Abstract
During in vitro broth culture, bacterial gene expression is typically dominated by highly expressed factors involved in protein biosynthesis, maturation, and folding, but it is unclear if this also applies to conditions in natural environments. Here, we used a promoter trap strategy with an unstable green fluorescent protein reporter that can be detected in infected mouse tissues to identify 21 Salmonella enterica promoters with high levels of activity in a mouse enteritis model. We then measured the activities of these and 31 previously identified Salmonella promoters in both the enteritis and a murine typhoid fever model. Surprisingly, the data reveal that instead of protein biosynthesis genes, disease-specific genes such as Salmonella pathogenicity island 1 (SPI-1)-associated genes and genes involved in anaerobic respiration (enteritis) or SPI-2-associated genes and genes of the PhoP regulon (typhoid fever), respectively, dominate Salmonella in vivo gene expression. The overall functional profile of highly expressed genes suggests a marked shift in major transcriptional activities to nutrient utilization during enteritis or to fighting against the host during typhoid fever. The large proportion of known and novel essential virulence factors among the identified genes suggests that high expression levels during infection may correlate with functional relevance.
Gene expression in microorganisms during in vitro culture is dominated by a small number of highly expressed genes (31, 45, 59) that are predominantly involved in protein biosynthesis, maturation, and folding (transcription/translation factors, ribosomal proteins, proteases, and chaperones). The essential functions of many of the gene products, the strongly biased codon usage that corresponds to abundant tRNAs, and the biased amino acid composition that might minimize the substantial biosynthesis energy costs (1) indicate the high biological significance of these genes.
Some of these genes could also dominate gene expression in pathogenic bacteria during infection (45). Numerous studies have investigated pathogenic in vivo gene expression by diverse techniques such as in vivo expression technology (IVET), differential fluorescence induction, in vivo-induced antigen technology, and microarray transcriptome analysis (9, 38, 54, 56, 58, 60, 62, 66, 72, 74), but these studies have mostly focused on genes that are specifically induced during infection, whereas genes with high levels of in vitro activity have been mostly disregarded. The in vivo significance of such “constitutive” genes thus remains largely unclear.
We have recently developed a modified differential fluorescence induction (72) technique that allowed identification of a large number of Salmonella enterica promoters with very high activity in infected spleen in a murine typhoid fever model, regardless of their in vitro activity (14, 61, 72). In this strategy, a Salmonella green fluorescent protein (GFP) promoter trap library is sorted for high fluorescence in infected mice by two-color flow cytometry, which suppresses interfering host autofluorescence (14). Utilization of unstable GFP variants such as GFP_OVA decreases sensitivity and thus allows selective identification of very strong promoters (61, 76).
In the present study, 21 Salmonella promoters with high levels of activity in a different disease model, the recently described murine enteritis model (3), were identified. The combination of these promoters and previously identified promoters with high activity in the typhoid fever model enabled us to analyze highly expressed Salmonella genes under two distinct diseases conditions in the same host species. The results indicate an unexpected dominance of disease-specific genes and a marked shift in the overall functional profiles during enteritis and systemic disease.
MATERIALS AND METHODS
Bacterial strains, Salmonella promoter trap library, and construction of mutants.
Streptomycin-resistant virulent Salmonella enterica serovar Typhimurium SL1344 (41) was used for all infection experiments. Salmonella organisms were cultured in LB medium containing 0.4% NaCl to minimize detrimental effects of high GFP levels (D. Bumann, unpublished observations). Appropriate antibiotics (streptomycin, 90 μg ml−1; ampicillin, 100 μg ml−1; and kanamycin, 50 μg ml−1) were added.
We used an episomal GFP_OVA promoter trap library in SL1344 with about 20-fold genome coverage that was previously described (61). In brief, sheared Salmonella enterica serovar Typhimurium SL1344 genomic DNA fragments (each, 500 to 700 bp) were inserted upstream of a promoterless gfp_ova gene on a medium-copy-number plasmid. The resulting plasmid library containing about 1.1 × 106 independent transcriptional fusions was transformed into strain SL1344, yielding 106 independent transformants.
Fluorescence-activated cell sorter (FACS)-sorted fluorescent clones were characterized by PCR amplification of the inserted DNA fragment with flanking primers up2 (5′-GTGATGTCGGCGATATAG) and do2 (5′-GAATTGGGACAACTCCAG), followed by restriction fragment fingerprinting using frequently cutting endonucleases Tsp509I, AluI, and HpaII. Nonredundant clones were sequenced using primer do (5′-TACTCATATGTATATCTCCTTCTTA). Associated Salmonella genes were identified by comparison with the genome sequence of Salmonella enterica serovar Typhimurium LT2. Both the primary annotation (53) and the somewhat different annotation of The Institute for Genomic Research (available at http://www.tigr.org) were used. Three transcriptional fusions were located within regions of the Salmonella genome sequence that lack annotated genes in the right orientation (fragment I, nucleotides 1656707 to 1656128 [nucleotides in the genome sequence]; fragment II, nucleotides 2807962 to 2807445; fragment III, nucleotides 1334351 to 1333800), and these were excluded from further analysis (see Results).
Strains with a chromosomal gfp insertion were constructed as previously described (61). In brief, gfp.mut2 was fused to a kanamycin resistance cassette by PCR, and products were integrated into the Salmonella chromosome at the predicted start codon of the respective gene using the temperature-sensitive helper plasmid pKD46 carrying a Lambda Red recombinase expression cassette (22). This recombinase protects the ends of linear DNA fragments and mediates homologous recombination with extraordinary efficiency, thus enabling directed chromosomal insertion of PCR-generated fragments with very short (40-bp) terminal homologous regions. Correct transformants were identified using appropriate flanking and internal primers, and pKD46 was cured during growth at 37°C, as verified by loss of resistance to ampicillin. For constructing deletion strains, target genes were replaced by the kanamycin-resistance cassette by Lambda Red recombinase-mediated homologous recombination (22) with the PCR primers described at http://falkow.stanford.edu/whatwedo/wanner/stmA.primers.
Mouse Salmonella infection models.
A recently described enteritis model has been developed for C57BL/6 mice (3). To allow direct comparison to our previous data set (61), we used BALB/c mice instead. For the enteritis model, mice were pretreated with 20 mg streptomycin; 1 day later, mice were intragastrically infected with 109 CFU Salmonella cells as previously described (3). At 1 day or 2 days postinfection, mice were killed, and ceca were prepared and homogenized. Each cecum generally contained 107 to 108 CFU Salmonella cells and showed typical signs of inflammation that are dependent on functional hilA (data not shown), as observed previously for C57BL/6 mice (3).
For the typhoid fever model, mice were systemically infected with 100 to 200 CFU by tail vain injection. After four days mice were killed and the spleen was removed for analysis. At this stage, spleen contained 10,000 to 40,000 CFU.
To determine virulence defects, groups of five streptomycin-pretreated mice were infected intragastrically (enteritis) or groups of five untreated mice were infected systemically (typhoid fever) with a mixture of wild-type and mutant Salmonella strains. Two days (enteritis) or 5 days (typhoid fever) postinfection, mice were killed and the CFU numbers in the cecum or spleen were determined for both wild-type and mutant strains by plating on selective media. The competitive index (CI) was calculated as (output mutant/wild-type ratio)/(input mutant/wild-type ratio). The significance of growth defects of the mutants compared to wild-type Salmonella was evaluated using the t test on logarithmically transformed CI data (expectation value CI = 1; log CI = 0).
Sorting of promoter trap library and single-clone analysis.
To identify clones carrying transcriptional fusions with high levels of activity in a murine enteritis model, three streptomycin-treated mice were intragastrically infected with the Salmonella promoter trap library (109 CFU). One day later, ceca were removed, homogenized in phosphate-buffered saline (PBS), and treated with 0.1% (final concentration) Triton X-100. After 100-fold dilution into PBS, GFP_OVA-expressing Salmonella cells were sorted by two-color flow cytometry using a biosafety level 2-equipped high-speed sorter (FacsDIVA; BD Biosciences) as previously described (14, 61). In brief, background fluorescence from other gut particles interfering with detection of GFP-expressing Salmonella was suppressed based on ratiometric comparison of green and orange emission intensities. Compared to background fluorescence, GFP-expressing Salmonella strains have a substantially higher green/orange emission ratio (14). In addition, forward and sideward scatter properties were used to further distinguish Salmonella cells from larger particles. An appropriate scatter gate was set using cecal contents of mice infected with a brightly fluorescent Salmonella strain that could be easily identified by green and orange emissions alone. The flow cytometer was calibrated each time with previously established calibration standards (76).
From the pooled cecal contents of three Salmonella library-infected mice, 40,000 Salmonella cells containing >20,000 GFP_OVA molecules were sorted and cultivated on agar plates. The clones were pooled, and a total of 109 CFU was administered to three streptomycin-treated mice. One day later, cecal contents were prepared as described above and pooled. A total of 4,000 Salmonella cells containing >100,000 GFP molecules were sorted and cultivated on agar plates.
To compare Salmonella gene expression during enteritis and typhoid fever, we also included some of our previously identified typhoid fever-associated promoters (61). In that study, many overlapping promoter-carrying inserts had been obtained in different sort cycles. For each promoter, only the activity of the fusion with the smallest distance between insert end and start codon of the respective gene was measured. In the present study, we used only those fusions that were obtained from a single sort cycle for high activity in vivo, while the other original sort cycles selecting for certain in vitro activities in LB culture were disregarded. This was done to avoid any in vitro bias in comparing data from the two disease models. As a result, for some promoters we had to measure fusions other than those of the previous study. The resulting combined data set contained 50 different promoters (Tables 1 and 2).
TABLE 1.
Salmonella operons with high expression during enteritis
| First locusa | Gene | Operonb | Function (classificationc) | Expression leveld
|
Previous evidence for expression/attenuatione (reference) | |
|---|---|---|---|---|---|---|
| Enteritis | Typhoid fever | |||||
| STM3668 | yiaK | yiaK-O-lyxK-sgbHUE | Uptake and utilization of lyxose ascorbate (P) | 470 | 30 | NA |
| STM4501 | STM4501-4500 | Putative cytoplasmic protein, putative SAM-dependent methyltransferase (U) | 380 | 40 | NA | |
| STM4315 | rtsA | rtsAB | Regulator of SPI-1 expression (V) | 290 | b.t. | NA |
| STM0974 | focA | focA-pflB | Formate transporter, pyruvate formiate lyase (P) | 300, 210 | b.t. | exp (78) |
| STM2283 | glpT | glpTQ | Glycerol transporter (P) | 240 | 30 | exp (15) |
| STM2344 | STM2344-2340 | PTS system, utilization of unknown carbohydrates (P) | 210 | 40 | NA | |
| STM0744 | ybgC | ybgC-tolQRAB-pal-ybgF | Maintenance of cell envelope integrity (P) | 210 | 35 | att* (tolB) (8) |
| STM3881 | rbsD | rbsDACBK | Ribose ABC transporter (P) | 230, 170 | 40, 35, 30, 25 | exp (15) |
| STM0608 | ahpC | ahpCF | Alkylhydroperoxide reductase (P) | 180 | b.t. | NA |
| STM2876 | hilA | hilA-iagB | Regulator of SPI-1 expression, cell invasion protein (V) | 270, 120 | b.t. | att (3, 69) |
| STM1328 | Putative outer membrane protein (U) | 170 | b.t. | NA | ||
| STM2328 | nuoA | nuoA-N | NADH dehydrogenase I (P) | 140 | 50 | NA |
| STM2875 | hilD | Regulator of SPI-1 expression (V) | 120, 100, 90, 85, 85, 80, 80 | 65, 45 | NA | |
| STM2330 | lrhA | Regulation of flagella, motility, chemotaxis genes (P) | 160, 130, 80, 70 | 15, b.t. | NA | |
| STM2886 | sicA | sicA-sipBCDA-iacP | SPI-1 effector proteins and chaperons (V) | 120, 90 | b.t. | att (3, 69) |
| STM1155 | htrB | Lipid A biosynthesis (P) | 105 | b.t. | NA | |
| STM0260 | dniR | Regulator of nitrite reductase (P) | 90 | 40 | NA | |
| STM0191 | fhuA | fhuACDB | Ferrichrome uptake system (P) | 130, 45 | b.t. | exp (38) |
| STM2867 | hilC | Regulator of SPI-1 expression (V) | 40 | b.t. | NA | |
| STM4423 | Putative AraC-type DNA-binding domain-containing protein (U) | 35 | b.t. | NA | ||
| STM2079 | wzzB | LPS synthesis (P) | 35, 20 | 15 | NA | |
First locus downstream of the identified promoter. Loci with the prefix STM were annotated by McClelland et al. (53).
Predicted operons according to published data and operon prediction at TIGR.
Classification into functional categories: P, physiological; V, virulence; U, unknown function.
GFP_OVA levels in ceca of infected streptomycin-pretreated mice or in spleens of intravenously infected mice in 1,000 copies per Salmonella cell (each value represents one infected mouse); b.t., below detection threshold of 10,000 GFP copies per cell in spleens of infected mice.
exp, expression in gut; att, inactivation of the operon leads to loss of enteritis phenotype in mice or cattle; att* (tolB), inactivation results in tolB-dependent attenuation in typhoid fever model; NA, no information available.
TABLE 2.
Salmonella operons with high expression during typhoid fever
| First locusa | Gene | Operonb | Function (classificationc) | Expression leveld
|
Previous evidence for expression/attenuatione (reference) | |
|---|---|---|---|---|---|---|
| Enteritis | Typhoid fever | |||||
| STM2781 | virK | PhoP-regulated gene (V) | Part | 720, 510, 480, 430, 370 | att (24) | |
| STM1602 | sifB | SPI-2 effector protein (V) | 10 | 300, 280, 380, 360 | NA | |
| STM1406 | ssaG | ssaG-L | SPI-2 TTSS (V) | b.t.; b.t.; b.t. (+) | 280, 270, 270, 220, 180, 120 | exp, att (73) |
| STM2080 | ugd | UDP-glucose/GDP-mannose dehydrogenase (V) | b.t.; b.t.; b.t. | 160, 160, 150, 120, 65 | exp (73) | |
| STM1393 | ssaB | ssaBC | SPI-2 TTSS (V) | b.t.; b.t.; b.t. | 170, 150, 150, 90, 65 | att (71) |
| STM4157 | sseK1 | SPI-2 effector protein (V) | 15 | 150, 120, 75 | NA | |
| ST64B | sb26 (sseK3) | Putative SPI-2 effector protein (V*) | Part | 120, 110, 75 | NA | |
| STM1397 | sseA | SPI-2 effector protein (V) | b.t. | 130, 70 | exp, att (40, 79) | |
| STM1231 | phoP | phoPQ | Virulence gene regulator (V) | b.t., b.t. (+) | 180, 65, 50 | exp, att (38, 55, 79) |
| STM1330 | Putative DNA/RNA nonspecific endonuclease (P) | Part | 85, 70 | NA | ||
| STM1672 | Putative cytoplasmic protein (U) | 5 | 90, 80, 60 | NA | ||
| PSLT046 | mig-5 | Putative carbonic anhydrase (V) | 17 | 90, 60 | exp, att (73) | |
| STM1633 | Putative periplasmic binding protein (V*) | 10 | 100, 45 | NA | ||
| STM1088 | pipB | SPI-2 effector protein (V) | b.t., b.t. (+) | 110, 65, 55, 45 | att* (47) | |
| STM0617 | rna | RNase I (P) | 10 | 70, 45 | exp (73) | |
| STM3195 | ribB | Riboflavin synthesis (P) | 190 | 60, 50, 45, 40 | att (this study) | |
| STM1275 | yaoF | Putative hemolysin (V) | 20, 10 | 55, 35 | att (this study) | |
| STM0701 | speF | speF-potE | Polyamine metabolism (P) | 40 | 45, 40 | NA |
| STM2666 | pheL | pheLA | Aromatic amino acid bisoynthesis (P) | 230, 195, 75 | 60, 45, 30 | NA |
| STM1224 | sifA | SPI-2 effector protein (V) | b.t., b.t. (+) | 45, 40 | att (6) | |
| STM0972 | sopD2 | SPI-2 effector protein (V) | b.t., b.t. (+) | 45, 40, 35 | att (11) | |
| STM1583 | Putative cytoplasmic protein (U) | b.t., b.t. | 50, 40, 30 | NA | ||
| STM2875 | hilD | Regulator of SPI-1 expression (V) | 40, 25 | 50, 35, 30 | NA | |
| STM2328 | nuoA | nuoA-N | NADH dehydrogenase (P) | 25, 20 | 40, 35 | NA |
| STM1269 | aroQ | Putative chorismate mutase (P) | b.t., b.t.; b.t. (+) | 35, 30 | Not attenuated (this study) | |
| STM2585A | pagK2 | Homologue of pagK (V) | b.t. (+) | 35, 30 | NA | |
| STM1333 | thrS | Threonine tRNA synthetase (P) | 125 | 40, 25 | NA | |
| STM0809 | Putative inner membrane protein (U) | b.t., b.t. | 30, 30 | NA | ||
| STM2780 | pipB2 | SPI-2 TTSS effector protein (V) | Part | 30, 25 | att (47) | |
| STM1444 | slyA | Transcriptional regulator for hemolysin (MarR family) (V) | b.t., b.t. (+) | 30, 25 | att (50) | |
| STM0474 | ybaJ | ybaJ-hha | Putative cytoplasmic protein, negative regulator of SPI-1 expression (V) | 30 | 25, 20 | NA |
First locus downstream of the identified promoter. Loci with prefixes STM and PSLT were annotated by McClelland et al. (53).
Predicted operons according to published data and operon prediction at TIGR.
Classification into functional categories: P, physiological; V, virulence; V*, virulence associated according to homology to known virulence factors or genomic context (see text); U, unknown function.
GFP_OVA levels in ceca of infected streptomycin-pretreated mice and in spleens of infected mice in 1,000 copies per Salmonella cell (each value represents one infected mouse); part, only a fraction of the total population has a GFP content higher than the detection limit of 4,000 copies per cell in ceca of infected streptomycin-pretreated mice; b.t., below detection threshold; (+), a minority of Salmonella cells (<10%) express detectable amounts of GFP_OVA.
exp, expression in infected mice; att, inactivation of the operon attenuates Salmonella for systemic virulence in mice; att*, inactivation results in attenuation, but the responsible gene has not yet been identified; NA, no information available.
GFP_OVA expression of individual clones was analyzed in the enteritis and typhoid fever models in mouse infection experiments by two-color flow cytometry using a FacsSORT (BD Biosciences) flow cytometer. For several fusions (35 in the typhoid fever model and 13 in the enteritis model), two to eight independent replicate measurements were done, and a median coefficient of variance of 25% was obtained. The results of independent measurements (where applicable) are shown in Tables 1 and 2. The activity of 10 identified promoters was also determined using single-copy chromosomal gfp fusions (Table 3; two independent replicates each). The flow cytometer was calibrated each time with previously established calibration standards (76). In contrast to the sorting of the libraries (see above), samples for these experiments were fixed with 2% formalin. This treatment decreased fluorescence intensities by about 25%, in agreement with previous observations (35), and this was taken into account for calculating GFP contents. The fluorescence intensity of fixed samples was constant over several hours of incubation at 4°C, thus permitting comparative measurements of different samples.
TABLE 3.
Salmonella in vivo GFP expression from chromosomal promotor fusions in cecum of streptomycin-pretreated mice
| Locus | Gene | Expression level in enteritis modela |
|---|---|---|
| STM0608 | ahpC | 12, 35 |
| STM2283 | glpT | 10, 13 |
| STM3881 | rbsD | 9, 15 |
| STM0974 | focA | 8, 11 |
| STM4315 | 8, 10 | |
| STM1328 | 7, 10 | |
| STM2330 | lrhA | b.t., b.t. |
| STM2344 | b.t., b.t. | |
| STM2876 | hilA | b.t., b.t. |
| STM4423 | b.t., b.t. |
GFP.mut2 levels in 1,000 copies per Salmonella cell in ceca of infected streptomycin-pretreated mice. b.t., below detection threshold of 4,000 GFP copies per cell. Each value represents one infected mouse.
Determination of GFP and GFP_OVA fluorophore formation and degradation.
Logarithmic- or stationary-phase in vitro cultures or freshly prepared cecal or splenic homogenates containing GFP- or GFP_OVA-expressing Salmonella strains were diluted 10 fold in prewarmed (37°C) and aerated LB medium containing chloramphenicol (170 μg ml−1) to block de novo protein synthesis. The mixtures were vigorously agitated at 37°C; after different time intervals, aliquots were removed and fixed in 2% formaldehyde in PBS. GFP or GFP_OVA fluorescence was then quantified by two-color flow cytometry.
RESULTS
GFP detection in the gut despite limited oxygen availability.
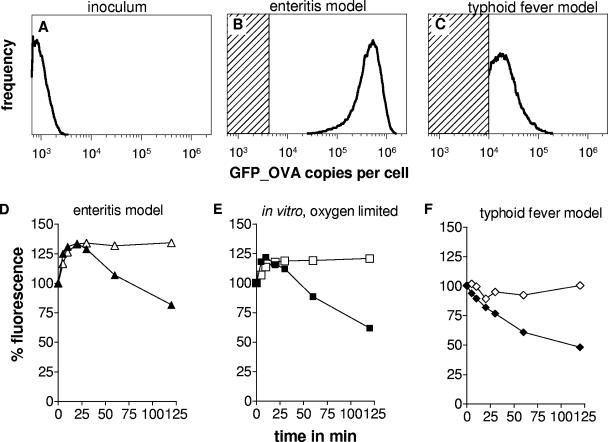
Our strategy to identify Salmonella promoters with high levels of activity during murine enteritis was based on GFP fluorescence detection. GFP fluorophore formation is oxygen dependent (37) and thus might be impaired in the anaerobic gut environment (36, 63). To evaluate this potential problem, streptomycin-treated mice (3) were infected with initially nonfluorescent stationary-phase Salmonella cells carrying a transcriptional gfp_ova fusion to the ribosomal promoter PrpsA (Fig. 1A). Two-color flow cytometry of cecal contents obtained 8, 24, or 48 h postinfection revealed Salmonella-like particles with high green fluorescence (Fig. 1B; data not shown) and emission ratios typical for GFP (14), indicating that GFP synthesis and at least partial fluorophore formation occurred in the gut. Background fluorescence emission of other gut particles including bacteria and tissue fragments could be spectrally separated from GFP fluorescence (14) by two-color flow cytometry (data not shown).
FIG. 1.
GFP_OVA content in Salmonella cells carrying a transcriptional PrpsA-gfp_ova fusion during enteritis and typhoid fever, as determined by flow cytometry. (A) Mice were infected with initially nonfluorescing Salmonella cells from a stationary in vitro culture. Salmonella were recovered 8 h after oral infection from infected ceca (Β) or 4 days after systemic infection from spleen (C). The shaded areas represent GFP_OVA levels below the detection threshold in the respective tissue homogenates. Similar data were obtained in three independent experiments. To assess GFP maturation and degradation, Salmonella cells expressing stable GFP (open symbols) or nonstable GFP_OVA (solid symbols) were freshly prepared from infected ceca (D), in vitro cultures with limited aeration (E), or infected spleen (F) and incubated in fully aerated LB medium containing chloramphenicol to block de novo synthesis. At different time intervals, GFP fluorescence was determined by flow cytometry.
Further incubation of freshly prepared cecal content in vigorously agitated LB broth containing 170-μg ml−1 chloramphenicol to block GFP de novo synthesis enhanced Salmonella fluorescence by 33% ± 4% (Fig. 1D, solid triangles), followed by a decline with a half-life of 88 ± 5 min that is typical for degradation of unstable GFP_OVA (61). Salmonella expressing stable GFP.mut2 (19) in the gut showed a similar initial fluorescence increase during in vitro incubation in LB broth containing 170 μg ml−1 chloramphenicol, but no degradation as expected (Fig. 1D, open triangles). Fluorescence enhancement was also observed when GFP_OVA- or GFP.mut2-expressing Salmonella cells grown under oxygen-limiting in vitro conditions were analyzed by this assay (Fig. 1E), suggesting that the early fluorescence increase might reflect oxygen-dependent fluorophore formation in a fraction of initially nonfluorescing GFP molecules. To account for this nonfluorescing GFP without laborious in vitro incubation steps, fluorescence intensities of immediately fixed cecal contents were multiplied by a normalization factor of 1.33. However, the major conclusions of this study would not be changed in the absence of applying this normalization factor (see below).
In contrast to this enteritis model, mice that had not received a streptomycin pretreatement had no fluorescent Salmonella in their ceca 1 day after intragastric infection with initially nonfluorescent Salmonella carrying the PrpsA-gfp_ova fusion, suggesting an absence of de novo GFP biosynthesis. Initially highly fluorescent Salmonella remained so in the cecum for 24 h, but aerobic incubation did not result in maturation of recently expressed GFP_OVA; the degradation assay showed very little turnover activity (data not shown), suggesting that the detected GFP_OVA may have been derived from the inoculum instead of being synthesized de novo in the gut. This apparent lack of ribosomal promoter activity is in agreement with the failure of Salmonella to colonize the gut of untreated mice or to cause intestinal pathology (3).
After systemic infection, initially nonfluorescent Salmonella carrying a transcriptional gfp_ova fusion to the ribosomal promoter PrpsA synthesized de novo GFP_OVA in the spleen (Fig. 1C), and fluorophore formation was already complete in vivo (Fig. 1F). GFP_OVA had a similar half-life (83 ± 5 min) in Salmonella in spleen (Fig. 1F) compared to Salmonella in gut lumen (Fig. 1D), suggesting that this degradable GFP variant allows detection of very strong Salmonella promoters with similar sensitivity (76) in both disease models.
Identification of Salmonella promoters with high levels of activity during enteritis.
To identify Salmonella promoters with high levels of activity in the gut, a diverse plasmid-based Salmonella GFP-promoter trap library was FACS sorted for high fluorescence in the cecal contents of streptomycin-pretreated, intragastrically infected mice. Ninety-six recovered clones were characterized by PCR restriction fragment length polymorphism, revealing 34 nonredundant clones. Sequencing of the corresponding inserts revealed 24 different promoters, 21 of which were associated with an appropriately oriented downstream open reading frame (Table 1). Three promoters were located within open reading frames or associated with open reading frames in the reverse direction (for sequence information, see Materials and Methods). Incomplete annotation by gene prediction programs, as well as transcription of noncoding genome sequences, is not uncommon (68). In particular, the identified promoters might drive expression of small RNAs or antisense RNAs, but we could not find clear evidence for that. Specific sequence properties of the insert-plasmid fusion might also have resulted in artificial promoter activities. We therefore excluded these promoters from further analysis.
High levels of in vivo activity in all identified promoters was confirmed by separate infection experiments (Table 1). To further validate the identified promoters, gfp was chromosomally integrated downstream of 10 selected promoters. For six of these constructs, GFP expression in the native genomic context and copy number was sufficiently high for detection in duplicate experiments (Table 3), confirming the successful identification of promoters with exceptionally high in vivo activity (35). The residual four promoters might be active but not strong enough to overcome the high detection threshold for single-copy reporter constructs. In addition, autoregulatory effects might occur for promoters PhilA, PlrhA, and Pstm4423 that drive expression of regulatory genes. For example, gfp inserted directly upstream of the hilA gene might interfere with HilA expression, which is required for full PhilA activity (23).
Previous data qualitatively confirm the in vivo activity of six of the identified Salmonella promoters, which further supports our data set. In particular, HilA (expressed from PhilA) and SipA and SipB (expressed from PsicA) are required for full Salmonella virulence in murine and/or bovine colitis models, suggesting that they are expressed in the gut (33, 69, 80). In addition, an IVET study has revealed intestinal fhuA expression (38). Indirect evidence for bacterial expression of rbsDACB, glpTQ, and pflB has been provided by transcriptome analysis of Escherichia coli growing on intestinal mucus (15) and Vibrio cholerae in a rabbit ileal loop model (78).
We restricted our analysis to the small minority of exceptionally highly active promoters by using the insensitive, degradable GFP_OVA reporter variant and very stringent sort criteria. The high redundancy of the sorted sublibrary (see above) might suggest substantial coverage of such promoters. On the other hand, the small set of 21 promoters obviously represents only a small part of all Salmonella activities during enteritis. Interestingly, only 2 out of the 21 promoters identified (PhilD and PnuoA-N) were previously found by our high-coverage analysis with the same Salmonella promoter trap library in a murine typhoid fever model (61).
Disease-dependent Salmonella promoter activities.
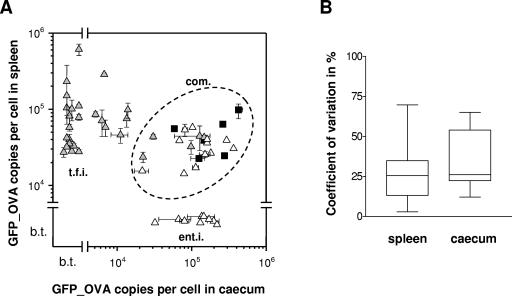
To compare Salmonella gene expression during different disease conditions in the same host species, we combined the enteritis-associated promoters with 31 previously identified typhoid fever-associated promoters that were sorted for high in vivo activity (61), resulting in a set of 50 different Salmonella promoters with high in vivo activity (for promoter selection, see Materials and Methods; two promoters were isolated from both models). We measured the activity of each fusion in the typhoid fever and enteritis models by two-color flow cytometry (Tables 1 and 2; Fig. 2A). We carried out two to eight replicate measurements for 35 fusions in the typhoid fever model and for 13 fusions in the enteritis model. All results from individual infection experiments are shown in Tables 1 and 2, and the observed ranges are shown in Fig. 2A. We were unable to perform full triplicate measurements for all 100 settings (50 fusions in two disease models) because of restrictions on animal experimentation. However, the available replicate data revealed a median coefficient of variation of 25% for promoter activity measurements in different mice (or 7% for the log-transformed values) (Fig. 2B), and this moderate level of variance may allow semiquantitative comparisons. This comparison is further limited by analyzing only a single time point per disease model. Pathogen gene expression during infection can considerably vary over time (74), but a detailed kinetic study was beyond the scope of the present study.
FIG. 2.
(A) Salmonella promoter activities in murine colitis (cecum) and typhoid fever (spleen) models. Promoter fusions were originally obtained from flow cytometric sorts of infected cecal lumen (open triangles) or spleen (gray triangles). For comparison, activities of five ribosomal promoters are also shown (black squares). b.t., below detection threshold; com, commonly active promoters; ent.i., enteritis induced; t.f.i., typhoid fever induced. Error bars indicate the range of repeated measurements. (B) Coefficient of variation of results obtained from repeated measurements of individual promoter-gfp_ova fusions in typhoid fever (spleen) and enteritis (cecum) models. The plots shows ranges (whiskers) and quartiles (the boxes extend from the 25th percentile to the 75th percentile, and the lines within boxes depict the medians).
Keeping these limitations in mind, an unexpected large proportion of the 50 characterized promoters showed activities that differed substantially (up to several orders of magnitude) between the two disease models (“disease-specific promoters”) (Fig. 2A). Some promoter fusions showed high levels of activity in both models, but most of them were still much more active during enteritis (median GFP_OVA ratio of enteritis/typhoid fever, around 5). The same trend of higher activity during enteritis was also observed with gfp_ova fusions to ribosomal promoters that we had previously identified in an unrelated project (Fig. 2A, black squares). We classified this group of promoters as “commonly active” (Fig. 2A).
For some promoter fusions with very high activity in the typhoid fever model, we also observed a small fraction of fluorescent Salmonella cells in the enteritis model (Table 2). From a quantitative comparison of volumetric FACS particle counts and plating results (14), we estimated that these subpopulations always represented <10% of the total Salmonella population in the gut lumen, while the large majority of Salmonella cells had an undetectably low GFP_OVA content. Some of these promoters are known to be active only within host cells (see below), suggesting that these small fluorescent populations could represent a few Salmonella cells that had been phagocytosed by host cells infiltrating the gut lumen or lamina propria cells that were later released from damaged villi (17, 34). To simplify the interpretation, we excluded these minor populations from further analysis.
Properties of disease-specific and commonly expressed genes.
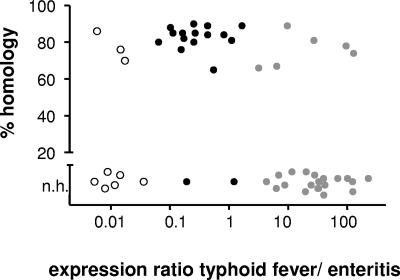
Twenty-seven of 50 Salmonella promoters (56%) with very high levels of activity during infection drive expression of genes with no homologue in the closely related species E. coli, which was more than expected (32% of all annotated Salmonella genes lack E. coli homologues). The absence of homologues in E. coli was particularly evident for disease-specific promoters (Fig. 3). Salmonella genes with no E. coli homologue also shared an atypically low GC content (data not shown), suggesting that they might have been acquired by horizontal gene transfer from an unknown microorganism. As a consequence of low GC content, these genes had a markedly different codon usage compared to “classical” highly expressed genes such as ribosomal proteins and chaperones, resulting in poor codon adaptation indices (reference 25 and data not shown). Moreover, the calculated metabolic costs for amino acid biosynthesis (1) of these genes were significantly higher than the genome average (data not shown), indicating that these genes were not yet optimized for low energy costs. Salmonella thus appeared to adapt to different disease conditions, mainly by using genes that lack properties of “typical” highly expressed genes.
FIG. 3.
Homology of highly in vivo-expressed Salmonella genes to E. coli K-12. A majority of Salmonella genes with disease-specific expression (enteritis, open circles; typhoid fever, gray circles; for classifications, see the legend to Fig. 2A) lack E. coli homologues, whereas most commonly expressed genes (black circles) are conserved in E. coli. Promoters that were only partially detected in one of the models were assumed to have median expression values close to the respective detection thresholds (6,000 molecules for typhoid fever and 4,000 molecules for enteritis) for calculating expression ratios. Promoters with undetectable expression in one of the disease models were assumed to have median expression values of about a quarter of the respective detection thresholds, based on the typical bandwidths of GFP measurements. n.h., homology below 5%.
In contrast to the disease-specific genes, most of the commonly expressed genes had an E. coli homologue (Fig. 3), but both codon usage and metabolic costs were not significantly different from the genome average (data not shown). Some “typical” highly expressed genes such as ribosomal genes (high conservation, highly biased codon usage, and low estimated biosynthetic costs) were also strongly expressed during infection (Fig. 2), but these genes did not appear to dominate Salmonella in vivo gene expression.
Functional profile of Salmonella gene expression in enteritis and typhoid fever.
The individual comparison of 50 different promoter activities revealed marked disease-specific Salmonella gene expression. To compare the overall functional distribution of in vivo highly expressed genes, promoters were classified as “virulence associated,” “physiological,” or “function unknown” (for detailed descriptions of individual genes, see Discussion), and the respective GFP-based promoter activities were calculated for both disease models (Fig. 4). These data represented only a first coarse estimate of the functional profile of highly in vivo-expressed Salmonella genes because of the limited number of promoter fusions and replicate measurements, as well as potentially distorting effects of copy number, topology, and position of the fusion junctions. With these limitations in mind, the overall analysis revealed a striking functional difference between the two different disease models. In the enteritis model, Salmonella expressed predominantly genes involved in general physiological functions such as nutrient utilization and energy conversion, while virulence genes that were mostly associated with SPI-1 accounted for only a minor fraction of gene expression (Fig. 4A). In marked contrast, Salmonella in the typhoid fever model predominantly expressed virulence genes that were mostly associated with Salmonella pathogenicity island 2 (SPI-2) or the PhoPQ regulon, whereas genes involved in general physiology were comparatively weakly expressed (Fig. 4B). These quantitative differences in gene expression between the two disease models were compatible with the predominant identification of physiology-associated promoters in the enteritis sort (Table 1) and virulence-associated promoters in the typhoid fever sort (Table 2) and also reflected the weaker expression of physiology-associated and ribosomal promoters in the typhoid fever model (Fig. 2).
FIG. 4.
Functional profile of highly expressed Salmonella genes during enteritis (A) or typhoid fever (B). Expression levels determined as the number of GFP_OVA copies per cell of promoters associated with physiology, virulence, or unknown function were separately summed. The cumulative activity of promoters associated with nutrient utilization and energy conversion (UTIL), SPI-1, SPI-2, or the PhoPQ regulon (PhoP) is also shown.
Disease-specific expression correlates with functional requirements.
Complementary sets of Salmonella virulence genes are known to be involved in enteritis induction (SPI-1-associated genes) and systemic typhoid fever (SPI-2-associated genes; PhoPQ regulon). Our expression analysis revealed that for most of these genes, the specific expression pattern reflected the differential relevance (i.e., SPI-2 genes were highly expressed during typhoid fever but not during enteritis; for detailed description of individual genes, see Discussion), in good agreement with our initial analysis (14).
To further explore the relationship between disease-specific expression and function, we selected four previously uncharacterized Salmonella genes that were not associated with SPI-1, SPI-2, or PhoPQ. In particular, we generated a mutant lacking the global anaerobiosis regulator fnr to test the combined relevance of several identified Fnr-dependent genes associated with anaerobic respiration and mixed acid fermentation that were selectively expressed during enteritis. We also tested mutants lacking the putative hemolysin yaoF or the periplasmic chorismate mutase aroQ, which were both selectively expressed during typhoid fever, and a mutant that lacked the riboflavin biosynthesis gene ribB with high expression in both disease models. All four Salmonella mutants were compared to the isogenic wild-type strain SL1344 by competitive infection experiments in both disease models.
Three of the four mutants had strong in vivo phenotypes (Table 4). The global anaerobiosis regulator fnr was essential during enteritis and had a weak role in typhoid fever. Limited oxygen availability in the gut (36), which was also reflected by delayed GFP fluorophore formation (Fig. 1D), appeared to result in a requirement for anaerobic metabolism. The yaoF mutant was attenuated in the typhoid fever model, where it was specifically expressed, but not during enteritis. The riboflavin biosynthesis gene ribB was essential in both disease models, in agreement with its expression pattern. In contrast to these three mutants, the aroQ mutant had no detectable phenotype even during typhoid fever, where aroQ was selectively expressed. A chorismate mutase isozyme, which is a part of pheA, was highly expressed during typhoid fever (Table 2) and might compensate for deficient aroQ. Taken together, these data confirmed and extended evidence for extensive Salmonella adaptation to various disease conditions by differential expression of specifically required genes.
TABLE 4.
Phentoype of Salmonella mutants in different disease models
| Mutation | Enteritis model
|
Typhoid fever model
|
||||
|---|---|---|---|---|---|---|
| CIa | log CIb | P valuec | CIa | log CIb | P valuec | |
| fnr | 0.025 | −1.8 ± 0.4 | 0.0007 | 0.251 | −0.7 ± 0.4 | 0.011 |
| yaoF | 0.47 | −0.36 ± 0.2 | NS | 0.035 | −1.69 ± 0.5 | 0.0003 |
| aroQ | 0.85 | −0.09 ± 0.2 | NS | 1.278 | −0.08 ± 0.2 | NS |
| ribB | 0.0030 | −2.7 ± 0.6 | 0.0031 | <0.0001 | Less than −4 | <0.0001 |
CI = competitive index (geometric mean of data from five mice). For calculations see Materials and Methods.
Arithmetic mean of log-transformed CI values ± standard deviation.
P values from two-tailed t test of log-transformed CI values. NS, not significant.
DISCUSSION
Highly in vivo-expressed pathogen genes could provide important targets for novel strategies for diagnosis, prevention, and treatment of infectious diseases. Moreover, major in vivo activities, as defined by gene expression patterns, could provide detailed insight into pathogen biology under relevant conditions. In vitro data for diverse bacterial species suggest that highly expressed genes encode predominantly highly conserved factors involved in protein biosynthesis and folding including transcription factors, ribosomal proteins, and chaperones. Many of these genes share a strongly biased codon usage, and this has been exploited to predict highly expressed genes in both environmental and pathogenic bacteria (44, 46). Extensive in vivo gene expression data have been obtained for various pathogens; these data could be used for testing the relevance of protein biosynthesis genes during infection, but most studies focused on genes specifically induced during infection, while constitutively expressed genes were mostly disregarded. To identify highly in vivo-expressed genes independently of whether they are expressed or not in vitro, we analyzed Salmonella promoter activities in murine models of enteritis and typhoid fever by a quantitative GFP reporter assay. As a critical precondition for this approach, we demonstrated that GFP fluorescence can be used to monitor Salmonella gene expression in the gut despite low oxygen availability.
In total, we identified 21 strong Salmonella promoters in the enteritis model and combined them with 31 previously identified strong promoters from the typhoid fever model (61). Interestingly, none of the associated Salmonella genes with high expression during infection belonged to the highly conserved core of transcription factors, ribosomal proteins, and chaperones that typically dominate gene expression in vitro. Moreover, most highly in vivo expressed genes did not share the “typical,” strongly biased codon usage of ribosomal genes, suggesting a limited utility of theoretical predictions based on codon adaptation indices. While protein biosynthesis is essential for bacterial growth under all conditions, these data suggested that it might not be the dominating activity in Salmonella during infection.
It might be expected that instead an activity related to host-pathogen interactions might dominate Salmonella gene expression in both intestinal and systemic infections. However, only 2 out of 50 identified promoters were commonly isolated from the two diseases models, despite substantial coverage of strong promoters. Moreover, semiquantitative activity measurements revealed that a surprisingly large proportion of the identified strong promoters were differentially regulated between enteritis and typhoid fever. Many of these differences might simply reflect different microenvironments in the spleen and the cecum. Indeed, for several identified promoters, previous in vitro evidence suggested differential regulation under conditions that are thought to mimic the gut lumen or intracellular compartments in infected host cells (51). Our data provided some in vivo confirmation of these data (see below). More importantly, the present study revealed that these and other disease-specific genes might actually dominate overall Salmonella in vivo gene expression.
Enteritis-specific genes.
Several genes with specific high expression during enteritis are known to be involved in virulence. In particular, four identified promoters were associated with the type III secretion system encoded on SPI-1 that is known to be important for enteritis induction and invasion. The PsicA-associated sicA-sipBCDA-iacP operon drives expression of the chaperone SicA that also has regulatory functions, the SPI-1 translocon components (and effectors) SipB and SipC, and the effector SipA (21). The rtsA, hilA, and hilC genes encode members of a complex regulatory cascade that positively regulates expression of various SPI-1-associated genes (26, 64).
The SPI-1-associated rtsA gene is cotranscribed with rtsB, which encodes a negative regulator of flagellum synthesis (26). Interestingly, we found a second negative regulator of flagellum genes (lrhA) with specific expression during enteritis, which can also downregulate motility and chemotaxis genes (49). However, the functional relevance of these negative flagella gene regulators in the murine colitis model remained unclear, since we also found high levels of enteric expression of the flhDC operon encoding the master regulator of class II flagellar genes and expression of the major flagellin gene fliC (data not shown). Indeed, direct examination of freshly prepared cecal contents of intragastrically infected mice revealed highly motile green fluorescent Salmonella cells (data not shown), in agreement with recent functional evidence for Salmonella motility and chemotaxis in the murine enteritis model (65). Interestingly, transcriptome analysis of Vibrio cholerae from stool samples revealed repression of chemotaxis genes, although Vibrio was also directly observed to be highly motile in the same samples (54). These apparently contradictory results might be related to posttranscriptional regulation, which is not revealed in promoter or transcriptome studies.
Several other identified genes had specific physiological functions. A formate transporter (FocA) and the pyruvate formiate lyase (PflB) are involved in mixed acid fermentation during anaerobic growth of Salmonella, which is consistent with oxygen limitation in the gut. PflB is also upregulated in Vibrio cholerae in a rabbit ileal loop model (78). The strong virulence defect of our regulator fnr mutant (Table 4) supported the high relevance of anaerobic Salmonella metabolism in the enteritis model.
The enterically expressed fhuACDB operon encodes a ferrichrome uptake system that utilizes ferrichrome siderophores produced by fungi (20). The expression of this operon confirmed a previous IVET result (38) and might reflect limited iron availability (7) in the gut during enteritis.
The ahpCF operon that encodes an alkylhydroperoxide reductase involved in detoxification of reactive oxygen and nitrogen intermediates was found to be specifically expressed in the colitis model, potentially reflecting antibacterial host responses during cecum inflammation. The enteric expression was verified with a GFP insertion in the native chromosomal locus (Table 3). AhpC is induced during interaction with macrophages, and the gene product is recognized as an antigen during systemic infection (29, 67); but in our hands, neither an episomal nor the chromosomal promoter fusion was sufficiently active for detection in infected spleen.
Finally, two genes with unknown function were specifically expressed in the enteritis model. STM4423 encodes an araC-type regulator and STM1328 encodes a conserved bacterial protein (COG3528).
Typhoid fever-specific genes.
The vast majority of genes that were found to be associated with typhoid fever-specific promoters are known or potential virulence factors and reside in clusters of presumably horizontally acquired genes (“genomic islands”) (data not shown). In particular, we identified nine operons that are associated with the type III secretion system encoded on SPI-2. This system is essential for systemic virulence and has been shown to interfere with microtubuli and actin networks, vesicle trafficking, and phagoendosome maturation resulting in the exclusion of inducible nitric oxide synthase and NADPH oxidase from the Salmonella-containing vacuole (75). Among the operons identified in our data set were previously characterized structural components expressed from PssaB and PssaG and secreted effectors/translocon components and their associated chaperones expressed from PsseA, PsifA, PsifB, PpipB, PpipB2, PsopD2, and PsseK1 (11, 12, 30, 40, 47, 48, 67). We also identified up to three novel candidate genes, STM1583, STM1633, and sb26 (carried on phage ST64B, which is absent in the sequenced strain LT2) (70), that are potentially associated with SPI-2, as they bear serine-rich N-terminal domains potentially acting as a signal for type III secretion (32). Indeed, sb26 is a close homologue to secreted sseK1 (STM4157) and sseK2 (STM2137) and has recently been renamed sseK3 (48). The association of STM1633 with virulence functions was suggested by its location in a pathogenicity islet that also includes the SPI-2 effector sseJ.
The key regulator phoP and five other genes of the PhoPQ regulon (slyA, ugd, pagK2, virK, and mig-5) were found to be selectively expressed in the typhoid fever model. SlyA itself is another transcriptional regulator (13). Members of the PhoPQ regulon regulate lipopolysaccharide structural modifications and changes in the composition of outer membrane proteins that render Salmonella more resistant to antimicrobial peptides (27). Finally, yaoF, which is weakly homologous to a Shigella hemolysin (57), represents a novel typhoid fever-specific virulence factor (Table 4).
In addition to the large number of known or putative virulence factors, three genes with putative housekeeping functions were found to be selectively expressed in the typhoid fever model, with products including a dispensable periplasmic chorismate mutase (expressed by aroQ) (Table 4), and the two endonucleases RNase I (rna) and the nonspecific DNA/RNA endonuclease STM1330, which is homologous to an extracellular nuclease of Serratia marcescens. Interestingly, another RNA-degrading enzyme, polynucleotide phosphorylase, profoundly influences the infection biology of Salmonella (16). Possibly, these nucleases modulate mRNA turnover for regulation of virulence gene expression, degrade antibacterial nets produced by neutrophils (10), and/or are involved in scavenging nucleic acids for nucleoside supply (5).
Finally, we identified the typhoid fever-associated hypothetical operon, STM0809-STM0810, which encodes putative inner membrane proteins that are exclusively present in S. enterica serovar Typhimurium but absent in S. enterica serovar Typhi, and STM1672, which is present in both serovars but absent in nonpathogenic E. coli.
Commonly expressed genes.
Most of the commonly highly expressed genes have a physiological function. In particular, we identified the two biosynthetic genes ribB and pheA. RibB is involved in the biosynthesis of riboflavin, an essential precursor of flavin adenine dinucleotide and flavin mononucleotide cofactors, and is required in both disease models (Table 4). pheA encodes the bifunctional chorismate mutase/prephenate dehydratase involved in phenylalanine biosynthesis. Four commonly expressed promoters drive the expression of operons involved in carbohydrate uptake and utilization. RbsDACBK and GlpTQ mediate the uptake of ribose and glycerol-3-phosphate, respectively. The yiaK operon and the putative operon STM2344-2340 encode carbohydrate uptake systems and utilization enzymes. The expression of glpTQ and the yiaK operons can be induced in vitro by anaerobiosis (42, 77), and their somewhat higher expression in the gut may reflect the oxygen limitation in this compartment compared to that in the spleen. Two identified promoters drive expression of proteins involved in respiration. NADH dehydrogenase I (nuoA-N) is expressed under both aerobic and anaerobic conditions (2), and DniR is a positive regulator of the formate-dependent respiratory nitrite reductase, which uses formate as electron donor to reduce nitrite to ammonia in anaerobic respiration (43). In addition, genes of lipopolysaccharide synthesis (htrB and wzzB) and maintenance of cell envelope integrity (ybgC-tolQRAB-pal-ybgF operon), the speF-potE operon whose gene products are involved in polyamine metabolism, and the gene of the threonine tRNA synthetase (thrS) were found to be commonly expressed. The commonly expressed putative operon STM4501-STM4500 encodes a protein broadly conserved in bacteria (COG3811) and a putative S-adenosylmethionine-dependent methyltransferase.
Surprisingly, two virulence-associated promoters were found to be active in both disease models, and one of these (PhilD) was independently identified in nonredundant clones in both disease models. HilD is a positive regulator in a regulatory cascade that leads to expression of SPI-1-associated genes, which are required for induction of enteritis but not during systemic infection. HilD expression is only modestly regulated under different in vitro conditions (52), and HilD activity is diminished by direct interaction with HilE (4) suggesting that intracellular hilD transcription might not correlate with SPI-1 activity. The second commonly expressed virulence promoter drives expression of the ybaJ-hha operon. While the function of YbaJ is unknown, Hha negatively regulates expression of HilA, which itself is a positive regulator of SPI-1-associated genes (28). The complex regulation of SPI-1 is still incompletely understood, and the functional relevance of enteric expression of the ybaJ-hha operon is presently unclear.
Global functional adaptations.
In addition to the differential regulation of individual operons, the data also suggested marked differences in the major functions of highly expressed genes during enteritis and typhoid fever. While nutrient utilization and energy conversion appeared to dominate Salmonella transcription during enteritis, virulence factor transcription seemed to dominate during typhoid fever. This reallocation of transcriptional activities to complementary functions closely corresponded to differential microenvironmental constraints, including competition with the host and other gut microorganisms for nutrients and low oxygen availability during enteritis and potent antimicrobial immune responses that impair intracellular Salmonella growth during typhoid fever. Indeed, the highly expressed genes play an outstanding role for Salmonella virulence, since at least 16 (32%) of the identified 50 operons with very high levels of expression during infection encoded genes with essential functions in one or both disease models (Tables 1, 2, and 4). This high proportion was remarkable, compared to the low frequency of essential genes among in vivo-expressed genes that had been previously identified with the nonquantitative IVET technology (18, 39) and an estimated 4% essential virulence genes in the entire Salmonella genome (8). Exceptionally high in vivo expression levels thus appeared to correlate with functional importance; this could reflect economical use of the substantial biosynthesis costs that are associated with highly expressed genes.
However, it is not yet clear whether these strong promoter activities correlate with high abundance of the corresponding proteins. Technical difficulties currently make it difficult to determine the protein composition of Salmonella in infected mouse tissues, but the atypically high AT content and the potentially suboptimal codon usage of many SPI-2-associated genes and genes of the PhoP regulon suggest that despite high levels of transcription of these virulence genes, the corresponding proteins might be less abundant than other well-translated proteins. Further studies are required to clarify this issue to more fully characterize Salmonella adaptation to different disease conditions on the proteome level.
Another issue that requires further study is the relevance of the mouse enteritis model for human disease. Several histological features show that some important aspects of human disease are reproduced by the mouse model. However, in mice this model leads eventually to lethal systemic infection, while in immunocompetent patients, gastroenteritis is not associated with detectable systemic dissemination. We utilized the mouse model, since it allowed comparison of systemic disease and enteritis in the same animal host species, but further studies are be needed to validate our findings in more realistic model systems.
Acknowledgments
We thank Thomas F. Meyer for critical discussion and generous support and Katharina Raba and Meike Sörensen for excellent technical assistance.
This work was supported in part by Deutsche Forschungsgemeinschaft (SFB621-A9).
Editor: A. D. O'Brien
REFERENCES
- 1.Akashi, H., and T. Gojobori. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA 99:3695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, C. D., and T. Elliott. 1995. Transcriptional control of the nuo operon which encodes the energy-conserving NADH dehydrogenase of Salmonella typhimurium. J. Bacteriol. 177:2335-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxter, M. A., T. F. Fahlen, R. L. Wilson, and B. D. Jones. 2003. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect. Immun. 71:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benedik, M. J., and U. Strych. 1998. Serratia marcescens and its extracellular nuclease. FEMS Microbiol. Lett. 165:1-13. [DOI] [PubMed] [Google Scholar]
- 6.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjarnason, J., C. M. Southward, and M. G. Surette. 2003. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowe, F., C. J. Lipps, R. M. Tsolis, E. Groisman, F. Heffron, and J. G. Kusters. 1998. At least four percent of the Salmonella typhimurium genome is required for fatal infection of mice. Infect. Immun. 66:3372-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyce, J. D., I. Wilkie, M. Harper, M. L. Paustian, V. Kapur, and B. Adler. 2002. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect. Immun. 70:6871-6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinkmann, V., U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D. S. Weiss, Y. Weinrauch, and A. Zychlinsky. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532-1535. [DOI] [PubMed] [Google Scholar]
- 11.Brumell, J. H., S. Kujat-Choy, N. F. Brown, B. A. Vallance, L. A. Knodler, and B. B. Finlay. 2003. SopD2 is a novel type III secreted effector of Salmonella typhimurium that targets late endocytic compartments upon delivery into host cells. Traffic 4:36-48. [DOI] [PubMed] [Google Scholar]
- 12.Brumell, J. H., P. Tang, S. D. Mills, and B. B. Finlay. 2001. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic 2:643-653. [DOI] [PubMed] [Google Scholar]
- 13.Buchmeier, N., S. Bossie, C. Y. Chen, F. C. Fang, D. G. Guiney, and S. J. Libby. 1997. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect. Immun. 65:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bumann, D. 2002. Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol. Microbiol. 43:1269-1283. [DOI] [PubMed] [Google Scholar]
- 15.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 101:7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clements, M. O., S. Eriksson, A. Thompson, S. Lucchini, J. C. Hinton, S. Normark, and M. Rhen. 2002. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. USA 99:8784-8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coburn, B., Y. Li, D. Owen, B. A. Vallance, and B. B. Finlay. 2005. Salmonella enterica serovar Typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 73:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conner, C. P., D. M. Heithoff, and M. J. Mahan. 1998. In vivo gene expression: contributions to infection, virulence, and pathogenesis. Curr. Top. Microbiol. Immunol. 225:1-12. [DOI] [PubMed] [Google Scholar]
- 19.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 20.Coulton, J. W., P. Mason, and M. S. DuBow. 1983. Molecular cloning of the ferrichrome-iron receptor of Escherichia coli K-12. J. Bacteriol. 156:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-960. [DOI] [PubMed] [Google Scholar]
- 22.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Keersmaecker, S. C., K. Marchal, T. L. Verhoeven, K. Engelen, J. Vanderleyden, and C. S. Detweiler. 2005. Microarray analysis and motif detection reveal new targets of the Salmonella enterica serovar Typhimurium HilA regulatory protein, including hilA itself. J. Bacteriol. 187:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Detweiler, C. S., D. M. Monack, I. E. Brodsky, H. Mathew, and S. Falkow. 2003. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48:385-400. [DOI] [PubMed] [Google Scholar]
- 25.dos Reis, M., L. Wernisch, and R. Savva. 2003. Unexpected correlations between gene expression and codon usage bias from microarray data for the whole Escherichia coli K-12 genome. Nucleic Acids Res. 31:6976-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3:1327-1334. [DOI] [PubMed] [Google Scholar]
- 28.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francis, K. P., P. D. Taylor, C. J. Inchley, and M. P. Gallagher. 1997. Identification of the ahp operon of Salmonella typhimurium as a macrophage-induced locus. J. Bacteriol. 179:4046-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freeman, J. A., M. E. Ohl, and S. I. Miller. 2003. The Salmonella enterica serovar Typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infect. Immun. 71:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 32.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 33.Hapfelmeier, S., K. Ehrbar, B. Stecher, M. Barthel, M. Kremer, and W. D. Hardt. 2004. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:795-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hapfelmeier, S., B. Stecher, M. Barthel, M. Kremer, A. J. Muller, M. Heikenwalder, T. Stallmach, M. Hensel, K. Pfeffer, S. Akira, and W. D. Hardt. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 174:1675-1685. [DOI] [PubMed] [Google Scholar]
- 35.Hautefort, I., M. J. Proenca, and J. C. Hinton. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He, G., R. A. Shankar, M. Chzhan, A. Samouilov, P. Kuppusamy, and J. L. Zweier. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. USA 96:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heim, R., D. C. Prasher, and R. Y. Tsien. 1994. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 91:12501-12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heithoff, D. M., C. P. Conner, P. C. Hanna, S. M. Julio, U. Hentschel, and M. J. Mahan. 1997. Bacterial infection as assessed by in vivo gene expression. Proc. Natl. Acad. Sci. USA 94:934-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heithoff, D. M., R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2000. In vivo gene expression and the adaptive response: from pathogenesis to vaccines and antimicrobials. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 41.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 42.Ibanez, E., E. Campos, L. Baldoma, J. Aguilar, and J. Badia. 2000. Regulation of expression of the yiaKLMNOPQRS operon for carbohydrate utilization in Escherichia coli: involvement of the main transcriptional factors. J. Bacteriol. 182:4617-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kajie, S., R. Ideta, I. Yamato, and Y. Anraku. 1991. Molecular cloning and DNA sequence of dniR, a gene affecting anaerobic expression of the Escherichia coli hexaheme nitrite reductase. FEMS Microbiol. Lett. 67:205-211. [DOI] [PubMed] [Google Scholar]
- 44.Karlin, S., M. J. Barnett, A. M. Campbell, R. F. Fisher, and J. Mrazek. 2003. Predicting gene expression levels from codon biases in alpha-proteobacterial genomes. Proc. Natl. Acad. Sci. USA 100:7313-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlin, S., J. Mrazek, A. Campbell, and D. Kaiser. 2001. Characterizations of highly expressed genes of four fast-growing bacteria. J. Bacteriol. 183:5025-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlin, S., J. Theriot, and J. Mrazek. 2004. Comparative analysis of gene expression among low G+C gram-positive genomes. Proc. Natl. Acad. Sci. USA 101:6182-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knodler, L. A., B. A. Vallance, M. Hensel, D. Jackel, B. B. Finlay, and O. Steele-Mortimer. 2003. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol. Microbiol. 49:685-704. [DOI] [PubMed] [Google Scholar]
- 48.Kujat Choy, S. L., E. C. Boyle, O. Gal-Mor, D. L. Goode, Y. Valdez, B. A. Vallance, and B. B. Finlay. 2004. SseK1 and SseK2 are novel translocated proteins of Salmonella enterica serovar Typhimurium. Infect. Immun. 72:5115-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 50.Libby, S. J., W. Goebel, A. Ludwig, N. Buchmeier, F. Bowe, F. C. Fang, D. G. Guiney, J. G. Songer, and F. Heffron. 1994. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc. Natl. Acad. Sci. USA 91:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lucas, R. L., and C. A. Lee. 2000. Unravelling the mysteries of virulence gene regulation in Salmonella typhimurium. Mol. Microbiol. 36:1024-1033. [DOI] [PubMed] [Google Scholar]
- 52.Lucas, R. L., and C. A. Lee. 2001. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:2733-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 54.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Motley, S. T., B. J. Morrow, X. Liu, I. L. Dodge, A. Vitiello, C. K. Ward, and K. J. Shaw. 2004. Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell. Microbiol. 6:849-865. [DOI] [PubMed] [Google Scholar]
- 57.Nagamune, K., K. Yamamoto, and T. Honda. 1995. Cloning and sequencing of a novel hemolysis gene of Vibrio cholerae. FEMS Microbiol. Lett. 128:265-269. [DOI] [PubMed] [Google Scholar]
- 58.Orihuela, C. J., J. N. Radin, J. E. Sublett, G. Gao, D. Kaushal, and E. I. Tuomanen. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 72:5582-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pedersen, S., P. L. Bloch, S. Reeh, and F. C. Neidhardt. 1978. Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell 14:179-190. [DOI] [PubMed] [Google Scholar]
- 60.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rollenhagen, C., M. Sorensen, K. Rizos, R. Hurvitz, and D. Bumann. 2004. Antigen selection based on expression levels during infection facilitates vaccine development for an intracellular pathogen. Proc. Natl. Acad. Sci. USA 101:8739-8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rollins, S. M., A. Peppercorn, L. Hang, J. D. Hillman, S. B. Calderwood, M. Handfield, and E. T. Ryan. 2005. In vivo induced antigen technology (IVIAT). Cell. Microbiol. 7:1-9. [DOI] [PubMed] [Google Scholar]
- 63.Ruiz-Perez, F., J. Sheikh, S. Davis, E. C. Boedeker, and J. P. Nataro. 2004. Use of a continuous-flow anaerobic culture to characterize enteric virulence gene expression. Infect. Immun. 72:3793-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 65.Stecher, B., S. Hapfelmeier, C. Muller, M. Kremer, T. Stallmach, and W. D. Hardt. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:4138-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taylor, P. D., C. J. Inchley, and M. P. Gallagher. 1998. The Salmonella typhimurium AhpC polypeptide is not essential for virulence in BALB/c mice but is recognized as an antigen during infection. Infect. Immun. 66:3208-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tjaden, B., R. M. Saxena, S. Stolyar, D. R. Haynor, E. Kolker, and C. Rosenow. 2002. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 30:3732-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsolis, R. M., L. G. Adams, T. A. Ficht, and A. J. Baumler. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tucker, C. P., and M. W. Heuzenroeder. 2004. ST64B is a defective bacteriophage in Salmonella enterica serovar Typhimurium DT64 that encodes a functional immunity region capable of mediating phage-type conversion. Int. J. Med. Microbiol. 294:59-63. [DOI] [PubMed] [Google Scholar]
- 71.Uchiya, K., M. A. Barbieri, K. Funato, A. H. Shah, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 73.Valdivia, R. H., and S. Falkow. 1998. Flow cytometry and bacterial pathogenesis. Curr. Opin. Microbiol. 1:359-363. [DOI] [PubMed] [Google Scholar]
- 74.Virtaneva, K., S. F. Porcella, M. R. Graham, R. M. Ireland, C. A. Johnson, S. M. Ricklefs, I. Babar, L. D. Parkins, R. A. Romero, G. J. Corn, D. J. Gardner, J. R. Bailey, M. J. Parnell, and J. M. Musser. 2005. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA 102:9014-9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 76.Wendland, M., and D. Bumann. 2002. Optimization of GFP levels for analyzing Salmonella gene expression during an infection. FEBS Lett. 521:105-108. [DOI] [PubMed] [Google Scholar]
- 77.Wong, K. K., and H. S. Kwan. 1992. Transcription of glpT of Escherichia coli K12 is regulated by anaerobiosis and fnr. FEMS Microbiol. Lett. 73:15-18. [DOI] [PubMed] [Google Scholar]
- 78.Xu, Q., M. Dziejman, and J. J. Mekalanos. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. USA 100:1286-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zaharik, M. L., B. A. Vallance, J. L. Puente, P. Gros, and B. B. Finlay. 2002. Host-pathogen interactions: host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proc. Natl. Acad. Sci. USA 99:15705-15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang, S., R. L. Santos, R. M. Tsolis, S. Stender, W. D. Hardt, A. J. Baumler, and L. G. Adams. 2002. The Salmonella enterica serotype Typhimurium effector proteins SipA, SopA, SopB, SopD, and SopE2 act in concert to induce diarrhea in calves. Infect. Immun. 70:3843-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]