Abstract
Seven cathepsin B-like cysteine proteases (CBLs) were identified from the immunoprotective excretory-secretory products of Haemonchus contortus. Two-dimensional (2-D) zymography and biotinylated inhibitors were employed to localize active CBLs in 2-D protein gels. Mass spectrometry provided the identification of AC-4, HMCP1, HMCP2, and GCP7 as well as three novel CBLs encoded by clustered expressed sequence tags.
Cysteine proteases are prime targets for vaccine development against parasitic nematodes (11, 14, 19). In Haemonchus contortus, a highly pathogenic parasite of ruminants, cathepsin B-like cysteine proteases (CBLs) are encoded by a family of at least 22 genes (7, 13, 21) and are abundantly expressed, representing 4% of all adult worm expressed sequence tags (ESTs) (6, 12). No function for any of the CBLs, nor their potential functional diversity or redundancy, has been resolved. Such knowledge is key to the design and evaluation of vaccination experiments, and it will be necessary to trace back immune protection to uniquely identified proteins. Specific inhibitor profiles of H. contortus CBL activity have mainly been determined in crude extracts of whole worms and gut tissue. These contain a mixture of CBL gene products (4, 8, 9, 10, 15), of which only a few have directly been identified by N-terminal amino acid sequencing (20).
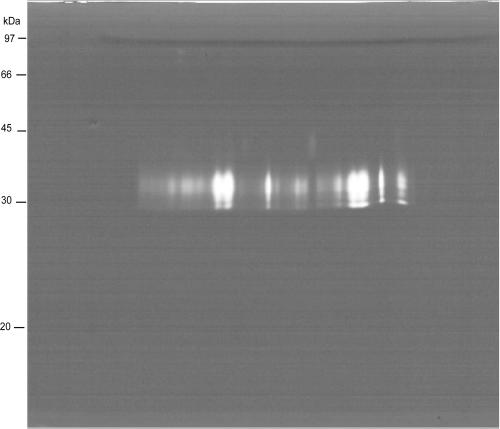
Cysteine proteases are the most active proteases of the excretory-secretory products (ES) of H. contortus (12) and are likely to be involved in induction of protective immunity (2). Proteomic analysis of 102 prominent spots present on a two-dimensional (2-D) protein gel of ES identified only members of three other protease classes (28). Therefore, the ability of CBLs to separate and migrate into 2-D protein gels was investigated by gel activity assay (zymogram), as commonly performed after 1-D electrophoresis in gelatin containing sodium dodecyl sulfate (SDS)-polyacrylamide gels. ES (200 μg) was submitted to isoelectric focusing on 13-cm immobilized pH gradient strips (pH 3 to 10 nonlinear [NL]) as described previously (28), but alkylation by iodoacetamide was omitted. Separation in the second dimension was performed on a SDS-12.5% polyacrylamide gel containing 0.1% gelatin in the absence of dithiothreitol (DTT). Under conditions favoring cysteine protease activity (18 h at 37°C in 10 mM Tris, 20 mM NaCl, 10 mM DTT, pH 5.0), abundant proteolytic activity was found in a region between 30 and 35 kDa (Fig. 1) corresponding to the position of cysteine proteases in 1-D zymograms of H. contortus ES (8). No proteolytic activity was detected at the higher molecular weight (MW) range, where serine proteases, metalloproteases, and aspartic proteases are expected to be located (28), possibly due to unfavorable experimental conditions.
FIG. 1.
Two-dimensional zymography (13-cm strips; pH 3 to 10 NL). Proteolysis is visualized as a clear area where gelatin has been digested against a blue background stained with Coomassie blue R250. Molecular masses were estimated from a standard molecular mass marker.
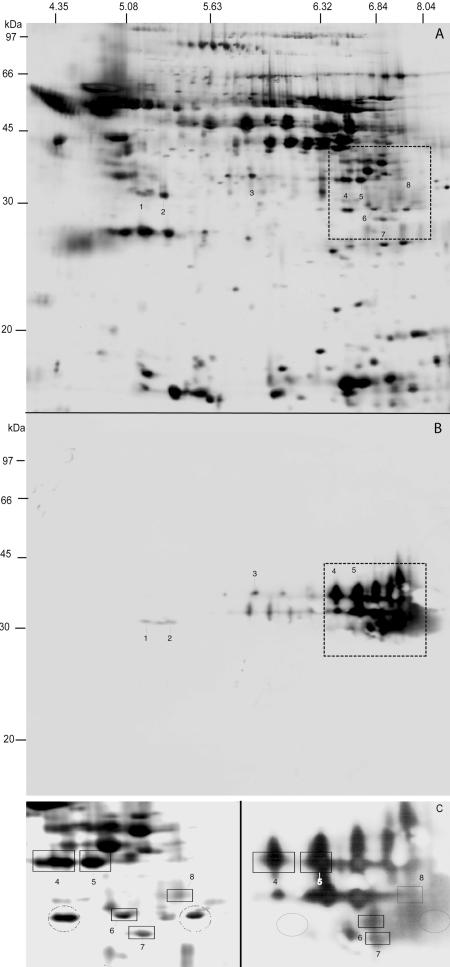
The observed wide pI range corresponds to the range of predicted pI values for individual CBLs (Table 1). However, reduced resolution due to the presence of gelatin and the absence of DTT prohibits precise colocalization with spots present in a silver-stained 2-D gel run in parallel. Therefore, spots representing putative CBLs were localized by affinity labeling with a biotinylated irreversible dipeptide inhibitor specific for cysteine proteases (20). ES (200 μg) was incubated with 5 μM biotin-phenylalanine-alanine-fluoromethylketone (Bt-FA-FMK) (Enzyme Systems Products) for 15 min at 37°C. After 2-D gel electrophoresis (immobilized pH gradient strips, pH 3 to 10 NL; 12.5% polyacrylamide gel electrophoresis ), proteins were blotted onto polyvinylidene difluoride membranes and blocked overnight with 5% nonfat dry milk in PBS-0.05% Tween (PBS-T). Membranes were incubated with streptavidin-horseradish peroxidase conjugate (GE Healthcare) diluted 1:500 in 2% nonfat dry milk in PBS-T, followed by detection by chemiluminescence (ECL plus; GE Healthcare). The noncharged and low-MW inhibitor Bt-FA-FMK binds covalently to a cysteine in the active site of the protease and is unlikely to cause changes in pI and MW in comparison to the silverstained gel, in which cysteines are blocked by alkylation with iodoacetamide. As on the zymogram, spots were detected between 30 and 35 kDa within a pI range of 5.0 to 8.3, a number of which could be colocalized with spots on a silver-stained gel (Fig. 2) using Phoretix Software (NonLinear). Mass spectrometry (ms) analysis by liquid chromatography (LC)/MS/MS allowed the identification of seven CBLs, an aspartic protease, and a metalloprotease by searching the GenBank protein database and a database of 21,791 clustered H. contortus ESTs with the obtained fragmentation spectra. CBL identifications are summarized in Table 1, and LC/MS/MS-derived peptide sequences are indicated in the alignment of Fig. 3. We obtained peptide sequences encoded by the cDNA sequences of four known CBLs (AC-4, spots 4 and 5; HMCP1, spot 8; HMCP2, spot 6; GCP7, spot 7) as well as the corresponding homologous EST clusters. However, the EST clusters most similar to HMCP1 and HMCP2 display a few remarkable sequence variations (Fig. 3). The gene clusters may encode allelic variants, but spots 8 and 6 could also derive from paralogous genes (represented by the EST clusters) most similar to HMCP1 and HCMP2.
TABLE 1.
Cathepsin B-like cysteine protease spots identified from H. contortus ESa
| Protease | Spot | Protein accession no. | EST accession no. | No. of ESTs in cluster | Predicted pI (−SP) | Predicted pI (−SP, −PR) | pI of spot | Predicted mass (−SP) | Predicted mass (−SP, −PR) | Mass of spot |
|---|---|---|---|---|---|---|---|---|---|---|
| AC4 | 4 and 5 | AAA29177 | 6.45 | 7.68 | 6.34 and 6.75 | 36.1 | 28.4 | 33.8 | ||
| GCP7 | 7 | AAC05262 | 8.08 | 8.09 | 7.32 | 37.4 | 27.8 | 28.9 | ||
| HMCP1 | 8 | CAA93275 | 7.88 | 8.27 | 8.29 | 38.3 | 28.7 | 32.0 | ||
| HMCP1-like | 8 | CA958902 | 94 | 7.99 | 8.34 | 8.29 | 38.3 | 28.6 | 32.0 | |
| HMCP2 | 6 | CAA93276 | 6.91 | 8.01 | 6.93 | 38.1 | 28.7 | 29.9 | ||
| HMCP2-like | 6 | CA958204 | 8 | Incomplete | NAb | 6.93 | NA | NA | 29.9 | |
| HMCP7 | 3 | CA869450 | 40 | 5.92 | 6.69 | 5.76 | 36.8 | 28.05 | 34.8 | |
| HMCP8 | 1 and 2 | CB019057 | 1 | Incomplete | NA | 5.05 and 5.34 | NA | NA | 32.5 and 32.4 | |
| HMCP9 | 8 | CA034108 | 3 | Incomplete | NA | 8.29 | NA | NA | 32.0 |
The predicted isoelectric points (pI) and molecular masses (in kDa) are indicated without signal peptide (−SP) and without the propeptide region (−PR). The observed pI and mass (in kDa) of the spots are also indicated.
NA, not applicable.
FIG.2.
(A) Silver-stained two-dimensional SDS-PAGE gel of H. contortus ES (13-cm strips; pH 3 to 10 NL). (B) Detection and localization of ES cysteine proteases of H. contortus by the biotinylated peptide inhibitor Bt-FA-FMK. C) Zoom of the region in panels A (gel on the left) and B (membrane on the right) marked with a square. Specificity is exemplified by two circled spots showing no binding of inhibitor (right) despite intense silver staining (left). The estimation of the molecular mass of stained proteins was done using a biotinylated marker (GE Healthcare).
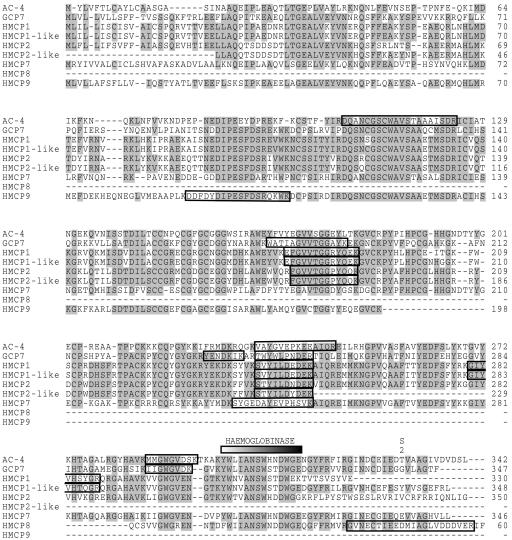
FIG. 3.
Alignment of the CBL sequences enclosing the identifications of CBLs made from ES of H. contortus. GenBank accession numbers of the displayed sequences have been indicated in Table 1. Positions having four or more identical residues have been shaded. The peptide sequences obtained from each spot by LC/MS/MS (as described in reference 28) are boxed in the alignment. The hemoglobinase motif and S2 substrate binding site described in the text are shown, and the propeptide region is indicated by a black bar.
Three novel CBLs, designated HMCP7 (spot 3), HMCP8 (spot 1 and 2), and HMCP9 (spot 8), are encoded by EST clusters with little similarity at the DNA level to any known CBL, thus excluding the possibility that they represent allelic variants of known CBLs.
A full-length protein sequence was obtained for HMCP7 by conceptual translation from the EST clusters. Sequence comparison (Fig. 3) reveals 71% amino acid identity to HMCP4. Remarkably, HMCP7 carries a glutamic acid at position 338, whereas most H. contortus CBLs have a hydrophobic residue at this position. Amino acids at the homologous position, lining the S2 substrate pocket (depicted in Fig. 3), in cathepsin B-like proteases of other organisms determine substrate specificity. Site-specific mutagenesis (3, 16) has demonstrated that a glutamic acid residue at this position supports the typical cathepsin B-like activity of hydrolyzing substrates Phe-Arg-AMC (FR) and Arg-Arg-AMC (RR), whereas a hydrophobic residue results in cathepsin L-like specificity leading to hydrolysis of only FR. Thus, the potential presence of abundantly expressed HMCP7 may well explain the previous observation that H. contortus intestinal extracts containing CBL activity seemed hardly more efficient in hydrolysis of FR compared to RR (15). Resolving the substrate specificity of individual CBLs, in ES as well as in intestinal extracts, is clearly important in determination of their function. With the identification and mapping of specific CBLs, predicted specificities can be tested by further fractionation from native extracts or characterization of recombinant proteins.
HMCP8 displays 57% identity to a Trichuris suis cysteine protease (accession no. AAC78691) and is represented by a single EST. Staining intensity of spots 1 and 2 suggests efficient secretion of this protease. A replacement of tyrosine by phenylalanine in the putative hemoglobinase domain (Fig. 3) is likely to modify substrate specificity. This domain is strictly conserved within most CBLs from blood feeding helminths (1), and detection of CBLs with and without this motif in the excretory/secretory products of H. contortus prompts for further testing of the potential functional diversity of the secreted CBLs.
CBLs are translated as preproteins harboring an N-terminal propeptide that blocks access to the active site. Activation results from autocleavage triggered by a drop in pH (25). All peptide sequences obtained by LC/MS/MS are localized after the cleavage site (Fig. 3), with the exception of the peptide mapping to the predicted propeptide (13, 20) region of HCMP9 (spot 8, colocalizing with HCMP1), indicating that it may be secreted as a nonactive protease. HCMP9 is represented by three ESTs and is 55% identical to GCP7. The observed MW and pI of AC 4 (spots 4 and 5) and HMCP7 match better with the values predicted for their precursor proteins still containing the propeptide (Table 1).
In addition to HCMP8, spots 1 and 2 provided simultaneous identifications with the aspartic protease PEP2 (CAE12199; spots 1 and 2) and the metalloprotease MEP3 (AAC31568; spot 2). Both are components of a galactose-containing glycoprotein complex (H-gal-GP) with immunoprotective properties located on the luminal surface of the intestine (22-24). PEP2 is cleaved into N- and C-terminal domains that are held together by disulfide bonds which are broken under the reducing conditions used for gel electrophoresis in the second dimension (28). The estimated molecular masses of spots 1 and 2 (32.4 and 32.5 kDa, respectively) correspond to the 31-kDa size reported for the C-terminal domain (22). Similarly, MEP3 (with a predicted size of 95.5 kDa) was shown to resolve in N- and C-terminal domains of 41 and 47 kDa under reducing conditions (24), and the size observed for MEP3 in spot 2 (32.5 kDa) indicates further processing. Previous proteomic analysis of H. contortus ES identified the presence of the N- and C-terminal domains obtained after cleavage of other metalloproteases (MEP1, MEP1B, and MEP2) and serine proteases (28). As for PEP2 and MEP3, the fragments may be kept together by disulfide bonds, possibly giving rise to proteolytically active complexes under native conditions. Colocalization with HCMP8 is considered to be coincidental, and labeling with Bt-FA-FMK is probably exclusively due to binding of this substrate to HCMP8.
Several CBLs induce protective immune responses (14), but a function has not been identified. A proposed (27) and partially reconstituted proteolytic cascade for the metabolism of hemoglobin, the major food source of blood-feeding parasites, encompasses the sequential cleavage by aspartic proteases, cysteine proteases, metalloproteases, and exopeptidases (26). Aspartic and metalloproteases are also involved in the activation of procathepsin B to cathepsin B (5). Immunohistology has demonstrated localization of many proteases at the surface of the microvilli (26). Their presence in ES suggests that hemoglobin digestion may take place not only at the cell surface but also in the lumen of the gut, thus greatly enhancing the rate of digestion. Proteases in ES may perform other essential functions, outside of the worm, in penetration of mucus layers, gaining access to blood vessels and intervention with host processes like blood-clotting and immune responses. The molecular identification of proteases in ES provides specific tools to explore these options.
The protective properties against infection induced by immunization with ES of H. contortus have inspired attempts to determine the molecular components involved (17, 18). A global analysis (28) of the most abundant proteins in ES already identified several known vaccine candidates (H11 and GA1) and demonstrated the complexity and variability of other immunologically relevant molecules (Hc-ASP1, Hc-ASP2, and Hc15). The identification in ES of new CBLs and proteases with previously demonstrated protective properties further expands this group of proteins, thus contributing to the challenging search for the minimal set of proteins required for induction of a protective immune response.
Acknowledgments
This work was supported by the European Union (Project QRLT-PL-1999-00565) and The Netherlands Proteomics Center.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Baig, S., R. T. Damian, and D. S. Peterson. 2002. A novel cathepsin B active site motif is shared by helminth bloodfeeders. Exp. Parasitol. 101:83-89. [DOI] [PubMed] [Google Scholar]
- 2.Bakker, N., L. Vervelde, K. Kanobana, D. P. Knox, A. W. Cornelissen, E. de Vries, and A. P. Yatsuda. 2004. Vaccination against the nematode Haemonchus contortus with a thiol-binding fraction from the excretory/secretory products (ES). Vaccine 22:618-628. [DOI] [PubMed] [Google Scholar]
- 3.Chan, V. J., P. M. Selzer, J. H. McKerrow, and J. A. Sakanari. 1999. Expression and alteration of the S2 subsite of the Leishmania major cathepsin B-like cysteine protease. Biochem. J. 340:113-117. [PMC free article] [PubMed] [Google Scholar]
- 4.Cox, G. N., D. Pratt, R. Hageman, and R. J. Boisvenue. 1990. Molecular cloning and primary sequence of a cysteine protease expressed by Haemonchus contortus adult worms. Mol. Biochem. Parasitol. 41:25-34. [DOI] [PubMed] [Google Scholar]
- 5.Ishidoh, K., and E. Kominami. 2002. Processing and activation of lysosomal proteinases. Biol. Chem. 383:1827-1831. [DOI] [PubMed] [Google Scholar]
- 6.Jasmer, D. P., M. D. Mitreva, and J. P. McCarter. 2004. mRNA sequences for Haemonchus contortus intestinal cathepsin B-like cysteine proteases display an extreme in abundance and diversity compared with other adult mammalian parasitic nematodes. Mol. Biochem. Parasitol. 137:297-305. [DOI] [PubMed] [Google Scholar]
- 7.Jasmer, D. P., J. Roth, and P. J. Myler. 2001. Cathepsin B-like cysteine proteases and Caenorhabditis elegans homologues dominate gene products expressed in adult Haemonchus contortus intestine. Mol. Biochem. Parasitol. 116:159-169. [DOI] [PubMed] [Google Scholar]
- 8.Karanu, F. N., F. R. Rurangirwa, T. C. McGuire, and D. P. Jasmer. 1993. Haemonchus contortus: identification of proteases with diverse characteristics in adult worm excretory-secretory products. Exp. Parasitol. 77:362-371. [DOI] [PubMed] [Google Scholar]
- 9.Knox, D. P., D. L. Redmond, and D. G. Jones. 1993. Characterization of proteinases in extracts of adult. Haemonchus contortus, the ovine abomasal nematode. Parasitology 106:395-404. [DOI] [PubMed] [Google Scholar]
- 10.Knox, D. P., S. K. Smith, and W. D. Smith. 1999. Immunization with an affinity purified protein extract from the adult parasite protects lambs against infection with Haemonchus contortus. Parasite Immunol. 21:201-210. [DOI] [PubMed] [Google Scholar]
- 11.Loukas, A., J. M. Bethony, A. L. Williamson, G. N. Goud, S. Mendez, B. Zhan, J. M. Hawdon, M. Elena Bottazzi, P. J. Brindley, and P. J. Hotez. 2004. Vaccination of dogs with a recombinant cysteine protease from the intestine of canine hookworms diminishes the fecundity and growth of worms. J. Infect. Dis. 189:1952-1961. [DOI] [PubMed] [Google Scholar]
- 12.Parkinson, J., C. Whitton, R. Schmid, M. Thomson, and M. Blaxter. 2004. NEMBASE: a resource for parasitic nematode ESTs. Nucleic Acids Res. 32:D427-D430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pratt, D., L. G. Armes, R. Hageman, V. Reynolds, R. J. Boisvenue, and G. N. Cox. 1992. Cloning and sequence comparisons of four distinct cysteine proteases expressed by Haemonchus contortus adult worms. Mol. Biochem. Parasitol. 51:209-218. [DOI] [PubMed] [Google Scholar]
- 14.Redmond, D. L., and D. P. Knox. 2004. Protection studies in sheep using affinity-purified and recombinant cysteine proteinases of adult Haemonchus contortus. Vaccine 22:4252-4261. [DOI] [PubMed] [Google Scholar]
- 15.Rhoads, M. L., and R. H. Fetterer. 1995. Developmentally regulated secretion of cathepsin L-like cysteine proteases by Haemonchus contortus. J. Parasitol. 81:505-512. [PubMed] [Google Scholar]
- 16.Sajid, M., and J. H. McKerrow. 2002. Cysteine proteases of parasitic organisms. Mol. Biochem. Parasitol. 120:1-21. [DOI] [PubMed] [Google Scholar]
- 17.Schallig, H. D., M. A. van Leeuwen, and A. W. Cornelissen. 1997. Protective immunity induced by vaccination with two Haemonchus contortus excretory secretory proteins in sheep. Parasite Immunol. 19:447-453. [DOI] [PubMed] [Google Scholar]
- 18.Schallig, H. D., M. A. van Leeuwen, B. E. Verstrepen, and A. W. Cornelissen. 1997. Molecular characterization and expression of two putative protective excretory secretory proteins of Haemonchus contortus. Mol. Biochem. Parasitol. 88:203-213. [DOI] [PubMed] [Google Scholar]
- 19.Selzer, P. M., S. Pingel, I. Hsieh, B. Ugele, V. J. Chan, J. C. Engel, M. Bogyo, D. G. Russell, J. A. Sakanari, and J. H. McKerrow. 1999. Cysteine protease inhibitors as chemotherapy: lessons from a parasite target. Proc. Natl. Acad. Sci. USA 96:11015-11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shompole, S., and D. P. Jasmer. 2001. Cathepsin B-like cysteine proteases confer intestinal cysteine protease activity in Haemonchus contortus. J. Biol. Chem. 276:2928-2934. [DOI] [PubMed] [Google Scholar]
- 21.Skuce, P. J., D. L. Redmond, S. Liddell, E. M. Stewart, G. F. Newlands, W. D. Smith, and D. P. Knox. 1999. Molecular cloning and characterization of gut-derived cysteine proteinases associated with a host protective extract from Haemonchus contortus. Parasitology 119:405-412. [DOI] [PubMed] [Google Scholar]
- 22.Smith, W. D., P. J. Skuce, G. F. Newlands, S. K. Smith, and D. Pettit. 2003. Aspartyl proteases from the intestinal brush border of Haemonchus contortus as protective antigens for sheep. Parasite Immunol. 25:521-530. [DOI] [PubMed] [Google Scholar]
- 23.Smith, W. D., S. K. Smith, and J. M. Murray. 1994. Protection studies with integral membrane fractions of Haemonchus contortus. Parasite Immunol. 16:231-241. [DOI] [PubMed] [Google Scholar]
- 24.Smith, W. D., S. K. Smith, D. Pettit, G. F. Newlands, and P. J. Skuce. 2000. Relative protective properties of three membrane glycoprotein fractions from Haemonchus contortus. Parasite Immunol. 22:63-71. [DOI] [PubMed] [Google Scholar]
- 25.Turk, B., D. Turk, and V. Turk. 2000. Lysosomal cysteine proteases: more than scavengers. Biochim. Biophys. Acta 1477:98-111. [DOI] [PubMed] [Google Scholar]
- 26.Williamson, A. L., P. J. Brindley, D. P. Knox, P. J. Hotez, and A. Loukas. 2003. Digestive proteases of blood-feeding nematodes. Trends Parasitol. 19:417-423. [DOI] [PubMed] [Google Scholar]
- 27.Williamson, A. L., P. Lecchi, B. E. Turk, Y. Choe, P. J. Hotez, J. H. McKerrow, L. C. Cantley, M. Sajid, C. S. Craik, and A. Loukas. 2004. A multi-enzyme cascade of hemoglobin proteolysis in the intestine of blood-feeding hookworms. J. Biol. Chem. 279:35950-35957. [DOI] [PubMed] [Google Scholar]
- 28.Yatsuda, A. P., J. Krijgsveld, A. W. Cornelissen, A. J. Heck, and E. de Vries. 2003. Comprehensive analysis of the secreted proteins of the parasite Haemonchus contortus reveals extensive sequence variation and differential immune recognition. J. Biol. Chem. 278:16941-16951. [DOI] [PubMed] [Google Scholar]