Abstract
Salmonella enterica serovar Typhi causes typhoid fever in humans. Central to the pathogenicity of serovar Typhi is its capacity to invade intestinal epithelial cells. The role of lipopolysaccharide (LPS) in the invasion process of serovar Typhi is unclear. In this work, we constructed a series of mutants with defined deletions in genes for the synthesis and polymerization of the O antigen (wbaP, wzy, and wzz) and the assembly of the outer core (waaK, waaJ, waaI, waaB, and waaG). The abilities of each mutant to associate with and enter HEp-2 cells and the importance of the O antigen in serum resistance of serovar Typhi were investigated. We demonstrate here that the presence and proper chain length distribution of the O-antigen polysaccharide are essential for serum resistance but not for invasion of epithelial cells. In contrast, the outer core oligosaccharide structure is required for serovar Typhi internalization in HEp-2 cells. We also show that the outer core terminal glucose residue (Glc II) is necessary for efficient entry of serovar Typhi into epithelial cells. The Glc I residue, when it becomes terminal due to a polar insertion in the waaB gene affecting the assembly of the remaining outer core residues, can partially substitute for Glc II to mediate bacterial entry into epithelial cells. Therefore, we conclude that a terminal glucose in the LPS core is a critical residue for bacterial recognition and internalization by epithelial cells.
Salmonella enterica serovar Typhi causes typhoid fever, a disease that annually affects more than 21 million people worldwide and is associated with about 200,000 deaths (7). Unlike the other serovars of S. enterica, serovar Typhi is restricted exclusively to humans (28, 56). The molecular mechanisms specifically involved in the pathogenesis of serovar Typhi are poorly understood in part due to the lack of an animal model. Most of the available information arises from studies using cell lines of human origin and from extrapolations of results obtained with Salmonella enterica serovar Typhimurium, which causes a typhoid-like disease in mice (13, 19). Since serovar Typhimurium causes only gastroenteritis of limited duration in immunocompetent patients, great care should be taken in extrapolating data from the mouse model to human typhoid (61). Moreover, genome sequence comparisons between representative serovar Typhi (42) and serovar Typhimurium (33) strains reveal that despite 97% identity between housekeeping genes, about 10% of their genes are different and unique to each serovar. These genes are likely to be involved in determining the differences in host specificity and pathogenesis between the two serovars (9).
An essential step in the pathogenesis of Salmonella is the ability of bacteria to penetrate the intestinal epithelium. The type III secretion system (TTSS) encoded within the Salmonella pathogenicity island 1 is critical for invasion of both serovars Typhi and Typhimurium into epithelial cells. The effector proteins injected by the TTSS into the host cells mediate actin cytoskeleton rearrangements and nuclear responses that ultimately facilitate entry of Salmonella into epithelial cells (1, 12, 16, 43). The full activation of TTSS requires an environmental signal, usually derived from contact with the host cell (12). Thus, early interactions of bacteria with epithelial cells are necessary before invasion can occur. The nature of these interactions for serovar Typhi is not clear, but it may involve the expression of adhesins or fimbria.
Multiple fimbrial operons in Salmonella species have been reported (52, 55). Genomic sequence comparisons and hybridization analyses revealed that serovar Typhi possesses a unique repertoire of fimbrial genes (52). Serovar Typhi also carries a type IV pilus operon, and studies show that wild-type (pil+) bacteria adhere to and enter intestinal epithelial cells better than the isogenic pil mutant (63). However, the nonpiliated mutant strain retains up to 25% of the invasion ability of the wild type, suggesting that other adhesin or adhesins are required to mediate serovar Typhi attachment to and/or entry into intestinal epithelial cells (63). Adherence and/or invasion of serovar Typhi but not of serovar Typhimurium was inhibited by soluble pre-PilS protein, suggesting that these two serovars enter intestinal epithelial cells by different mechanisms (63). Indeed, serovar Typhi but not serovar Typhimurium employs the cystic fibrosis transmembrane conductance regulator (CFTR) as a receptor to facilitate the invasion of intestinal epithelial cells (44).
Various studies have identified other surface determinants required for invasion of epithelial cells by Salmonella. For example, flagella and intact motility are necessary for entry of serovar Typhimurium (21, 22, 26) and serovar Typhi (29) into cultured epithelial cells. However, the role of motility appears to be different in the two species. In serovar Typhimurium, motility would merely facilitate contact of the bacteria with the cell (20), whereas in serovar Typhi, motility and entry appear to be functionally linked (29).
Lipopolysaccharide (LPS) has also been implicated in the adherence and/or invasion of epithelial cells (17, 50). LPS, a major component of the outer membrane of gram-negative bacteria, consists of lipid A, core oligosaccharide, and in many bacteria O-antigen polysaccharide (46). Early reports demonstrated that colonization of the mouse intestine is impaired in LPS-defective mutants of serovar Typhimurium (40) and that mutants lacking O antigen are attenuated in mice (34). Not only the presence but also the proper distribution of the O-antigen chain length is required for the full virulence of serovar Typhimurium (38). However, O antigen is not required for invasion of epithelial cells in vitro (23).
In serovar Typhi, the role of the O antigen in the invasion process is unclear. An early report indicated that O-antigen-defective mutants of serovar Typhi are unable to adhere to and invade HeLa cells (37), but more recently it has been suggested that the bacterial ligand for the receptor in intestinal epithelial cells is located on or near the outer core oligosaccharide (31). The experiments carried out in these studies were performed using chemical or transposon mutants isolated by screening for the loss of O antigen after random mutagenesis. Therefore, many of these mutants lack genetic definition, and a relation between invasion and the presence of O antigen has not been directly confirmed.
In this work, we have investigated the role of LPS components in the invasion of serovar Typhi into epithelial cells using a series of mutants with defined deletions of genes for the synthesis and polymerization of the O antigen and the assembly of the outer core. Our results indicate that the presence and proper chain length distribution of the O antigen are not essential for invasion of epithelial cells but are absolutely required for serum resistance. We also demonstrate that the outer core terminal glucose residue is necessary for efficient entry of serovar Typhi into epithelial cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Table 1 summarizes the properties of the bacterial strains and plasmids used in this study. Bacteria were grown in Luria-Bertani medium (LB) (10 g/liter Bacto tryptone, 5 g/liter Bacto yeast extract, 5 g/liter NaCl). Solid medium contained 1.5% (wt/vol) agar. Ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (20 μg/ml), and novobiocin (25 μg/ml) were added when appropriate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| S. enterica serovar Typhi strains | ||
| Ty2 | Wild type | ISPb |
| M985 | Ty2 Δwzz | This study |
| M1224 | Ty2 Δwzy | This study |
| MSS1 | Ty2 ΔwbaP::cat Cmr | 49 |
| M8 | Ty2 ΔrfaH::cat Cmr | 47 |
| M115 | Ty2 ΔwaaK | This study |
| M102 | Ty2 ΔwaaJ | This study |
| M1015 | Ty2 ΔwaaI | This study |
| M109 | Ty2 ΔwaaB | This study |
| M108 | Ty2 ΔwaaB::aph Kanr | This study |
| M113 | Ty2 ΔwaaG | This study |
| Plasmids | ||
| pKD46 | bla PBADgam bet exo pSC101 oriTS | 8 |
| pKD3 | bla FRT cat FRT PS1 PS2 oriR6K | 8 |
| pKD4 | bla FRT aph FRT PS1 PS2 oriR6K | 8 |
| pCP20 | bla cat cI857 λPRflp pSC101 oriTS | 5 |
| pGEM-T Easy | TA cloning vector | Promega |
| pJC102 | waaJ cloned into pGEM-T Easy | This study |
| pJC1015 | waaI cloned into pGEM-T Easy | This study |
| pJC109 | waaB cloned into pGEM-T Easy | This study |
| pJC113 | waaG cloned into pGEM-T Easy | This study |
Cmr, chloramphenicol resistant; Kanr, kanamycin resistant.
ISP, Institute of Public Health, Santiago, Chile.
Mutagenesis of the LPS biosynthesis genes from serovar Typhi.
Mutagenesis was performed by the method of Datsenko and Wanner (8) to create chromosomal mutations by homologous recombination using PCR products. To disrupt the LPS biosynthesis genes, serovar Typhi Ty2 cells were first transformed with the thermosensitive plasmid pKD46, which expresses the λ Red recombinase system. These cells were transformed with PCR products that were generated using plasmids pKD3 and pKD4 as templates, which contain FRT-flanked kanamycin resistance (aph) or chloramphenicol resistance (cat) genes, respectively. Each primer pair also carried 25 bases that were homologous to the edge of the gene targeted for disruption. The sequences of the oligonucleotide primers used in this study are available on request. In the presence of the Red recombinase system, the integration of the amplicons resulted in the targeted replacement of the wild-type gene by the antibiotic resistance cassette. The Kanr and Cmr transformants were replica plated in the absence of antibiotic selection at 42°C and finally assayed for ampicillin sensitivity to confirm the loss of pKD46. To obtain nonpolar deletion mutants, the antibiotic resistance gene was removed by transforming the gene replacement mutants with pCP20, which encodes the FLP recombinase (5). Transformants were plated on LB agar plates containing ampicillin and kanamycin or ampicillin and chloramphenicol at 37°C. Individual colonies were replica plated on LB agar plates, LB agar plates containing ampicillin, and LB agar plates containing kanamycin or chloramphenicol. The plates were incubated at 42°C. Transformants that had lost the resistance gene (aph or cat) and plasmid pCP20 were selected as those colonies that were able to grow only on LB agar plates. Correct insertional gene replacements and the deletion of the antibiotic gene cassettes were confirmed by PCR analysis.
To complement the mutations, the intact gene was amplified from the chromosome by PCR, cloned into pGEM-T, and electroporated into the corresponding mutant. The sequences of the oligonucleotide primers used are available on request.
Adhesion and invasion assays.
HEp-2 cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal calf serum (FCS). Monolayers for infection were prepared by seeding 8 × 104 cells into each well of a 48-multiwell plate and incubating at 37°C for 20 h in 5% CO2 and 95% air. Bacteria were grown aerobically to an optical density at 600 nm of 0.7, washed twice with phosphate-buffered saline (PBS), suspended in DMEM-FCS (100 μl), and added to confluent HEp-2 cells at a multiplicity of infection of approximately 50. Adhesion assays were performed by allowing bacteria to adhere at 4°C for 1 h, and then each well was rinsed three times with 100 μl of ice-cold PBS. Adherent bacteria were released by incubation with 100 μl of sodium deoxycholate (0.5% in PBS) for 10 min. LB (900 μl) was then added, and each sample was vigorously mixed. Adherent bacteria were quantified by plating for CFU on LB agar plates. Adhesion was calculated as follows: percent adhesion = 100 × (number of cell-associated bacteria/initial number of bacteria added). For invasion assays, HEp-2 cells were infected as described above, but bacteria were incubated with the monolayers at 37°C for 1 h. Then, cells were washed three times with prewarmed PBS and incubated for an additional 2 h in DMEM-FCS containing gentamicin (250 μg/ml) to kill extracellular bacteria. Cells were then washed three times with PBS and lysed with sodium deoxycholate (0.5% in PBS). Intracellular bacteria were diluted and plated on LB agar plates. In agreement with a previous report (35), no multiplication of bacteria occurred during the course of the assay. Invasion was calculated as follows: percent invasion = 100 × (number of bacteria resistant to gentamicin/initial number of bacteria added). Data were calculated from at least three independent experiments performed in triplicate and are expressed as means ± standard errors. The statistical significance of differences in the data was determined using the one-way analysis of variance (ANOVA) test and the Tukey post test.
LPS analysis.
Culture samples were adjusted to an optical density at 600 nm of 2.0 in a final volume of 100 μl. Then, proteinase K-digested whole-cell lysates were prepared as described previously (15), and LPS was separated on 14% acrylamide gels using a Tricine-sodium dodecyl sulfate (SDS) buffer system (27). Gel loadings were normalized so that each sample represented the same number of cells. Gels were silver stained by a modification of the procedure of Tsai and Frasch (53). LPS electrophoretic profiles in all samples were quantified by densitometric analysis with the UN-SCAN-IT gel software (Silk Scientific).
Motility assays.
Phenotypic assays for swimming motility were performed as described by Bengoechea et al. (2). Briefly, 2 μl of an overnight culture was stabbed into the center of a plate containing 20 ml of soft agar (0.3% agar and 1% tryptone). Plates were analyzed after 24 h of incubation at 37°C, and the halo that migrated from the inoculation point for each strain was measured in centimeters. Experiments were run in duplicate in three independent occasions. Data are expressed as means ± standard errors. The statistical significance of differences in the data was determined using the one-way ANOVA test and the Tukey post test.
Serum resistance.
Serum bactericidal activity was determined as described by Bengoechea et al. (2). Human serum samples containing no antibodies against Salmonella group D1 (provided by the blood bank of the University of Chile Clinical Hospital) were divided into small aliquots and stored at −70°C until use. An overnight bacterial culture was diluted in PBS to 104 CFU/ml, and 20 μl of the dilution was mixed with 20 μl of 20% serum and incubated at 37°C for 0, 5, 10, 20, 30, and 60 min. Complement function was stopped by the addition of 60 μl of brain heart infusion broth. Tubes were kept on ice until the full contents of each tube were plated onto LB agar plates to determine the number of viable bacteria. The same experiments were carried out using serum inactivated at 56°C for 30 min. Resistance to serum was calculated as percent survival, taking the bacterial counts obtained with heat-inactivated serum as 100%. Each experiment was carried out in duplicate in three independent assays. Data are expressed as means ± standard errors. The statistical significance of differences in the data was determined using the one-way ANOVA test and the Tukey post test.
RESULTS
Construction of serovar Typhi strains with chromosomal LPS synthesis gene mutations.
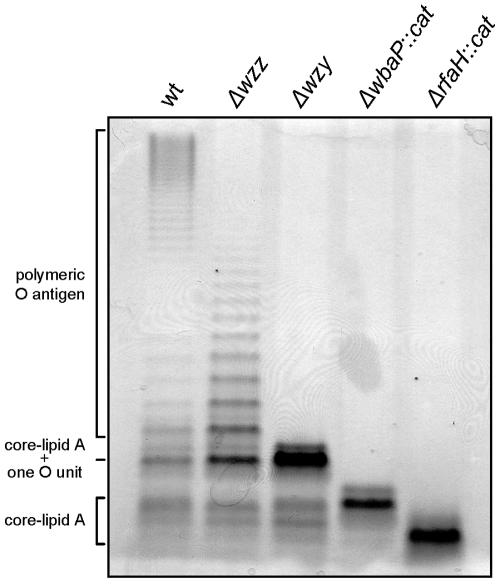
Defined mutations in various O-antigen and core oligosaccharide synthesis genes of serovar Typhi strain Ty2 were constructed to dissect the LPS components contributing to invasion of epithelial cells. Mutagenesis was carried out by the one-step PCR method of Datsenko and Wanner (8) as described above in Materials and Methods, which allowed us to obtain defined, nonpolar deletions of each targeted gene. Furthermore, nonpolar deletions of the O-antigen modal chain regulator wzz gene (strain M985) and the O-antigen polymerase wzy gene (strain M1224) were obtained. We also constructed a mutant strain with a deletion in the wbaP gene (strain MSS1), which encodes WbaP, the enzyme that initiates O-antigen synthesis in serovar Typhi by catalyzing the transfer of galactosyl-1-phosphate from UDP-galactose to undecaprenyl-phosphate (49, 57). In addition, we included in this study strain M8, which carries a deletion in the rfaH gene. RfaH is a transcriptional antiterminator that regulates the expression of the O-antigen and core operons in enterobacteria (10, 32, 45, 58). In previous work, we demonstrated that wild-type expression of LPS in serovar Typhi requires RfaH (47). Figure 1 shows the LPS patterns of the wild-type Ty2 strain and the O-antigen mutants. Strain M985 (Δwzz) produced O-antigen chains of random length, whereas strain M1224 (Δwzy) showed a semirough phenotype with a very prominent band of the core substituted with one O-antigen unit. The mutant in which the wbaP gene was disrupted (strain MSS1) did not express O antigen but produced a complete core, while mutant M8 produced LPS with an incomplete core region.
FIG. 1.
Analysis of LPS from serovar Typhi strain Ty2 and O-antigen mutants. LPS samples from equal numbers of bacterial cells (1 × 107 CFU) were loaded in each lane and were analyzed by Tricine/SDS-polyacrylamide gel electrophoresis on a 14% acrylamide gel followed by silver staining. The strains are Ty2 (wild type [wt]), M985 (Ty2 Δwzz), M1224 (Ty2 Δwzy), MSS1 (Ty2 ΔwbaP::cat), and M8 (Ty2 ΔrfaH::cat).
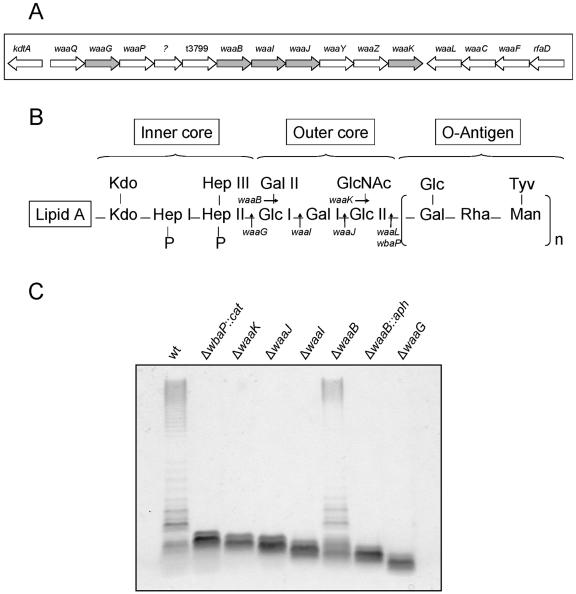
To our knowledge, the structure of the LPS core from serovar Typhi has not been elucidated. The genes for the biosynthesis of the core oligosaccharide are in the waa gene cluster. A comparison of the sequences of the waa genes in serovar Typhi Ty2 with those of serovar Typhimurium LT2 revealed 99% identity and the same gene organization. The high level of sequence conservation and the identical gene organization (Fig. 2A) strongly support the notion that the LPS core structures in both serovars are identical, as depicted in Fig. 2B. We constructed precise, nonpolar deletions in the genes waaK (strain M115), waaJ (strain M102), waaI (strain M1015), waaB (strain M109), and waaG (strain M113). Analysis of the LPS profiles of these mutants revealed that strain M115 (ΔwaaK) produced a core region that migrated slightly faster than that of the wbaP mutant that synthesizes a complete core (Fig. 2C). This is consistent with the lack of the GlcNAc residue in the outer core (Fig. 2B). Because this GlcNAc residue is essential for the recognition of core oligosaccharide acceptor by the O-antigen ligase WaaL (14), the ΔwaaK mutant does not express O antigen. Deletion of waaJ and waaI genes resulted in additional truncations of the core LPS region (Fig. 2C). The waaJ gene product adds the outer core terminal glucose residue (Glc II), while the waaI product adds the Gal I residue to the outer core (Fig. 2B). Mutant M109, which is due to a deletion of the waaB gene lacks the Gal II residue in the outer core, showed a heterogeneous core region. The major LPS band migrated slightly faster than the core band observed in the ΔwaaI mutant. In addition, minor LPS bands, which migrated as though they have larger cores as well as complete LPS molecules containing O antigen were observed (Fig. 2C). This phenotype is consistent with previous work conducted in serovar Typhimurium, which demonstrated that the waaI gene product cannot efficiently add Gal I in the absence of the branch galactose, and as a consequence, the majority of the LPS molecules contain only Glc I. However, the requirement of WaaI for the branch galactose is not absolute, and thus, the waaB mutant has a small proportion of complete core molecules, which can be substituted with O antigen (60). In contrast, strain M108, which carries an insertion of an aph cassette in waaB, produced a fast-migrating truncated core (Fig. 2C). We constructed this mutant because the insertion of the antibiotic resistance gene causes a polar effect on the expression of the genes located downstream in the operon, and thus, this mutant has a LPS structure that contains only the Glc I residue of the outer core (Fig. 2B). Finally, deletion of waaG (strain M113) resulted in major truncation of the LPS core. Accordingly, this strain synthesized only the inner core region (Fig. 2C).
FIG. 2.
(A) Genetic organization of genes for core biosynthesis in serovar Typhi. The targets of the mutations obtained in this study are shown in gray. (B) Proposed structures of the inner core, outer core, and O antigen of serovar Typhi. Kdo, 3-deoxy-d-manno-octulosonic acid. (C) Analysis of LPS from serovar Typhi strain Ty2 and core mutants. LPS samples from equal numbers of bacterial cells (2.5 × 107 CFU) were loaded in each lane and were analyzed by Tricine/SDS-polyacrylamide gel electrophoresis on a 14% acrylamide gel followed by silver staining. The strains are Ty2 (wild type [wt]), MSS1 (Ty2 ΔwbaP::cat), M115 (Ty2 ΔwaaK), M102 (Ty2 ΔwaaJ), M1015 (Ty2 ΔwaaI), M109 (Ty2 ΔwaaB), M108 (Ty2 ΔwaaB::aph), and M113 (Ty2 ΔwaaG).
The O antigen does not play a direct role in the invasion of epithelial cells by serovar Typhi.
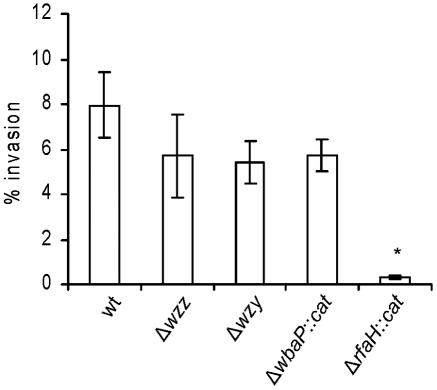
To address the role of the O antigen in the entry of serovar Typhi into epithelial cells, we compared the abilities of mutants and the wild-type Ty2 to invade HEp-2 cells in vitro. The invasion levels of strains M985 (Δwzz) and M1224 (Δwzy) were similar to that of the wild type (Fig. 3), suggesting that neither the length of O antigen nor the proper modal distribution of O-antigen chains plays a direct role in invasion of epithelial cells. Consistent with this conclusion, the invasion level of strain MSS1 (ΔwbaP::cat), which produces a complete lipid A-core and lacks O antigen, was not significantly different from that of the wild type (Fig. 3). In contrast, the rfaH mutant M8, which produces an incomplete core, displayed a 10-fold reduction (P < 0.05) in invasion level compared to those of the wild type and the O-antigen mutant strains (Fig. 3). Together, these results suggest that the outer core region, but not the O antigen, is required for entry of serovar Typhi into epithelial cells.
FIG. 3.
Invasion of HEp-2 epithelial cells by serovar Typhi Ty2 and O-antigen mutants. The strains are Ty2 (wild type [wt]), M985 (Ty2 Δwzz), M1224 (Ty2 Δwzy), MSS1 (Ty2 ΔwbaP::cat), and M8 (Ty2 ΔrfaH::cat). Invasion assays were performed in triplicate on at least three independent occasions. Averages ± standard errors (error bars) are shown. The value that was significantly different from that of the wild type (P < 0.05) by the one-way ANOVA test and Tukey posttest is indicated by an asterisk.
The terminal glucose residue of the outer core is necessary for efficient entry of serovar Typhi into epithelial cells.
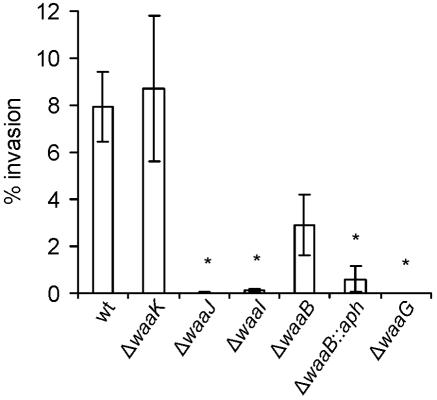
Results from a previous study (31) suggested that the serovar Typhi LPS core mediates the internalization of bacteria by epithelial cells. Our findings described in the above section are in support of this hypothesis. To define more precisely the ligand responsible for interaction with the HEp-2 cells, we assessed the ability of core LPS-defective mutants to enter HEp-2 cells. The results indicated that deletions in waaG, waaI, and waaJ (strains M113, M1015, and M102, respectively) abrogated the ability of serovar Typhi to enter epithelial cells (Fig. 4). All these strains synthesize truncated outer core regions (Fig. 2C). On the contrary, strain M115 (ΔwaaK), which lacks a terminal GlcNAc residue, invaded epithelial cells as efficiently as the wild-type strain, indicating that this branching residue is not needed for entry. Strain M109 (ΔwaaB) showed a reproducibly lower invasive capacity than the wild type, but the difference was not statistically significant. This mutant produces a heterogeneous core region (Fig. 2C), which contains some complete molecules that lack the Gal II residue but can attach O antigen, and a major fraction of truncated core molecules that contain only the Glc I residue. This heterogeneity may account for the slightly lower invasion level exhibited by this mutant compared to that of the wild-type strain.
FIG. 4.
Invasion of HEp-2 epithelial cells by serovar Typhi Ty2 and core mutants. The strains are Ty2 (wild type [wt]), M115 (Ty2 ΔwaaK), M102 (Ty2 ΔwaaJ), M1015 (Ty2 ΔwaaI), M109 (Ty2 ΔwaaB), M108 (Ty2 ΔwaaB::aph), and M113 (Ty2 ΔwaaG). Invasion assays were performed in triplicate on at least three independent occasions. Averages ± standard errors (error bars) are shown. Values that were significantly different from that of the wild-type Ty2 strain (P < 0.05) by the one-way ANOVA test and Tukey posttest are indicated by asterisks.
Strain M108 (ΔwaaB::aph), which produces a core LPS structure where the Glc I residue is terminal (Fig. 2B), invaded HEp-2 cells with an efficiency of approximately 8% of that of the wild-type Ty2 strain. Although this invasion level is significantly lower than Ty2, it is worth noting that this mutant consistently invaded HEp-2 cells.
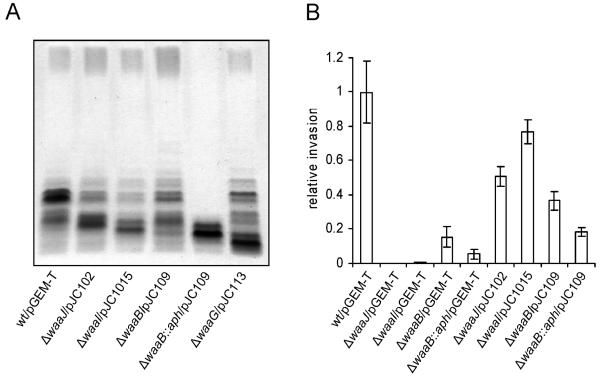
To confirm that the defect in invasion exhibited by mutants M102, M1015, M109, and M113 is indeed due to the deletion of the corresponding gene, each of the intact genes (waaJ, waaI, waaB, and waaG, respectively) was cloned into pGEM-T and transformed into the appropriate mutant. Analysis of LPS revealed that introduction of the intact gene restored the mutants' ability to synthesize a complete core and to attach O antigen (Fig. 5A). However, in all cases a significant amount of truncated core region was still present. This was probably due to the high copy number of the cloning vector and high expression levels. A similar phenomenon has been reported with other cloned LPS genes, such as wzy, and it was attributed to limiting pools of rare tRNAs, which impair transcription and translation of cloned genes containing abundant rare codons in highly efficient expression vectors (30). We also complemented strain M108 (ΔwaaB::aph) with the waaB gene. Because the insertion of the antibiotic resistance gene causes a polar effect on the expression of the genes located downstream in the operon, introduction of waaB did not restore the normal LPS phenotype (Fig. 5A), as expected. We assessed the ability of the complemented strains to enter HEp-2 cells and compared it with the invasion of the respective mutant transformed with the cloning vector alone. The complemented mutants ΔwaaJ/pJC102, ΔwaaI/pJC1015, and ΔwaaB/pJC109 showed invasion levels similar to that of the wild-type Ty2 strain. In contrast, invasion of ΔwaaB::aph/pJC109 was similar to that of mutant ΔwaaB::aph complemented with pGEM-T (Fig. 5B). Strain ΔwaaG/pJC113 did not show better invasion levels than the mutant alone, but the growth of this strain was seriously impaired (data not shown), probably accounting for the defect in invasion.
FIG. 5.
(A) Analysis of LPS from serovar Typhi Ty2 and complemented core mutants. LPS samples from equal numbers of bacterial cells (2.5 × 107 CFU) were loaded in each lane and were analyzed by Tricine/SDS-polyacrylamide gel electrophoresis on a 14% acrylamide gel followed by silver staining. (B) Invasion of HEp-2 epithelial cells by serovar Typhi Ty2 and complemented core mutants. Invasion assays were performed in triplicate on at least three independent occasions. Values represent invasion of each strain relative to invasion of Ty2/pGEM-T. The strains are Ty2 (wild type [wt])/pGEM-T, M102 (Ty2 ΔwaaJ)/pGEM-T, M1015 (Ty2 ΔwaaI)/pGEM-T, M109 (Ty2 ΔwaaB)/pGEM-T, M108 (Ty2 ΔwaaB::aph)/pGEM-T, M102 (Ty2 ΔwaaJ)/pJC102, M1015 (Ty2 ΔwaaI)/pJC1015, M109 (Ty2 ΔwaaB)/pJC109, and M108 (Ty2 ΔwaaB::aph)/pJC109.
Together, our results indicate that the outer core structure composed of Glc I-Gal I-Glc II is required for optimal entry of serovar Typhi into epithelial cells. The terminal Glc II residue is essential for invasion, but Glc I in mutant M108 can partially substitute for Glc II as the ligand responsible for interaction with the epithelial cell receptor to mediate entry of serovar Typhi.
Adherence of LPS-defective mutants to epithelial cells.
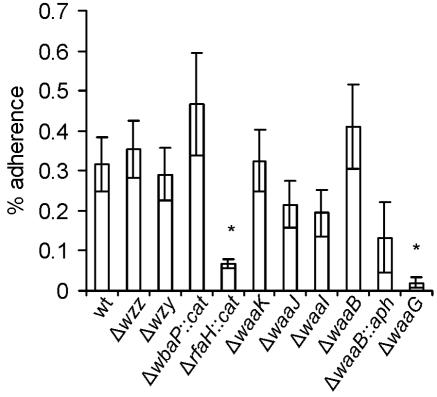
Because in a previous work it was suggested that the serovar Typhi O antigen is required for invasion and adherence to epithelial cells (37), we also investigated the adherence to HEp-2 cells of the O-antigen-defective mutants M985 (Δwzz), M1224 (Δwzy), and MSS1 (ΔwbaP::cat). As expected, we did not find any significant differences in the adherence of these mutants compared to that of the wild-type Ty2 strain (Fig. 6). Therefore, we conclude that the O antigen is not required either for invasion or adherence of serovar Typhi to epithelial cells.
FIG. 6.
Adherence of serovar Typhi Ty2 and mutants on HEp-2 epithelial cells. The strains are Ty2 (wild type [wt]), M985 (Ty2 Δwzz), M1224 (Ty2 Δwzy), MSS1 (Ty2 ΔwaaP::cat), M8 (Ty2 ΔrfaH::cat), M115 (Ty2 ΔwaaK), M102 (Ty2 ΔwaaJ), M1015 (Ty2 ΔwaaI), M109 (Ty2 ΔwaaB), M108 (Ty2 ΔwaaB::aph), and M113 (Ty2 ΔwaaG). Adhesion assays were performed in triplicate on at least three independent occasions. Averages ± standard errors (error bars) are shown. Values that are significantly different from that of the wild-type Ty2 (P < 0.05) by the one-way ANOVA test and Tukey posttest are indicated by asterisks.
We also tested the adhesion properties of the mutants with core truncations to investigate whether the defects in invasion capacity of some of these mutants could be due to reduced bacterial adherence to the epithelial cells. As shown in Fig. 6, strains M102 (ΔwaaJ), M1015 (ΔwaaI), and M108 (ΔwaaB::aph), which were invasion defective, displayed no significant differences in adhesion compared with the invasion-proficient strains MSS1 (ΔwbaP::cat), M115 (ΔwaaK), M109 (ΔwaaB), and the wild-type Ty2. We can conclude from these results that the differences observed in the invasion capacity of the core mutants cannot be attributed to defects in adhesion to epithelial cells. The mutants with more truncated LPS core, such as M8 (ΔrfaH::cat) and M113 (ΔwaaG), were the only strains with a significant reduction in adherence.
Motility of LPS-defective mutants and sensitivity to novobiocin.
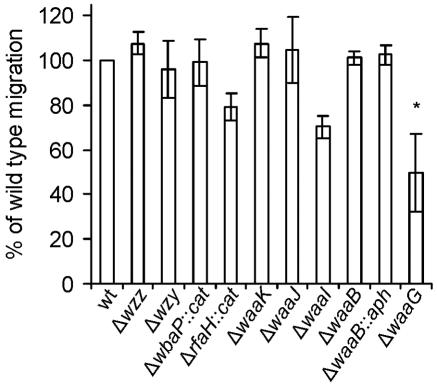
Previously, it has been reported that intact motility is required for the invasion of HeLa cells by serovar Typhi (29). To test the possibility that the impaired invasiveness exhibited by mutants M102 (ΔwaaJ), M1015 (ΔwaaI), M113 (ΔwaaG), M108 (ΔwaaB::aph), and M8 (ΔrfaH::cat) was due to an altered motility that could result in an inability to reach the monolayer, we quantified the migration of the wild type and LPS mutants in semisolid medium. All the strains were as motile as the wild type, with the exception of the ΔwaaG mutant M113 (Fig. 7), which produces an LPS lacking the outer core region. The loss of cell motility associated with the waaG deletion may be due to the absence of flagella. A similar phenotype has been well documented in Escherichia coli K-12, where it has been suggested that the lack of flagella is due specifically to the absence of Glc I in the LPS core and not with the LPS truncation (41). This is also in agreement with the results of Komeda et al. (24), who found that E. coli galU mutants could not synthesize flagella and exhibited a decrease in flagellin mRNA. Thus, it may be possible that rather than a problem in the export or assembly of flagellar structures, a down-regulation of flagellar genes may be triggered by the change in LPS.
FIG. 7.
Motility phenotype of serovar Typhi Ty2 and mutants. Swimming motility on soft agar was performed as described in Materials and Methods. Bars represent the migration of each mutant relative to the migration of the wild-type strain. The strains are Ty2 (wild type [wt]), M985 (Ty2 Δwzz), M1224 (Ty2 Δwzy), MSS1 (Ty2 ΔwaaP::cat), M8 (Ty2 ΔrfaH::cat), M115 (Ty2 ΔwaaK), M102 (Ty2 ΔwaaJ), M1015 (Ty2 ΔwaaI), M109 (Ty2 ΔwaaB), M108 (Ty2 ΔwaaB::aph), and M113 (Ty2 ΔwaaG). Averages ± standard errors (error bars) are shown. Experiments were performed in duplicate on three independent occasions. The value that is significantly different from that of the wild-type Ty2 (P < 0.05) by the one-way ANOVA test and Tukey posttest is indicated by an asterisk.
We assayed the susceptibility of the core-defective mutants to the hydrophobic antibiotic novobiocin as a means to determine whether the overall permeability of the outer membrane was compromised. Strain M113 (ΔwaaG) was unable to grow in LB containing 25 μg/ml novobiocin. Mutant M8 (ΔrfaH::cat), which synthesizes an incomplete core migrating slightly slower than that of M113, was also sensitive to novobiocin (data not shown).
Together, our results suggest that the low invasive capacity of the core mutants, except for mutants with more severe core truncation (M8 and M113), is not attributable to a defect in motility or to a decreased stability of the outer membrane, but it is indeed a result of their altered LPS structures.
Serum resistance of LPS-defective mutants.
The previous results suggested that the O antigen has no direct role in invasion of epithelial cells. However, the presence of O antigen has been associated with pathogenicity in serovar Typhimurium by mediating resistance to complement-mediated serum killing (38, 39). We examined the role of serovar Typhi O antigen in serum resistance by incubating wild-type Ty2 and the LPS mutants in 10% human serum at different times during a 1-hour period as described in Materials and Methods. Table 2 shows the percent survival after 10 min of incubation with serum relative to the survival in heat-inactivated serum. All the mutants lacking O antigen were highly susceptible to the serum bactericidal action, with percentages of survival ranging from 0% to 3.5%. Mutant M985 (Δwzz) was more sensitive than the wild type (approximately 50% survival), indicating that O antigen with a proper distribution of chain lengths is required for serum resistance. The semirough mutant M1224 (Δwzy) was as sensitive as the rough mutants, suggesting that the presence of high-molecular-weight molecules, rather than the extent to which the core is capped by O antigen, is crucial for survival in serum. This notion is supported by the resistance of mutant M109; although it produces a smaller amount of complete O antigen, it exhibited a survival level similar to that of the wild-type strain.
TABLE 2.
Serum resistance of serovar Typhi Ty2 and mutants
| Strain | Mutation | LPS phenotype | % Serum resistancea (10 min) |
|---|---|---|---|
| Ty2 | Wild type | Normal | 55.05 ± 3.14 |
| M985 | Δwzz | No modal distribution | 27.1 ± 6.2 |
| M1224 | Δwzy | Only one O-antigen unit | 0.2 ± 0.1 |
| MSS1 | ΔwbaP::cat | No O antigen; complete outer core | 3.5 ± 0.56 |
| M8 | ΔrfaH::cat | No O antigen; truncated outer core | 0 ± 0 |
| M115 | ΔwaaK | No O antigen; truncated outer core | 0 ± 0 |
| M102 | ΔwaaJ | No O antigen; truncated outer core | 2.74 ± 0.37 |
| M1015 | ΔwaaI | No O antigen; truncated outer core | 3.1 ± 3.1 |
| M109 | ΔwaaB | Fewer O-antigen molecules; truncated outer core | 77.95 ± 4.34 |
| M108 | ΔwaaB::aph | No O antigen; truncated outer core | 0 ± 0 |
| M113 | ΔwaaG | No O antigen; no outer core | 0 ± 0 |
Serum resistance is expressed as percent survival, taking as 100% the bacterial counts obtained with heat-inactivated serum. Each sample was tested at least three times; the averages and standard errors were calculated from these values.
DISCUSSION
This study has examined the importance of the LPS O-antigen and core structures in the ability of serovar Typhi to enter epithelial cells and to resist the bacteriolytic action of serum complement. The role of LPS in invasion of epithelial cells varies in different species of enterobacteria. In Salmonella, the role of the O antigen in invasion of epithelial cells is dependent on the serotype. Rough mutants of Salmonella enterica serovar Choleraesuis are defective for entry (11), while rough mutants of serovar Typhimurium are not (23) and rough mutants of serovar Enteritidis are only slightly defective for entry (51). To our knowledge, only two reports from the literature have examined the role of LPS on invasion of epithelial cells by serovar Typhi, and their results were contradictory. Mroczenski-Wildey et al. (37) observed that rough mutants of serovar Typhi are unable to adhere to and invade HeLa cells, but more recently, Lyczak et al. (31) reported that loss of expression of the LPS O antigen does not affect the efficiency with which serovar Typhi strain Ty2 is internalized by epithelial cells.
We found that the presence and the length of the O-antigen chains do not affect invasion of HEp-2 cells, since mutants that do not synthesize O antigen (ΔwbaP::cat), that produce only one O-antigen unit (Δwzy), or that have an altered modal chain distribution (Δwzz) can enter epithelial cells as efficiently as the wild-type strain. Thus, our results agree with previous work that suggested that the bacterial ligand responsible for interaction with the epithelial cells is located on or near the LPS core (31). However, that study could not clearly define the bacterial ligand responsible for the interaction with epithelial cells, since transposon mutants were used in which polar defects could not be ruled out. Also, none of the mutants previously employed had defects in the core biosynthesis gene cluster.
The experiments carried out in this work, using defined mutations in genes for the biosynthesis of the O-antigen and core regions, allowed us to determine that the terminal glucose residue of the outer core is necessary for the interaction and subsequent entry of serovar Typhi into epithelial cells. An intact core is not required, since mutant ΔwaaK, which cannot add the GlcNAc substituent, and mutant ΔwaaB, which lacks the Gal II residue, are able to invade HEp-2 cells. Our results also indicate that Glc I, when it becomes a terminal residue, can partially substitute for Glc II as the ligand for interaction with epithelial cells although with reduced efficiency.
Our findings are consistent with results obtained by Zaidi et al. (62) indicating that a complete outer core with an exposed terminal glucose residue is necessary for maximal adherence and entry of Pseudomonas aeruginosa into corneal epithelial cells. Their work was carried out with mutants of known chemical structure, differing from one another in the number of sugar residues in the LPS core (6, 18, 25, 48). These mutants were selected after spontaneous or chemical mutagenesis and were genetically undefined, so the impaired invasiveness observed by Zaidi et al. (62) in mutants with truncated cores could be due not only to the LPS defect but also to unknown secondary mutations. However, their results and those from this study stress the importance of terminal glucose residues in the bacterium-epithelial cell interaction.
It is puzzling that the presence of O antigen cannot prevent interactions with epithelial cells. Moreover, the mutants defective in O-antigen production did not show an increased invasion, indicating that the absence of O antigen does not represent an advantage, probably due to sufficient amounts of ligand already present to interact with epithelial cell receptors. In support of this notion, densitometry analysis (not shown) of the gel in Fig. 1 revealed that approximately 40% of the LPS core in serovar Typhi Ty2 is not capped with O antigen, and therefore, it is exposed and can interact with epithelial cell receptors. Alternatively, it is possible that the O antigen in serovar Typhi has nonstoichiometric substitutions with terminal glucose that could serve as a ligand for invasion. Analysis of the genome of serovar Typhi Ty2 revealed the presence of genes for a putative gtr system for the glucosylation of O antigen, highly similar to gtr genes from Shigella flexneri. It has recently been reported that glucosylation of Shigella O antigen promotes invasion of cultured epithelial cells, as well as evasion of innate immunity (59). We constructed a mutant strain with a deletion in both the gtrA and gtrB genes of serovar Typhi Ty2 and assayed its ability to enter HEp-2 cells. The mutant strain did not show any significant reduction in invasion (unpublished results), suggesting that either this putative gtr system is not functional in serovar Typhi or the glucosylation of the O antigen is not relevant to invasion of epithelial cells by this pathogen.
Our results also demonstrate that, although the LPS core is required for an efficient entry of serovar Typhi into epithelial cells, it does not mediate bacterial adhesion. We found that mutants that produce truncated cores (M102, M1015, and M108), which are invasion defective, displayed no significant differences in adhesion compared with invasion-proficient strains. The mutants that were unable to synthesize the outer core (M8 and M113) were defective in both adherence and invasion. Thus, we suggest that adhesion of serovar Typhi to epithelial cells, at least in tissue culture, is mediated by one or more adhesins different than the LPS core. Zhang et al. (63) have shown that the type IVB pilus expressed by serovar Typhi acts as an adhesin to mediate bacterium-intestinal cell attachment. Furthermore, these authors demonstrated that the PilS protein of the type IVB pili binds to CFTR (54), a human epithelial cell receptor for serovar Typhi (44), and have proposed that, after mediating bacterial self-association (36), the pili then act to attach the bacterial aggregates to the epithelial cell membrane (54). We propose that, after this initial adhesion, the LPS core would interact with the intestinal cell receptor, probably CFTR, to facilitate bacterial invasion. It is worth noting that by using the electron microscope, we visualized fimbrial appendages in all our mutants, except for the adhesion-defective strains M8 and M113 (not shown).
Previous work suggested that, upon interaction with epithelial cells, the LPS of serovar Typhi undergoes a phenotypic alteration that unmasks a ligand present in the LPS outer core (31). One structural alteration of the LPS observed in that study was a diminished amount of O-antigen chains. Work from our laboratory has demonstrated that the production of serovar Typhi O antigen increases at the onset of stationary phase and correlates with a growth-regulated expression of the rfaH gene under the control of the alternative sigma factors RpoN and RpoS (3, 4). It is tempting to speculate that in the gut, where the environment is rich in nutrients, the expression of the O antigen may be down-regulated (as in exponential phase of growth), facilitating the exposure of the terminal glucose of the outer core for interaction with epithelial cell receptor. Thus, a fine regulation of O-antigen expression may be of advantage at different stages of serovar Typhi infection: in the intestinal mucosa, a smaller amount of this polysaccharide would be produced to promote invasion, whereas in the bloodstream and tissues, a larger amount of O antigen would be required to confer resistance to the bactericidal action of serum complement.
Acknowledgments
This work was supported by grant 1040562 from FONDECYT (to I.C.) and grant 10206 from the Canadian Institutes of Health Research (to M.A.V.). M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Editor: V. J. DiRita
REFERENCES
- 1.Alonso, A., and F. García-del Portillo. 2004. Hijacking of eukaryotic functions by intracellular bacterial pathogens. Int. Microbiol. 7:181-191. [PubMed] [Google Scholar]
- 2.Bengoechea, J. A., H. Najdenski, and M. Skurnik. 2004. Lipopolysaccharide O antigen status of Yersinia enterocolitica O:8 is essential for virulence and absence of O antigen affects the expression of the other Yersinia virulence factors. Mol. Microbiol. 52:451-469. [DOI] [PubMed] [Google Scholar]
- 3.Bittner, M., S. Saldías, C. Estévez, M. Zaldívar, C. L. Marolda, M. A. Valvano, and I. Contreras. 2002. O-antigen expression in Salmonella enterica serovar Typhi is regulated by nitrogen availability through RpoN-mediated transcriptional control of the rfaH gene. Microbiology 148:3789-3799. [DOI] [PubMed] [Google Scholar]
- 4.Bittner, M., S. Saldías, F. Altamirano, M. A. Valvano, and I. Contreras. 2004. RpoS and RpoN are involved in the growth-dependent regulation of rfaH transcription and O antigen expression in Salmonella enterica serovar Typhi. Microb. Pathog. 36:19-24. [DOI] [PubMed] [Google Scholar]
- 5.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalized excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 6.Coyne, M. J., Jr., K. S. Russell, C. L. Coyle, and J. A. Goldberg. 1994. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176:3500-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crump, J. A., S. P. Luby, and E. D. Mintz. 2004. The global burden of typhoid fever. Bull. W. H. O. 8:346-353. [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, R. A., G. J. Olsen, and S. R. Maloy. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 10.Farewell, A., R. Brazas, E. Davie, J. Mason, and L. I. Rothfield. 1991. Suppression of the abnormal phenotype of Salmonella typhimurium rfaH mutants by mutations in the genes for transcription termination factor Rho. J. Bacteriol. 173:5188-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlay, B. B., M. N. Starnbach, C. L. Francis, B. A. D. Stocker, S. Chatfield, G. Dougan, and S. Falkow. 1998. Identification and characterization of TnphoA mutants of Salmonella that are unable to pass through a polarized MDCK epithelial cell monolayer. Mol. Microbiol. 2:757-766. [DOI] [PubMed] [Google Scholar]
- 12.Galán, J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53-86. [DOI] [PubMed] [Google Scholar]
- 13.Groissman, E. A., and C. Mouslim. 2000. Molecular mechanisms of Salmonella pathogenesis. Curr. Opin. Infect. Dis. 13:519-522. [DOI] [PubMed] [Google Scholar]
- 14.Heinrichs, D. E., M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. The assembly system for the lipopolysaccharide R2 core-type of Escherichia coli is a hybrid of those found in Escherichia coli K12 and Salmonella enterica. J. Biol. Chem. 273:8849-8859. [DOI] [PubMed] [Google Scholar]
- 15.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.House, D., A. Bishop, C. Parry, G. Dougan, and J. Wain. 2001. Typhoid fever: pathogenesis and disease. Curr. Opin. Infect. Dis. 14:573-578. [DOI] [PubMed] [Google Scholar]
- 17.Jacques, M. 1996. Role of lipo-oligosaccharides and lipopolysaccharides in bacterial adherence. Trends Microbiol. 4:408-409. [DOI] [PubMed] [Google Scholar]
- 18.Jarrell, K., and A. M. Kropiski. 1977. The chemical composition of the lipopolysaccharide from Pseudomonas aeruginosa strain PAO and a spontaneously derived rough mutant. Microbios 19:103-116. [PubMed] [Google Scholar]
- 19.Jones, B. D., and S. Falkow. 1996. Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 14:533-561. [DOI] [PubMed] [Google Scholar]
- 20.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, G. H., L. A. Richardson, and D. Uhlman. 1981. The invasion of HeLa cells by Salmonella typhimurium: reversible and irreversible bacterial attachment and the role of bacterial motility. J. Gen. Microbiol. 127:351-360. [DOI] [PubMed] [Google Scholar]
- 22.Khoramina-Falsafi, T., S. Harayama, K. Kutsukake, and J. C. Pechere. 1990. Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb. Pathog. 9:47-53. [DOI] [PubMed] [Google Scholar]
- 23.Kihlstrom, E., and L. Edebo. 1976. Association of viable and inactivated Salmonella typhimurium 395 MS and MR 10 with HeLa cells. Infect. Immun. 14:851-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komeda, Y., T. Icho, and T. Iino. 1977. Effects of galU mutation on flagellar formation in Escherichia coli. J. Bacteriol. 129:908-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kropiski, A. M., L. C. Chan, and F. H. Milazzo. 1979. The extraction and analysis of lipopolysaccharide from Pseudomonas aeruginosa strain PAO, and three rough mutants. Can. J. Microbiol. 25:390-398. [DOI] [PubMed] [Google Scholar]
- 26.Lee, C. A., B. D. Jones, and S. Falkow. 1992. Identification of a Salmonella typhimurium locus by selection of hyperinvasive mutants. Proc. Natl. Acad. Sci. USA 87:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesse, A., A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 28.Levine, M. M., and B. Sztein. 2000. Shigella, Salmonella typhi, and Escherichia coli, p. 171-194. In M. W. Cunningham and R. S. Fujinami (ed.), Effects of microbes on the immune system. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 29.Liu, S.-L., T. Ezaki, H. Miura, K. Matsui, and E. Yabuuchi. 1988. Intact motility as a Salmonella typhi invasion-related factor. Infect. Immun. 56:1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukomski, S., R. A. Hull, and S. I. Hull. 1996. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J. Bacteriol. 178:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyczak, J. B., T. S. Zaidi, M. Grout, M. Bittner, I. Contreras, and G. B. Pier. 2001. Epithelial cell contact-induced alterations in Salmonella enterica serovar Typhi lipopolysaccharide are critical for bacterial internalization. Cell. Microbiol. 3:763-772. [DOI] [PubMed] [Google Scholar]
- 32.Marolda, C. L., and M. A. Valvano. 1998. Promoter region of the Escherichia coli O:7-specific lipopolysaccharide gene cluster: structural and functional characterization of an upstream untranslated mRNA sequence. J. Bacteriol. 180:3070-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 34.Miller, I., D. Maskell, C. Hormaeche, K. Johnson, D. Pickard, and G. Dougan. 1989. Isolation of orally attenuated Salmonella typhimurium following TnphoA mutagenesis. Infect. Immun. 57:2758-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills, S. D., and B. B. Finlay. 1994. Comparison of Salmonella typhi and Salmonella typhimurium: invasion, intracellular growth and localization in cultured human epithelial cells. Microb. Pathog. 17:409-423. [DOI] [PubMed] [Google Scholar]
- 36.Morris, C., C. M. C. Yip, I. S. M. Tsui, D. K-H. Wong, and J. Hackett. 2003. The shufflon of Salmonella enterica serovar Typhi regulates type IVB pilus-mediated bacterial self-association. Infect. Immun. 71:1141-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mroczenski-Wildey, M. J., J. L. Di Fabio, and F. C. Cabello. 1989. Invasion and lysis of HeLa cell monolayers by Salmonella typhi: the role of lipopolysaccharide. Microb. Pathog. 6:143-152. [DOI] [PubMed] [Google Scholar]
- 38.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence: identification of FepE as a second Wzz. Mol. Microbiol. 47:1395-1406. [DOI] [PubMed] [Google Scholar]
- 39.Murray, G. L., S. R. Attridge, and R. Morona. 2005. Inducible serum resistance in Salmonella typhimurium is dependent on wzzfepE-regulated very long O antigen chains. Microbes Infect. 7:1296-1304. [DOI] [PubMed] [Google Scholar]
- 40.Nevola, J., B. A. D. Stocker, D. Laux, and P. Cohen. 1985. Colonization of a mouse intestine by a virulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect. Immun. 50:152-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. Mungall, S. D. Bentley, M. T. G. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. The complete genome sequence of a multidrug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 43.Patel, J. C., and J. E. Galán. 2005. Manipulation of the host actin cytoskeleton by Salmonella—all in the name of entry. Curr. Opin. Microbiol. 8:10-15. [DOI] [PubMed] [Google Scholar]
- 44.Pier, G., M. Grout, T. Zaidi, G. Meluleni, S. S. Mueschenborn, G. Banting, R. Ratcliff, M. J. Evans, and W. H. Colledge. 1998. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature 393:79-82. [DOI] [PubMed] [Google Scholar]
- 45.Pradel, E., and C. A. Schnaitman. 1991. Effect of rfaH (sfrB) and temperature on expression of rfa genes of Escherichia coli K-12. J. Bacteriol. 173:6428-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rojas, G., S. Saldías, M. Bittner, M. Zaldívar, and I. Contreras. 2001. The rfaH gene, which affects lipopolysaccharide synthesis in Salmonella enterica Serovar Typhi, is differentially expressed during the bacterial growth phase. FEMS Microbiol. Lett. 204:123-128. [DOI] [PubMed] [Google Scholar]
- 48.Rowe, P. S. N., and P. M. Meadow. 1983. Structure of the core oligosaccharide from the lipopolysaccharide of Pseudomonas aeruginosa and its defective mutants. Eur. J. Biochem. 132:329-337. [DOI] [PubMed] [Google Scholar]
- 49.Saldías, M. S. 2004. Functional analysis of WbaP, essential enzyme in the biosynthesis of O-antigen in Salmonella enterica. Ph.D. thesis. Universidad de Chile, Santiago, Chile.
- 50.St. Geme, J. W., III. 2002. Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell. Microbiol. 4:191-200. [DOI] [PubMed] [Google Scholar]
- 51.Stone, B. J., C. M. García, J. L. Badger, T. Hassett, R. I. Smith, and V. L. Miller. 1992. Identification of novel loci affecting entry of Salmonella enteritidis into eukaryotic cells. J. Bacteriol. 174:3945-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Townsend, S. M., N. Kramer, R. Edwards, S. Baker, N. Hamlin, M. Simmonds, K. Stevens, S. Maloy, J. Parkhill, G. Dougan, and J. Bäumler. 2001. Salmonella enterica serovar Typhi possesses a unique repertoire of fimbrial gene sequences. Infect. Immun. 69:2894-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 54.Tsui, I. S. M., C. M. C. Yip, J. Hackett, and C. Morris. 2003. The type IVB pili of Salmonella enterica serovar Typhi bind to the cystic fibrosis transmembrane conductance regulator. Infect. Immun. 71:6049-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wain, J., D. House, D. Pickard, G. Dougan, and G. Frankel. 2001. Acquisition of virulence-associated factors by the enteric pathogens Escherichia coli and Salmonella enterica. Philos. Trans. R. Soc. London B 356:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wain, J., D. House, J. Parkhill, C. Parry, and G. Dougan. 2002. Unlocking the genome of the human typhoid bacillus. Lancet Infect. Dis. 2:163-170. [DOI] [PubMed] [Google Scholar]
- 57.Wang, L., and P. R. Reeves. 1994. Involvement of the galactosyl-1-phosphate transferase encoded by the Salmonella enterica rfbP gene in O-antigen subunit processing. J. Bacteriol. 176:4348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, L., S. Jensen, R. Hallman, and P. R. Reeves. 1998. Expression of the O antigen gene cluster is regulated by RfaH through the JUMPstart sequence. FEMS Microbiol. Lett. 165:201-206. [DOI] [PubMed] [Google Scholar]
- 59.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M.-C. Prévost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313-1317. [DOI] [PubMed] [Google Scholar]
- 60.Wollin, R., S. Greeger, L. I. Rothfield, B. A. D. Stocker, and A. A. Lindberg. 1983. Salmonella typhimurium mutants defective in UDP d-galactose: lipopolysaccharide α-1,6-d-galactosyl-transferase. J. Biol. Chem. 258:3769-3774. [PubMed] [Google Scholar]
- 61.Young, D., T. Hussell, and G. Dougan. 2002. Chronic bacterial infections: living with unwanted guests. Nat. Immunol. 3:1026-1032. [DOI] [PubMed] [Google Scholar]
- 62.Zaidi, T. S., S. M. J. Fleiszig, M. J. Preston, J. B. Goldberg, and G. Pier. 1996. Lipopolysaccharide outer core is a ligand for corneal cell binding and ingestion of Pseudomonas aeruginosa. Investig. Ophthmal. Vis. Sci. 37:976-986. [PubMed] [Google Scholar]
- 63.Zhang, X.-L., I. S. M. Tsiu, C. M. C. Yip, A. W. Y. Fung, D. K.-H. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]