Abstract
Coccidioides posadasii and Coccidioides immitis are dimorphic, soil-dwelling pathogenic ascomycetes endemic to the southwestern United States. Infection can result from inhalation of a very few arthroconidia, but following natural infection, long-lived immunity is the norm. Previous work in the field has shown that spherule-derived vaccines afford more protection than those from mycelia. We have used two-dimensional differential in-gel electrophoresis coupled with nano-high-performance liquid chromatography-tandem mass spectrometry to directly assess both absolute abundance and differential expression of proteins in the spherule and the mycelial phases of C. posadasii with the intent to identify potential vaccine candidates. Peptides derived from 40 protein spots were analyzed and a probable identity was assigned to each. One spherule-abundant protein, identified as Pmp1, showed homology to allergens from Aspergillus fumigatus and other fungi, all of which exhibit similarity to yeast thiol peroxidases. Recombinant Pmp1 was reactive with serum from individuals with both acute and protracted disease, and evoked protection in two murine models of infection with C. posadasii. These results demonstrate the utility of proteomic analysis as a point of discovery for protective antigens for possible inclusion in a vaccine candidate to prevent coccidioidomycosis.
Coccidioides posadasii and Coccidioides immitis are dimorphic, soil-dwelling pathogenic ascomycetes endemic to the southwestern United States. In the soil the fungus grows as saprobic mycelium, resulting in the formation of single-celled arthroconidia. Infection in humans and animals is initiated when an arthroconidium is inhaled into the respiratory tract. Subsequently, in the lung, arthroconidia develop into thick- walled, spore-filled structures known as spherules, presumably with significant changes in the protein expression profile of the organism (12, 24, 49). Spherule development can be replicated in vitro by growing arthroconidia under conditions of elevated temperature and CO2 (8).
It is estimated that about 150,000 otherwise healthy individuals are infected with Coccidioides spp. each year, up to 30% of whom may develop symptoms including protracted pulmonary or systemic syndromes (17), miss school or work due to their illness, and seek repeated medical attention (6). A small percentage of patients suffer life-threatening illness. In most affected persons, however, illness is self-limited, spherule proliferation is arrested, and markers of cellular immunity against antigens of Coccidioides spp. become evident (4, 9, 18). Recovery from an initial infection nearly always produces long-lived and complete immunity to illness from a second infection, a fact which has motivated extensive efforts to discover specific coccidioidal antigens which may induce protection when administered as vaccines (1, 10, 13, 30, 34). Despite this progress, it is possible that a more effective vaccine candidate could be developed by discovering additional immunoprotective coccidioidal proteins.
Antigens that produce protective immunity against coccidioidomycosis have exhibited several common characteristics. The most protective coccidioidal vaccines to date have been spherule-derived (32, 33, 56), or from recombinant proteins that are abundantly, though not necessarily exclusively, represented in the spherule phase (27). Moreover, protective antigens derived from Coccidioides spp. have been either cell wall associated or actively secreted from spherules (1, 13), similar to effective subcellular immunogens in vaccines against other pathogens (23, 32, 49). An additional theoretical characteristic of a protective antigen for any pathogen is that it be directly virulence associated, e.g., an adhesin or immunomodulatory protein, specifically expressed during the infectious phase (2, 25, 31, 35, 48).
We chose to use two-dimensional differential in-gel electrophoresis (2D-DIGE) coupled with nano-high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) to directly assess both absolute abundance and differential expression of proteins in the spherule and the mycelial phases of C. posadasii. This approach led to the identification of a protein, Pmp1, which showed protection in murine models of infection with C. posadasii.
(This work was presented in part at the 105th Annual Meeting of the American Society for Microbiology, 5 to 9 June 2005.)
MATERIALS AND METHODS
Strains and culture conditions.
Coccidioides posadasii strain Silveira stock cultures were maintained on 2XGYE agar plates at room temperature. For growth in the spherule phase, arthroconidia were harvested from 4-week-old cultures by the spin bar technique of Sun and Huppert (47); 7 × 108 arthroconidia were inoculated into 1 liter modified Converse medium (7) supplemented with 0.05% NZ-Amine (Sigma, St. Louis, MO) and cultures were incubated at 39°C, 8% CO2, and 160 rpm, for 96 h. Mycelia for protein extraction were grown in liquid 2XGYE at 37°C and 180 rpm, for 48 h. All manipulations of potentially viable C. posadasii were accomplished under biosafety level 3 conditions in laboratories registered with the Centers for Disease Control and Prevention for possession of this select agent.
Protein extraction.
Spherules were harvested by centrifugation and the carbohydrate rich outer wall material removed from the surface of the cell pellet. The cell pellet was washed twice with water, then frozen at −80°C. Mycelia were harvested by filtration, washed extensively with water, and frozen at −80°C. The pellets were thawed on ice, and mixed with 5 ml per g (wet weight) lysis buffer [20 mM Tris, pH 7.9, 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 0.3 M (NH4)2SO4, 1× protease inhibitors (Calbiochem, La Jolla, CA), 1 mM phenylmethylsulfonyl fluoride]. The resuspended fungal material was aliquoted into 15-ml conical centrifuge tubes and silica-zirconium beads (0.5 mm; Biospec, Bartlesville, OK) were added (∼2 ml beads and ∼2.5 ml suspension). The cells were disrupted by vortexing eight times for 1 min separated by 1 min on ice and the cell lysate was collected. The beads were washed three times with 1 ml lysis buffer. The washes combined with the cell lysate were centrifuged at 4,000 rpm for 30 min at 4°C. Supernatants were precipitated with 10% trichloroacetic acid in the presence of 0.7% β-mercaptoethanol, and the pellets were washed with cold acetone, air dried, and resuspended in 200 μl solubilization buffer (8 M urea, 4% NP-40, 2% β-mercaptoethanol, pH 9.5).
Two-dimensional differential in-gel electrophoresis.
The protein concentration for each extract was determined using the 2-D Quant kit (Amersham Biosciences, Piscataway, NJ). Excess salts and lipids were removed with the Plus One 2D-Clean-Up kit and samples resuspended in sample buffer containing 7 M urea, 2 M thiourea, 2% dithiothreitol, and 4% CHAPS (cholamidopropyldimethylammoniopropanesulfonate); 1 μl of CyDye (Amersham) working solution (400 pmol/μl in 99% anhydrous dimethyl formamide) was used to label protein aliquots for 30 min on ice in the dark. The reactions were quenched with an excess of lysine (1 μl of a 10 mM stock) and further incubated on ice for 10 min.
For the first experiment, 25 μg spherule extract labeled with Cy3, 25 μg mycelial extract labeled with Cy5, and an additional 25 μg mycelial extract labeled with Cy2 were mixed with an unlabeled pool of mycelial and spherule proteins (112.5 μg each). For the second experiment, spherule extracts labeled with Cy3 and Cy2 and mycelial extract labeled with Cy5 were combined with unlabeled protein.
For first-dimension isoelectric focusing, samples were applied to duplicate 24-cm pH 3 to 7 nonlinear immobilized pH gradient strips (Amersham) in the presence of 0.012% Destreak solution (Amersham) and 0.5% IPG buffer (Amersham), rehydrated for 12 h at 50 V in the dark, and focused in the first dimension (69,111 V · h). The focused strips were reequilibrated following established protocols, applied to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and electrophoresed at 30 mA until the bromophenol blue eluted from the gel. Images were obtained by scanning the gels on a Typhoon 9400 fluorescent laser scanner, at the appropriate emissions, filters, and wavelengths for each label used: Cy3 with a 580 BP emission filter, green laser, 532 nm; Cy5 with a 670 BP emission filter, red laser, 633 nm; and Cy2 with a 520 BP emission filter, blue laser, 488 nm. Initial scan parameters were press sample, depth plus 3 mm, 600-V photomultiplier tube setting and 500-μm pixel size, before repeating at 50 to 100-μm pixel size for a high-resolution scan. Images were acquired using the Typhoon scanner control software, and saved in a format compatible with the DIA (differential in-gel analysis) module of DeCyder software (Amersham Biosciences).
Forty protein spots revealed as differentially expressed were manually excised at the center of the visible spot from one of the gels after poststaining of the unlabeled protein with colloidal Coomassie stain. Proteins in each spot were digested in gel with trypsin as described (5, 54) and the resulting peptides subjected to nano-HPLC-MS/MS using an LCQ-Deca XP Plus ion trap mass spectrometer (Thermo-Finnigan, San Jose, CA) as described (38). MS/MS data were analyzed with the SEQUEST program (55), which allows the correlation of experimental data with theoretical spectra generated from known protein sequences. Criteria for preliminary peptide identification were as described (38), searching against a locally created database consisting of an unannotated GENESCAN file (Theo Kirkland, personal communication) created from eightfold coverage of the C. posadasii genome (TIGR release of September 2003) and a collection of publicly available protein sequences from fungi, Saccharomyces cerevisiae, Escherichia coli, and other microorganisms (38). The dominant protein in each visible spot was taken to be the one represented by at least twice as many peptides as all others combined. In cases in which there was not a clearly dominant protein, the two most abundant were considered to be equally likely.
MS/MS and bioinformatics.
Protein identities based on peptide sequences deduced by the SEQUEST program were verified in several ways. First, peptides were manually validated against the GENESCAN database, and then the GENESCAN-generated deduced protein sequences were analyzed by BLASTP (3) against the nr protein database at NCBI to obtain a putative identity by homology, with an arbitrary maximum value of e < 10−5. Second, sets of peptide fragments which did not appear in the C. posadasii GENESCAN data set, but were identified with other fungal sequences, were localized to individual contigs of the eightfold coverage version of the C. posadasii genome sequence (TIGR) using the TFASTS algorithm (36).
During the course of this work, sequencing of the genome of C. posadasii was substantially completed (August 2004), and this version was used in later analyses. The DNA sequence 1,000 bp up- and downstream from the open reading frames for matching peptides was identified using TBLASTX against the nr database at NCBI. Homology and gene structure were validated manually. Putative protein sequences homologous to “hypothetical” proteins were tentatively identified as possible using the Conserved Domain Database and CDART at NCBI (37) and protein sequence analysis tools available online at ExPASy (http://au.expasy.org).
Cloning and protein expression.
One protein, identified by SEQUEST as a peroxisomal membrane protein from C. posadasii, and highly expressed in the spherule phase, was selected for further study (see Results and Discussion). First-strand cDNA produced from mycelial RNA as previously described (14) was amplified using primers designed from the genomic sequence of C. posadasii (TIGR) up- and downstream from the coding region of pmp1. Three separate 5′ primers were used in conjunction with a single 3′ primer to deduce the transcription start site for mRNA (Table 1).
TABLE 1.
Primers used in this study
| Orientation | Primer sequence | Purpose | Vector | Product |
|---|---|---|---|---|
| Forward | CTTCACCGCAGCCTTGCTTAG | Locate 5′ end of mRNA | None | |
| Forward | GTCTGCCAACGCTCGATTCAC | Locate 5′ end of mRNA | Perfectly blunt | 675 bp |
| Forward | AACCTCACCTCGCTACTAC | Locate 5′ end of mRNA | 591 bp | |
| Reverse | GGCCGTTTCCCTTGATTCTCC | 3′ end of mRNA | Perfectly blunt | |
| Forward | GAATTC-ATGGCCTCTCTCAAAGCTGGAGAC | Expression | YEp-FLAG-1 | 512 bp |
| Reverse | GTCGACTTA-CAACTTGGAAATGACGGCGTC | Expression | Yep-FLAG-1 |
After confirming the cDNA sequence, primers were designed to include convenient restriction sites, and the resultant amplimer was cloned into the yeast expression vector YEp-FLAG-1 (Sigma) to create pTP73415 (Table 1). Recombinant protein was produced from Saccharomyces cerevisiae strain BJ3505/pTP73415 by inoculating a single colony into 30 ml of synthetic complete medium containing 1.0 g/liter yeast synthetic drop-out medium supplement without tryptophan (Sigma), 6.7 g/liter yeast nitrogen base without amino acids (Difco/BD, Sparks, MD) and 2% glucose and incubating at 30°C at 175 rpm, for 48 to 72 h; 25 ml of this starter culture was diluted into 500 ml of YP High Stability Expression Medium (YPHSM) containing 1% yeast extract, 8% peptone, 1% glucose, 3% glycerol, and 20 mM CaCl2 and grown for an additional 96 h. Secreted recombinant Pmp1 (rPmp1) was purified from the culture supernatant with anti-FLAG-M2 Affinity Gel (Sigma) by elution with 0.1 M glycine, dialyzed against phosphate-buffered saline, and stored at −70°C until use. Protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL). rPmp1 was visualized as a 17.9-kDa protein by Coomassie stain following SDS-PAGE and immunoblotting using a pool of serum from patients with high titers to the diagnostic complement fixation antigen.
ELISA and source of serum.
To assess whether Pmp1 was recognized by the immune system of naturally infected patients, we assayed for anti-Pmp1 immunoglobulin G (IgG) antibody in several patient populations. Ninety-six-well plates (Polysorp; Nunc, Rochester, NY) were coated with 10 ng rPmp1 in Dulbecco's phosphate-buffered saline (PBS) containing 0.2 M MgCl2. Enzyme-linked immunosorbent assays (ELISAs) were performed as previously described (53).
Serum from individuals with complicated coccidioidal disease was from patients enrolled in a study to examine the effectiveness of new antifungal agents, and represents the earliest available sample from each patient. Sera from patients with primary coccidioidal pneumonia were obtained during a prospective study of coccidioidomycosis as a cause of community-acquired pneumonia. Serum samples from patients with no apparent coccidioidal disease were deidentified samples selected randomly from the clinical chemistry laboratory at a hospital in Tucson. The titer was the highest dilution with an absorbance of >2 times the background optical density at a 1:80 dilution for the individual serum sample. Sera were also evaluated for immunodiffusion-complement fixation (IDCF) and immunodiffusion-tube precipitin antibodies used for diagnosis of coccidioidomycosis, either in a clinical laboratory or by standard methods in our laboratory (52). University of Arizona and Southern Arizona Veterans Administration Health Care System Institutional Review Board approval was obtained in all cases.
Protection studies.
Female, 6-week-old C57BL/6 mice were purchased from Harlan-Sprague-Dawley (Indianapolis, IN) and maintained according to National Institutes of Health guidelines. Groups of 10 mice were immunized subcutaneously with 1, 5, or 50 μg rPmp1 with monophosphoryl lipid A-stable emulsion (MPL-SE) adjuvant (Corixa, Hamilton, MT) on day 0 and day 14. Control mice received adjuvant only. Four weeks after boosting (day 42), the mice were infected intraperitoneally with 310 arthroconidia and sacrificed 2 weeks later. The right lung and spleen were quantitatively cultured as previously described (1) and results were expressed as log10 CFU per organ. Cultures without growth at the lowest dilution (1:100) were recorded as one-half the lower limit of detection, or 50 CFU/organ. Results were analyzed using the Wilcoxon signed ranks test.
In a separate study, 10 mice were immunized intradermally with 1 μg rPmp1 and immunostimulatory oligonucleotide sequences (1018 ISS; Dynavax, Berkeley, CA) (10 μg/mouse) as adjuvant on days 0 and 14. Another group received adjuvant alone. The groups were challenged on day 42 with 82 arthroconidia intranasally. Survival was assessed for 56 days after infection and the differences in survival between groups were analyzed by the Mantel-Haenzel test of significance.
All studies were conducted with the approval of the Institutional Animal Care and Use Committee.
Nucleotide sequence accession number.
The accession number for the cDNA sequence of pmp1 is DQ225176.
RESULTS
Proteomic analysis.
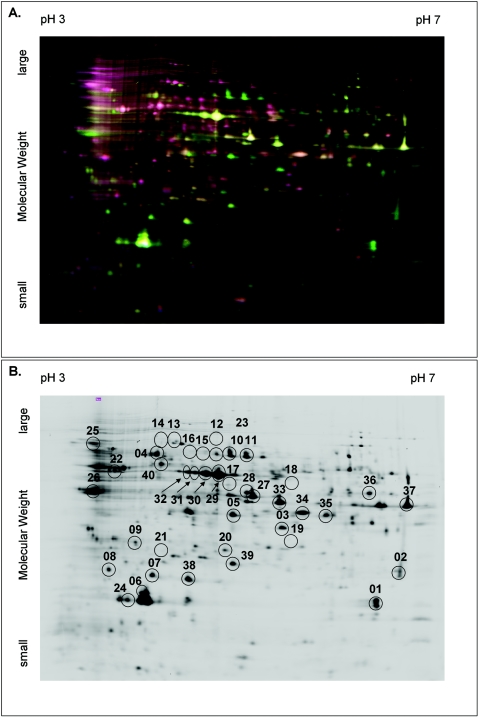
We used 2D-DIGE to compare the relative abundance of highly expressed proteins in the mycelial and the spherule phases of C. posadasii. De-Cyder software was used to compare Cy3 and Cy5 image intensity against a Cy2-labeled control and against each other. Thirty-one protein spots were identified which differed in abundance between the two phases either by more than two standard deviations from the mean spot intensity or threefold on two cross-labeled gels. Another nine spots were picked for further evaluation based on simple abundance in the spherule phase (Fig. 1). Proteins could be identified in all 40 spots examined (Table 2), although several discrete spots were dominated by the same protein for a total of 32 different proteins identified as dominant.
FIG. 1.
A. Two-dimensional differential in-gel electrophoresis of spherule and mycelial extracts. Spherule extract labeled with Cy3 (green), mycelial extract labeled with Cy5 (red) and Cy2 (cyan). B. Spots selected for analysis are identified on the Cy3-spherule virtual image. Circles enclosing apparently blank areas of the image indicate the positions of mycelium dominant protein.
TABLE 2.
Protein identificationa
| Spot | Tentative identification | Phase | No. of peptides | Accession no. | e value |
|---|---|---|---|---|---|
| 1 | HSP30 | S | 13 | XP_660134 | 4 e-32 |
| 2 | Benzoquinone reductase | S | 10 | AF452883 | 1 e-101 |
| 2 | HSP30 | S | 7 | XP_660134 | 4 e-32 |
| 3 | Short-chain alcohol dehydrogenase | S | 33 | EAL87182 | 3 e-67 |
| 4 | Short-chain alcohol dehydrogenase | S | 31 | EAL87182 | 3 e-67 |
| 5 | Aldehyde reductase | S | 20 | AAL27089 | 3.9 e-133 |
| 6 | Peroxisomal protein | S | 60 | AAQ84041 | 4 e-61 |
| 7 | Fructose 1,6-bisphosphate aldolase | S | 19 | AAG00613 | 0 |
| 8 | Hypothetical | S | 19 | ZP_00302283 | 1 e-21 |
| 9 | Peroxisomal protein | m | 8 | AAQ84041 | 4 e-61 |
| 10 | Pyruvate decarboxylase | S | 47 | EAL92474 | 0 |
| 11 | Pyruvate decarboxylase | S | 60 | EAL92474 | 0 |
| 12 | Pyruvate decarboxylase | M | 50 | EAL92474 | 0 |
| 13 | HSP70 (mitochondrial) | M | 28 | AAP05987 | 0 |
| 14 | HSP70 (mitochondrial) | M | 25 | AAP05987 | 0 |
| 15 | Leucine aminopeptidase | M | 19 | AAS76669 | 2 e-151 |
| 16 | Leucine aminopeptidase | M | 22 | AAS76669 | 2 e-151 |
| 17 | Isovaleryl-coenzyme A DH | M | 19 | EAL91706 | 0 |
| 17 | Peptidyl-prolyl cis-trans isomerase | M | 15 | EAL87302 | 5 e-99 |
| 18 | Ornithine aminotransferase | M | 21 | EAL89873 | 0 |
| 19 | Cytochrome c peroxidase | M | 11 | EAL89876 | 4 e-139 |
| 19 | Orotidine 5′-monophosphate decarboxylase (pyrG) | M | 10 | AAQ16206 | 0 |
| 20 | GDSL motif lipase | S | 8 | EAL93188 | 9.6 e-46 |
| 21 | HSP70 (mitochondrial) | M | 4 | AAP05987 | 0 |
| 22 | 1,2-α-d-Mannosidase | S | 13 | P31723 | 2.7 e-107 |
| 23 | Leucine aminopeptidase | M | 13 | AAS76669 | 2 e-151 |
| 24 | Peroxisomal protein | S | 8 | AAQ84041 | 4 e-61 |
| 25 | HSP70 | = | 35 | AAQ83701 | 0 |
| 26 | HSP70 | $ | 44 | AAQ83701 | 0 |
| 27 | Fructose 1,6-bisphosphate aldolase | = | 48 | AAG00613 | 0 |
| 28 | HSP70 (mitochondrial) | $ | 23 | AAP05987 | 0 |
| 29 | Enolase | = | 47 | Q12560 | 0 |
| 30 | Enolase | $ | 33 | Q12560 | 0 |
| 31 | Enolase | $ | 26 | Q12560 | 0 |
| 32 | Enolase | $ | 17 | Q12560 | 0 |
| 33 | Ketol-acid reductoisomerase (ilv2) | = | 23 | EAL92139 | 0 |
| 34 | Zinc-binding alcohol DH | = | 18 | EAL88173 | 5 e-83 |
| 35 | G-beta-like protein | = | 7 | AAF98065 | 8 e-160 |
| 35 | Glyceraldehyde-3-phosphate DH | = | 5 | EAL88988 | 0 |
| 35 | Zinc-binding alcohol DH | = | 5 | EAL88173 | 5 e-83 |
| 36 | Formate DH | = | 14 | AAV67970 | 9 e-151 |
| 37 | Glyceraldehyde-3-phosphate DH | = | 26 | AAN76496 | 1 e-165 |
| 38 | Enolase | = | 6 | Q12560 | 0 |
| 38 | Hypothetical | = | 4 | EAL89506 | 3 e-54 |
| 39 | GDSL motif fungal lipase | $ | 1 | EAL93188 | 9.6 e-46 |
| 40 | Arginosuccinate synthetase | m | 17 | EAA65048 | 3 e-101 |
| 40 | Glutamate carboxypeptidase | m | 12 | EAL93018 | 0 |
Phase refers to relative intensity of a given spot as follows: S, higher than twice the standard deviation of mean spot intensity in spherule phase; $, more than threefold higher in spherule phase; M, higher than twice the standard deviation of mean spot intensity in mycelial phase; m, more than threefold higher in mycelial phase; =, similar in intensity. DH, dehydrogenase. Accession number refers to the closest homolog in the nonredundant databases at NCBI. e value refers to the alignment BLASTP score using the deduced amino acid sequence as the query.
The most highly represented category of proteins in these extracts was that of enzymes active in energy production (12), followed by heat shock and stress related proteins (6), and hydrolases (4). Two proteins mapping to different contigs were identified as homologous to HSP70. Eight proteins were identified only in spherule extract, and eight others only in mycelial extract.
Analysis and expression of an abundant spherule protein.
Peptides derived from one protein that appeared upregulated in 96-h spherules (spot 6; Fig. 1) were identified by SEQUEST as derived from a 99-amino-acid peroxisomal membrane protein from C. posadasii (partial sequence in GenBank AY007646). We chose to examine this protein for its immunologic properties based on the apparent absolute abundance of the protein and on preliminary data from serial analysis of gene expression (29), which showed that mRNA for the protein was highly expressed. The genomic DNA sequence (TIGR) was searched for open reading frames corresponding to the peptides generated from this spot and a single locus on one contig encoded 53 of the peptides generated, including several peptides initially assigned a different identity. Analysis of the DNA sequence and comparison of the deduced amino acid sequence to several closely related fungal proteins predicted that the complete protein contained 166 amino acids. The presence of a C-terminal peroxisomal sorting signal (Ser-Lys-Leu) and conserved domain architecture also suggested that the protein in spot 6 is likely a peroxisomal matrix protein (Pmp1) related to thioredoxin peroxidases.
Pmp1 was found to be highly homologous to allergens derived from several fungi, including Asp f3 of Aspergillus fumigatus (68% identity) (11) and Pen c3 of Penicillium citrinum (66% identity) (45). Of the five Saccharomyces cerevisiae thiol peroxidases thus far identified, Pmp1 and the homologous allergenic proteins showed the greatest similarity to TSA2/Ahp1/YLR109w, which has been identified as an alkyl hydroperoxide peroxidase (43). Pmp1 was found to contain two cysteine residues, at positions 31 and 61, similar to Ahp1. C-61 has been shown to be part of the catalytic domain in the S. cerevisiae protein (44), and this residue is highly conserved in homologous proteins from several fungi closely related to C. posadasii, suggesting it could be the catalytic site in these proteins as well. In contrast, Pmp1 is no more than 36% identical to mammalian thiol peroxidases.
ELISA.
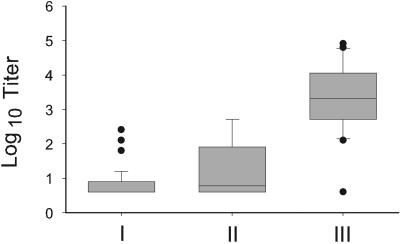
Individual sera from three groups of patients were evaluated by quantitative ELISA (Fig. 2) for anti-Pmp1 IgG antibodies. A panel of serum from 20 patients with complicated disease showed a median titer of 1:20,480. Only one of these patients showed an undetectable level (<1:80) of anti-Pmp1 antibody. In comparison, in a panel of serum from 18 patients with primary coccidioidal pneumonia, half showed an undetectable level of antibody. Of those with a detectable antibody response, the median titer was 1:640. Of serum samples from 98 patients with no evidence of coccidioidal disease, 59% had undetectable levels of anti-Pmp1 antibody. The median titer of those with detectable levels of antibody was 1:160.
FIG. 2.
Comparison of anti-Pmp1 IgG titer among three groups of patients by ELISA. Group I, no evidence of infection; II, primary coccidioidal pneumonia; III, complicated disease.
The anti-Pmp1 antibody titers were not concordant with results from either the clinically useful quantitative complement fixation test (IDCF) or the qualitative immunodiffusion-tube precipitin test (42) (data not shown).
Protection by vaccination with rPmp1.
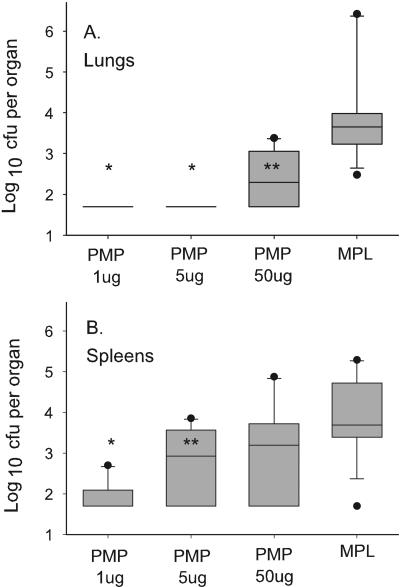
An initial study assessed protection of mice infected intraperitoneally with 310 arthroconidia (Fig. 3) following subcutaneous vaccination with rPmp1 at three doses. Lungs of mice vaccinated with 1 or 5 μg of rPmp1 exhibited mean log fungal CFU per organ approaching the lower limits of detection. Less protection was observed using 50 μg rPmp1, but reduction in CFU for all doses was significant compared to MPL-SE adjuvant alone (P < 0.01 for all). Spleens of vaccinated mice also exhibited a significant reduction in CFU compared to controls when lower doses of antigen were used (P < 0.005), but protection was lost when the immunizing dose was increased to 50 μg protein.
FIG. 3.
CFU in the spleens and lungs of immunized mice. Groups of 10 C57BL/6 mice were immunized subcutaneously with rPmp1 or MPL-SE adjuvant only and challenged intraperitoneally with 310 arthroconidia. Organs were quantitatively cultured 2 weeks later and results were expressed as CFU per organ. A. Lungs. *, P = 0.005; **, P = 0.008 compared to adjuvant-only control. B. Spleens. *, P < 0.01; **, P = 0.053 compared to the adjuvant control.
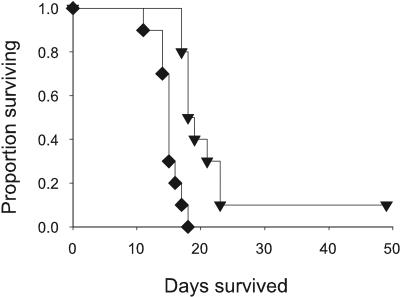
Survival following a more stringent intranasal challenge was assessed subsequent to intradermal vaccination with 1 μg rPmp1 using immunostimulatory sequences (1018 ISS) as the adjuvant. Although the change in survival following IN challenge appeared modest, it was significant (P = 0.005) compared to adjuvant alone (Fig. 4).
FIG. 4.
Prolonged survival following immunization with Pmp1. Groups of 10 C57BL/6 mice were immunized intradermally with 1 μg rPmp1 or ISS adjuvant only and challenged intranasally with 81 arthroconidia. Survival was assessed for 56 days. ▾, Pmp1; ⧫, ISS adjuvant alone (P < 0.005).
DISCUSSION
We used 2D-DIGE to search for proteins more abundantly expressed in the spherule phase of C. posadasii than the mycelial phase. Forty dominant proteins identified in this study, which analyzed whole-cell extracts, were examined in silico for export signal sequences, other localization and functional motifs, and homology to mammalian proteins. Of the 32 different proteins identified in the analysis of 40 spots, none met all three postulated criteria for an optimally protective antigen: abundance and preferential expression in the spherule phase, motifs indicative of secreted or cell wall localization, and lack of similarity to host (self) antigens. Cellular localization often cannot be deduced solely by in silico analysis, and a fourth criterion of immunogenicity, functional contribution to virulence (48), must be established experimentally.
It should be noted that proteins with dual function can be localized to the cell surface (26) and that similarity to host antigens has not precluded their ability to produce protection in some models (19). For example, after these studies were completed, Urban et al. (50) showed that a thiol peroxidase from Candida albicans, having no apparent signal sequence, was found to be differentially expressed on the cell surface in the hyphal phase compared to the yeast phase. Thus, proteomic analysis as employed in these studies may provide an initial point of discovery for potential vaccine candidate antigens but would likely not supplant the need for further characterization by a variety of other approaches.
Of the proteins we identified, Pmp1 appeared to be abundantly and preferentially expressed in mature spherules. In other studies of gene expression in C. posadasii, serial analysis of gene expression showed that mRNA for this protein was abundant in 48-h spherules, although it was even more abundant in mycelium (29). This apparent discrepancy may be explained in several ways. In the present study, we analyzed 96-h spherules, which are developmentally and metabolically different from 48-h spherules (20, 24). Further, it has been demonstrated that mRNA levels do not necessarily predict protein levels for all proteins (21).
By homology, Pmp1 appears to be related to a thiol-specific antioxidant protein of S. cerevisiae (Ahp1/TSA2/YLR109w) which is reported to be an alkyl hydroperoxide reductase. These enzymes are thought to play a role in oxidative homeostasis, stress response and resistance to heavy metal toxicity (15, 41), but a specific role in C. posadasii cannot be confidently deduced based on the functions of homologs in other organisms (16). Recently, a different member of the thiol peroxidase family (TSA1) was shown to be essential for growth at 37°C in the pathogenic fungus Cryptococcus neoformans, and deletion mutants were shown to be reduced in virulence (39). A thiol peroxidase from Leishmania major, also with similarity to TSA1, has been shown to be protective (51), and a related protein from Mycobacterium paratuberculosis is antigenic in an in vitro model (40). Although lacking clear signal sequences, the Leishmania and Mycobacterium TSAs and related thiol peroxidases from Candida albicans (50), Dirofilaria immitis and Entamoeba histolytica are all secreted either to culture medium or the cell wall and are all highly antigenic (51). Our studies have not determined if Pmp1 is similarly present on the cell surface, but if so, its serological activity and immunogenicity might be more easily understood.
In addition to its abundance in spherules, Pmp1 shows homology to an Aspergillus fumigatus antigen, Asp f3, and related fungal allergens (11, 22), and this similarity encouraged us to evaluate further the immunogenicity of Pmp1.
In our studies, the group of patients with complicated coccidioidal disease had a higher mean IgG titer against Pmp1 than those without infection or with less severe disease. However, for individual patients, anti-Pmp1 antibody levels did not uniformly discriminate between uninfected persons and those with early infections as effectively as do currently available clinical serological tests (42). Although the antibody titer against one of these clinical tests, the IDCF test, is also negatively correlated to disease status (42), there was no apparent concordance between IDCF and Pmp1 titers. Our findings, therefore, demonstrate humoral responses in patients to Pmp1 after naturally acquired infection but do not suggest a diagnostic role for this antigen for clinical purposes.
When used as a vaccine, rPmp1 evoked significant protection in mice against both intraperitoneal and intranasal infection. The degree of protection after the more stringent intranasal infection was modest compared to other coccidioidal vaccine antigens reported previously (13, 28, 34, 46). Protection was diminished following high-dose immunization (50 μg) with rPmp1compared to an intermediate dose (1 to 2 μg), a finding similar to the “inverted U” dose-response pattern described by Ibrahim et al. when immunizing against Candida albicans with a single recombinant antigen, and may be typical of immunogens which elicit cell-mediated immunity (25). Nonetheless, the demonstration of significant protection by rPmp1 corroborates the utility of the strategy employed in these studies. Our findings suggest that differential proteomic analysis coupled with cell fractionation techniques and augmented by transcriptional analysis is a promising approach to identify protective antigens for vaccine candidates against Coccidioides spp. and other complex pathogens in future and more comprehensive studies.
Acknowledgments
This work was supported in part by the U.S. Office of Veteran's Affairs, NIH-NAIAD 1PO1AI061310, and the California Health Care Foundation.
We thank Leah Bennett of the Proteomics Analysis Laboratory, Bio5 Institute, for excellent technical assistance, and Susan Miller of the Biotechnology Computing Facility, Arizona Research Labs, for developing scripts for bioinformatic analysis.
Editor: A. Casadevall
REFERENCES
- 1.Abuodeh, R. O., L. F. Shubitz, E. Siegel, S. Snyder, T. Peng, K. I. Orsborn, E. Brummer, D. A. Stevens, and J. N. Galgiani. 1999. Resistance to Coccidioides immitis in mice after immunization with recombinant protein or a DNA vaccine of a proline-rich antigen. Infect. Immun. 67:2935-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida, S. R., C. S. Unterkircher, and Z. P. Camargo. 1998. Involvement of the major glycoprotein (gp43) of Paracoccidioides brasiliensis in attachment to macrophages. Med. Mycol. 36:405-411. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ampel, N. M., L. A. Kramer, L. Li, D. S. Carroll, K. M. Kerekes, S. M. Johnson, and D. Pappagianis. 2002. In vitro whole-blood analysis of cellular immunity in patients with active coccidioidomycosis by using the antigen preparation T27K. Clin. Diagn. Lab Immunol. 9:1039-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breci, L., E. Hattrup, M. Keeler, J. Letarte, R. Johnson, and P. A. Haynes. 2005. Comprehensive proteomics in yeast using chromatographic fractionation, gas phase fractionation, protein gel electrophoresis, and isoelectric focusing. Proteomics 5:2018-2028. [DOI] [PubMed] [Google Scholar]
- 6.Cole, G. T., J. M. Xue, C. N. Okeke, E. J. Tarcha, V. Basrur, R. A. Schaller, R. A. Herr, J. J. Yu, and C. Y. Hung. 2004. A vaccine against coccidioidomycosis is justified and attainable. Med. Mycol. 42:189-216. [DOI] [PubMed] [Google Scholar]
- 7.Converse, J. L. 1959. Nutrition of the parasitic phase of Coccidioides immitis in a chemically defined liquid medium. J. Bacteriol. 78:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Converse, J. L. 1956. Effect of physicochemical environment of spherulation of Coccidioides immitis in a chemically defined medium. J. Bacteriol. 72:784-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corry, D. B., N. M. Ampel, L. Christian, R. M. Locksley, and J. N. Galgiani. 1996. Cytokine production by peripheral blood mononuclear cells in human coccidioidomycosis. J. Infect. Dis. 174:440-443. [DOI] [PubMed] [Google Scholar]
- 10.Cox, R. A., and D. M. Magee. 2004. Coccidioidomycosis: host response and vaccine development. Clin. Microbiol. Rev. 17:804-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crameri, R. 1998. Recombinant Aspergillus fumigatus allergens: from the nucleotide sequences to clinical applications. Int. Arch. Allergy Immunol. 115:99-114. [DOI] [PubMed] [Google Scholar]
- 12.Delgado, N., C. Y. Hung, E. Tarcha, M. J. Gardner, and G. T. Cole. 2004. Profiling gene expression in Coccidioides posadasii. Med. Mycol. 42:59-71. [DOI] [PubMed] [Google Scholar]
- 13.Delgado, N., J. Xue, J. J. Yu, C. Y. Hung, and G. T. Cole. 2003. A recombinant β-1,3-glucanosyltransferase homolog of Coccidioides posadasii protects mice against coccidioidomycosis. Infect. Immun. 71:3010-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dugger, K. O., K. M. Villareal, A. Ngyuen, C. R. Zimmermann, J. H. Law, and J. N. Galgiani. 1996. Cloning and sequence analysis of the cDNA for a protein from Coccidioides immitis with immunogenic potential. Biochem. Biophys. Res. Commun. 218:485-489. [DOI] [PubMed] [Google Scholar]
- 15.Farcasanu, I. C., D. Hirata, E. Tsuchiya, K. Mizuta, and T. Miyakawa. 1999. Involvement of thioredoxin peroxidase type II (Ahp1p) of Saccharomyces cerevisiae in Mn2+ homeostasis. Biosci. Biotechnol. Biochem. 63:1871-1881. [DOI] [PubMed] [Google Scholar]
- 16.Flohe, L., T. Jaeger, S. Pilawa, and H. Sztajer. 2003. Thiol-dependent peroxidases care little about homology-based assignments of function. Redox Rep. 8:256-264. [DOI] [PubMed] [Google Scholar]
- 17.Galgiani, J. N. 1993. Coccidioidomycosis. West. J. Med. 159:153-171. [PMC free article] [PubMed] [Google Scholar]
- 18.Galgiani, J. N., N. M. Ampel, A. Catanzaro, R. H. Johnson, D. A. Stevens, and P. L. Williams. 2000. Practice guideline for the treatment of coccidioidomycosis. Infectious Diseases Society of America. Clin. Infect. Dis. 30:658-661. [DOI] [PubMed] [Google Scholar]
- 19.Gomez, F. J., R. Allendoerfer, and G. S. Deepe, Jr. 1995. Vaccination with recombinant heat shock protein 60 from Histoplasma capsulatum protects mice against pulmonary histoplasmosis. Infect. Immun. 63:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guevara-Olvera, L., C. Y. Hung, J. J. Yu, and G. T. Cole. 2000. Sequence, expression and functional analysis of the Coccidioides immitis ODC (ornithine decarboxylase) gene. Gene 242:437-448. [DOI] [PubMed] [Google Scholar]
- 21.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemmann, S., K. Blaser, and R. Crameri. 1997. Allergens of Aspergillus fumigatus and Candida boidinii share IgE-binding epitopes. Am. J. Respir. Crit. Care Med. 156:1956-1962. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz, M. A., B. W. Lee, B. J. Dillon, and G. Harth. 1995. Protective immunity against tuberculosis induced by vaccination with major extracellular proteins of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 92:1530-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huppert, M., S. H. Sun, and J. L. Harrison. 1982. Morphogenesis throughout saprobic and parasitic cycles of Coccidioides immitis. Mycopathologia 78:107-122. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahim, A. S., B. J. Spellberg, V. Avenissian, Y. Fu, S. G. Filler, and J. E. Edwards, Jr. 2005. Vaccination with recombinant N-terminal domain of Als1p improves survival during murine disseminated candidiasis by enhancing cell-mediated, not humoral, immunity. Infect. Immun. 73:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeffery, C. J. 2003. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 19:415-417. [DOI] [PubMed] [Google Scholar]
- 27.Jiang, C., D. M. Magee, and R. A. Cox. 1999. Coadministration of interleukin-12 expression vector with antigen 2 cDNA enhances induction of protective immunity against Coccidioides immitis. Infect. Immun. 67:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, C., D. M. Magee, F. D. Ivey, and R. A. Cox. 2002. Role of signal sequence in vaccine-induced protection against experimental coccidioidomycosis. Infect. Immun. 70:3539-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellner, E. M., M. A. Mandel, J. N. Galgiani, and M. J. Orbach. 2005. Serial analysis of gene expression in Coccidioides posadasii, abstr. 171. Abstr. 23rd Fungal Genet. Conf.
- 30.Kirkland, T. N., P. W. Thomas, F. Finley, and G. T. Cole. 1998. Immunogenicity of a 48-kilodalton recombinant T-cell-reactive protein of Coccidioides immitis. Infect. Immun. 66:424-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein, B. S. 1997. Role of cell surface molecules of Blastomyces dermatitidis in the pathogenesis and immunobiology of blastomycosis. Semin. Respir. Infect. 12:198-205. [PubMed] [Google Scholar]
- 32.Knudsen, G. M., K. F. Medzihradszky, K. C. Lim, E. Hansell, and J. H. McKerrow. 2005. Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol. Cell Proteomics 4:1862-1875. [DOI] [PubMed]
- 33.Kong, Y. C., H. B. Levine, and C. E. Smith. 1963. Immunogenic properties of nondisrupted and disrupted spherules of Coccidioides immitis in mice. Sabouraudia 2:131-142. [PubMed] [Google Scholar]
- 34.Li, K., J. J. Yu, C. Y. Hung, P. F. Lehmann, and G. T. Cole. 2001. Recombinant urease and urease DNA of Coccidioides immitis elicit an immunoprotective response against coccidioidomycosis in mice. Infect. Immun. 69:2878-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long, K. H., F. J. Gomez, R. E. Morris, and S. L. Newman. 2003. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J. Immunol. 170:487-494. [DOI] [PubMed] [Google Scholar]
- 36.Mackey, A. J., T. A. J. Haystead, and W. R. Pearson. 2002. Getting more from less: algorithms for rapid protein identification with multiple short peptide sequences. Mol. Cell Proteomics 1:139-147. [DOI] [PubMed] [Google Scholar]
- 37.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medina, M. L., P. A. Haynes, L. Breci, and W. A. Francisco. 2005. Analysis of secreted proteins from Aspergillus flavus. Proteomics 5:3153-3161. [DOI] [PubMed] [Google Scholar]
- 39.Missall, T. A., M. E. Pusateri, and J. K. Lodge. 2004. Thiol peroxidase is critical for virulence and resistance to nitric oxide and peroxide in the fungal pathogen, Cryptococcus neoformans. Mol. Microbiol. 51:1447-1458. [DOI] [PubMed] [Google Scholar]
- 40.Mullerad, J., A. H. Hovav, R. Nahary, Y. Fishman, and H. Bercovier. 2003. Immunogenicity of a 16.7 kDa Mycobacterium paratuberculosis antigen. Microb. Pathog. 34:81-90. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen-nhu, N., and B. Knoops. 2002. Alkyl hydroperoxide reductase 1 protects Saccharomyces cerevisiae against metal ion toxicity and glutathione depletion. Toxicol. Lett. 135:219-228. [DOI] [PubMed] [Google Scholar]
- 42.Pappagianis, D., and B. L. Zimmer. 1990. Serology of coccidioidomycosis. Clin. Microbiol. Rev. 3:247-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park, S. G., M. K. Cha, W. Jeong, and I. H. Kim. 2000. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 275:5723-5732. [DOI] [PubMed] [Google Scholar]
- 44.Rhee, S. G., S. W. Kang, T. S. Chang, W. Jeong, and K. Kim. 2001. Peroxiredoxin, a novel family of peroxidases. IUBMB Life 52:35-41. [DOI] [PubMed] [Google Scholar]
- 45.Shen, H. D., C. W. Wang, H. Chou, W. L. Lin, M. F. Tam, M. H. Huang, M. L. Kuo, S. R. Wang, and S. H. Han. 2000. Complementary DNA cloning and immunologic characterization of a new Penicillium citrinum allergen (Pen c3). J Allergy Clin. Immunol. 105:827-833. [DOI] [PubMed] [Google Scholar]
- 46.Shubitz, L., T. Peng, R. Perrill, J. Simons, K. Orsborn, and J. N. Galgiani. 2002. Protection of mice against Coccidioides immitis intranasal infection by vaccination with recombinant antigen 2/PRA. Infect. Immun. 70:3287-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, S. H., and M. Huppert. 1976. A cytological study of morphogenesis in Coccidioides immitis. Sabouraudia 14:185-198. [PubMed] [Google Scholar]
- 48.Tavares, D., P. Ferreira, and M. Arala-Chaves. 2003. Increased resistance in BALB/c mice to reinfection with Candida albicans is due to immunoneutralization of a virulence-associated immunomodulatory protein. Microbiology 149:333-339. [DOI] [PubMed] [Google Scholar]
- 49.Tonui, W. K., J. S. Mejia, L. Hochberg, M. L. Mbow, J. R. Ryan, A. S. Chan, S. K. Martin, and R. G. Titus. 2004. Immunization with Leishmania major exogenous antigens protects susceptible BALB/c mice against challenge infection with L. major. Infect. Immun. 72:5654-5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urban, C., X. Xiong, K. Sohn, K. Schroppel, H. Brunner, and S. Rupp. 2005. The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans. Mol. Microbiol. 57:1318-1341. [DOI] [PubMed] [Google Scholar]
- 51.Webb, J. R., A. Campos-Neto, P. J. Ovendale, T. I. Martin, E. J. Stromberg, R. Badaro, and S. G. Reed. 1998. Human and murine immune responses to a novel Leishmania major recombinant protein encoded by members of a multicopy gene family. Infect. Immun. 66:3279-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wieden, M. A., J. N. Galgiani, and D. Pappagianis. 1983. Comparison of immunodiffusion techniques with standard complement fixation assay for quantitation of coccidioidal antibodies. J. Clin. Microbiol. 18:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wieden, M. A., L. L. Lundergan, J. Blum, K. L. Delgado, R. Coolbaugh, R. Howard, T. Peng, E. Pugh, N. Reis, J. Theis, and J. N. Galgiani. 1996. Detection of coccidioidal antibodies by 33-kDa spherule antigen, Coccidioides EIA, and standard serologic tests in sera from patients evaluated for coccidioidomycosis. J. Infect. Dis. 173:1273-1277. [DOI] [PubMed] [Google Scholar]
- 54.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 55.Yates, J. R., III, J. K. Eng, A. L. McCormack, and D. Schieltz. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67:1426-1436. [DOI] [PubMed] [Google Scholar]
- 56.Zimmermann, C. R., S. M. Johnson, G. W. Martens, A. G. White, B. L. Zimmer, and D. Pappagianis. 1998. Protection against lethal murine coccidioidomycosis by a soluble vaccine from spherules. Infect. Immun. 66:2342-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]