuvette in a Jasco J-710 spectropolarimeter, and 0.3 U of either cobalt or zinc allantoinase was added and mixed. The room temperature reaction was followed by taking spectra at various times or by continuous monitoring at a wavelength of 220 nm.
RESULTS
Effects of metal ions on recombinant allantoinase activity in cell cultures.
Using the genomic DNA sequence of E. coli, primers were designed to PCR amplify the segment containing the allantoinase gene and incorporate restriction sites for optimal placement in the T7 polymerase-controlled expression plasmid pET11a. The resulting construct, designated pEA14, was used for all subsequent experiments. When cultures were grown and induced by IPTG at 37°C, SDS-PAGE analysis of cell extracts revealed the production of some soluble allantoinase along with a larger fraction of insoluble enzyme (data not shown). Yields of total allantoinase were examined at IPTG concentrations of 0 to 0.5 mM with optimal amounts of protein observed at 50 μM inducer. Growth at lower temperatures resulted in a larger fraction of soluble enzyme.
The TB medium used for cell growth contains metal-binding components that reduce the available concentrations of metal ions suspected to be required for allantoinase activity; thus, the effects of added metal ions were examined. At metal ion concentrations of 1 mM, only zinc, cobalt, and nickel provided activities (28, 35, and 6 U ml−1, respectively) that were significantly above that of the no-supplemental-metal control (1.3 U/ml). Specific activities of cultures grown in a range of zinc concentrations from 0 to 5 mM revealed maximal activity levels at 2.5 mM metal ion. Zinc concentrations above 2.5 mM and cobalt or nickel concentrations above 1 mM were inhibitory to cell growth. The conditions selected for routine purification of allantoinase from zinc-amended media used 2.5 mM zinc acetate. Similarly, 1 mM cobalt chloride or nickel chloride was added to cultures to obtain the cobalt or nickel forms of the enzyme. In all cases, cells were grown at 30°C until they reached an A600 of 0.1 to 0.2, at which time, IPTG was added to a final concentration of 50 μM, and growth continued at room temperature overnight (22 to 25°C). Under these conditions, typical cultures contained 34,000 to 83,000 U (zinc supplemented), 35,000 to 90,000 U (cobalt supplemented), or 2,800 to 7,300 U (nickel supplemented) of allantoinase per liter.
Nonrecombinant E. coli allantoinase was previously reported to be synthesized only when cells were grown under anaerobic conditions (10), suggesting that the enzyme may be oxygen labile or that its regulation is controlled by the oxidation state of the cells. To test the effects of oxygen and added ferrous ions on recombinant allantoinase activity, extracts were prepared from cells grown anaerobically with various concentrations of ferrous ions and assays performed under anaerobic conditions. No significant activity was detected for any of these samples. In contrast, control experiments with cells grown in 2.5 mM zinc acetate under aerobic conditions and lysed by the anaerobic lysozyme method gave an activity of 21 U ml−1, while a parallel sample of cells disrupted by sonication yielded 34 U ml−1.
Allantoinase purification.
Recombinant allantoinase was purified from sonicated extracts of cells grown in the presence of added zinc, cobalt, or nickel and from cultures with no metal supplementation. Each sample was purified by successive DEAE-Sepharose and Mono-Q chromatography steps. Typical purifications of allantoinases from zinc- and cobalt-amended cultures are shown in Table 1. Purifications were performed with buffers containing 50 mM HEPES (pH 8.1), and the pure enzyme samples were stored as pooled fractions directly off the Mono-Q column (estimated KCl concentration of 250 mM in 50 mM HEPES [pH 8.1]) or in 100 mM KCl-50 mM HEPES (pH 8.1). The allantoinase purified from zinc-amended cultures could be frozen and thawed at least four times without significant reduction of specific activity. The enzyme purified from cobalt-supplemented cultures was labile to freezing and lost about 50% of the initial activity within 1 week when stored at 4 to 8°C, with subsequent losses occurring at a somewhat slower rate. Purified allantoinase from either source was routinely stored on ice. In this way, the zinc form has been stored as long as 5 months and the cobalt form has been stored as long as 1 month without significant loss of activity. Short-term incubation with metal chelators, such as EDTA and dipicolinic acid, had negligible effects on either form of allantoinase activity at the pH of the storage buffer. Furthermore, long-term incubation of the enzyme isolated from zinc-amended cultures with 10 μM cobalt did not lead to any increase in activity.
TABLE 1.
Purification of E. coli allantoinase from cultures supplemented with zinc and cobalt
| Purification step | Allantoinase
|
|||||
|---|---|---|---|---|---|---|
| Protein (mg)a | Total Ub | Sp act (U mg−1) | Recovery (%) | |||
| Zinc-amended culture | ||||||
| Sonicated cells | 985 | 21,600 | 22.0 | 100 | ||
| Streptomycin SO4 | 643 | 23,300 | 36.1 | 100 | ||
| DEAE-Sepharose | 323 | 11,600 | 35.9 | 53.6 | ||
| Mono-Q | 53.7 | 4,090 | 76.1c | 18.9 | ||
| Cobalt-amended culture | ||||||
| Sonicated cells | 670 | 44,300 | 66.0 | 100 | ||
| Streptomycin SO4 | 330 | 46,500 | 142 | 100 | ||
| DEAE-Sepharose | 127 | 14,300 | 113 | 32.3 | ||
| Mono-Q | 20.0 | 8,200 | 410c | 18.5 | ||
From 250 ml of culture.
Measured at 50 mM allantoin.
Concentration of final protein determined by A280 with a calculated extinction coefficient of 35,700 M−1 cm−1.
Allantoinase purified from cultures grown without added metal ions had a final specific activity of 0.7 μmol min−1 mg of protein−1 (9.4 mg from a 250-ml culture). This form of the enzyme showed small, gradual increases in activity when incubated in buffers containing 2 or 20 mM cobalt, yielding enzyme with 3.9 and 8.1 μmol min−1 mg of protein−1, respectively, after 10 days at room temperature. No increase in activity was detected in samples incubated with 100 μM zinc, even in the presence of mild concentrations of urea. Concentrations of zinc above 100 μM could not be tested in this experiment due to precipitation of the protein. Inclusion of 10 mM bicarbonate to test the possible requirement that lysine carbamylation is required for enzyme activation as in urease (36) (and likely to be needed for dihydroorotase and hydantoinase) (1, 2, 8, 47) did not affect the allantoinase activity. For comparison, the activities of allantoinases purified from zinc- or cobalt-supplemented cultures were unaffected by incubation with these additives.
Metal content.
The metal contents of representative allantoinase preparations are illustrated in Table 2. Growth conditions had a large impact on the metal content of the enzyme, with zinc, cobalt, nickel, and iron being the only metals found in significant quantities. Allantoinase prepared from cells grown in excess zinc contained 0.62 to 1.45 mol of metal per mol of monomer, as well as significant amounts of iron. No cobalt or nickel was detected in this form of the enzyme. Enzyme purified from cells grown in cobalt-supplemented medium contained approximately stoichiometric amounts of this metal (ranging from 0.81 to 1.0 mol per mol of monomer), but also had small amounts of zinc and iron. Unexpectedly, enzyme prepared from nickel-amended cultures contained only small amounts of this metal (0.13 mol of metal per mol of monomer) and larger amounts of iron (0.62 mol). Similarly, allantoinase purified from cultures grown without added metals had more than stoichiometric levels of iron and only trace amounts of zinc.
TABLE 2.
Properties of allantoinase purified from zinc-, cobalt-, and nickel-amended cultures and nonamended cultures
| Culture | Allantoin Km (mM) | kcat (min−1) | DTT Ki (μM) | No. of mol of metal/mol of monomera
|
Isomer preference | |||
|---|---|---|---|---|---|---|---|---|
| Zn | Co | Fe | Ni | |||||
| Zinc amended | 17.0 ± 2.7 | 5,000 ± 250 | 237 | 1.44 | 0.00 | 0.38 | 0.00 | S |
| Cobalt amended | 19.5 ± 1.3 | 28,200 ± 540 | 795 | 0.12 | 1.00 | 0.17 | 0.00 | S (fast), R (slow) |
| Nickel amended | 79.9 ± 5.5 | 200 ± 5 | NDb | 0.00 | 0.00 | 0.62 | 0.13 | ND |
| Nonamended | ND | 34.7c | ND | 0.03 | 0.00 | 1.08 | 0.00 | ND |
Values are shown for representative samples.
ND, not determined.
Measured at 50 mM allantoin.
Absorbance spectroscopy.
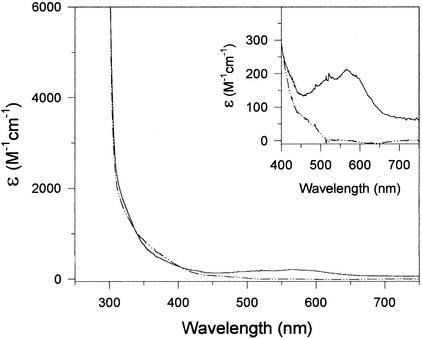
As described in the Discussion section, several cobalt-containing enzymes possess characteristic absorbance bands in the 500- to 700-nm-wavelength range. When examined by UV-visible spectroscopy, the cobalt-containing allantoinase showed a similar absorption band centered at 570 nm with an ɛ570 of 209 M−1 cm−1 (Fig. 3).
FIG. 3.
Absorbance spectra of allantoinases prepared from cobalt (solid line), or zinc (dashed-dotted line)-amended cultures. (Inset) Expanded absorbance scale of the 400- to 750-nm region. The concentration of enzymes were 20 μM for the zinc form and 15 μM for the cobalt form in 50 mM HEPES (pH 7.8) buffer. The concentrations of enzymes in the inset were 400 μM for the zinc form and 300 μM for the cobalt form.
Kinetic parameters.
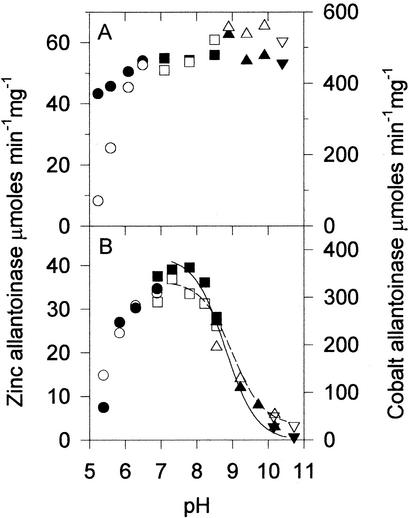
Allantoinases purified from zinc-, cobalt-, and nickel-supplemented cultures were found to have significant differences in their kinetics, as shown in Table 2. The kcat values were 5,000 ± 250, 28,200 ± 540, and 200 ± 5 μmol min−1 for the three species, respectively, while the corresponding Km values were 17.0, 19.5, and 80 mM. Because the enzyme isolated from nonsupplemented medium exhibited only very low levels of activity, detailed kinetic analyses were not performed. Tests of the pH stability of the metal-supplemented allantoinase samples showed that the cobalt form of the enzyme (Fig. 4A, open symbols) is less stable at acidic pH than the zinc (Fig. 4A, closed symbols) or nickel (data not shown) forms. Each of the enzymes exhibited optimal activity at pH 7 to 8, with the high-pH end of the plots fitting nicely to theoretical curves yielding pKa values of 8.8 ± 0.1 for the zinc form and 9.0 ± 0.1 for the cobalt form (Fig. 4B).
FIG. 4.
Effects of pH on allantoinase stability and activity. Open symbols represent results for the cobalt form of the enzyme (corresponding to the right axis), and solid symbols represent those for the zinc form (corresponding to the left axis). The buffers used were MES (circles), HEPES (squares), CHES (triangles), and CAPS (inverted triangles). (A) Stability of allantoinases for 1 h at room temperature as a function of pH. (B) Activity as a function of pH. The curves are fit to the data in the range of pH 7.3 to 10.5 yielding pKa values of 9.0 for the cobalt form of allantoinase (dashed line) and 8.8 for the zinc form (solid line).
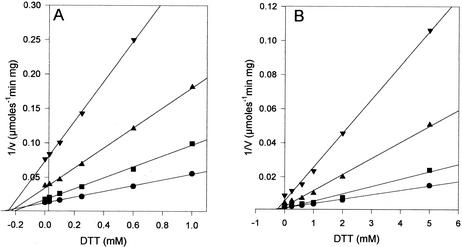
Alternative substrates and potential inhibitors of allantoinase activity were examined. No hydrolytic activity was observed with hydantoin or hydantoin-5-acetic acid, and no significant inhibition of allantoin hydrolysis was detected when assays were performed under standard conditions plus 20 mM hydantoin. The thiols β-mercaptoethanol and DTT were identified as inhibitors during tests to determine whether additives would alter allantoinase stability and activity. Since β-mercaptoethanol inhibited only weakly, inhibition of the zinc and cobalt forms of allantoinase by DTT was examined in detail. This compound exhibited competitive inhibition of both allantoinases with Ki values of 237 μM for the zinc form and 795 μM for the cobalt form (Fig. 5).
FIG. 5.
DTT inhibition of allantoinase. (A) Zinc form of allantoinase. Allantoin concentrations were 50 mM (•), 30 mM (▪), 15 mM (▴), and 7 mM (▾). (B) Cobalt form of allantoinase. The allantoin concentrations were 80 mM (•), 50 mM (▪), 25 mM (▴), and 10 mM (▾).
Enantioselectivity of allantoinase.
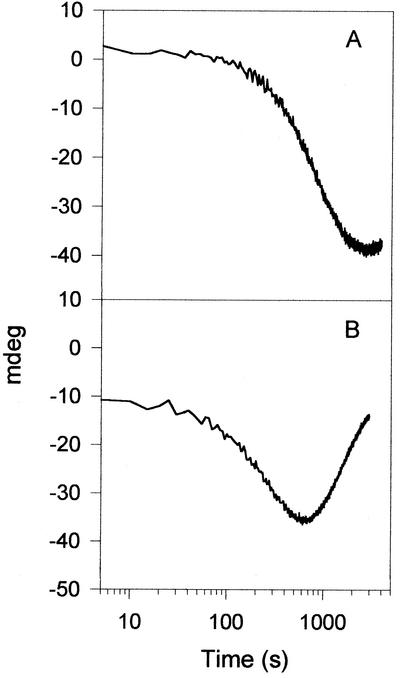
When reaction progress curves were determined with the zinc form of allantoinase, only half of the allantoin was consumed, suggesting hydrolysis of a single isomer. The enantiomeric selectivity of the zinc- and cobalt-containing allantoinases was examined by CD spectropolarimetry, monitoring changes in the spectral range of 200 to 400 nm (data not shown) and at a fixed wavelength of 220 nm (Fig. 6). The reaction progress curves clearly show that the zinc form of allantoinase is enantioselective for the (+) or S allantoin. In contrast, the cobalt form of allantoinase consumes both isomers, but with the (−) or R enantiomer hydrolyzed at about 10% the rate of the (+) enantiomer.
FIG. 6.
Allantoinase assays monitored by CD spectropolarimetry. (A) Allantoinase prepared from zinc-amended cultures. (B) Allantoinase from cobalt-amended cultures.
DISCUSSION
Allantoinase has been proposed to belong to a superfamily of metal-containing amidohydrolases related by evolution and predicted to have a common three-dimensional fold (17). More specifically, sequence comparisons versus crystallographically characterized proteins (Fig. 2) allow us to predict that allantoinase contains a dinuclear metal site bridged by a carbamylated lysine residue. To date, no report has appeared on the metal content of any purified allantoinase; however, stimulation of partially or completely purified allantoinases by incubation with various metal ions has been reported (24, 53). Below, we discuss the effect of supplementing the cultures with various metal ions on the activity and metal content of recombinant E. coli allantoinase.
Our results confirm that E. coli allantoinase is a metalloenzyme. The highest levels of allantoinase activities are observed in cell extracts from cultures supplemented with zinc or cobalt to concentrations slightly below those found to be toxic to the cells. Of other added metals examined, only nickel yields activities above those of cells grown in medium without metal supplementation. Notably, supplementation of the medium with ferrous ions, even for anaerobically grown cultures, fails to enhance cellular allantoinase activity. Metal quantitation results for allantoinase species purified under the various culture conditions are consistent with the presence of a dinuclear metal active site in fully active enzyme. For example, protein purified from zinc-amended cultures typically contains more than stoichiometric levels of zinc, as well as significant amounts of iron. The high level of production of recombinant allantoinase (up to 1,000 mg liter−1 or 29% of the soluble protein) is likely to result in enzyme with a low stoichiometry of bound metal, especially as we observed in protein isolated from the cobalt- and nickel-supplemented cultures and the nonsupplemented sample. This low stoichiometry may be due to factors such as limiting metal ion transport or the requirement for an accessory protein that is present in low abundance (described below). Thus, our purified allantoinase species likely represent populations of enzyme with dinuclear centers and populations with less than a full complement of active site metal ions.
The identity of the metal in nonrecombinant allantoinase is most likely to be zinc ions. Both the zinc and cobalt forms of the enzyme are active, with the cobalt form exhibiting nearly sixfold higher activity, but it is unlikely that cobalt has a biological role in vivo. For example, Walker and Bradshaw (54) have argued that yeast methionine aminopeptidase is a zinc-dependent enzyme, rather than cobalt dependent as previously reported, on the basis of the predicted low bioavailability of cobalt ions resulting in cytoplasmic concentrations that are 1,000-fold lower than that of zinc. Cobalt's ability to substitute for zinc and produce hyperactive enzymes is well known. For example, reconstitution of the apoenzyme form of hydantoinase with cobalt results in a 1.5-fold increase in activity compared to the level in the native zinc enzyme (28). Similarly, replacement of zinc with cobalt in untreated or N-ethylmaleimide-treated dihydroorotase results in an ∼50% increase in hydrolytic activity (6, 18). In the case of adenosine deaminase, however, the activity of the cobalt-substituted enzyme is the same as that of the native zinc enzyme (9). Although not a member of the amidohydrolase superfamily, it is worth noting that cobalt-substituted alcohol dehydrogenase from Saccharomyces cerevisiae has 100-fold more activity than the native zinc enzyme (49). We conclude that cobalt substitutes for the native zinc in allantoinase to provide hyperactive enzyme when provided in the medium at high concentrations. If zinc and cobalt are not abundant, the allantoinase apoprotein appears to exhibit high affinity for iron, as shown by both the presence of stoichiometric amounts of this metal in enzyme purified from cultures grown in the absence of added metal ions and the detectable iron levels in other preparations. If not displaced or competed against by zinc or cobalt, this metal becomes locked into the enzyme. Perhaps related to this finding, E. coli cytosine deaminase is an iron-containing enzyme (19). Similarly, ferrous ions are found at the active sites of methionine aminopeptidase (12, 30) and peptide deformylase (38), whereas a zinc(II)/iron(III) dinuclear site is found in purple acid phosphatase (45), calcineurin (58), or serine/threonine phosphatase-1 (13). Thus, we cannot exclude the possibility that iron may play a significant role in allantoinase; however, we find that protein containing only iron is inactive or possesses very low levels of activity and could not be activated by incubation under any conditions we could identify.
The UV/visible absorbance spectrum of the cobalt form of allantoinase provides insight into the coordination geometry of at least one of the metal sites. An absorption band centered at 570 nm with a shoulder at 520 nm resembles the absorption bands in cobalt-substituted dihydroorotase (6, 18), carboxypeptidase A (32), alcohol dehydrogenase, and sorbitol dehydrogenase (26). Such spectra, with ɛ = 50 to 250 M−1 cm−1 as observed for allantoinase, are indicative of cobalt(II) d-d transitions associated with a pentacoordinate site (5). In contrast, no distinct ∼350-nm transition, as found in the thiolate-liganded cobalt sites of β-lactamase II (11) or alcohol and sorbitol dehydrogenases (26), is observed in the cobalt form of allantoinase. We suggest that cobalt in the allantoinase active site is coordinated in a five-coordinate environment lacking cysteine ligation.
Two lines of evidence address the mechanism of assembly of the allantoinase metal center. Activity is not affected at pH 8.1 by incubation with EDTA or dipicolinic acid, suggesting that the metal is inaccessible to these metal chelators and deeply buried in the protein. Second, efforts to activate protein isolated from cultures not amended with metal ions proved unsuccessful, even in the presence of added bicarbonate (needed for lysine carbamylation) or mild concentrations of denaturant. This situation differs from those of several amidohydrolase family members, including hydantoinase (28), dihydroorotase (6, 18, 37), and adenosine deaminase (9). Bound metals are readily removed from and restored to these enzymes at neutral pH, consistent with an accessible active site. Our results also contrast with those from earlier studies of allantoinase in which chelators and metal additions exhibited a larger effect on activity (24, 52, 53). On the other hand, the properties of the apparently buried metallocenter of allantoinase somewhat resemble those of urease. Nickel ions bound at the urease active site cannot be removed by dialysis against various chelators, and formation of this buried metallocenter is a complex process requiring accessory proteins that appear to act as protein chaperones (15) and a metallochaperone (44). Thus, we speculate that allantoinase activation may also require the participation of an accessory protein. A requirement for an accessory protein may explain the observed low levels of activation in the overproducing recombinant, where the single-copy accessory gene is expressed at insufficient levels. Possible functions of such a protein include action in metal delivery or in the incorporation of CO2/bicarbonate, which is suspected to occur as part of the activation step. The E. coli gene cluster containing the allantoinase gene does not include obvious candidates for such an accessory gene (10); however, it may be encoded elsewhere on the chromosome. Further efforts to identify an allantoinase accessory protein are warranted; however, the experimental observations noted above are also compatible with metal incorporation occurring during the protein folding process.
The kinetic parameters for E. coli allantoinase reported here differ from previously reported values. Working with partially purified allantoinase from E. coli, Vogels et al. (52) reported a specific activity of 6 μmol min−1 mg of protein−1. More recently, Kim et al. (24, 25) examined purified recombinant E. coli allantoinase and found specific activities of 5.7 to 19.9 μmol min−1 mg of protein−1. These values are significantly lower than the Vmax of 101 μmol min−1 mg of protein−1 presented here for the zinc form of the enzyme. In addition, the Km of 17.0 mM for zinc allantoinase in this study is higher than the 4.2 and 7.4 mM values reported by Kim et al. (24, 25). Two possible explanations for the disparate kinetic parameters between our results and those reported earlier for purified E. coli enzyme relate to differences in enzyme metal content and assay conditions. No metal content of the purified enzyme was reported in the earlier studies and, perhaps significantly, no metal ions were added to the LB medium used for cell growth. Thus, the lower activity observed in the prior studies may be due to a large proportion of inactive enzyme arising from low levels of metal ions in their growth medium. In addition, the earlier assays were performed in the presence of 0.5 mM DTT, which we have shown to be a competitive inhibitor. The presence of a competitive inhibitor also would be expected to lead to a reduction in activity.
The most intriguing finding in this study was the observed difference in substrate stereospecificities between the zinc and cobalt allantoinases when examined by CD spectropolarimetry. The zinc enzyme utilized only the (+) S isomer, resulting in an initial negative phase (Fig. 6). A short upward phase at the end of the progress curve most likely results from spontaneous racemization, a process that occurs at neutral pH with a half-life of 10 h at room temperature (23). The cobalt enzyme reaction profile initially proceeded in the same direction, but faster because of the greater kcat. The change in optical activity then reversed direction almost until the baseline was reached. This result clearly indicates that both isomers are utilized, but at rates that differ by about 10-fold. Precise kinetic parameters were not derived from these reaction profiles, since the spectropolarimeter response was unknown and the reactions were run at 1 mM allantoin, far below the Km. Our results imply that cobalt allantoinase can hydrolyze both isomers of allantoin; however, the possibility exists that only one isomer is hydrolyzed and the enzyme possesses a slow racemase activity. Previous studies have suggested that crude preparations or partially isolated allantoinases from various sources are aspecific or favor the (+) isomer (14, 34, 50, 52). Care must be taken when interpreting these earlier studies, because allantoin is a substrate for d-hydantoinase (unpublished observations), a not surprising result considering that hydantoinases can hydrolyze a wide variety of 5-substituted hydantoins (46). Thus, contamination of partially purified allantoinases with hydantoinase could lead to incorrect conclusions about stereospecificity. The ability to affect the stereospecificity of allantoinase by simply replacing the metal cofactor could have important biotechnology implications. Illustrating this potential, studies have been undertaken to change the enantiomeric preference of hydantoinase by using directed evolution (27). The findings presented here indicate that metal substitution may be an alternative method to achieve this desired result.
Acknowledgments
We thank Vladimir Romanov for initiating these studies, Sarah Ward for help in characterizing the Ni-containing enzyme, Rafik Momin and Muralee Nair for assistance with CD spectroscopy, and Lina Patino for some metal analyses.
This study was supported by U.S. Department of Agriculture grant 0003496.
REFERENCES
- 1.Abendroth, J., K. Niefind, O. May, M. Siemann, C. Syldatk, and D. Schomburg. 2002. The structure of L-hydantoinase from Arthrobacter aurescens leads to an understanding of dihydropyrimidinase substrate and enantio specificity. Biochemistry 41:8589-8597. [DOI] [PubMed] [Google Scholar]
- 2.Abendroth, J., K. Niefind, and D. Schomburg. 2002. X-ray structure of a dihydropyrimidinase from Thermus sp. at 1.3 Å resolution. J. Mol. Biol. 320:143-156. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M. 1994. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Benning, M. M., J. M. Kuo, F. M. Raushel, and H. M. Holden. 1995. 3-Dimensional structure of the binuclear metal center of phosphotriesterase. Biochemistry 34:7973-7978. [DOI] [PubMed] [Google Scholar]
- 5.Bertini, I., and C. Luchinat. 1994. The reaction pathways of zinc enzymes and related biological catalysts, p. 37-106. In I. Bertini, H. B. Gray, S. J. Lippard, and J. S. Valentine (ed.), Bioinorganic chemistry. University Science Books, Mill Valley, Calif.
- 6.Brown, D. C., and K. D. Collins. 1991. Dihydroorotase from Escherichia coli: substitution of Co(II) for the active site Zn(II). J. Biol. Chem. 266:1597-1604. [PubMed] [Google Scholar]
- 7.Buchbinder, J. L., R. C. Stephenson, M. J. Dresser, J. W. Pitera, T. S. Scanlan, and R. J. Fletterick. 1998. Biochemical characterization and crystallographic structure of an Escherichia coli protein from the phosphotriesterase gene family. Biochemistry 37:5096-5106. [DOI] [PubMed] [Google Scholar]
- 8.Cheon, Y.-H., H.-S. Kim, K.-H. Han, J. Abendroth, K. Niefind, D. Schomburg, J. Wang, and Y. Kim. 2002. Crystal structure of D-hydantoinase from Bacillus stearothermophilus: insight into the stereochemistry of enantioselectivity. Biochemistry 41:9410-9417. [DOI] [PubMed] [Google Scholar]
- 9.Cooper, B. F., V. Sideraki, D. K. Wilson, D. Y. Dominguez, S. W. Clark, F. A. Quiocho, and F. B. Rudolph. 1997. The role of divalent cations in structure and function of murine adenosine deaminase. Protein Sci. 6:1031-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cusa, E., N. Obradors, L. Baldoma, J. Badia, and J. Aguilar. 1999. Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 181:7479-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, R. B., and E. P. Abraham. 1974. Metal cofactor requirements of beta-lactamase II. Biochem. J. 143:129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'souza, V. M., and R. C. Holz. 1999. The methionyl aminopeptidase from Escherichia coli can function as an iron(II) enzyme. Biochemistry 38:11079-11085. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg, J., H.-B. Huang, Y.-G. Kwon, P. Greengard, A. C. Nairn, and J. Kuriyan. 1995. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376:745-753. [DOI] [PubMed] [Google Scholar]
- 14.Gravenmade, E. J., G. D. Vogels, and C. van Pelt. 1969. Preparation, properties, and absolute configuration of (−)-allantoin. Recl. Trav. Chim. 88:929-936.
- 15.Hausinger, R. P., G. J. Colpas, and A. Soriano. 2001. Urease: a paradigm for protein-assisted metallocenter assembly. ASM News 67:78-84. [Google Scholar]
- 16.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holm, L., and C. Sander. 1997. An evolutionary treasure: unification of a broad set of amidohydrolases related to urease. Proteins Struct. Funct. Genet. 28:72-82. [PubMed] [Google Scholar]
- 18.Huang, D. T. C., M. A. W. Thomas, and R. I. Christopherson. 1999. Divalent metal derivatives of the hamster dihydroorotase domain. Biochemistry 38:9964-9970. [DOI] [PubMed] [Google Scholar]
- 19.Ireton, G. C., G. McDermott, M. E. Black, and B. L. Stoddard. 2002. The structure of Escherichia coli cytosine deaminase. J. Mol. Biol. 315:687-697. [DOI] [PubMed] [Google Scholar]
- 20.Jabri, E., M. B. Carr, R. P. Hausinger, and P. A. Karplus. 1995. The crystal structure of urease from Klebsiella aerogenes. Science 268:998-1004. [PubMed] [Google Scholar]
- 21.Janssen, D. B., R. Smits, and C. Vanderdrift. 1982. Allantoinase from Pseudomonas aeruginosa. Purification, properties and immunochemical characterization of its in vivo inactivation. Biochim. Biophys. Acta 718:212-219. [DOI] [PubMed] [Google Scholar]
- 22.Kahn, K., and P. A. Tipton. 1997. Kinetic mechanism and cofactor content of soybean root nodule urate oxidase. Biochemistry 36:4731-4738. [DOI] [PubMed] [Google Scholar]
- 23.Kahn, K., and P. A. Tipton. 2000. Kinetics and mechanism of allantoin racemization. Bioorg. Chem. 28:62-72. [Google Scholar]
- 24.Kim, G. J., D. E. Lee, and H.-S. Kim. 2000. Functional expression and characterization of the two cyclic amidohydrolase enzymes, allantoinase and a novel phenylhydantoinase, from Escherichia coli. J. Bacteriol. 182:7021-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, G. J., D. E. Lee, and H. S. Kim. 2001. High-level expression and one-step purification of cyclic amidohydrolase family enzymes. Protein Expr. Purif. 23:128-133. [DOI] [PubMed] [Google Scholar]
- 26.Maret, W. 1989. Cobalt(II)-substituted class III alcohol and sorbitol dehydrogenases from human liver. Biochemistry 28:9944-9949. [DOI] [PubMed] [Google Scholar]
- 27.May, O., P. T. Nguyen, and F. H. Arnold. 2000. Inverting enantioselectivity by directed evolution of hydantoinase for improved production of L-methionine. Nat. Biotechnol. 18:317-320. [DOI] [PubMed] [Google Scholar]
- 28.May, O., M. Siemann, M. G. Siemann, and C. Syldatk. 1998. Catalytic and structural function of zinc for the hydantoinase from Arthrobacter aurescens DSM 3745. J. Mol. Catal. B Enzym. 4:211-218. [Google Scholar]
- 29.McClure, P. R., and D. W. Israel. 1979. Transport of nitrogen in the xylem of soybean plants. Plant Physiol. 64:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng, L., S. Ruebush, V. M. D'souza, A. J. Copik, S. Tsunasawa, and R. C. Holz. 2002. Overexpression and divalent metal binding properties of the methionyl aminopeptidase from Pyrococcus furiosus. Biochemistry 41:7199-7208. [DOI] [PubMed] [Google Scholar]
- 31.Miroux, B., and J. E. Walker. 1996. Overproduction of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 32.Moratal, J. M., J. Castells, A. Donaire, J. Salgado, H. R. Jimenez, and R. Domingo. 1994. Interaction of cobalt ions with carboxypeptidase A. J. Inorg. Biochem. 53:1-11. [DOI] [PubMed] [Google Scholar]
- 33.Noguchi, T., S. Fujiwara, and S. Hayashi. 1986. Evolution of allantoinase and allantoicase involved in urate degradation in liver peroxisomes. A rapid purification of amphibian allantoinase and allantoicase complex, its subunit locations of the 2 enzymes, and its comparison with fish allantoinase and allantoicase. J. Biol. Chem. 261:4221-4223. [PubMed] [Google Scholar]
- 34.Okumura, I., K. Kondo, Y. Miyaki, K. Itaya, and T. Yamamoto. 1976. Stereospecificity of conversion of uric acid into allantoic acid by enzymes of Candida utilis. J. Biochem. 79:1013-1019. [DOI] [PubMed] [Google Scholar]
- 35.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park, I.-S., and R. P. Hausinger. 1995. Requirement of carbon dioxide for in vitro assembly of the urease nickel metallocenter. Science 267:1156-1158. [DOI] [PubMed] [Google Scholar]
- 37.Pettigrew, D. W., R. R. Bidigare, B. J. Mehta, M. I. Williams, and E. G. Sander. 1985. Dihydroorotase from Clostridium oroticum. Purification and reversible removal of essential zinc. Biochem. J. 230:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopalan, P. T. R., X. C. Yu, and D. Pei. 1997. Peptide deformylase: a new type of mononuclear iron protein. J. Am. Chem. Soc. 119:12418-12419. [Google Scholar]
- 39.Reynolds, P. H. S., M. J. Boland, D. G. Blevins, D. D. Randall, and K. R. Schubert. 1982. Ureide biogenesis in leguminous plants. Trends Biochem. Sci. 7:366-368. [Google Scholar]
- 40.Romanov, V., M. T. Merski, and R. P. Hausinger. 1999. Assays for allantoinase. Anal. Biochem. 268:49-53. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Sarma, A. D., P. Serfozo, K. Kahn, and P. A. Tipton. 1999. Identification and purification of hydroxyisourate hydrolase, a novel ureide-metabolizing enzyme. J. Biol. Chem. 274:33863-33865. [DOI] [PubMed] [Google Scholar]
- 43.Schubert, K. R. 1981. Enzymes of purine biosynthesis and catabolism in Glycine max. I. Comparison of activities with N2 fixation and composition of xylem exudate during nodule development. Plant Physiol. 68:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song, H. K., S. B. Mulrooney, R. Huber, and R. P. Hausinger. 2001. Crystal structure of Klebsiella aerogenes UreE, a nickel-binding metallochaperone for urease activation. J. Biol. Chem. 276:49359-49364. [DOI] [PubMed] [Google Scholar]
- 45.Sträter, N., T. Klabunde, P. Tucker, H. Witzel, and B. Krebs. 1995. Crystal structure of a purple acid phosphatase containing a dinuclear Fe(III)-Zn(II) active site. Science 268:1489-1492. [DOI] [PubMed] [Google Scholar]
- 46.Syldatk, C., O. May, J. Altenbuchner, R. Mattes, and M. Siemann. 1999. Microbial hydantoinases—industrial enzymes from the origin of life? Appl. Microbiol. Biotechnol. 51:293-309. [DOI] [PubMed] [Google Scholar]
- 47.Thoden, J. B., G. N. Phillips, T. M. Neal, F. M. Raushel, and H. M. Holden. 2001. Molecular structure of dihydroorotase: a paradigm for catalysis through the use of a binuclear metal center. Biochemistry 40:6989-6997. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, R. J., and L. E. Schrader. 1981. The assimilation of ureides in shoot tissues of soybeans. 1. Changes in allantoinase activity and ureide contents of leaves and fruits. Plant Physiol. 67:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanni, A., E. Pessione, L. Anfossi, C. Baggiani, M. Cavaletto, M. Gulmini, and C. Giunta. 2000. Properties of a cobalt-reactivated form of yeast alcohol dehydrogenase. J. Mol. Catal. B Enzym. 9:283-291. [Google Scholar]
- 50.Vogels, G. D. 1969. Stereospecificity in the allantoin metabolism. Antonie Leeuwenhoek 35:236-238. [DOI] [PubMed] [Google Scholar]
- 51.Vogels, G. D., F. E. de Windt, and W. Bassie. 1969. Hydrolysis and racemization of allantoin. Recl. Trav. Chim. 88:940-950. [Google Scholar]
- 52.Vogels, G. D., F. Trijbels, and A. Uffink. 1966. Allantoinases from bacterial, plant and animal sources. I. Purification and enzymatic properties. Biochim. Biophys. Acta 122:482-496. [Google Scholar]
- 53.Vogels, G. D., and C. van der Drift. 1966. Allantoinase from bacterial, plant and animal sources. II. Effect of bivalent cations and reducing substances on the enzymatic activity. Biochim. Biophys. Acta 122:497-509. [Google Scholar]
- 54.Walker, K. W., and R. A. Bradshaw. 1999. Yeast methionine aminopeptidase. I. Alteration of substrate specificity by site-directed mutagenesis. J. Biol. Chem. 274:13403-13409. [DOI] [PubMed] [Google Scholar]
- 55.Webb, M. A., and J. S. Lindell. 1993. Purification of allantoinase from soybean seeds and production and characterization of anti-allantoinase antibodies. Plant Physiol. 103:1235-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson, D. K., and F. A. Quiocho. 1993. A pre-transition-state mimic of an enzyme. X-ray structure of adenosine deaminase with bound 1-deazaadenosine and zinc-activated water. Biochemistry 32:1689-1694. [DOI] [PubMed] [Google Scholar]
- 57.Yang, C., D. Carlow, R. Wolfenden, and S. A. Short. 1992. Cloning and nucleotide sequence of the Escherichia coli cytidine deaminase (ccd) gene. Biochemistry 31:4168-4174. [DOI] [PubMed] [Google Scholar]
- 58.Yu, L., A. Haddy, and F. Rusnak. 1995. Evidence that calcineurin accommodates an active site binuclear metal center. J. Am. Chem. Soc. 117:10147-10148. [Google Scholar]