Abstract
GdhR is a meningococcal transcriptional regulator that was previously shown to positively control the expression of gdhA, encoding the NADP-specific l-glutamate dehydrogenase (NADP-GDH), in response to the growth phase and/or to the carbon source. In this study we used reverse transcriptase-PCR-differential display (to identify additional GdhR-regulated genes. The results indicated that GdhR, in addition to NADP-GDH, controls the expression of a number of genes involved in glucose catabolism by the Entner-Doudoroff pathway and in l-glutamate import by an unknown ABC transport system. The genes encoding the putative periplasmic substrate-binding protein (NMB1963) and the permease (NMB1965) of the ABC transporter were genetically inactivated. Uptake experiments demonstrated an impairment of l-glutamate import in the NMB1965-defective mutant in the absence or in the presence of a low sodium ion concentration. In contrast, at a sodium ion concentration above 60 mM, the uptake defect disappeared, possibly because the activity of a sodium-driven secondary transporter became predominant. Indeed, the NMB1965-defective mutant was unable to grow at a low sodium ion concentration (<20 mM) in a chemically defined medium containing l-glutamate and four other amino acids that supported meningococcal growth, but it grew when the sodium ion concentration was raised to higher values (>60 mM). The same growth phenotype was observed in the NMB1963-defective mutant. Cell invasion and intracellular persistence assays and expression data during cell invasion provided evidence that the l-glutamate ABC transporter, tentatively named GltT, was critical for meningococcal adaptation in the low-sodium intracellular environment.
Neisseria meningitidis (meningococcus) is a commensal bacterium of the human nasopharynx that occasionally provokes life-threatening or other severe diseases including meningitis and sepsis. A crucial factor in the commensal and pathogenic behavior of this bacterium is its capacity to obtain and synthesize nutrients essential for its survival in the different microenvironments within the human host during the course of a natural infection. A genomewide analysis of the attributes of N. meningitidis required for disseminated infection strongly supported this view. Using signature-tagged mutagenesis, 73 genes that are essential for systemic infection in an infant rat model have been recently identified. Remarkably, about half of the 73 genes encode enzymes that are involved in metabolism and transport of nutrients (42).
Although the study of the meningococcal metabolism was begun more than 40 years ago and genomic technology has become a fundamental tool for investigating the meningococcal metabolome (44) (http://www.sciencemag.org/feature/data/1046515.shl), our knowledge about the metabolic pathways and transport systems in this microorganism is far from being exhaustive. In addition, only a few regulatory pathways have been studied in some detail with respect to the meningococcal metabolism and host cell interaction (7, 8, 10, 15).
In a previous work, limited transcriptional analysis of invasive and commensal meningococcal isolates led to discovery of a transcriptional regulator, GdhR, differentially expressed among the strains (32). GdhR belongs to the GntR family of bacterial helix-turn-helix regulators (34), and it was shown to positively control the expression of gdhA, encoding the NADP-specific l-glutamate dehydrogenase (NADP-GDH), in response to the growth phase and/or to the carbon source. The negative effector 2-oxoglutarate, a product of the catabolic reaction of the NADP-GDH, and an intermediate of the tricarboxylic acid (TCA) cycle, plays a key role in this control by preventing the binding of GdhR to the stronger gdhA promoter (32). The existence of this regulatory circuit led us to speculate that the major role of NADP-GDH in meningococci might be catabolic, to supply the TCA with 2-oxoglutarate from l-glutamate under certain growth conditions, a hypothesis that is supported by the natural auxotrophy for l-glutamate of this microorganism (5). Indeed, studies on the closely related species N. gonorrhoeae indicate that intracellular levels of 2-oxoglutarate may range considerably in response to the carbon source, being higher in the presence of lactate (or pyruvate) than in the presence of glucose (17, 19, 20, 22, 31, 38). Glucose is the predominant carbon source in blood; by contrast, lactate is the major carbon source during growth in the cerebrospinal fluid (13), as well as in the saliva and in mucosal environments that are colonized by lactic bacteria, such as the nasopharynx (25, 26). Lactate and pyruvate tend to be used as major carbon (energy) sources within phagocytic cells (46). These arguments support the hypothesis that the role of the NADP-GDH and the regulatory mechanisms concerned might have wider implications and that GdhR might control other genes in addition to gdhA.
To test this hypothesis, a reverse transcriptase-PCR differential display (RT-PCR-DD) strategy was developed in this study. The strategy relied on the presence of highly and moderately repetitive transcribed DNA sequences in the meningococcal genome. In particular, the meningococcal genome is characterized by the presence of more than 1,900 copies of 10-bp-long DNA uptake signal sequences (DUS) often found in the base-paired stem of transcription terminators (33, 39, 44). In addition, the neisserial miniature insertion sequences (nemis or Correia elements) are 154 to 158 bp (unit-length) or 104 to 108 bp (internally rearranged) elements carrying terminal inverted repeats (TIRs) present in about 250 copies (28, 33, 44). The subfamilies of 26L/26R, 26L/27R, 27L/27R, and 27L/26R elements originate from the combination of TIRs, which vary in length (26 to 27 bp) as well as in sequence content (L and R types). More than two-thirds of nemis are interspersed with single-copy DNA and are cotranscribed with cellular genes serving as portable promoters (2, 4, 35) and/or as RNase III processing sites (9, 28).
The results of the RT-PCR-DD, independently confirmed by Northern and slot blot analyses, indicate that GdhR, in addition to NADP-GDH, controls the expression of a number of genes involved in glucose catabolism by the Entner-Doudoroff pathway and the l-glutamate import by an unknown ABC transport system, tentatively named GltT, evolutionarily conserved among gram-negative bacteria and plant chloroplasts. Several phenotypes of knocked-out ABC transport mutants were tested, including the ability (i) to grow on complex or chemically defined media, (ii) to import l-glutamate, (iii) to invade HeLa cells, and (iv) to survive and multiply within the host cell. Finally, expression of the GltT operon by intracellular meningococci was evaluated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The meningococcal strain used in this study as a wild type was H44/76, a serogroup B isolate belonging to the ET-5 hypervirulent lineage, kindly provided by IRIS, Chiron SpA, Siena, Italy. Meningococci were cultured on complex media chocolate agar (Becton-Dickinson) or on gonococcus agar or broth supplemented with 1% (vol/vol) Polyvitox (OXOID) at 37°C in 5% CO2. When required, Bacto agar was added at a 1.5% (wt/vol) final concentration. Meningococci were also cultured in the chemically defined media MCDA (5), MCDA-1, and MCDA-2, which were formulated on the basis of MCDA. The final concentration of each component of the MCDA is as follows: 100 mM NaCl, 2.5 mM KCl, 7.5 mM NH4Cl, 7.5 mM Na2HPO4, 1.25 mM KH2PO4, 2.2 mM Na3C6H5O7 · 2 H2O, 2.5 mM MgSO4 · 7 H20, 0.0075 mM MnSO4 · H2O, 8.0 mM l-glutamic acid, 0.5 mM l-arginine, 2.0 mM glycine, 0.2 mM l-serine, 0.06 mM l-cysteine · HCl · H2O, 0.5% (vol/vol) glycerin, 0.25 mM CaCl2 · 2H2O, 0.01 mM Fe2(SO4)3, 1% glucose. In MCDA-1 the NaCl was omitted; in MCDA-2 the NaCl concentration was decreased to 40 mM. The pHs of all media were adjusted to 7.4.
Escherichia coli strain DH5α[F− Φ80d lacZΔM15 endA1 recA1 hsdR17 supE44 thi-1 λ− gyrA96 Δ(lacZYA-argF) U169] was used in cloning procedures. This strain was grown in Luria-Bertani (LB) medium. To allow plasmid selection, LB medium was supplemented with ampicillin (50 μg ml−1).
DNA procedures.
High-molecular-weight genomic DNA from N. meningitidis strains was prepared as described previously (3). DNA fragments were isolated through acrylamide slab gels and recovered by electroelution as described previously (36).
Oligonucleotides used in this study as primers in PCRs are listed in Table 1. Oligonucleotide synthesis was performed as a service by MWG-Biotech AG Oligo Production. The amplification reactions generally consisted of 35 cycles including 45 s of denaturation at 94°C, 45 s of annealing at 65°C, and 45 s of extension at 72°C. They were carried out in a Perkin-Elmer Cetus DNA Thermal Cycler 2400. The DNA from strain H44/76 was used as a template.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| NMB1966-1 | 5′-GACCCTGAAATTATGTTGTACGACGAG-3′ |
| NMB1965-1 | 5′-GTTGCCGGATCCGGGCTGTTTGTCGGC ATGGTCTTGGG-3′ |
| NMB1965-2 | 5′-GCAATGGGATCCCTGATGCACGGCAAT CAGCGTTAC-3′ |
| NMB1964-1 | 5′-TTGGGTCGGACTGTTCGTCCTGATTGG-3′ |
| NMB1964-2 | 5′-TTATTCGGCGGCTTTTTCCGCATTGCC-3′ |
| NMB1964-f | 5′-TTGGGTCGGACTGTTCGTCCTG-3′ |
| NMB1964-r | 5′-GACCAATACGCCTGCGGATTTG-3′ |
| NMB1963-1 | 5′-GCCAAAGGATCCAAAACGCCACTCAAG TATTGAGC-3′ |
| NMB1963-2 | 5′-CGAATTGGATCCGGTACACGGTAACCA GGCTCGC-3′ |
| NMB1961-1 | 5′-TTTTCAGGATCCACGACCAAGCCGACC GCTACATTTTCG-3′ |
| NMB1961-2 | 5′-GCGGCTGGATCCAGACTGTCGGTCAAA TCGAGCAG-3′ |
| NMB1960-1 | 5′-TTCTGTCGGAACGGACAGTCCGAAACC-3′ |
| NMB1960-2 | 5′-GTAGAAAGCTGGATGTCGAACCATTCG-3′ |
| NMB1958-2 | 5′-AATCAGGGTTACTTTGCCTTGCAGGTC-3′ |
| NMB2147-1 | 5′-TATTTTGATAACCGCCTGCAGCACACC-3′ |
| NMB2147-2 | 5′-TAATGTCCCGGACAGGATGCTCATCGG-3′ |
| Zwf-1 | 5′-TGAGGCTTGGGCATCGTTTGTCGAACG-3′ |
| Zwf-2 | 5′-GGTCAGCGGCTTCAATGACTTGATGAC-3′ |
| DUS-IN | 5′-GCCGTCTGAA-3′ |
| DUS-OUT | 5′-TTCAGACGGC-3′ |
| 26L-IN | 5′-GTGGATTAACAAAAATCAGGAC-3′ |
| 26L-OUT | 5′-GTCCTGATTTTTGTTAATCCAC-3′ |
| 27L-IN | 5′-GTGGATTAAATTTAAATCAGGAC-3′ |
| 27L-OUT | 5′-GTCCTGATTTAAATTTAATCCAC-3′ |
| 16Suniv-1 | 5′-CAGCAGCCGCGGTAATAC-3′ |
| 16Suniv-2 | 5′-CCGTCAATTCCTTTGAGTTT-3′ |
| 16S-r | 5′-CTACGCATTTCACTGCTACACG-3′ |
Southern blot hybridizations were carried out according to standard protocols (36). 32P labeling of the DNA fragments was performed by random priming using the Klenow fragment of the E. coli DNA polymerase I and [α-32P]dATP and [α-32P]dGTP (3,000 Ci mmol−1) (36).
DNA sequencing was performed as a service by the MWG Biotech Custom Sequencing Service. Processing of the DNA sequences was performed with the software GeneJockey Sequence Processor (published and distributed by Biosoft). Deduced amino acid sequence similarity searching was performed with the BLAST program using the Conserved Domain Database available at the NCBI. Smith-Waterman alignments (40) were performed with the SSEARCH program using the NPSA 3DSEQ or the NPSA SWISSPROT databases available at the Pôle Bioinformatique Lyonnaise.
Plasmids and cloning procedures.
The Neisseria-E. coli shuttle vector pDEX and the derivative plasmid pDEΔgdhR harboring a 540-bp Sau3AI DNA fragment containing the central part of gdhR have been described previously (32). To construct pDEΔNMB1965, the DNA corresponding to the central segment of the open reading frame (ORF) NMB1965 was amplified using the primers NMB1965-1 and NMB1965-2, and the BamHI-restricted 482-bp PCR product was cloned into the BamHI site of pDEX. An analogous strategy was used to obtain pDEΔNMB1963 with the primers NMB1963-1 and NMB1963-2 amplifying a BamHI-restricted 417-bp PCR product and pDEΔNMB1961 with the primers NMB1961-1 and NMB1961-2 amplifying a BamHI-restricted 481-bp PCR product.
Knockout of gdhR, NMB1965, NMB1963, and NMB1961.
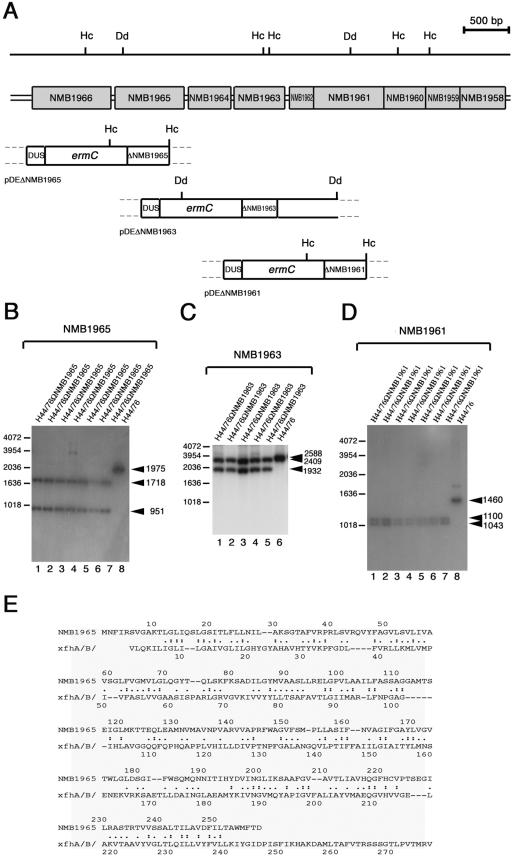
The gdhR gene was inactivated in H44/76 by single crossing-over event using pDEΔgdhR originating the strain H44/76ΩgdhR as described previously (32). The ORFs NMB1965, NMB1963, and NMB1961 were genetically knocked out by the same procedure using the plasmids pDEΔNMB1965, pDEΔNMB1963, and pDEΔNMB1961, respectively. Transformations were performed by using 0.1 to 1 μg of plasmid DNA. Transformants were selected on GC agar medium supplemented with erythromycin (7 μg ml−1). Successful gene inactivation was demonstrated by Southern blot hybridization. NMB1965-, NMB1963- and NMB1961-specific probes (see Fig. 4B to D) were obtained by 32P labeling of BamHI-restricted PCR fragments. The sizes of these fragments were 482 bp (primers NMB1965-1 and NMB1965-2), 417 bp (primers NMB1963-1 and NMB1963-2), and 481 bp (primers NMB1961-1 and NMB1961-2).
FIG.4.
Knockout of NMB1965, NMB1963, and NMB1961 in H44/76 and alignment of deduced amino acid sequences. (A) Experimental design for NMB1965, NMB1963, and NMB1961 disruption by single crossing-over. The genetic and physical map of the NMB1966-to-NMB1958 region is indicated above the map of the genetic determinants of pDEΔNMB1965, pDEΔNMB1963, and pDEΔNMB1961 used for the gene disruption. Hc, HincII restriction sites; Dd, DdeI restriction sites. The genetic determinants of pDEΔNMB1965, pDEΔNMB1963, and pDEΔNMB1961 are as follows: (i) ermC, the erythromycin resistance gene used as a selective marker for transformation; (ii) ΔNMB1965 (in pDEΔNMB1965), a BamHI-restricted 482-bp DNA fragment spanning the central part of NMB1965; (iii) ΔNMB1963 (in pDEΔNMB1963), a BamHI-restricted 417-bp DNA fragment spanning the central part of NMB1963; (iv) ΔNMB1961 (in pDEΔNMB1961), a BamHI-restricted 481-bp DNA fragment spanning the central part of NMB1961; (v) a DNA fragment containing a DUS, required for efficient DNA uptake during transformation. (B to D) Southern blot analysis demonstrating the inactivation of NMB1965 (B), NMB1963 (C), and NMB1961 (D). Chromosomal DNA was extracted from the parental strain, H44/76 (lane 8 in B and D and lane 6 in C), and from several recombinant strains transformed either with pDEΔNMB1965 (lanes 1 to 7, B), pDEΔNMB1963 (lanes 1 to 5, C) or with pDEΔNMB1961 (lanes 1 to 7, D). The HincII-restricted (B and D) or DdeI-restricted (C) chromosomal DNAs were analyzed by Southern blotting using as probes the 32P-labeled NMB1965-specific (B), the NMB1963-specific (C), or the NMB1961-specific (D) DNA fragments cloned in pDEΔNMB1965, pDEΔNMB1963, or pDEΔNMB1961, respectively. Arrows on the right of each panel indicate NMB1965-specific (B), NMB1963-specific (C), or NMB1961-specific (D) fragments whose sizes were deduced on the basis of the relative migration of DNA ladders (bars on the left of each panel). (E) Smith-Waterman alignment between the NMB1965 deduced protein sequence and the proton/glutamate symport protein from Pyrococcus horikoshii. Identities (:) and synonym substitutions (.) are reported.
Standard RNA procedures.
Total bacterial RNA was extracted from logarithmically growing cells in GC medium by the guanidine hydrochloride procedure previously described (32) or by the RNeasy Midi kit according to the manufacturer's instructions (QIAGEN). Both of these procedures gave comparable results in terms of RNA integrity using the gdhA transcript (32) in Northern blot experiments as a reference mRNA. Electrophoretic analysis was performed by fractionating the total RNA on 1% agarose gels containing formaldehyde (36). RNA transfer to Hybond-N+ nylon membranes (Amersham) and hybridization with 32P-labeled fragments were according to standard procedures (36).
Slot blot experiments were performed as described previously (36). Briefly, total bacterial RNA was extracted as described above, heated at 80°C for 5 min, and then immobilized onto Hybond-N+ nylon filters by slow filtration in a slot blot apparatus (Schleicher & Schuell). The immobilized RNA was then hybridized to denatured 32P-labeled fragments. The hybridization reactions were carried out in 5× SSC (1× SSC: 0.015 M NaCl, 15 mM Na citrate [pH 7.0]), 0.5% sodium dodecyl sulfate, 0.5× Denhardt (36) at 68°C for 6 h. 32P labeling of NMB1965- and NMB1961-specific probes, used in Northern blot and slot blot experiments (see Fig. 2 and 6), was performed by random priming as described above (36). The NMB1964-, NMB2147-, and zwf-specific probes were obtained by 32P labeling of PCR fragments. The sizes of these fragments were as follows: 471 bp (primers NMB1964-1 and NMB1964-2), 345 bp (primers NMB2147-1 and NMB2147-2), and 603 bp (primers Zwf-1 and Zwf-2). The semiquantitative analysis of the different transcripts was performed by densitometry using a Scanmaster 3 (Howtek, Inc., Hudson, NH), a high-performance desktop flatbed color scanner equipped with the RFLPrint (Pdi, Huntington Station, NY) software package, or by directly counting the radioactivity bands by using a PhosphorImager SI (Molecular Dynamics, Inc., Sunnyvale, CA). Slot blot experiments were carried out with triplicate samples in each assay. To evaluate the statistical significance of the results, each experiment was repeated at least three times with different RNA preparations, and means and standard deviations were determined.
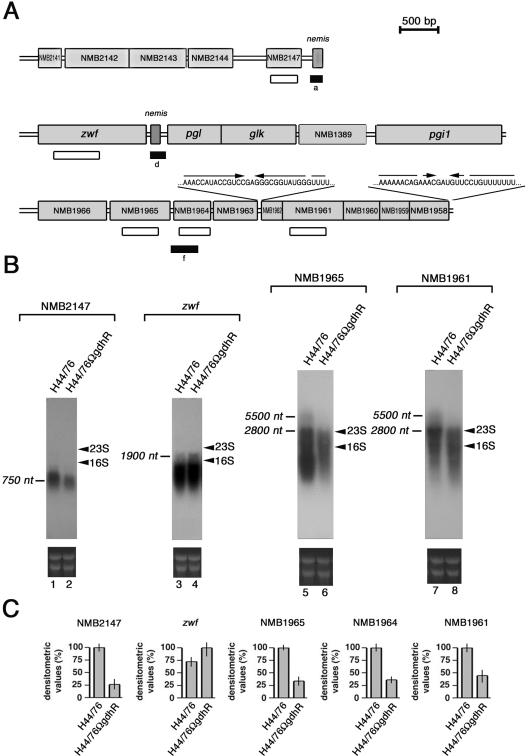
FIG. 2.
Genetic map of GdhR-regulated loci and Northern blot analysis. (A) The position of the cDNA (closed rectangles) corresponding to either up-regulated (d) or down-regulated (a and f) genes in the GdhR-defective strain is indicated with respect to the available genetic map of MC58 (44), an ET-5 strain phylogenetically related to H44/76 (35). Open rectangles locate the NMB2147-, the zwf-, the NMB1965-, the NMB1964-, and the NMB1961-specific probes used in Northern blot (B) and slot blot (C) experiments. The RNA sequences of the palindromic elements mapping in the intercistronic NMB1963-NMB1962 region and at the 3′ end of NMB1958 are reported. (B) Northern blot analysis of GdhR-regulated transcripts in isogenic strains H44/76 and H44/76ΩgdhR. Top, H44/76 (lanes 1, 3, 5, and 7) and H44/76ΩgdhR (lanes 2, 4, 6, and 8) were grown to late logarithmic phase (OD550 of 0.8) in complex GC medium. Total RNAs (10 μg) were extracted and analyzed by Northern blotting using the 32P-labeled NMB2147-, zwf-, NMB1965-, and NMB1961-specific probes shown in panel A. Bars indicate GdhR-regulated transcripts whose approximate sizes were deduced on the basis of the relative migration of 23S and 16S rRNA (arrowheads). Bottom, Ethidium bromide staining of ribosomal RNAs is reported as a loading control. (C) Relative levels of NMB2147-, zwf-, NMB1965-, NMB1964,- and NMB1961-specific mRNAs in strains H44/76 and H44/76ΩgdhR determined by slot blot experiments. For NMB2147, NMB1965, NMB1964, and NMB1961, transcript levels in H44/76 are arbitrarily assumed to be equal to 100%. For zwf the transcript level in H44/76ΩgdhR is assumed to be equal to 100%. Values represent means from three independent experiments, each with triplicate samples. Bars indicate standard deviations.
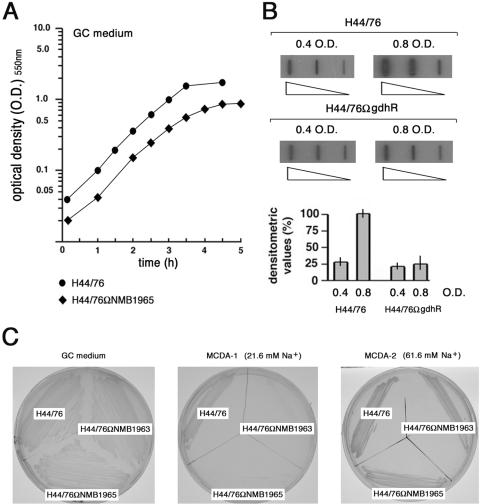
FIG. 6.
Effects of NMB1965 genetic inactivation on growth and growth phase-dependent expression of NMB1965. (A) Growth curves of H44/76 and H44/76ΩNMB1965 (permease defective) in GC-rich broth. The lag phase is omitted. (B) Slot blot analysis of NMB1965-specific transcripts. Top, total RNAs were extracted from H44/76 and H44/76ΩgdhR grown in GC-rich broth to middle (OD550 of 0.4) or to late logarithmic phase (OD550 of 0.8). The RNAs (0.01, 0.1, and 1 μg) were fixed onto Hybond-N+ nylon membranes and hybridized to the 32P-labeled NMB1965-specific probe used in Northern blot experiments (Fig. 2B). Bottom, densitometry analysis of the slot blot. The transcript level in H44/76 grown to an OD550 of 0.8 is arbitrarily assumed to be equal to 100%. Values represent means from three independent experiments, each with triplicate samples. Bars indicate standard deviations. (C) Strains H44/76, H44/76ΩNMB1963 (periplasmic substrate-binding protein defective), and H44/76ΩNMB1965 were streaked on GC medium, MCDA-1, or MCDA-2 agar plates.
RT-PCR-DD analysis.
Total bacterial RNA was extracted from meningococci grown in GC-rich medium to late logarithmic phase (optical density at 550 nm [OD550] of 0.8) as described above. Twelve-microliter mixtures consisting of 100 ng of total RNA from H44/76 or H44/76ΩgdhR, 0.9 mM deoxynucleoside triphosphates, 0.04 μg μl−1 primer DUS-IN, DUS-OUT, 26L-IN plus 27L-IN, or 26L-OUT plus 27L-OUT were heated at 65°C and immediately cooled on ice. Then, first-strand cDNA synthesis was carried out with 5 U μl−1 Superscript reverse transcriptase (RT) (Invitrogen) in the presence of 0.01 M dithiothreitol and 2 U μl−1 RNase inhibitor at 37°C for 50 min. Reactions were stopped by heat inactivation at 70°C for 15 min. Two microliters of the synthesized cDNA was used as templates in PCRs that were carried out in the presence of 0.2 mM deoxynucleoside triphosphates, 0.2 μM of the corresponding primers, and 0.5 μM random hexamers as reverse primers. The amplification reactions consisted of 35 cycles including 20 s of denaturation at 94°C, 20 s of annealing at 37°C, and 1 min of extension at 65°C. They were carried out in a Perkin-Elmer Cetus DNA Thermal Cycler 2400. Samples were subjected to phenol and chloroform-isoamylic alcohol (24:1) extraction and ethanol precipitation before loading onto 6% acrylamide-bisacrylamide gel in TBE buffer (1× TBE is 8.9 mM Tris-borate, 89 mM boric acid, 2 mM EDTA). Electrophoresis was performed at room temperature. The gel was stained by the silver staining method (36). The slices of gel containing the differentially expressed transcripts were cut with a sharp scalpel, and the DNA fragments were allowed to diffuse into 100 μl of sterile water at 37°C for 4 h under stirring. The eluted DNA was ethanol precipitated using glycogen as a carrier. Then, it was used as a template in a PCR with the corresponding primers. The positive amplicons were cloned in a pGEM easy vector (Promega) and sequenced as described above.
Whole-cell l-glutamate uptake assay.
l-Glutamate transport assay was performed essentially as described previously (47) with some modification. Meningococci were grown in 10 ml GC broth supplemented with 1% (vol/vol) Polyvitox (OXOID) at 37°C with shaking. Cells were harvested at an OD550 of 0.7 to 0.8 by centrifugation at 5,000 × g for 10 min at room temperature. The cell pellets were washed by three resuspensions with 5 ml of buffer A (50 mM potassium phosphate, pH 6.9, and 0.5 mM MgCl2). Following the final centrifugation, cell pellets were resuspended in 1 ml of buffer B (buffer A containing chloramphenicol [300 μg ml−1]). For whole-cell transport assay, mixtures (60 μl) were prepared at room temperature as follows: 20 μl of the cell resuspension was added to 20 μl of buffer A containing 3% glycerol and, where indicated, NaCl at twice final concentration. Glycerol was used as an energizer. When present, sodium orthovanadate (Sigma) was added at concentrations ranging between 0 and 1 μM. Samples were allowed to incubate for 15 min, and the assays were then initiated by the addition of 20 μl of buffer A containing a mixture of l-[3,4-3H]glutamic acid (specific activity, 52 Ci/mmol) (provided by Perkin-Elmer Life Science) and unlabeled l-glutamic acid at a 3 times final concentration and, where indicated, unlabeled amino acids l-glutamine, l-aspartic acid, l-asparagine, or l-alanine at a 3 times final concentration, and NaCl at final concentration. The assay mixtures were mixed with a Vortex mixer, and the assays were terminated after different times by vacuum filtration onto a 0.45-μm-pore-size White HAWP Millipore membrane. Filters were washed with 0.5 ml of buffer A and dried. The radioactivity was determined by scintillation counting in 20-mm (inside diameter) vials containing 5 ml of pseudocumene scintillation solution. Background binding of l-[3,4-3H]glutamic acid to membrane or bacteria (energy-independent binding) was determined in control samples lacking bacteria or glycerol (as an energizer), and it was subtracted from all values. The protein concentration was determined by the Bradford method, with bovine serum albumin as a standard. All transport data are expressed as nanomoles of substrate transported per mg of cell protein min−1.
Invasion and intracellular viability assays.
For standard invasion and intracellular viability assays, HeLa cells were grown for 1 day to near-confluence in 24-well tissue culture plates (Falcon) in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (GIBCO) and 2 mM l-glutamine. Meningococci were grown overnight on GC agar supplemented with 1% (vol/vol) Polyvitox (OXOID) at 37°C in 5% CO2. HeLa cells were infected at a multiplicity of infection of 100 and incubated for 1 h at 37°C in 5% CO2. The infection was initiated by a centrifugation step (600 × g for 5 min at room temperature). Noninternalized bacteria were killed by the addition of 100 μg ml−1 of gentamicin (Sigma) to the culture medium. After 30 min, HeLa cells were washed three times with phosphate-buffered saline (to remove gentamicin and dead bacteria) and reincubated in DMEM medium for various times. The medium was then collected, and HeLa cells were washed extensively and lysed by 0.1% saponin in phosphate-buffered saline for 5 min to release internalized bacteria. Lysates and medium were then plated onto GC agar plates in order to determine numbers of CFU. Alternatively, in order to check that the infection protocol was working successfully, after infection and reincubation times, cells were processed for immunofluorescence to distinguish between adherent and internalized bacteria (27).
Slot blot analysis of meningococcal transcripts in infected cells.
In analyzing the meningococcal transcripts in infected cells, an obvious obstacle was represented by the small number of recoverable bacteria, making inappropriate the application of any standard procedures for RNA analysis. To overcome this limitation, an original slot blot procedure was ad hoc developed. For this purpose, total RNA was extracted from about 5 × 107 to 5 × 108 intracellular meningococci following saponin lysis of infected HeLa cell 8 h after internalization (see above) by the RNeasy Mini kit according to the manufacturer's instructions (QIAGEN). The extracted RNA was subjected to reverse transcription using the oligonucleotide NMB1965-2, NMB1964-2, NMB1961-2, or 16Suniv-2 (loading control) as primers, according to the above-described procedure (see RT-PCR-DD analysis). The labeling procedure consisted of a PCR, where 2 μl of cDNA was used as a template in a 50-μl mixtures containing 0.2 mM (each) of dCTP and dTTP, 2.5 μM (each) of dATP and dGTP, 0.12 μM [α-32P]ATP (3,000 Ci mmol−1), and [α-32P]GTP (3,000 Ci mmol−1), 0.2 μM forward (NMB1965-1, NMB1964-1, NMB1961-1, or 16Suniv-1) and reverse (NMB1965-2, NMB1964-2, NMB1961-2, or 16Suniv-2) primers, and 0.05 U μl−1 of Taq polymerase (Perkin-Elmer SpA). The amplification reaction consisted of 20 to 25 cycles including 45 s of denaturation at 94°C, 45 s of annealing at 55°C, and 45 s of extension at 72°C. The number of cycles in the amplification reaction was critical to operate in the linear range of the PCR, and it was determined in preliminary experiments with different primer pairs. In these experiments the amplification reaction was terminated after 5, 10, 15, 20, 25, 30, and 35 cycles. The 32P-labeled cDNA probes were hybridized to different amounts (0.05 to 100 ng) of denatured NMB1965-, NMB1964-, NMB1961-, or 16S rRNA gene-specific DNA fragments generated by PCR with the corresponding primers NMB1965-1 and NMB1965-2, NMB1964-1 and NMB1964-2, NMB1961-1 and NMB1961-2, and 16Suniv-1 and 16Suniv-2, respectively. The denatured fragments were fixed on positively charged Hybond-N+ nylon membranes by using a hybrid-slot manifold (Bethesda Research Laboratories) according to standard procedures (36). The hybridization reactions were carried out in Church buffer (36) at 63°C overnight. Assays were carried out with triplicate samples. Each experiment was repeated at least five times using RNA preparations from distinct infection assays, and means and standard deviations were determined.
RT real-time PCR analysis.
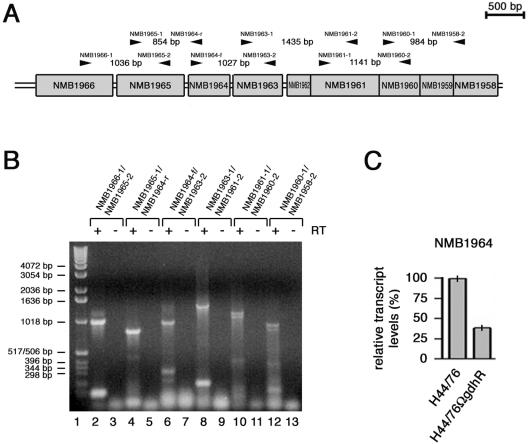
Semiquantitative analysis of the NMB1964 transcript, normalized to 16S rRNA, was performed by RT real-time PCR with the iQ SYBRgreen Supermix (Bio-Rad) on a Bio-Rad iCycler iQ instrument. For RT real-time PCR experiments, total RNA was isolated with the procedure described above either from exponentially growing meningococci in GC medium or from intracellular meningococci. Total RNA (1 μg) was then reverse transcribed by using random hexamer (2.5 μM) with Superscript RT (Invitrogen) as previously described. For each RNA sample, retro-transcription was performed five times. About 0.1 to 1% of each RT reaction was used to run real-time PCR with the primer pairs 16Suniv-1/16S-r, specific for 16S rRNA, and NMB1964-f/NMB1964-r, specific for NMB1964. PCR products were 185 bp long for 16Suniv-1/16S-r and 175 bp long for NMB1964-f/NMB1964-r primer pairs. Real-time PCRs were run in triplicate. The real-time PCR conditions were as follows: 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C for 45 cycles. Data were analyzed by the Bio-Rad iCycler iQ real-time PCR detection system software, version 3.1. Each experiment was repeated five times using distinct cDNA preparations for each RNA sample, and means and standard deviations were determined.
RESULTS
RNA differential display analysis of isogenic GdhR-proficient and GdhR-defective meningococcal strains.
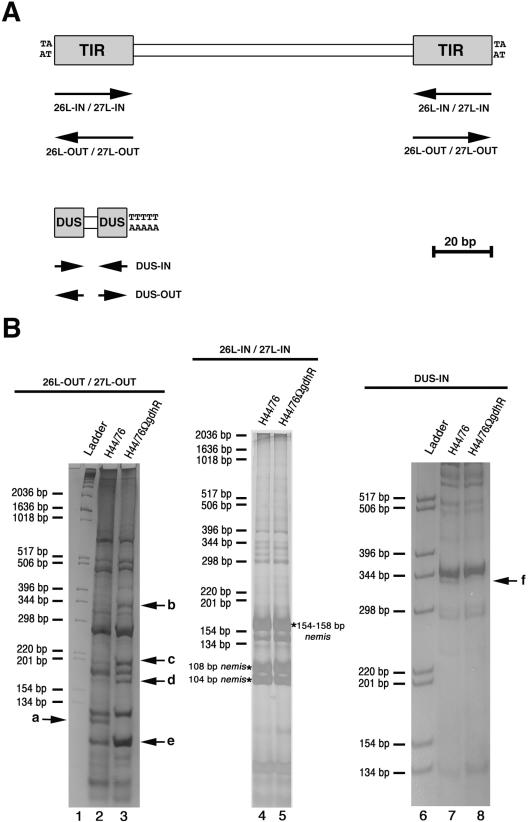
In an attempt to isolate GdhR-regulated genes, an RT-PCR-DD strategy was developed that relied on the presence of highly and moderately repetitive transcribed DNA sequences in the meningococcal genome. Oligonucleotides designed on the basis of the DUS (DUS-IN and DUS-OUT) or the 26L, 27L nemis sequences (26L-IN, 26L-OUT, 27L-IN, 27L-OUT) (Fig. 1A) were used as primers in RT assays to prepare cDNAs from H44/76 (GdhR proficient) and the isogenic derivative H44/76ΩgdhR (GdhR defective) grown in GC-rich medium to late logarithmic phase. The cDNAs were then amplified by PCR using the corresponding oligonucleotides and a mixture of random hexamers as primers, and the PCRs were analyzed by polyacrylamide gel electrophoresis. By this approach, several bands corresponding to either up-regulated (b, c, d, and e) or down-regulated (a and f) genes were identified in the GdhR-defective strain (Fig. 1B). The bands a, d, and f were then excised from the gels, and the corresponding cDNAs were cloned and subjected to nucleotide sequences analysis. The bands of b-, c- and e-associated cDNAs, when subjected to cloning, each gave rise to more than one clone, and therefore they were not further analyzed in this study.
FIG. 1.
RT-PCR-DD analysis of transcript in H44/76 and H44/76ΩgdhR. (A) Schematic representation of the neisserial nemis (also known as Correia element) and DNA uptake sequence (DUS) repeats with the relative positions (arrows) of the oligonucleotides 26L-IN, 26L-OUT, 27L-IN, 27L-OUT, DUS-IN, and DUS-OUT used as primers in RT-PCR-DD analysis in panel B. TIR, terminal inverted repeat. (B) The oligonucleotides 26L-OUT and 27L-OUT (left), 26L-IN and 27L-IN (middle), or DUS-IN (right) were used as primers in RT assays to prepare cDNAs from H44/76 (lanes 2, 4, and 7) and the isogenic derivative H44/76ΩgdhR (lanes 3, 5, and 8) grown in GC-rich medium to late logarithmic phase. The cDNAs were then amplified by PCR using the corresponding oligonucleotides and a mixture of random hexamers as primers, and the PCRs were analyzed by polyacrylamide gel electrophoresis. In lanes 1 and 6, molecular weight ladders, whose sizes are indicated on the left of each panel, were run in parallel. Arrows indicate bands corresponding to either up-regulated (b, c, d, and e) or down-regulated (a and f) genes in the GdhR-defective strain. The asterisks mark the position of the 104-bp-, the 108 bp-, the 154-bp-, or the 158-bp-long subfamily of the nemis element.
Identification of genes differentially expressed in GdhR-proficient and GdhR-defective strains.
The nucleotide sequence analysis demonstrated that the band d-associated cDNA (up-regulated in the GdhR-defective strain) corresponded to an operon region between the zwf and pgl cistrons encoding, respectively, the glucose 6-phosphate 1-dehydrogenase and the 6-phosphogluconolactonase, two key enzymes of the Entner-Doudoroff pathway (Fig. 2A and Table 2). The band a-associated cDNA (down-regulated) was localized immediately downstream from NMB2147, coding for a probable lipoprotein, whereas the band f-associated cDNA (down-regulated) corresponded to NMB1964, encoding a possible outer membrane transport protein on the basis of the available gene annotations (Fig. 2A and Table 2). The inspection of the genetic map suggested that the ORFs NMB1966 to NMB1958 might constitute an operon of unknown function (Fig. 2A).
TABLE 2.
Annotations of meningococcal loci identified by RT-PCR-DD
| ORF name (from MC58) | ORF name (from Z2491) | Gene name | Annotation |
|---|---|---|---|
| NMB2141 | NMA0227 | Probable periplasmic protein | |
| NMB2142 | NMA0228 | Hypothetical protein | |
| NMB2143 | NMA0229 | Hypothetical protein | |
| NMB2144 | NMA0230 | Probable ECF-family RNA polymerase sigma factor | |
| NMB2147 | NMA0233 | Probable lipoprotein | |
| NMB1392 | NMA1609 | zwf | Glucose 6-phosphate 1-dehydrogenase |
| NMB1391 | NMA1608 | pgl | 6-Phosphogluconolactonase |
| NMB1390 | NMA1607 | glk | Glucokinase |
| NMB1389 | NMA1605 | Probable transcriptional regulator; LacI family signature | |
| NMB1388 | NMA1604 | pgil | Glucose-6-phosphate isomerase |
| NMB1958 | NMA0493 | Possible periplasmic protein; thioredoxin, putative | |
| NMB1959 | NMA0492 | Conserved hypothetical protein; thioesterase superfamily | |
| NMB1960 | NMA0491 | Hypothetical protein | |
| NMB1961 | NMA0490 | Possible periplasmic/outer membrane protein; similar to VacJ lipoprotein required for intercellular spreading of S. flexneri | |
| NMB1962 | NMA0489 | Hypothetical protein containing NTP-Binding domain (STAS) | |
| NMB1963 | NMA0488 | Possible periplasmic transport protein | |
| NMB1964 | NMA0487 | Possible outer membrane transport protein | |
| NMB1965 | NMA0486 | Possible ABC transporter inner membrane subunit | |
| NMB1966 | NMA0485 | Probable ABC transporter ATP-binding subunit |
In order to confirm the results of the RT-PCR-DD analysis, Northern blot, slot blot, RT-PCR, and RT real-time PCR experiments were performed (Fig. 2 and 3). An about 750-nucleotide (nt)-long transcript was detected with an NMB2147-specific probe in the wild-type strain H44/76 (Fig. 2B, lane 1). As expected, this transcript was less abundant in the GdhR-defective H44/76ΩgdhR by densitometry analysis (Fig. 2B, lane 2). In contrast, the about 1,900-nt-long zwf-specific transcript was slightly more copious in H44/76ΩgdhR than in the wild type (Fig. 2B, lanes 4 and 3, respectively). The length of this transcript was compatible with a processing event possibly operated by the RNase III at the level of the nemis element in the zwf-pgl intercistronic region.
FIG. 3.
Linked RT-PCR and RT real-time PCR analyses of the NMB1966-to-NMB1958 region. (A) Genetic map of the NMB1966-to-NMB1958 region with the positions of the primer pairs (arrowheads) used in linked RT-PCR experiments. (B) Linked RT-PCR experiment. Total RNA from H44/76 grown to late logarithmic phase (OD550 of 0.8) in complex GC medium was analyzed by RT-PCR using the following primer pairs: NMB1966-1/NMB1965-2 (lanes 2 and 3), NMB1965-1/NMB1964-r (lanes 4 and 5), NMB1964-f/NMB1963-2 (lanes 6 and 7), NMB1963-1/NMB1961-2 (lanes 8 and 9), NMB1961-1/NMB1960-2 (lanes 10 and 11), and NMB1960-1/NMB1958-2 (lanes 12 and 13). In lanes 3, 5, 7, 9, 11, and 13, Superscript RT was omitted as a control. The relative migration of DNA molecular weight ladders (lane 1) is indicated (bars on the left). (C) Semiquantitative analysis of the NMB1964-specific transcript in H44/76 and H44/76ΩgdhR by RT real-time PCR experiment. The RNA was extracted from meningococci grown to late logarithmic phase (OD550 of 0.8) in GC medium. Results were normalized to 16S rRNA levels. Transcript levels in H44/76 are arbitrarily assumed to be equal to 100%. Values represent means from five independent experiments with triplicate samples using distinct cDNA preparations for each RNA sample ± standard deviations.
Semiquantitative analysis by slot blot experiments demonstrated that the NMB2147-specific RNA was about threefold less abundant in H44/76ΩgdhR than in the wild type. However, the same analysis, when extended to the zwf-specific RNA, failed to reveal statistically significant differences between the two strains because of its inability to distinguish between full-length and processed transcripts (Fig. 2C).
The result of the Northern blot analysis was consistent with the hypothesis that the genes NMB1966 to NMB1958 may constitute an operon subjected to positive control by GdhR. In fact, although barely detectable, an about 5,500-nt-long transcript, down-regulated in H44/76ΩgdhR, hybridized to either NMB1965- or NMB1961-specific probes (Fig. 2B, lanes 5 and 6 and lanes 7 and 8, respectively). The transcript length and the occurrence of (i) an ORF (NMB1967) in the opposite orientation upstream of NMB1966, (ii) a Rho-independent transcription terminator structure downstream from NMB1958, and (iii) translational coupling between the distal five cistrons (NMB1962 to NMB1958) supported this hypothesis.
It should be noted, however, that the Northern blot pattern was not particularly clean, although the experiments were repeated many times with different RNA preparations that were tested for mRNA integrity using a reference transcript (Materials and Methods). The Northern blot demonstrated two major transcripts about 2,800 nt in length with the two probes, much more abundant than the 5,500-nt long one. The levels of these two transcripts were greatly reduced in H44/76ΩgdhR compared to those in the wild-type strain. This transcript pattern was suggestive for the occurrence of mRNA processing regulating the segmental stability of the full-length 5,500-nt-long transcript. The presence of a stem-loop structure in the intercistronic region between NMB1963 and NMB1962 may be functional for the processing event (Fig. 2A), although we cannot exclude that it may also serve as a Rho-independent intercistronic transcription terminator. Therefore, to support the Northern blot data, we used linked RT-PCRs as an alternative strategy (Fig. 3A and B). The results of this analysis confirmed that the ORFs NMB1966 to NMB1958 are in the same transcriptional unit.
Semiquantitative analysis by slot blot experiments demonstrated that the NMB1965-, NMB1964-, and NMB1961-specific RNAs were significantly less abundant in H44/76ΩgdhR than in the wild-type strain (Fig. 2C). The result of RT real-time PCR experiments confirmed that NMB1964-specific transcript was almost threefold less abundant (38%) in H44/76ΩgdhR than in the wild-type strain (Fig. 3C).
In silico analysis of the NMB1966-to-NMB1958 gltT operon.
When the NMB1966 to NMB1962 deduced protein sequences were challenged in the data bank (Conserved Domain Database at the NCBI) in an attempt to gain information about their function, the ttg2 (ttg2A-ttg2B-ttg2C-ttg2D-ttg2E) operon of Pseudomonas putida GM73 gave the best score (Table 3). This operon was presumed to encode a second efflux pump responsible for toluene resistance in this microorganism, alternative to the SrpA-, SrpB-, SrpC-dependent pump (23). The possible existence of such a function in N. meningitidis, a narrow-host-range parasite, was rather puzzling. Moreover, the results of BLAST analysis demonstrated a high degree of conservation of the NMB1966 to NMB1962 deduced protein sequences in different Proteobacteria (Table 3). The BLAST analysis also revealed significant homology (32% identity, 55% similarity) between the NMB1966 deduced protein sequence and GluA of Corynebacterium glutamicum, the cytoplasmic ATP-binding component of the ABC four-member l-glutamate uptake porter.
TABLE 3.
Percent identities of related NMB1966 to NMB1958 ORFs in gram-negative bacteriaa
| Organism | ORF (% identity)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ATP-binding protein | Inner membrane permease | Outer membrane substrate binding protein | Periplasm transport protein | NTP-binding protein (STAS) | Lipoprot. VacJ-like | Hypothet. protein | Thioesterase | Peroxiredoxine | |
| β-Proteobacteria | |||||||||
| N. meningitidis | NMB1966 | NMB1965 | NMB1964 | NMB1963 | NMB1962 | NMB1961 | NMB1960 | NMB1959 | NMB1958 |
| N. gonorrhoeae | NGO2116 (99) | NGO2117 (98) | NGO2118 (100) | NGO2119 (97) | NGO2120 (91) | NG2121 (97) | NG2122 (98) | NG2123 (99) | NG2124 (99) |
| N. lactamica | (99) | (100) | (97) | (96) | (89) | (96) | (98) | (100) | (99) |
| C. violaceum | CV0449 (84) | CV0448 (76) | CV0447 (81) | CV0446 (65) | CV0445 (55) | CV0444 (60) | // CV4255 (67) | // CV4256 (56) | // CV4257 (58) |
| Bordetella pertussis | BP3757 (69) | BP3758 (68) | BP3759 (67) | BP3761 (56) | BP3760 (49) | ||||
| γ-Proteobacteria | |||||||||
| E. coli | yrbF (71) | yrbE (64) | yrbD (63) | yrbC (54) | yrbB (56) | // vacJ (42) | |||
| S. flexneri | ypsA (71) | vpsB (64) | ypsC (63) | yrbC (54) | yrbB (56) | // vacJ (43) | |||
| Yersinia pestis | YP03575 (72) | YP03574 (64) | YP03573 (65) | YP03572 (52) | YP03571 (57) | // YP02743 (44) | |||
| P. putida | ttg2A (71) | ttg2B (64) | ttg2C (63) | ttg2D (46) | ttg2E (61) | // PP2163 (46) | |||
| Haemophilus influenzae | HI1087 (78) | HI1086 (70) | HI1085 (63) | HI1084 (44) | HI1083 (57) | // HI0718 (45) | |||
| X. fastidiosa | XF0421 (61) | XF0420 (56) | XF0419 (59) | XF0418 (43) | XF0417 (50) | XF0416 (46) | |||
| α-Proteobacteria | |||||||||
| B. melitensis | BME10964 (60) | BME10965 (49) | BME10965 (49) | ||||||
| ɛ-Proteobacteria | |||||||||
| Helicobacter pylori | HP1465 (60) | HP1466 (46) | HP1464 (49) | ||||||
| Campylobacter jejunii | CJ1647 (59) | CJ1646 (47) | CJ1648 (45) | // CJ1372 (48) | // CJ1371 (43%) | ||||
The symbol // marks ORFs that are split with respect to the ABC transporter-encoding cluster. Lipoprot., lipoprotein; Hypothet., hypothetical.
In addition, Smith-Waterman alignment demonstrated homology (22% identity, 56% similarity) between the NMB1965 deduced protein sequence and the proton-glutamate symport protein from Pyrococcus horikoshii (Fig. 4E), whose structure has been determined by X-ray diffraction (48). These searches led us to hypothesize that the NMB1966-to-NMB1958 operon may code for an import system eventually involved in l-glutamate import rather than a toluene exporter, as previously annotated. The evidence that GdhR is responsible for coordinate regulation of both NADP-GDH and this operon strongly supported this hypothesis.
Functional characterization of the NMB1966-to-NMB1958 gltT operon: whole-cell l-glutamate uptake assays.
To substantiate this hypothesis with experimental data, isogenic mutant strains were constructed. In the strain H44/76ΩNMB1965, the gene encoding the putative permease of the four-member uptake system was insertionally inactivated by pDEΔNMB1965 (Fig. 4A). Southern blot analysis confirmed the insertion of the erythromycin-resistant circular cassette by a single crossing-over event in NMB1965. As a result of the recombination event, two HincII DNA fragments of the expected sizes (1,718 bp and 951 bp) were detected by NMB1965-specific probe in the transformed strains H44/76ΩNMB1965 (Fig. 4B, lanes 1 to 7) in place of the 1,975-bp HincII fragment of the parental strain H44/76 (Fig. 4B, lane 8). A similar strategy was used to inactivate NMB1963 encoding a putative periplasmic substrate-binding protein. Southern blot analysis demonstrated the insertional inactivation of the ORF by pDEΔNMB1963 (Fig. 4A). Indeed, two DdeI DNA fragments of the expected sizes (2,409 bp and 1,932 bp) were detected by a NMB1963-specific probe in the transformed strains H44/76ΩNMB1963 (Fig. 4C, lanes 1 to 5) in place of the 2,588-bp DdeI fragment of H44/76 (Fig. 4C, lane 6).
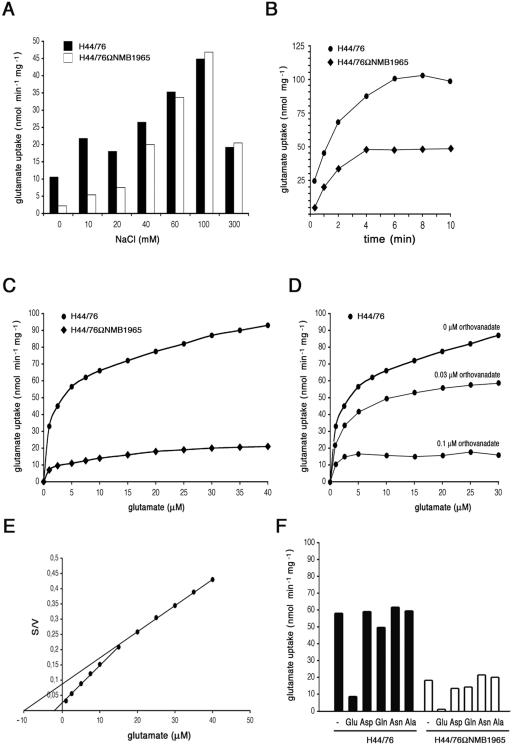
The uptake of l-glutamate was determined in wild-type H44/76 and in H44/76ΩNMB1965 under different assay conditions (Fig. 5). As in other microorganisms, the efficiency of l-glutamate import was largely affected by the sodium ion concentration (Fig. 5A). When compared to the wild-type H44/76, H44/76ΩNMB1965 exhibited severe impairment of l-glutamate uptake in the range between 0 and 40 mM NaCl. In contrast, at a sodium ion concentration above 40 mM, the uptake defect became milder, and it disappeared above 60 mM NaCl (Fig. 5A), suggesting that the activity of a sodium-driven, secondary transporter was predominant under these assay conditions. Time course experiments confirmed the reduced ability of H44/76ΩNMB1965 to import l-glutamate uptake in the presence of a low sodium ion concentration (20 mM) (Fig. 5B). In a buffer containing 20 mM NaCl, the velocity of l-glutamate uptake was reduced more than fourfold for H44/76ΩNMB1965 in a range of substrate concentration between 1 and 40 μM (Fig. 5C). The ATPase inhibitor sodium orthovanadate demonstrated dose-dependent inhibition of l-glutamate uptake in the wild-type strain (Fig. 5D) without affecting the residual uptake in the permease-defective strain (data not shown). In the wild type, this inhibition leveled off at values similar to those of the mutant strain (Fig. 5C). This result was consistent with impairment of an ATPase-driven l-glutamate transport in H44/76ΩNMB1965. Saturation data for the import of l-glutamate by the wild-type H44/76 indicated two major components with apparent Km values for l-glutamate of 2 μM and 10 μM (Fig. 5E). The component with the higher Km value was saturated at a higher l-glutamate concentration and was affected in H44/76ΩNMB1965. To test the specificity of the import system, l-glutamate uptake was determined in the presence of a 100-fold molar excess of l-glutamate, l-aspartate, l-glutamine, l-asparagine, or l-alanine (Fig. 5F). The results demonstrated that the import system was specific for l-glutamate in the wild-type strain. l-Glutamine scarcely inhibited (17%) l-glutamate uptake. The residual uptake of l-glutamate by H44/76ΩNMB1965 was inhibited by l-aspartate (28%) and by l-glutamine (22%). On the basis of these results, we assigned the name gltT (for l-glutamate transport) operon to the ORFs NMB1966 to NMB1958.
FIG. 5.
Effects of NMB1965 genetic inactivation on l-glutamate import. (A) Effects of sodium ion concentration on l-glutamate uptake by H44/76 and H44/76ΩNMB1965 (permease defective). The assay was performed as detailed in Materials and Methods using a final l-glutamate concentration of 0.5 μM and the indicated NaCl concentration. The transport assays were terminated after 20 s. (B) Time course of l-glutamate uptake. The assay was performed in the presence of 20 μM l-[3,4-3H]glutamic acid and a final NaCl concentration of 20 mM. (C) Concentration dependence of l-glutamate import by strains H44/76 and H44/76ΩNMB1965. The assay (20 s) was performed using a final NaCl concentration of 20 mM. (D) Effects of sodium orthovanadate addition on l-glutamate uptake. Transport assays were carried out as in panel C. (E) Reciprocal transformation of the saturation data of panel C relative to H44/76; S is expressed as micromolar concentrations, and V is expressed as nanomoles min−1 mg cell protein−1. (F) l-glutamate uptake (20 s) was determined using a final l-glutamate concentration of 5 μM and a final NaCl concentration of 20 mM, either in the absence or in the presence of 100-fold molar excess of l-glutamate, l-aspartate, l-glutamine, l-asparagine, or l-alanine.
Functional characterization of the NMB1966-to-NMB1958 gltT operon: effects on growth in complex or chemically defined media.
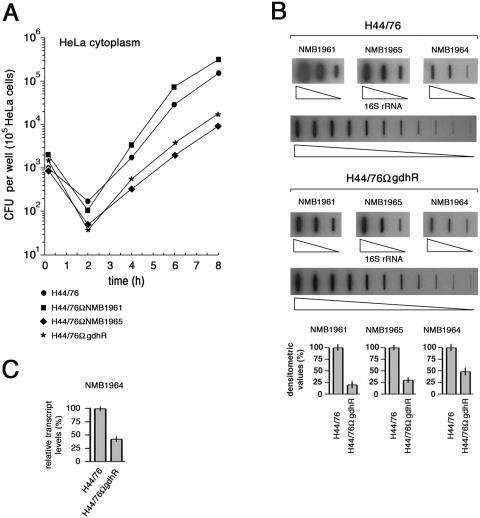
In liquid GC-rich medium, compared to the wild-type strain, H44/76ΩNMB1965 exhibited a prolonged lag phase, and the growth defect, only modest during the early logarithmic phase (OD550 of 0.02 to 0.2), increased during the middle and the late logarithmic phase (OD550, 0.2 to 0.8). The generation time was about 30 min for H44/76 and 60 min for the mutant strain in the growth interval between OD550s of 0.2 and 0.8. In addition, H44/76ΩNMB1965 entered the stationary phase at a biomass concentration significantly lower than that of H44/76 (Fig. 6A). Slot blot experiments demonstrated that the amount of NMB1965-specific mRNA increased about fourfold during the late logarithmic phase in the wild-type strain, whereas it did not change in H44/76ΩgdhR (Fig. 6B), confirming a major role for the gltT operon during the late logarithmic growth. It is relevant to note that transcription of gdhA also exhibited a similar timing (31).
The growth impairment of the NMB1965-defective mutant was much more marked in chemically defined media. At variance with the wild-type H44/76, this mutant failed to grow at a low sodium ion concentration (21.6 mM) on a chemically defined solid medium containing citrate, l-glutamate, and four other amino acids (l-arginine, l-glycine, l-serine, and l-cysteine) that are essential for meningococcal growth (5), but it grew on the same medium when the sodium ion concentration was raised to 61.6 mM (Fig. 6C). The same growth phenotype was observed for the NMB1963 (periplasmic substrate-binding protein)-defective mutant.
Functional characterization of the NMB1966-to-NMB1958 gltT operon: cell invasion and intracellular persistence assays.
A distinctive feature of the gltT operon of N. meningitidis is the presence of four additional ORFs (NMB1961 to NMB1958) that are located downstream from the ORFs (NMB1966 to NMB1962) coding for the ABC transporter. An identical gene arrangement is present in the closely related bacteria Neisseria gonorrhoeae (strain FA 1090) and Neisseria lactamica (ST-640). In the more distant neisseriacea Chromobacterium violaceum (strain ATCC 12472), the homologous ORFs coding for the ABC transporter (CV0449 to CV0445) precede a single ORF, CV0444 (NMB1961 in N. meningitidis), whereas the NMB1960-to-NMB1958 homologues map outside. A similar organization is present in Xylella fastidiosa but not in the other γ-Proteobacteria (Table 3). The gene product of NMB1961 is related to VacJ of Shigella flexneri, a lipoprotein of unknown function that is exposed on the bacterial surface and that has been involved in the intercellular spreading of this invasive microorganism (43). These searches led us to investigate the possible role of the meningococcal gltT operon in cell invasion and intracellular survival.
For this purpose, the ORF NMB1961, encoding the VacJ-like protein, was insertionally inactivated by pDEΔNMB1961 (Fig. 4A). The insertion of the erythromycin-resistant circular cassette by single crossover in NMB1961 was verified by Southern blot analysis (Fig. 4D). Two HincII DNA fragments of the expected sizes (1,100 bp and 1,043 bp) were detected by the NMB1961-specific probe in the transformed strains H44/76ΩNMB1961 (Fig. 4D, lanes 1 to 7) in place of the 1,460-bp HincII fragment of the parental strain, H44/76 (Fig. 4D, lane 8).
Strains H44/76, H44/76ΩgdhR, H44/76ΩNMB1965 (permease defective), and H44/76ΩNMB1961 (VacJ-like defective) were used in cell invasion and intracellular persistence assays. After allowing meningococci to invade HeLa cells for 1 h, intracellular viability was assessed immediately after gentamicin treatment and every 2 h post-gentamicin treatment (Fig. 7A). Results demonstrated the absence of statistically relevant differences in the ability to invade the HeLa cells by the four strains. At 2 h post-gentamicin treatment, a difference occurred in the intracellular viability of the wild type and the mutants. Normally, at this time, a decrease occurs in intracellular viable bacteria, which is also characteristic of gonococcal invasion (37, 46). In the experiment shown in Fig. 7A, the decrease was about sixfold for the wild-type H44/76, 20-fold for the permease-defective strain H44/76ΩNMB1965, 18-fold for the VacJ-like-defective strain H44/76ΩNMB1961, and 43-fold for H44/76ΩgdhR. Similar results were obtained in other tests. Between 2 and 8 h, fast bacterial duplication occurred with the following apparent generation times: 24 min, H44/76; 48 min, H44/76ΩNMB1965; 18 min, H44/76ΩNMB1961; 42 min, H44/76ΩgdhR. Thus, generation times appeared to be greatly increased for both the GdhR- and permease-defective strains and slightly decreased for the VacJ-like-defective strain. It should be noted that intracellular generation times are only apparent, since they reflect the sum of intracellular growth, intracellular death, and eventually bacterial egression from HeLa cells and reinvasion. Therefore, different processes may be affected in the three mutants.
FIG. 7.
Effects of genetic inactivation of NMB1965, NMB1961, or gdhR on HeLa cell invasion and intracellular survival. (A) After allowing meningococci to invade for 1 h, intracellular viability was assessed immediately after gentamicin treatment (0 h) and every 2 h post-gentamicin treatment. Intracellular viability is expressed as the average number of viable bacteria (CFU) per epithelial cell for H44/76 and derivative strains H44/76ΩgdhR, H44/76ΩNMB1965 (permease defective), and H44/76ΩNMB1961 (VacJ-like defective). (B) Slot blot analysis of NMB1965-, NMB1964-, and NMB1961-specific transcripts in the intracellular environment. Top, total RNAs were extracted from about 5 × 107 to 5 × 108 intracellular meningococci (strains H44/76 and H44/76ΩgdhR) following saponin lysis of infected HeLa cells at different times after 8 h of gentamicin treatment and were used to generate gene-specific 32P-labeled cDNA probes as detailed in the Materials and Methods section. The 32P-labeled cDNA probes were hybridized to different amounts (4, 20, and 100 ng) of denatured NMB1965-, NMB1964-, or NMB1961-specific fragments. For the 16S rRNA gene-specific fragment, twofold serial dilutions (from 0.05 to 50 ng) were used. Bottom, densitometry analysis of the slot blot. For NMB1965, NMB1964, and NMB1961, the relative transcript levels in H44/76 are arbitrarily assumed to be equal to 100%. Values represent means from five independent experiments, each with triplicate samples, using RNA preparations from distinct infection assays. Bars indicate standard deviations. (C) Semiquantitative analysis of the NMB1964-specific transcript in H44/76 and H44/76ΩgdhR by RT real-time PCR experiment. The RNA was extracted from intracellular meningococci as described above. Results were normalized to 16S rRNA levels. Transcript levels in H44/76 are arbitrarily assumed to be equal to 100%. Values represent means from five independent experiments with triplicate samples using distinct cDNA preparations for each RNA sample ± standard deviations.
Transcript levels of NMB1961 (VacJ-like), NMB1965 (l-glutamate putative permease), and NMB1964 (putative outer membrane substrate binding protein) in the intracellular milieu were evaluated by slot blot (Fig. 7B) and by semiquantitative RT real-time PCR (Fig. 7C) experiments. Reduction in the amounts of these transcripts in H44/76ΩgdhR compared to levels in the wild-type strain might account for the lower fitness of this strain in the intracellular environment.
DISCUSSION
In this study we demonstrate that GdhR, a positive transcriptional regulator of gdhA, encoding the NADP-GDH (32), controls the expression of a number of genes involved in glucose metabolism and l-glutamate import by an ABC transporter whose genetic counterpart was so far unexplored.
GdhR acts as a negative (direct or indirect) regulator for transcription of the zwf-pgl region, where two genes map encoding key enzymes of the Entner-Doudoroff pathway, the glucose 6-phosphate 1-dehydrogenase and the 6-phosphogluconolactonase, respectively (Fig. 1 and Fig. 2). The Entner-Doudoroff pathway is crucial for glucose metabolism in pathogenic Neisseriae. These narrow-host-range parasites are able to utilize only a few compounds as energy sources efficiently: lactate, pyruvate, glucose, and (the meningococcus) maltose.
Glucose is catabolized largely along the Entner-Doudoroff pathway, which generates relatively small amounts of energy, with a little contribution from the pentose phosphate pathway (19). At neutral pH values, the catabolism of glucose results in the accumulation of acetate, which is not further catabolized until glucose is depleted or growth becomes limiting. Indeed, growth on glucose markedly reduces the levels of TCA cycle enzymes in gonococci (17, 31).
Lactate metabolism is accomplished by at least three lactic dehydrogenases. The most important are two electron transport-linked lactic dehydrogenases that are associated with the cytoplasmic membrane (12, 38). Lactate provides energy for growth by being a substrate for electron transport when it is oxidized to pyruvate. Pyruvate then generates acetyl-coenzyme A, the precursor of fatty acid synthesis, and constituents and products of the TCA cycle, which is entirely functional under these growth conditions (20, 22, 38). Fueling of the TCA cycle by acetyl-coenzyme A accounts for lactate stimulation of neisserial metabolism in media containing glucose (38). A similar effect may be produced by the activity of the NADP-GDH, which supplies the TCA cycle with 2-oxoglutarate (32). Indeed, there is evidence that exogenous l-glutamate stimulates citrate catabolism in N. meningitidis (18). Interestingly, the extent of such stimulation is strain dependent (18) as is expression of NADP-GDH (32). On the basis of these arguments, the negative regulation of the glucose metabolism by GdhR may be unsurprising. In fact, 2-oxoglutarate is a negative effector for GdhR in N. meningitidis, and GdhR in turn regulates the Entner-Doudoroff pathway in response to this TCA intermediary. In this scheme, high levels of 2-oxoglutarate might be permissive for stimulation of glucose metabolism via GdhR inhibition by 2-oxoglutarate.
The main point of this study is the identification and the partial characterization of a GdhR-regulated operon encoding an ABC transporter evolutionarily conserved in eubacteria (Table 3) and also present in plastid genomes (see below). Bioinformatics provided the following evidence that this system might be responsible for l-glutamate uptake.
(i) The NMB1966 deduced protein sequence exhibits similarity to that corresponding to gluA of Corynebacterium glutamicum, the cytoplasmic ATP-binding component of the ABC four-member glutamate uptake porter. Similar homology was previously observed between GluA and the deduced protein sequence of XF0421 (NMB1966 homolog) in the plant pathogen Xylella fastidiosa (29). (ii) NMB1965 encodes an integral membrane protein belonging to the duf140 family and harboring six transmembrane domains, a typical architecture of the inner-membrane permease component of the ABC transport system. Significant homology exists between the NMB1965 deduced protein sequence and the proton-glutamate symport protein from eubacteria and archaea (Fig. 4E). The duf140 family is widespread over the evolution of eubacteria and plastid genomes and is absent in Firmicutes and in Archaea. (iii) NMB1964 encodes a protein of unknown function that is annotated as an outer membrane substrate-binding protein (Tables 2 and 3).
(iv) On the basis of structural features, NMB1963 may code for a periplasmic substrate-binding transport protein (Tables 2 and 3). (v) The small protein encoded by NMB1962 contains a STAS (for sulfate transporter and antisigma factor antagonist) domain. This domain is found in the C-terminal region of sulfate transporters and bacterial antisigma factor antagonists (1). Noteworthily, in the α-proteobacterium Brucella melitensis, the STAS domain is translationally fused to the putative permease encoded by the ORF BME10965 (Table 3). It has been suggested that the STAS domain may have a general NTP binding function. The presence of a predicted NTP-binding domain in the cytoplasmic portions of anion transporters suggested that anion transport could be regulated by intracellular concentrations of GTP and/or ATP (1). A similar function may be hypothesized for the product of the ORF NMB1962.
The function of the ORF NMB1961 is obscure. The gene product of NMB1961 is related to VacJ of S. flexneri, a lipoprotein of unknown function that is exposed on the bacterial surface. Interestingly, vacJ, as well as the S. flexneri unlinked genes vpsA and vpsC (NMB1966 and NMB1964 in N. meningitidis), is required for the intercellular spreading of this invasive microorganism, an essential prerequisite for shigellosis (21, 43). The VacJ-like-defective strain H44/76ΩNMB1961 did not exhibit decreased intracellular persistence and/or growth with respect to the wild-type strain (Fig. 7A). Intriguingly, the apparent intracellular generation time of this strain was slightly lower than that of the wild type. Because the intracellular generation time reflects the sum of intracellular growth, intracellular death, and eventually bacterial egression from HeLa cells and reinvasion, this difference might, in principle, be due to variation in any of these processes. A possibility to be explored is that the VacJ-like product of the ORF NMB1961 might be involved in bacterial egression from the host cells, a hypothesis that is compatible with the role of the related ORF in S. flexneri.
Functional data support the hypothesis that the ORFs NMB1966 to NMB1962 may code for an ABC transporter devoted to l-glutamate import and that this system maximally operates at a low sodium ion concentration (in a range around 10 mM NaCl) (Fig. 5). In contrast, at a high sodium ion concentration an alternative l-glutamate uptake system is predominant. Indeed, a sodium/glutamate symporter (GltS), encoded by the ORF NMB0085, is annotated in the genome of the serogroup B MC58 strain (44). The operation of two major, sodium-dependent and sodium-independent l-glutamate transporters is supported by the growth phenotype of the NMB1963- and NMB1965-defective mutants (Fig. 6).
The occurrence of two major systems operating at different sodium ion concentrations may be functional for meningococcal adaptation to different (intracellular versus extracellular) microenvironments. Although the question about the subcellular location of pathogenic Neisseriae is controversial (30), there is evidence that these parasites may have access to carbon sources in the host cell cytoplasm (46). In principle, there is little available intracellular glucose, since glucose entering cells is rapidly phosphorylated to glucose-6-phosphate (41), a substrate that, along with the other glycolytic intermediates, cannot be assimilated by meningococci. The best intracellular carbon sources are pyruvate and lactate or/and several amino acids, such as l-glutamate, which stimulates the citrate catabolism (18). Under physiological conditions, the extracellular concentration of this amino acid ranges from 30 to 90 μM in the plasma depending on feeding, exercise, and circadian rhythm (14, 45). The intracellular concentration may be considerably higher depending on the cell type and metabolism (6, 24, 45). Indeed, the l-glutamate metabolism in mammalian cells is strictly linked to that of l-glutamine, the most abundant amino acid in plasma (14). l-glutamine is an essential amino acid for growth of cells in culture (11) as well as an important oxidative fuel for cells grown in culture and in intact functioning organs (6, 24). In vivo l-glutamate may be generated from l-glutamine extracellularly by the action of the γ-glutamyltransferase and intracellularly by the action of the glutaminase (24). The intracellular milieu is characterized by low sodium ion concentrations, ranging between 15 and 20 mM; in contrast, the sodium ion concentration is much higher in the extracellular fluids and plasma (130 to 150 mM) (16). Thus, coregulation by GdhR of l-glutamate uptake via the ABC transporter operating at a low sodium ion concentration and l-glutamate oxidation via the NADP-GDH might be functional to support the activity of the TCA in the intracellular environment of the host cell. This hypothesis is supported by the results of the cell invasion and intracellular persistence assays. In these assays, HeLa cells were infected in the DMEM containing 2 mM l-glutamine as a source of the intracellular l-glutamate pool. Strains H44/76ΩNMB1965 (permease defective) and H44/76ΩgdhR (GdhR defective) exhibited normal cell invasion efficiency but a reduced ability to persist and/or grow in the intracellular environment (Fig. 7A). These experiments also demonstrated that the GdhR regulon is activated in the intracellular milieu (Fig. 7B), reflecting a limitation of the TCA cycle.
Acknowledgments
This work was partially supported by a grant from Progetto MIUR Cofin 2004 (area 06—scienze mediche, no. 24).
Editor: J. N. Weiser
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2000. The STAS domain—a link between anion transporters and antisigma-factor antagonists. Curr. Biol. 10:R53-R55. [DOI] [PubMed] [Google Scholar]
- 2.Black, C. G., J. A. M. Fyfe, and J. K. Davies. 1995. A promoter associated with the neisserial repeat can be used to transcribe the uvrB gene from Neisseria gonorrhoeae. J. Bacteriol. 177:1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucci, C., A. Lavitola, P. Salvatore, L. Del Giudice, D. R. Massardo, C. B. Bruni, and P. Alifano. 1999. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol. Cell 3:435-445. [DOI] [PubMed] [Google Scholar]
- 4.Cantalupo, G., C. Bucci, P. Salvatore, C. Pagliarulo, V. Roberti, A. Lavitola, C. B. Bruni, and P. Alifano. 2001. Evolution and function of the neisserial dam-replacing gene. FEBS Lett. 495:178-183. [DOI] [PubMed] [Google Scholar]
- 5.Catlin, B. W. 1973. Nutritional profiles of Neisseria gonorrhoeae, Neisseria meningitidis, and Neisseria lactamica in chemically defined media and the use of growth requirements for gonococcal typing. J. Infect. Dis. 128:178-194. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, H. N. 1990. Role of amino acid transport and countertransport in nutrition and metabolism. Physiol. Rev. 70:43-77. [DOI] [PubMed] [Google Scholar]
- 7.Deghmane, A. E., D. Giorgini, M. Larribe, J. M. Alonso, and M. K. Taha. 2002. Down-regulation of pili and capsule of Neisseria meningitidis upon contact with epithelial cells is mediated by CrgA regulatory protein. Mol. Microbiol. 43:1555-1564. [DOI] [PubMed] [Google Scholar]
- 8.Deghmane, A. E., D. Giorgini, L. Maigre, and M. K. Taha. 2004. Analysis in vitro and in vivo of the transcriptional regulator CrgA of Neisseria meningitidis upon contact with target cells. Mol. Microbiol. 53:917-927. [DOI] [PubMed] [Google Scholar]
- 9.De Gregorio, E., C. Abrescia, M. S. Carlomagno, and P. P. Di Nocera. 2002. The abundant class of nemis repeats provides RNA substrates for ribonuclease III in Neisseriae. Biochim. Biophys. Acta 1576:39-44. [DOI] [PubMed] [Google Scholar]
- 10.Delany, I., R. Rappuoli, and V. Scarlato. 2004. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52:1081-1090. [DOI] [PubMed] [Google Scholar]
- 11.Eagle, H., V. Oyama, M. Levy, C. L. Horton, and R. Fleischman. 1956. The growth response of mammalian cells in tissue culture to L-glutamine and L-glutamic acid. J. Biol. Chem. 218:607-617. [PubMed] [Google Scholar]
- 12.Erwin, A. L., and E. C. Gotschlich. 1993. Oxidation of d-lactate and l-lactate by Neisseria meningitidis: purification and cloning of meningococcal d-lactate dehydrogenase. J. Bacteriol. 175:6382-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Exley, R. M., J. Shaw, E. Mowe, Y. H. Sun, N. P. West, M. Williamson, M. Botto, H. Smith, and C. M. Tang. 2005. Available carbon source influences the resistance of Neisseria meningitidis against complement. J. Exp. Med. 201:1637-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernstrom, J. D., R. J. Wurtman, B. Hammarstrom-Wiklund, W. M. Rand, H. N. Munro, and C. S. Davidson. 1979. Diurnal variations in plasma concentrations of tryptophan, tryosine, and other neutral amino acids: effect of dietary protein intake. Am. J. Clin. Nutr. 32:1912-1922. [DOI] [PubMed] [Google Scholar]
- 15.Grifantini, R., S. Sebastian, E. Frigimelica, M. Draghi, E. Bartolini, A. Muzzi, R. Rappuoli, G. Grandi, and C. A. Genco. 2003. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 100:9542-9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guyton, A. C., and J. E. Hall. 1998. Pocket companion to textbook of medical physiology, 10th ed. W.B. Saunders Company, Philadelphia, Pa.
- 17.Hebeler, B. H., and S. A. Morse. 1976. Physiology and metabolism of pathogenic neisseria: tricarboxylic acid cycle activity in Neisseria gonorrhoeae. J. Bacteriol. 128:192-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill, J. C. 1971. Effect of glutamate on exogenous citrate catabolism of Neisseria meningitidis and of other species of Neisseria. J. Bacteriol. 106:819-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holten, E. 1974. 6-Phosphogluconate dehydrogenase and enzymes of the Entner-Doudoroff pathway in Neisseria. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. Immunol. 82:207-213. [DOI] [PubMed] [Google Scholar]
- 20.Holten, E. 1976. Radiorespirometric studies in genus Neisseria. 3. The catabolism of pyruvate and acetate. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. Immunol. 84:9-16. [PubMed] [Google Scholar]
- 21.Hong, M., Y. Gleason, E. E. Wyckoff, and S. M. Payne. 1998. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect. Immun. 66:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jyssum, K. 1960. Intermediate reactions of the tricarboxylic acid cycle in meningococci. Acta Pathol. Microbiol. Scand. Sect. B Microbiol. Immunol. 48:121-132. [DOI] [PubMed] [Google Scholar]
- 23.Kim, K., S. Lee, K. Lee, and D. Lim. 1998. Isolation and characterization of toluene-sensitive mutants from toluene-resistant bacterium Pseudomonas putida GM73. J. Bacteriol. 180:3629-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovacevic, Z., and J. D. McGivan. 1983. Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol. Rev. 63:547-605. [DOI] [PubMed] [Google Scholar]
- 25.Lentner, C. 1981. Geigy scientific tables, vol. 1. Units of measurements, body fluids, composition of the body, nutrition. Ciba Geigy Ltd., Basel, Switzerland.
- 26.Lentner, C. 1984. Geigy scientific tables, vol. 3. Physical chemistry, composition of the blood, hematology, somatometric data. Ciba Geigy Ltd., Basel, Switzerland.
- 27.Makino, S., J. P. van Putten, and T. F. Meyer. 1991. Phase variation of the opacity outer membrane protein controls invasion by Neisseria gonorrhoeae into human epithelial cells. EMBO J. 10:1307-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazzone, M., E. De Gregorio, A. Lavitola, C. Pagliarulo, P. Alifano, and P. P. Di Nocera. 2001. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene 278:211-222. [DOI] [PubMed] [Google Scholar]
- 29.Meidanis, J., M. D. Braga, and S. Verjovski-Almeida. 2002. Whole-genome analysis of transporters in the plant pathogen Xylella fastidiosa. Microbiol. Mol. Biol. Rev. 66:272-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merz, A. J., and M. So. 2000. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16:423-457. [DOI] [PubMed] [Google Scholar]
- 31.Morse, S. A., and B. H. Hebeler. 1978. Effect of pH on the growth and glucose metabolism of Neisseria gonorrhoeae. Infect. Immun. 21:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagliarulo, C., P. Salvatore, L. R. De Vitis, R. Colicchio, C. Monaco, M. Tredici, A. Talà, M. Bardaro, A. Lavitola, C. B. Bruni, and P. Alifano. 2004. Regulation and differential expression of gdhA encoding NADP-specific glutamate dehydrogenase in Neisseria meningitidis clinical isolates. Mol. Microbiol. 51:1757-1772. [DOI] [PubMed] [Google Scholar]
- 33.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 34.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 35.Salvatore, P., C. Pagliarulo, R. Colicchio, P. Zecca, G. Cantalupo, M. Tredici, A. Lavitola, C. Bucci, C. B. Bruni, and P. Alifano. 2001. Identification, characterization, and variable expression of a naturally occurring inhibitor protein of IS1106 transposase in clinical isolates of Neisseria meningitidis. Infect. Immun. 69:7425-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Shaw, J. H., and S. Falkow. 1988. Model for invasion of human tissue culture cells by Neisseria gonorrhoeae. Infect. Immun. 56:1625-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, H., E. A. Yates, J. A. Cole, and N. J. Parsons. 2001. Lactate stimulation of gonococcal metabolism in media containing glucose: mechanism, impact on pathogenicity, and wider implications for other pathogens. Infect. Immun. 69:6565-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, H. O., M. L. Gwinn, and S. L. Salzberg. 1999. DNA uptake signal sequences in naturally transformable bacteria. Res. Microbiol. 150:603-616. [DOI] [PubMed] [Google Scholar]
- 40.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 41.Stryer, L. 1988. Biochemistry, 3rd ed. WH Freeman, New York, N.Y.
- 42.Sun, Y. H., S. Bakshi, R. Chalmers, and C. M. Tang. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6:1269-1273. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, T., T. Murai, I. Fukuda, T. Tobe, M. Yoshikawa, and C. Sasakawa. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 11:31-41. [DOI] [PubMed] [Google Scholar]
- 44.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 45.Tsai, P. J., and P. C. Huang. 1999. Circadian variations in plasma and erythrocyte concentrations of glutamate, glutamine, and alanine in men on a diet without and with added monosodium glutamate. Metabolism 48:1455-1460. [DOI] [PubMed] [Google Scholar]
- 46.Williams, J. M., G.-C. Chen, L. Zhu, and R. F. Rest. 1998. Using the yeast two-hybrid system to identify human epithelial cell proteins that bind gonococcal Opa proteins: intracellular gonococci bind pyruvate kinase via their Opa proteins and require host pyruvate for growth. Mol. Microbiol. 27:171-186. [DOI] [PubMed] [Google Scholar]
- 47.Willis, R. C., and C. E. Furlong. 1975. Interactions of a glutamate-aspartate binding protein with the glutamate transport system of Escherichia coli. J. Biol. Chem. 250:2581-2586. [PubMed] [Google Scholar]
- 48.Yernool, D., O. Boudker, Y. Jin, and E. Gouaux. 2004. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 431:811-818. [DOI] [PubMed] [Google Scholar]