Abstract
The ability to invade and grow in macrophages is necessary for Mycobacterium tuberculosis to cause disease. We have found a Mycobacterium marinum locus of two genes that is required for both invasion and intracellular survival in macrophages. The genes were designated iipA (mycobacterial invasion and intracellular persistence) and iipB. The iip mutant, which was created by insertion of a kanamycin resistance gene cassette at the 5′ region of iipA, was completely avirulent to zebra fish. Expression of the M. tuberculosis orthologue of iipA, Rv1477, fully complemented the iip mutant for infectivity in vivo, as well as for invasion and intracellular persistence in macrophages. In contrast, the iipB orthologue, Rv1478, only partially complemented the iip mutant in vivo and restored invasion but not intracellular growth in macrophages. While IipA and IipB differ at their N termini, they are highly similar throughout their C-terminal NLPC_p60 domains. The p60 domain of Rv1478 is fully functional to replace that of Rv1477, suggesting that the N-terminal sequence of Rv1477 is required for full virulence in vivo and in macrophages. Further mutations demonstrated that both Arg-Gly-Asp (RGD) and Asp-Cys-Ser-Gly (DCSG) sequences in the p60 domain are required for function. The iip mutant exhibited increased susceptibility to antibiotics and lysozyme and failed to fully separate daughter cells in liquid culture, suggesting a role for iip genes in cell wall structure and function. Altogether, these studies demonstrate an essential role for a p60-containing protein, IipA, in the pathogenesis of M. marinum infection.
Mycobacterium tuberculosis is an extraordinarily successful human pathogen, with 2 to 3 billion people infected worldwide (12). Its success likely reflects its complex parasitic lifestyle with sophisticated mechanisms for combating host defense. For example, M. tuberculosis inhibits acidification of the bacterium-containing phagosome and its fusion to lysosomes (6, 29, 33). Within the “maturation-arrested” phagosome, M. tuberculosis proliferates and ultimately kills the host cells by apoptosis and/or necrosis (11, 13, 22). The ability to invade and grow inside host cells is an important virulence property for pathogenic mycobacteria, and infection of macrophages by M. tuberculosis plays a key role in initiating a primary infection in the lung (29). In recent years, some progress has been made toward understanding the M. tuberculosis molecules involved both in invasion and in intracellular growth. For example, the heparin-binding hemagglutinin adhesin has been shown to mediate M. tuberculosis adherence to epithelial cells and to potentiate extrapulmonary dissemination of M. tuberculosis (23). An exported repetitive protein (Erp) of M. tuberculosis and a Mycobacterium marinum homologue of M. tuberculosis Rv3881c both are required for intracellular growth in cultured macrophages and virulence in vivo (3, 16). However, as M. tuberculosis is known to utilize multiple mechanisms for invasion (14), there must be additional mycobacterial molecules that mediate invasion through discrete mechanisms. In fact, the M. tuberculosis molecule(s) required for invasion of macrophages has not yet been identified. In addition, much remains to be understood about how M. tuberculosis has transformed the macrophage phagosome into a replicative niche.
We initiated studies aimed at identifying Mycobacterium genes involved in invasion and intracellular growth in macrophages. Our strategy is to screen an M. marinum mutant library to identify mutants defective for invasion and intracellular growth in macrophages. M. marinum is a natural pathogen of fish and amphibians and an opportunistic pathogen of humans, which in recent years has been used increasingly as a model to study the pathogenesis of mycobacterial infections (10, 15-17, 26, 27, 31). M. marinum is genetically closely related to M. tuberculosis (32), and many aspects of the diseases it causes in fish and frogs are similar to those of tuberculosis in humans (8, 27, 31). Like M. tuberculosis, M. marinum grows and persists in host macrophages both in vivo and in vitro (4, 25). This screen led to identification of a putative operon of two genes, iipA (mycobacterial invasion and intracellular persistence) and iipB, homologous to Rv1477 and Rv1478, respectively, of M. tuberculosis. This paper examines in detail the virulence function of iipA and iipB in vivo, as well as the physiological function of the two gene products.
MATERIALS AND METHODS
Bacterial strains and media.
M. marinum strain M (ATCC BAA-535) and the DH5α strain of Escherichia coli were cultured and genetically manipulated as described previously (15, 17). The temperature range for optimal growth of M. marinum is 28 to 33°C. Unless otherwise specified, M. marinum strains were cultured at 32°C in 7H9 liquid medium (Difco) supplemented with ADC (Difco), 0.01% Tween 80, and 200 mM d-sorbitol or on 7H10 agar (Difco) supplemented with ADC, 0.001% oleic acid, and 200 mM d-sorbitol. Growth of M. marinum in vitro was examined by inoculating the mid-log-phase culture into fresh 7H9 at an optical density at 600 nm (OD600) of ∼0.1, followed by multiple measurements of OD600 at defined time points. To ensure an accurate reading of OD600, the bacterial clumps were disrupted by passage through a 26-gauge needle three times before measurement of optical density. In our practice, multiple passages through the needle disrupt large cell clumps to create significantly smaller ones that sediment at a rate low enough for consistent measurements of OD600, and, therefore, measurement of optical density becomes a reliable method for determining the growth status of each of the various M. marinum strains.
Culture, maintenance, and infection of macrophages.
J774 cells (ATCC TIB67) and bone marrow-derived macrophages (BMDMs) (C57BL/6J) were cultured and maintained at 37°C as described previously (15). Six-well plates were seeded with J774 cells and BMDMs to reach 1 × 106 and 5 × 105 cells/well, respectively, at the time of infection. M. marinum strains grown to the late log phase were processed to isolate single bacteria (16, 17). The inocula were determined by both microscopic counting of the single bacterial cells of the inocula and enumeration of the CFU of the inocula on 7H10 agar plates. The bacteria were added to the cell monolayers at a multiplicity of infection (MOI) of 5 in the absence of opsonization and incubated for 2 h at 32°C. When necessary, amikacin (200 μg/ml) was added to the cells and incubated for another 2 h at 32°C to kill extracellular bacteria. The infected cells were further incubated at 32°C, and the bacterial CFU were evaluated either at the end of amikacin treatment (to evaluate invasion) or after 4 days (to evaluate intracellular growth) by hypotonic lysis of the infected cells and plating on 7H10 agar plates, as described previously (15, 17). For detection of M. marinum bacteria associated with macrophages, the green fluorescent protein (GFP)-expressing bacteria were used and the infected cells were examined by fluorescent microscopy (16).
Isolation of M. marinum mutant with transposon insertion in the iip locus.
An M. marinum transposon mutant library was generated by the M4 procedure as previously described (15). A total of ∼1,000 mutants were screened to identify the ones with defective growth in J774 cells and BMDMs. The transposon insertion site was retrieved and sequenced (15), and sequence similarity searches were performed using BLASTp and BLASTn at the NCBI site (http://www.ncbi.nlm.nih.gov/BLAST) and the newly finished (unpublished) M. marinum genome database at the Sanger Center site (http://www.sanger.ac.uk/Projects/M_marinum/).
Generation of M. marinum iipA::kan mutant by homologous recombination.
A 3.6-kb DNA fragment (IIP) was PCR amplified from the M. marinum genome using primers IIPA::kan-F1 (5′ GCCACGACCTGCCTACTGTCAGGCTAGGG 3′) and IIPA::kan-R1 (5′ CCATCGATACGGGTACCGACGATGTCGGTCG 3′) (Invitrogen), as described previously (15). The IIP fragment cut with EcoRI and ClaI was ligated to the EcoRI/ClaI-cut pBluescript (Stratagene) to generate pBS.IIP. The cloned pBS.IIP plasmid was digested with AgeI that cut in the 5′ coding region of iipA. The AgeI-cut plasmid was blunt ended and ligated to the kanamycin resistance (Kanr) gene cassette cut from pUC4K (Pharmacia) with SalI and blunt ended, generating the pBS.iipA::kan plasmid. The blunt-ended ClaI-EcoRI fragment of pBS.iipA::kan was ligated to pLYG304.Zeo (16), which was cut with XbaI and blunt ended to generate the plasmid for homologous recombination. Correct construction of the plasmid was confirmed by sequencing the junctions between the iipA and Kanr genes and between the entire insert sequence and the plasmid backbone. The plasmid was introduced into the M. marinum wild type (WT) by electroporation (15), and the transformants were first selected for resistance to kanamycin (40 μg/ml). The Kanr colonies were then plated on plates containing kanamycin and sucrose (10%). The Kanr and sucrose-resistant colonies were further confirmed for homologous recombination by sensitivity to Zeocin (50 μg/ml) (Invitrogen), as described previously (16). Replacement of the WT iipA gene with iipA::kan on the chromosome was confirmed by PCR, in which the forward primer anneals to sequence upstream from the IIP fragment (5′ CATCTCGGTGACGGTGAGCACCACGTCG 3′) and the reverse primer is in the Kanr gene (5′ CTGGCAGTGTTCCTGCGCCGGTTGCATTCG 3′ or 5′ CGAATGCAACCGGCGCAGGAACACTGCCAG 3′, depending on orientation of the Kanr gene). Double crossing-over was confirmed by a second PCR using a forward primer in the Kanr gene (5′ CTGGCAGTGTTCCTGCGCCGGTTGCATTCG3′ or 5′ CGAATGCAACCGGCGCAGGAACACTGCCAG 3′, depending on orientation of the Kanr gene) and a reverse primer that anneals to a sequence downstream from the IIP fragment. The sizes of the PCR products were confirmed by agarose gel electrophoresis, and the sequences were confirmed by sequencing.
Quantitative PCR analysis.
Mid-log-phase bacterial cells were broken by bead beating in the presence of lysozyme and the QIAGEN RNAProtect bacterial reagent (QIAGEN), and RNA was isolated using the RNeasy kit from QIAGEN. After treatment of the resulting RNA preparations with RNase-free DNase, cDNA was generated using SuperScript III RNase H− reverse transcriptase (Invitrogen). Quantitative PCR was performed with the Stratagene SYBR green master mix kit on a Stratagene MX4000 PCR machine. The oligonucleotides used to amplify iipA were 5′ CAACAAGCGGTCAAAGACGCCAAT-3′ (forward) and 5′-TCCGACGGGCCATTCATATAGGT-3′ (reverse). The oligonucleotides used to amplify iipB were 5′-GCAAATAGGATCCCGCGCGTTTAT-3′ (forward) and 5′ TCGAAACCGACGGTGTTGGCAC-3′ (reverse).
Examination of maturation of M. marinum-containing phagosomes.
BMDMs infected by the M. marinum WT (MOI, 5) or the iip mutants (MOI, 20) for 24 h were incubated for 40 min with Lysotracker (100 nM; Molecular Probes), washed to remove residual Lysotracker, and further incubated for 1 h before imaging. A higher MOI was used for the iipA::kan mutant than for the WT, because of less efficient invasion by the mutant.
Complementation of the M. marinum iipA::kan mutant.
The coding sequences of iipA, iipB, and iipA plus iipB were PCR amplified from the M. marinum genome using primer pairs 1, 2, and 3, respectively (Table 1). The fragments were cloned individually into pLYG206.Zeo, as described previously (17). Similarly, the coding sequences of Rv1477, Rv1478, and Rv1477 plus Rv1478 were PCR amplified from the M. tuberculosis H37Rv genome (7) using primer pairs 4, 5, and 6, respectively, and cloned individually into pLYG206.Zeo. To delete the p60 domain from Rv1477, the fragment Rv1477Δp60 was PCR amplified from the M. tuberculosis H37Rv genome using primer pair 7. To replace the p60 domain of Rv1477 with that of Rv1478, a “sewing PCR” technique was used. In brief, the sequence of Rv1477 upstream from the p60 domain was PCR amplified from the M. tuberculosis genome using primer pair 8, and the p60 domain of Rv1478 was amplified using primer pair 9. Both sequences were “sewn” together using primer pair 10. The same technique was used to generate point mutations in the p60 domain of Rv1477. To make point mutations from GD to AA, primer pairs 11 and 12 were first used to generate the upstream and downstream sequences and primer pair 13 was used to “sew” together the two sequences. To mutate DCSG to AAAA, primer pairs 14 and 15 were first used to generate the upstream and downstream sequences, and primer pair 16 was used to “sew” together the two sequences. These fragments were cloned individually into pLYG206.Zeo. The above complementation plasmids were introduced into the iipA::kan mutant by electroporation as described previously (15).
TABLE 1.
Primers used for complementation study
| Primer pair | Gene(s) to amplify | Primer sequence
|
|
|---|---|---|---|
| Forward | Reverse | ||
| 1 | iipA | GGTACGTAGACGGACCCGCCGTAGCTCAG | GGCGAATTCTAGTACTCGATGTAGCGGACCACGAAGG |
| 2 | iipB | GCCCTACGTACCCGATACAAGCGATTTCGCCTACTC | GCGGAATTCAGTACTCGATGATCCTAGTCACGAAGGG |
| 3 | iipA + iipB | GGTACGTAGACGGACCCGCCGTAGCTCAG | GCGGAATTCAGTACTCGATGATCCTAGTCACGAAGGG |
| 4 | Rv1477 | GATATCTGAGACGGAATCGCCGTGGCTCGC | CAAGCTTCGCGTGTGGCGCATGAATCCTCG |
| 5 | Rv1478 | TGGCCATACGCCACACGCGTTTTCACCCGA | GAAGCTTGTGCCCGGCAAATCACACCTGGC |
| 6 | Rv1477 + Rv1478 | GATATCTGAGACGGAATCGCCGTGGCTCGC | GAAGCTTGTGCCCGGCAAATCACACCTGGC |
| 7 | Rv1477Δp60 | GATATCTGAGACGGAATCGCCGTGGCTCGC | GGCTCGAGTCAGCGGCGGATCACGTATTCAGAAG |
| 8 | Rv1477Δp60 + 2p60 | GATATCTGAGACGGAATCGCCGTGGCTCGC | CCCATCTGCGACCCGGCGCGGCGGATCACGTATTC |
| 9 | Rv1477Δp60 + 2p60 | GAATACGTGATCCGCCGCGCCGGGTCGCAGATGGG | CAAGCTTCGCGTGTGGCGCATGAATCCTCG |
| 10 | Rv1477Δp60 + 2p60 | GATATCTGAGACGGAATCGCCGTGGCTCGC | CAAGCTTCGCGTGTGGCGCATGAATCCTCG |
| 11 | Rv1477/GD→AA | GATATCTGAGACGGAATCGCCGTGGCTCGC | CGTAGAAGATGACGGCGGCGCGGCGCATCTGC |
| 12 | Rv1477/GD→AA | GCAGATGCGCCGCGCCGCCGTCATCTTCTACG | CAAGCTTCGCGTGTGGCGCATGAATCCTCG |
| 13 | Rv1477/GD→AA | GATATCTGAGACGGAATCGCCGTGGCTCGC | CAAGCTTCGCGTGTGGCGCATGAATCCTCG |
| 14 | Rv1477/DCSG→AAAA | GATATCTGAGACGGAATCGCCGTGGCTCGC | GAGTACAACACCAGGGCTGCGGCGGCGAAGCCGACGGTG |
| 15 | Rv1477/DCSG→AAAA | CACCGTCGGCTTCGCCGCCGCAGCCCTGGTGTTGTACTC | CAAGCTTCGCGTGTGGCGCATGAATCCTCG |
| 16 | Rv1477/DCSG→AAAA | GATATCTGAGACGGAATCGCCGTGGCTCGC | CAAGCTTCGCGTGTGGCGCATGAATCCTCG |
Zebra fish infection and virulence determination.
The zebra fish AB strain was maintained as described previously (18). Infection of zebra fish (intraperitoneally), determination of virulence, and bacterial growth in fish organs were performed as described previously (16), in compliance with the University of California, San Francisco, animal research policies.
Examination of M. marinum cell separation and cording.
To examine M. marinum cell separation, late-log-phase cultures were processed for isolation of single bacteria, which were then grown for 4 days without shaking in 7H9 in a 12-well plate at a starting density of 107 bacteria/ml. Bacterial morphologies were observed by fluorescent microscopy or transmission electron microscopy. To examine M. marinum cording, the bacteria were grown in 7H9 with shaking to reach mid-log phase before imaging.
Examination of M. marinum resistance to antibiotics and lysozyme.
Susceptibility of M. marinum to antibiotics was determined on 7H10 agar as described previously (17), and the MIC was determined as the minimal concentration of an antibiotic that completely inhibited appearance of colonies on agar. For testing susceptibility of M. marinum to lysozyme, a suspension of single bacteria was inoculated into 7H9 (107/ml) containing various concentrations of lysozyme and incubated for 24 h, followed by enumeration of CFU on 7H10, similarly to what we previously described (17).
RESULTS
Identification of iip genes required for M. marinum invasion and intracellular survival in macrophages.
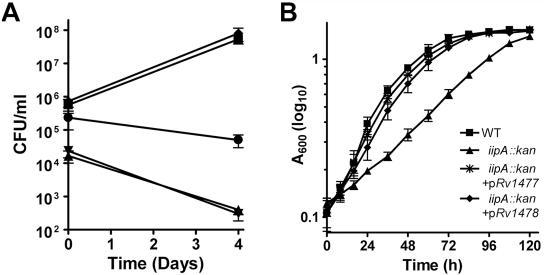
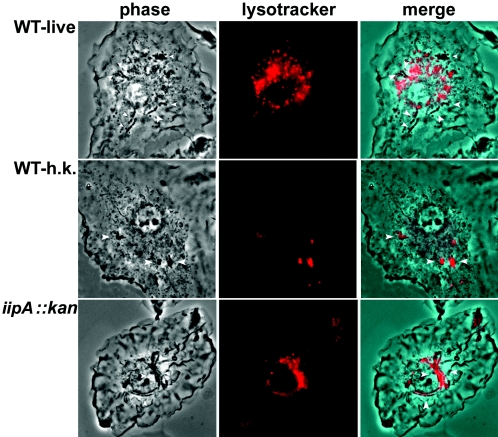
Our screen of an M. marinum transposon mutant library developed by the M4 procedure (15, 17) identified one mutant, 6H5, that exhibited a significantly reduced ability to invade and survive in J774 cells and BMDMs. This mutant had two transposon insertions: one in the promoter of a two-gene operon homologous to Rv1477 and Rv1478 of M. tuberculosis and the other one in a gene without an M. tuberculosis homologue. Initial complementation analysis suggested that the defects in invasion and intracellular survival of 6H5 were due to disruption of the Rv1477/Rv1478 homologues (data not shown), so we created an M. marinum mutant (iipA::kan) by inserting a kanamycin resistance gene cassette at the 5′ region of the Rv1477 gene homologue by homologous recombination. Infections of BMDMs by iipA::kan and WT strains were compared, in which prepared single bacterial cells were incubated with macrophages at an MOI of 5 (Fig. 1). The numbers of iipA::kan CFU associated with BMDMs were 80-fold lower than those of the WT immediately after 2 h of infection and another 2 h of antibiotic treatment to remove extracellular bacteria (Fig. 1A). The lower number of CFU for the mutant bacteria associated with BMDMs was not due to an inoculum lower than that of the WT, because the same inoculum sizes for both strains were ensured by (i) microscopic counting of the single bacterial cells of the inocula and (ii) enumeration of the CFU of the inocula on 7H10 agar plates (data not shown). By 4 days after infection, WT had multiplied in the cells ∼100-fold, while the iipA::kan mutant showed a drop in CFU to less than 5% of the level present at 4 h of infection (Fig. 1A). In striking contrast to this inability to survive in macrophages, iipA::kan cells grew in 7H9, although somewhat slower than WT cells (Fig. 1B). The loss of viability for the iipA::kan strain in macrophages correlated with fusion of the bacterium-containing phagosomes to lysosomes (Fig. 2). While the majority of WT bacteria did not colocalize with the late endosome/lysosome marker, Lysotracker, the iipA::kan bacteria predominantly colocalized with Lysotracker at a level similar to that of heat-killed WT bacteria.
FIG. 1.
(A) Invasion and intracellular survival of M. marinum iip mutants in macrophages. BMDMs (5 × 105/well) were infected by the WT strain (▪), strain 6H5 (▾), the iipA::kan mutant (▴), and the iipA::kan mutant complemented by M. tuberculosis Rv1477 (⧫) or Rv1478 (•). The cells were infected at an MOI of 5 for 2 h, followed by a 2-h treatment with amikacin to kill extracellular bacteria. The CFU were determined either immediately after amikacin treatment (day 0) or at 4 days after infection (day 4). (B) Growth of M. marinum strains in 7H9 liquid medium, starting with inocula of an OD600 of approximately 0.1, followed by multiple measurements. The bacterial clumps were disrupted by passage of the culture suspensions through a 26-gauge needle three times.
FIG. 2.
Detection of maturation of M. marinum-containing phagosomes. BMDMs were incubated with WT live (WT-live) or heat-killed (WT-h.k.) bacteria at an MOI of 5 or the iipA::kan mutant bacteria at an MOI of 20. After 2 h of incubation, extracellular bacteria were washed off and the cells were further incubated for 24 h prior to incubation with Lysotracker (red) for detection of the late endosome/lysosome compartments. The phase and Lysotracker images were merged to determine colocalization of M. marinum with late endosome/lysosomes. Compared to WT live bacteria, the WT heat-killed bacteria and the iipA::kan mutant bacteria both showed predominant colocalization with late endosomes/lysosomes.
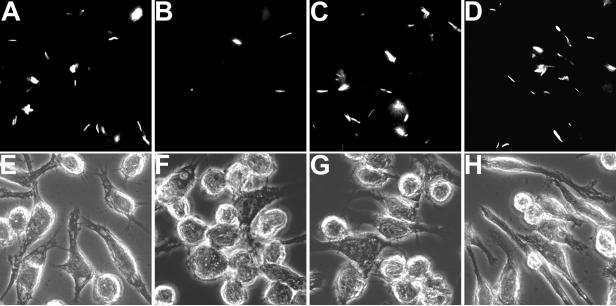
To determine whether the low number of macrophage-associated iipA::kan CFU immediately after infection was due to rapid death of the bacteria after invasion or to failure of bacterial uptake into the cells, we examined the association of WT and mutant bacteria with macrophages at 2 h of infection using GFP-expressing bacteria. As heat-killed GFP-expressing WT or iipA::kan mutant bacteria maintained GFP fluorescence in BMDMs for more than 3 h (data not shown), we set a criterion that all bacteria (WT or mutant) that were taken up by the macrophages for up to 2 h would exhibit detectable GFP fluorescence. The WT had an average of ∼3 bacteria/cell, whereas the iipA::kan mutant had an average of only ∼0.1 bacterium/cell, based on counting the numbers of bacteria by fluorescence and the numbers of cells by phase channels (Fig. 3). Thus, the iipA::kan mutant is defective at an early step for entry into BMDMs, as well as for intracellular survival (or persistence) in macrophages. For this reason, we have named the putative M. marinum operon homologous to Rv1477/Rv1478 the iip (macrophage invasion and intracellular persistence) locus and the two genes iipA and iipB. The encoded proteins, IipA and IipB, are very similar to the M. tuberculosis Rv1477 and Rv1478 products (7). Both genes are highly conserved among Mycobacterium species, including M. marinum, M. tuberculosis, M. bovis, M. leprae, M. avium, and M. smegmatis.
FIG. 3.
Detection of invasion of macrophages by M. marinum strains. BMDMs were infected for 2 h at an MOI of 5 by the WT (A and E), the iipA::kan mutant (B and F), or the mutant complemented by Rv1477 (C and G) or Rv1478 (D and H) expressing GFP. The infected cells were washed to remove extracellular bacteria and observed for cell-associated bacteria using a fluorescent microscope. Panels E, F, G, and H are the phase images of the fluorescent images in panels A, B, C, and D, respectively.
Quantitative PCR measurements demonstrated that iipB transcription was disrupted in the iipA::kan mutant (data not shown), indicating that iipA and iipB are in an operon and that the iipA::kan insertion mutation disrupted expression of iipB.
Complementation of the iipA::kan mutant by M. tuberculosis genes.
To determine whether the M. tuberculosis locus had a similar function and to determine the roles for the two genes in the putative operon, we performed complementation of the iipA::kan mutant with Rv1477 and Rv1478 genes. Expression of Rv1477 alone fully complemented the iipA::kan mutant for invasion and intracellular survival (Fig. 1A) and the in vitro growth lag (Fig. 1B), which was indistinguishable from complementation by both Rv1477 and Rv1478. However, while Rv1478 alone complemented the iipA::kan mutant for invasion, it did not rescue the intracellular survival defect (Fig. 1). Similarly, the iipA gene of M. marinum completely corrected the invasion and intracellular growth defects of the iipA::kan mutant and the iipB gene recovered invasion only (data not shown), emphasizing the functional homology of the M. marinum and M. tuberculosis genes. These results indicate that iipA and iipB have overlapping functions for invasion, while iipA has an additional role in intracellular persistence.
M. marinum iip genes are essential for virulence in zebra fish.
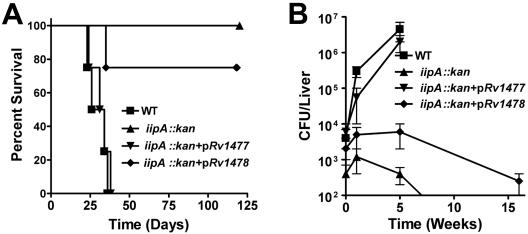
To determine whether the iip locus and the individual genes were necessary for in vivo virulence, we infected zebra fish with the WT, the iipA::kan mutant, and complemented strains. As shown in Fig. 4A, at a dose of 105 bacteria/fish, the WT caused 100% mortality within 3 to 5 weeks. In contrast, the iipA::kan strain was completely attenuated, with none of the infected fish dying during the entire 16 weeks of the experiment. As in macrophage infection, complementation by Rv1477 completely restored the in vivo virulence of the iipA::kan strain. However, the Rv1478 strain had little effect, as only one of the four fish infected by this complementation strain died, which was not statistically different from the iipA::kan strain. By 5 weeks postinfection, the CFU of both the WT and Rv1477-complemented mutant increased about 1,000-fold in the livers (Fig. 4B). In contrast, at 1 week of infection, the iipA::kan mutant already showed 2 logs fewer CFU than the WT, and by 8 weeks of infection, CFU were undetectable in the livers. Although the Rv1478-complemented mutant persisted in the livers for 5 weeks, it was significantly killed by the fish at late stage. These results demonstrated that iipA has a critical and nonredundant role in the virulence of M. marinum to zebra fish.
FIG. 4.
Role of iip genes in M. marinum infection of zebra fish. (A) Death curve for zebra fish infected (intraperitoneally) by the M. marinum strains listed in the inset (105/fish). For each M. marinum strain, 4 fish were infected, and fish were observed for 16 weeks. (B) Growth of the M. marinum strains in the livers of zebra fish infected under the same conditions as in panel A. Bacterial CFU were determined as described previously (16). Fish infected by the WT and the Rv1477-complemented mutant strain died before 10 weeks, and therefore no further CFU counts are shown.
IipA structural requirements for invasion and intracellular growth.
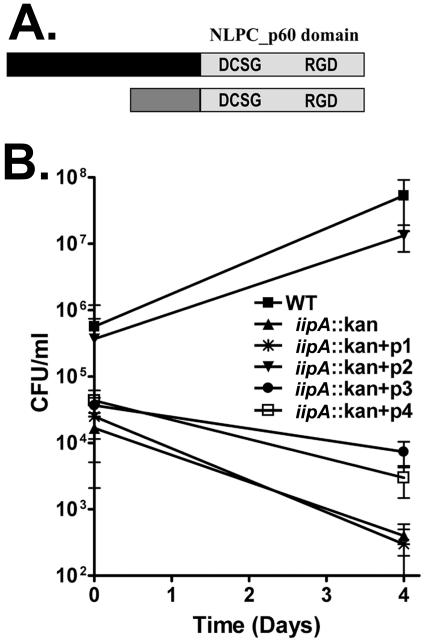
Alignment of the sequences of M. marinum IipA and IipB showed that they differed in their N termini, but were highly similar throughout their C termini, where an NLPC_p60 domain was identified (http://smart.embl-heidelberg.de/) for each (Fig. 5A). NLPC_p60 is a domain conserved through a variety of bacterial lineages. Although the function of this domain is not completely understood, some members of this family have been shown to act as murein hydrolases that cleave peptide linkages within peptidoglycan (1). As shown in Table 2, the p60 domains are 71% identical between M. marinum IipA and IipB and 69% identical between Rv1477 and Rv1478. Higher similarities were found between the p60 domains of the M. marinum IipA and Rv1477 (93% identical) and between the p60 domains of M. marinum IipB and Rv1478 (92% identical). To determine whether the p60 domain was required for Rv1477 function, we attempted to complement the iipA::kan strain with Rv1477 lacking the p60 domain (p1 in Fig. 5B). This strain was no different from the iipA::kan strain for invasion or persistence, demonstrating that the p60 domain is required for both functions. To determine whether the difference between the abilities of Rv1477 and Rv1478 to complement the mutant was due to differences in their p60 domains, we tested a hybrid molecule in which the p60 domain of Rv1478 was fused to the N-terminal domain of Rv1477 (p2 in Fig. 5B). This hybrid molecule completely restored both invasion and persistence of the iipA::kan strain, demonstrating that the p60 domains are functionally interchangeable in association with the Rv1477 N-terminal domain.
FIG. 5.
Requirement for NLPC_p60 motifs in Iip function. (A) Schematic illustrations of iipA and iipB and the signature sequences in the NLPC_p60 domain, DCSG and RGD. (B) Examination of the functions of the p60 domains and their signature sequences by complementation studies. BMDMs were infected by M. marinum WT, the iipA::kan mutant, or various complemented strains transduced with the following plasmids: p1, plasmid expressing Rv1477 with the p60 domain deleted; p2, plasmid expressing Rv1477 with the native p60 domain replaced by the p60 domain of Rv1478; p3, plasmid expressing Rv1477 with the RGD motif altered to RAA; and p4, plasmid expressing Rv1477 with the DCSG sequence altered to AAAA.
TABLE 2.
Similarities between the p60 domains of M. marinum and M. tuberculosis iip homologues
| p60 domain | % identitya
|
||
|---|---|---|---|
| IipA | IipB | Rv1477 | |
| IipB | 71 | ||
| Rv1477 | 93 | 72 | |
| Rv1478 | 69 | 92 | 69 |
The numbers indicate percentage of identity between the two amino acid sequences.
The RGD motif and the murein hydrolase activity site of the p60 domain of Rv1477 are necessary for invasion and intracellular persistence.
Examination of the p60 domains of Rv1477, Rv1478, and M. marinum Iips revealed that they all contain an RGD motif that is conserved among Rv1477/1478 homologues in all mycobacteria (Fig. 5A). The RGD sequence is well known to mediate binding of proteins to cell surface integrin receptors (2). As the predicted sequences of M. marinum and M. tuberculosis iip genes all have N-terminal signal peptides for protein secretion, and some p60 family proteins are known cell wall-associated proteins (5), we hypothesized that the RGD sequences of the Mycobacterium Iip proteins bind to host cell receptors for invasion. To begin to test this hypothesis, we mutated the glycine and aspartate residues (GD) of Rv1477 to alanines (AA) and expressed the mutant form of Rv1477 (p3 in Fig. 5B) to determine the effect of this mutation on function. The RGD mutation completely abolished the ability of Rv1477 to complement either phenotype, indicating that the RGD motif is necessary not only for invasion but also for intracellular persistence.
Several NLPC_p60 family proteins, including Listeria p60, have murein hydrolase activity. All of the p60 family murein hydrolases have a conserved DCSG sequence, in which the C residue has been demonstrated to be essential for activity (34) (Fig. 5A). To understand whether the function of iipA might involve peptidoglycanase activity, we mutated the DCSG residues of Rv1477 to AAAA and expressed the mutant form of Rv1477 in the iipA::kan mutant (p4 in Fig. 5B). Like the mutation of RGD, the mutation of DCSG completely abrogated the ability of Rv1477 to complement the iipA::kan mutant for invasion and intracellular growth.
The p60 domains of Rv1477 and Rv1478 are required for separation of dividing bacterial cells.
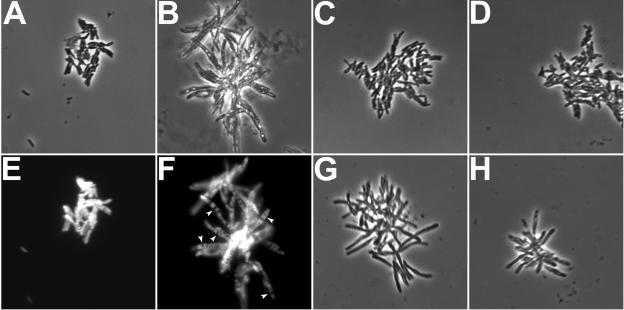
The p60 protein in Listeria and p60 domain-containing protein in Streptococcus have been shown to possess peptidoglycanase activity and are required for separation of daughter cells after cell division. Because our mutation analysis suggested that the putative peptidoglycan peptidase active site was required for virulence of M. marinum, we examined bacterial separation during in vitro growth. Isolated single bacterial cells were grown in 7H9 liquid medium without shaking for 4 days prior to imaging. As seen in Fig. 6A and B, the iipA::kan mutant frequently formed filaments longer than the WT. When the morphology of the GFP-expressing bacteria was examined, the WT showed predominantly short rods with evenly distributed GFP (Fig. 6E). In contrast, the iipA::kan strain formed longer filaments with a beaded pattern of GFP expression (Fig. 6F). Expression of either Rv1477 or Rv1478 in the iipA::kan mutant completely recovered the morphological defect of the mutant (Fig. 6C). The ability of Rv1478 to correct the morphological abnormalities suggests that the virulence defect of the mutant is not strictly dependent on its morphological problems. Moreover, in late log phase, the morphology of the mutant bacteria was much closer to WT (data not shown), as has been reported for Listeria (24); in fact, these are the bacteria that were used for the infection assays, emphasizing that the morphological and virulence defects are not strictly correlated. Expression of Rv1477 with the GD→AA or DCSG→AAAA mutation failed to complement the morphological abnormalities of the iipA::kan mutant (Fig. 6G and H), suggesting that in addition to DCSG, the RGD sequence is required for murein hydrolase activity and bacterial cell separation.
FIG. 6.
Requirement of Iips for normal bacterial morphology. M. marinum strains expressing GFP were grown to the late log phase, processed to isolate single bacteria, and subcultured without shaking for 4 days before imaging. Panels A and B are the phase images for the fluorescent images in panels E and F, respectively. (A and E) WT strain. (B and F) iipA::kan mutant. (C) iipA::kan mutant complemented by Rv1477. (D) iipA::kan mutant complemented by Rv1478. (G) iipA::kan mutant complemented by Rv1477/GD→AA. (H) iipA::kan mutant complemented by Rv1477/DCSG→AAAA.
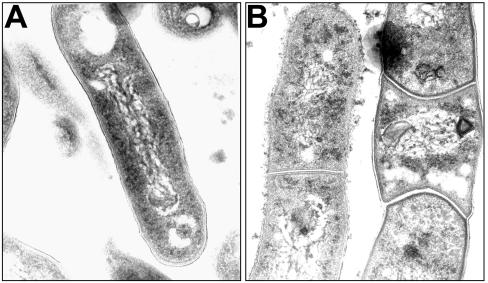
The morphology of the WT and iipA::kan strains was examined in greater detail by transmission electron microscopy. The bacteria were cultured under the same conditions described above. As seen in Fig. 7A, the WT showed a well-separated rod morphology. In contrast, the iipA::kan mutant frequently formed chains of shorter cells with an enlarged cross section, although septa were developed (Fig. 7B), consistent with a defect in separation of daughter cells.
FIG. 7.
Transmission electron microscopy of M. marinum. M. marinum WT (A) or iipA::kan mutant (B) single cells were grown for 4 days, vortexed vigorously in 7H9 liquid medium containing 0.05% Tween 80, and processed for transmission electron microscopy.
The p60 domains of Rv1477 and Rv1478 are required for cording of mycobacteria.
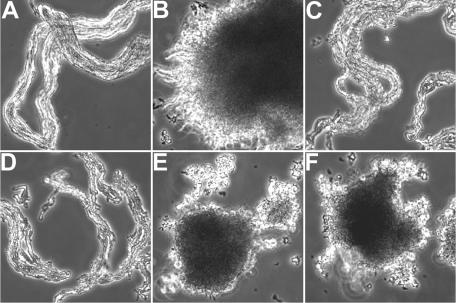
Cording is a major morphological feature of virulent mycobacteria in culture that is dependent on lipids and perhaps other components of the cell wall. Since the Iip proteins likely are enzymes involved in cell wall biosynthesis, we examined whether these proteins were required for cording. Different from the studies of bacterial cell separation described above, in which the bacterial cultures were grown without shaking, cording was assessed in cells grown with agitation, which facilitates cord formation. As seen in Fig. 8A, the WT formed cords, as indicated by ordered arrays of the bacteria in bundles. In contrast, the iipA::kan mutant formed disordered aggregates that lacked obvious cord formation (Fig. 8B). Expression of either Rv1477 or Rv1478 completely recovered the cording defect of the iipA::kan mutant (Fig. 8C and D, respectively). Expression of either Rv1477/GD→AA or Rv1477/DCSG→AAAA failed to restore cording of the iipA::kan mutant, demonstrating the role for the p60 domain of Rv1477 in cording of mycobacteria. Not surprisingly, mutations of either the GD or DCSG residues of Rv1477 failed to restore the somewhat low growth rate of the iipA::kan mutant in 7H9 liquid culture medium (data not shown), indicating that the normal cording morphology is in favor of mycobacterial growth in vitro.
FIG. 8.
Cording of the M. marinum strains grown to mid-log phase under shaking conditions. (A) WT. (B) iipA::kan mutant. (C) iipA::kan mutant complemented by Rv1477. (D) iipA::kan mutant complemented by Rv1478. (E) iipA::kan mutant complemented by Rv1477/RGD→RAA. (F) iipA::kan mutant complemented by Rv1477/DCSG→AAAA.
Rv1477 and Rv1478 have different effects on resistance to antibiotics and lysozyme.
We have previously shown that resistance to certain antibiotics is a sensitive assay for M. marinum cell wall impermeability (17). Since the Streptococcus p60 family protein (PcsB) is required for resistance to an array of antibiotics (28), we examined the antibiotic sensitivity of the WT, iipA::kan mutant, and complemented strains. As shown in Table 3, the iipA::kan mutant was 24- to 60-fold more sensitive to rifampin, ciprofloxacin, and erythromycin than the WT strain. Expression of Rv1477 restored the defect of the iipA::kan mutant essentially to WT levels. Rv1478 had a much smaller effect than Rv1477 on complementation of antibiotic susceptibility of the mutant. Expression of Rv1477 and Rv1478 together fully restored the iipA::kan strain. Thus, the antibiotic susceptibility patterns support the hypothesis that the iipA::kan strain has a defective cell wall. Interestingly, the effects of Rv1477 and Rv1478 on antibiotic susceptibility correlate with intracellular survival and virulence rather than bacterial morphology and cording.
TABLE 3.
MICs of antibiotics for M. marinum strains
| M. marinum strain | MIC (μg/ml)
|
||
|---|---|---|---|
| Rifampin | Ciprofloxacin | Erythromycin | |
| WT | 0.6 | 0.06 | 30 |
| iipA::kan | 0.025 | 0.001 | 0.5 |
| iipA::kan + pRv1477 | 0.4 | 0.02 | 15 |
| iipA::kan + pRv1478 | 0.1 | 0.008 | 3 |
| iipA::kan + pRv1477-1478 | 0.6 | 0.06 | 30 |
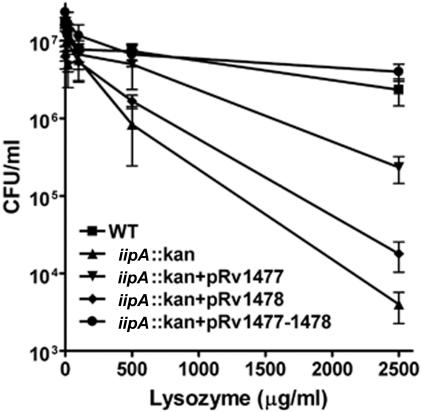
An important function of the mycobacterial cell wall is to confer resistance to host defense molecules such as lysozyme or defensin (17). As shown in Fig. 9, the iipA::kan mutant was much more susceptible than the WT to lysozyme. Resistance of the mutant was restored to a much greater extent by expression of Rv1477 than by expression of Rv1478. However, only expression of both Rv1477 and Rv1478 completely overcame susceptibility of the mutant to lysozyme. Thus, this assay reveals a requirement for both genes of the putative operon. Susceptibility to lysozyme, a hydrolytic enzyme present in macrophage lysosomes, may in part account for the failure of the iipA::kan mutant to survive after intracellular infection and for its marked attenuation during in vivo infections.
FIG. 9.
Susceptibility of M. marinum strains to lysozyme. M. marinum strains were grown to late log phase, processed for isolation of single bacteria, and incubated (107/ml) in the presence of various concentrations of lysozyme (0, 20, 100, 500, or 2,500 μg/ml) for 24 h prior to enumeration of CFU on 7H10 agar plates.
DISCUSSION
In this paper, we provide evidence that the iip locus is essential for M. marinum invasion and intracellular persistence in macrophages and for virulence to zebra fish. Since the iip locus is highly conserved between M. marinum and M. tuberculosis and the iip mutant can be fully complemented by the M. tuberculosis homologues, it is likely that the homologous Rv1477/1478 locus of M. tuberculosis has a similar essential role in M. tuberculosis virulence. This raises the possibility that targeting the iip locus may have great potential for novel drug and vaccine design. Because the locus, and presumably the function of Iips, is conserved in other mycobacteria, a drug targeting Iips' function might also have value in human diseases caused by other mycobacteria, such as M. leprae and M. avium. Indeed, a Listeria mutant in which p60 has been deleted is severely attenuated in vivo (19), suggesting the possibility that targeting p60 enzymatic activity could have relevance for diseases caused by a wide variety of bacterial pathogens.
The iip locus consists of two genes in tandem in all Mycobacterium species. The predicted amino acid sequences of iipA (or Rv1477) and iipB (or Rv1478) contain highly conserved C-terminal NLPC_p60 domains. Proteins containing NLPC_p60 domains in Listeria monocytogenes and Streptococcus have been shown to act as peptidoglycanases, and a highly conserved DCSG sequence has been implicated in the enzyme activity (20, 30). Many characteristics of the iipA::kan mutant are consistent with a similar function for the Iip proteins, as is the fact that mutation of DCSG in Rv1477 completely abolishes its ability to complement the separation defect of the iipA::kan mutant. Especially significant are the mutant bacterium's morphological defects, which are reminiscent of the abnormalities of the p60-deficient Listeria. We hypothesize that lack of this peptidoglycanase activity leads to the multiple cell wall defects of the iipA::kan strain, including separation defects, decreased cording, and decreased resistance to antibiotics and to lysozyme. Unfortunately, we have been unable so far to demonstrate enzymatic activity of recombinant Iip proteins made in E. coli to test this hypothesis further. Both cording and antibiotic resistance are known to depend on lipids in the mycobacterial cell wall, and it is intriguing to speculate that one role for the peptidoglycanase is to allow appropriate assembly of the complex mycobacterial cell wall lipid structures. However, this cannot be a global deficit, since electron microscopy demonstrates an apparently normal cell wall structure distant from the separation sites. Our data also suggest that decreased resistance to lysozyme may contribute to the failure of the iipA::kan mutant to survive in macrophages. The requirement for both Rv1477 and Rv1478 to fully complement lysozyme resistance is intriguing and suggests that increased in vivo survival of the bacteria may be the selection pressure that has conserved the two genes of the locus throughout the evolution of mycobacteria. Our failure to see a role for Rv1478 in vivo may reflect the small numbers of infections studied or the nonphysiologic route of fish infection.
Despite the similarity of their NLPC_p60 domains, Rv1477 complements the iipA::kan mutant almost completely, while Rv1478 is much less able to compensate for the mutant's defects. The fact that a hybrid molecule consisting of the Rv1477 N terminus and the Rv1478 p60 domain is able to complement as completely as Rv1477 clearly demonstrates that the difference between the two gene products lies in their N termini. There are several possible models for how these different N termini affect function. It is possible that the distinct N termini lead to different localizations of the peptidoglycanase activity of the Iip proteins and that IipA (Rv1477) localizes to a site more important for growth in fish, intracellular survival in macrophages, and resistance to lysozyme than does IipB (Rv1478). Alternatively, the N termini may affect the fine specificity of the Iip peptidoglycanases, and the IipA (Rv1477) substrates may be more significant for pathogenesis. Finally, the IipA (Rv1477) N terminus may have an additional unrecognized enzymatic activity that is required for pathogenesis. Further studies will be required to test those possibilities.
An intriguing aspect of the Iip proteins and their homologues in other Mycobacterium species is the presence of an RGD sequence, which is absent from all other NLPC_p60 family members. This RGD motif is required both for macrophage invasion and for intracellular growth of M. marinum. It is clear that this sequence is required for Iip activity on the cell wall, because mutation of the RGD leads to abnormal bacterial morphology and loss of cording. Many other members of the NLPC_p60 family, including Listeria monocytogenes and Streptococcus, also have the GD dipeptide in a homologous site of the domain, so it is likely that this sequence, like DCSG, is required for activity. However, RGD is also a canonical integrin binding motif. It is intriguing to note that the listerial p60 protein, lacking RGD, although required for virulence, is not required for invasion of cells (24), suggesting that this may be an additional role for the RGD sequence of the Iips.
There are three additional genes in the M. tuberculosis genome that encode proteins with the conserved p60 domain, including Rv0024, Rv1566c, and Rv2190c, none of which have RGD (7). Homologues of all three of these genes are present in the M. marinum genome. Functions for these additional mycobacterial p60 proteins have not been described. Interestingly, two earlier studies reported that Rv1477 and Rv1478 (21) and their homologues in M. avium (9) are highly expressed upon infection of macrophages. It is possible that the other mycobacterial p60 genes are expressed under different conditions and for different functions.
Acknowledgments
We thank Lalita Ramakrishnan (University of Washington, Seattle) for the GFP-expressing plasmid pmsp12.GFP.Apr. We thank Su Guo and Susie Lee (University of California, San Francisco) for assistance with some of the zebra fish work. We also thank members of the Brown laboratory (University of California, San Francisco) and members of the Gao laboratory (University of Maryland, College Park) for helpful suggestions and comments.
This work was supported by RO1AI055614; L.Y.G. was supported in part by T32AI07334.
Editor: J. T. Barbieri
REFERENCES
- 1.Anantharaman, V., and L. Aravind. 3. February 2003, posting date. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4:R11. [Online.] doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaout, M. A., S. L. Goodman, and J. P. Xiong. 2002. Coming to grips with integrin binding to ligands. Curr. Opin. Cell Biol. 14:641-651. [DOI] [PubMed] [Google Scholar]
- 3.Berthet, F. X., M. Lagranderie, P. Gounon, C. Laurent-Winter, D. Ensergueix, P. Chavarot, F. Thouron, E. Maranghi, V. Pelicic, D. Portnoi, G. Marchal, and B. Gicquel. 1998. Attenuation of virulence by disruption of the Mycobacterium tuberculosis erp gene. Science 282:759-762. [DOI] [PubMed] [Google Scholar]
- 4.Bouley, D. M., N. Ghori, K. L. Mercer, S. Falkow, and L. Ramakrishnan. 2001. Dynamic nature of host-pathogen interactions in Mycobacterium marinum granulomas. Infect. Immun. 69:7820-7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 6.Clemens, D. L., and M. A. Horwitz. 1995. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J. Exp. Med. 181:257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Cosma, C. L., D. R. Sherman, and L. Ramakrishnan. 2003. The secret lives of the pathogenic mycobacteria. Annu. Rev. Microbiol. 57:641-676. [DOI] [PubMed] [Google Scholar]
- 9.Danelishvili, L., M. J. Poort, and L. E. Bermudez. 2004. Identification of Mycobacterium avium genes up-regulated in cultured macrophages and in mice. FEMS Microbiol. Lett. 239:41-49. [DOI] [PubMed] [Google Scholar]
- 10.Davis, J. M., H. Clay, J. L. Lewis, N. Ghori, P. Herbomel, and L. Ramakrishnan. 2002. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17:693-702. [DOI] [PubMed] [Google Scholar]
- 11.Dobos, K. M., E. A. Spotts, F. D. Quinn, and C. H. King. 2000. Necrosis of lung epithelial cells during infection with Mycobacterium tuberculosis is preceded by cell permeation. Infect. Immun. 68:6300-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolin, P. J., M. C. Raviglione, and A. Kochi. 1994. Global tuberculosis incidence and mortality during 1990-2000. Bull. W. H. O. 72:213-220. [PMC free article] [PubMed] [Google Scholar]
- 13.Duan, L., H. Gan, D. E. Golan, and H. G. Remold. 2002. Critical role of mitochondrial damage in determining outcome of macrophage infection with Mycobacterium tuberculosis. J. Immunol. 169:5181-5187. [DOI] [PubMed] [Google Scholar]
- 14.Ernst, J. D. 1998. Macrophage receptors for Mycobacterium tuberculosis. Infect. Immun. 66:1277-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, L.-Y., R. Groger, J. S. Cox, S. M. Beverley, E. H. Lawson, and E. J. Brown. 2003. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 71:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, L. Y., S. Guo, B. McLaughlin, H. Morisaki, J. N. Engel, and E. J. Brown. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53:1677-1693. [DOI] [PubMed] [Google Scholar]
- 17.Gao, L. Y., F. Laval, E. H. Lawson, R. K. Groger, A. Woodruff, J. H. Morisaki, J. S. Cox, M. Daffe, and E. J. Brown. 2003. Requirement for kasB in Mycobacterium mycolic acid biosynthesis, cell wall impermeability and intracellular survival: implications for therapy. Mol. Microbiol. 49:1547-1563. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. A., E. L. Shen, A. Fiser, A. Sali, and S. Guo. 2003. The zebrafish forkhead transcription factor Foxi1 specifies epibranchial placode-derived sensory neurons. Development 130:2669-2679. [DOI] [PubMed] [Google Scholar]
- 19.Lenz, L. L., S. Mohammadi, A. Geissler, and D. A. Portnoy. 2003. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci. USA 100:12432-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lenz, L. L., and D. A. Portnoy. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol. Microbiol. 45:1043-1056. [DOI] [PubMed] [Google Scholar]
- 21.Mariani, F., G. Cappelli, G. Riccardi, and V. Colizzi. 2000. Mycobacterium tuberculosis H37Rv comparative gene-expression analysis in synthetic medium and human macrophage. Gene 253:281-291. [DOI] [PubMed] [Google Scholar]
- 22.Perskvist, N., M. Long, O. Stendahl, and L. Zheng. 2002. Mycobacterium tuberculosis promotes apoptosis in human neutrophils by activating caspase-3 and altering expression of Bax/Bcl-xL via an oxygen-dependent pathway. J. Immunol. 168:6358-6365. [DOI] [PubMed] [Google Scholar]
- 23.Pethe, K., S. Alonso, F. Biet, G. Delogu, M. J. Brennan, C. Locht, and F. D. Menozzi. 2001. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412:190-194. [DOI] [PubMed] [Google Scholar]
- 24.Pilgrim, S., A. Kolb-Mäurer, I. Gentschev, W. Goebel, and M. Kuhn. 2003. Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect. Immun. 71:3473-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramakrishnan, L., and S. Falkow. 1994. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect. Immun. 62:3222-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 27.Ramakrishnan, L., R. H. Valdivia, J. H. McKerrow, and S. Falkow. 1997. Mycobacterium marinum causes both long-term subclinical infection and acute disease in the leopard frog (Rana pipiens). Infect. Immun. 65:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinscheid, D. J., B. Gottschalk, A. Schubert, B. J. Eikmanns, and G. S. Chhatwal. 2001. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J. Bacteriol. 183:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell, D. G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2:569-577. [DOI] [PubMed] [Google Scholar]
- 30.Schubert, K., A. M. Bichlmaier, E. Mager, K. Wolff, G. Ruhland, and F. Fiedler. 2000. p45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21-28. [DOI] [PubMed] [Google Scholar]
- 31.Talaat, A. M., R. Reimschuessel, S. S. Wasserman, and M. Trucksis. 1998. Goldfish, Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect. Immun. 66:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tønjum, T., D. B. Welty, E. Jantzen, and P. L. Small. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vergne, I., J. Chua, S. B. Singh, and V. Deretic. 2004. Cell biology of Mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 20:367-394. [DOI] [PubMed] [Google Scholar]
- 34.Wuenscher, M. D., S. Köhler, A. Bubert, U. Gerike, and W. Goebel. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J. Bacteriol. 175:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]