Volume 70, no. 11, p. 6444-6447, 2002. Page 6444: The title should appear as shown above.
Pages 6444-6447: We had previously reported the characterization of 12 mutagenized CHO-K1 cell lines that were resistant to chlamydial infectivity by Chlamydia trachomatis and C. pneumoniae. We recently determined that the parental CHO-K1 cell line was resistant to infection by C. pneumoniae, leading us to question our published data for CHO-K1 and its derived mutants. It was determined that the strain used in this study was not C. pneumoniae but that the tube was mislabeled and contained C. psittaci. The strain used in these experiments was determined by sequencing of the ompA gene to be C. psittaci (6BC-type) strain (6BCPF). To validate the retrospective identification of the strain used in these studies as C. psittaci, the results were repeated with concurrently sequence-verified C. psittaci. The data published are accurate for each cell line, but the strain designation was not correct (see corrected Fig. 1 on following page). The use of C. psittaci rather than C. pneumoniae does not fundamentally change the conclusions of the study, as the purpose of using C. pneumoniae was to employ a strain that was phylogenetically distant from C. trachomatis, thereby permitting broad characterization of the mutant cell phenotypes. The use of C. psittaci fulfills this aim by demonstrating the spectrum of attachment and uptake deficiencies and supports the key conclusion that attachment and uptake involve separate molecular mechanisms specific for host cells.
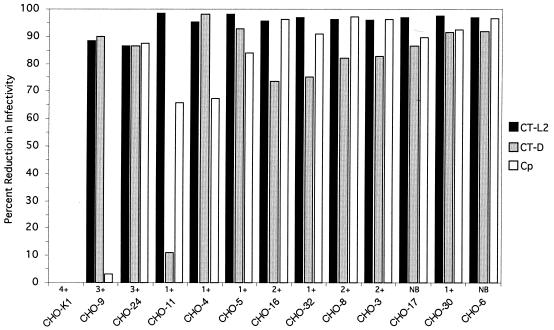
FIG. 1.
Parallel assessments of infectivity of C. trachomatis serovar L2 (CT-L2), C. trachomatis serovar D (CT-D), and C. psittaci (Cp) for each CHO cell mutant. Infectivity for monolayers of each cell line was tested in triplicate for each chlamydial strain. At approximately 48 h postinfection, chlamydial inclusions were detected by immunofluorescence and enumerated by counting inclusions. Similar data were obtained by using sequence-confirmed C. psittaci 6BCPF. Qualitative estimates of attachment for C. trachomatis serovar L2 are indicated as no binding (NB) by a scale of 1+ to 4+ when cell-associated binding was detected, where 1+ represents approximately 10 elementary bodies apparently associated with cell surfaces and 4+ represents many elementary bodies bound with entire staining of the cells.
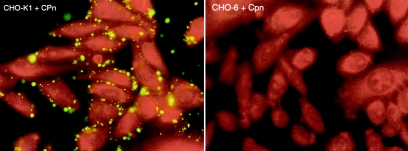
Although infectivity of the mutant cell line by C. pneumoniae cannot be addressed because of the nascent resistance to infection of the parental CHO-K1 line, we discovered that sequence-confirmed C. pneumoniae attached to CHO-K1 cells. CHO-6, a mutant cell line resistant to attachment by C. trachomatis and C. psittaci, was tested and was found to be resistant to attachment by C. pneumoniae (corrected Fig. 2 on following page). Other mutagenized CHO cell lines that were determined to have an attachment defect, such as CHO-17, CHO-30, and CHO-5, were tested and also found to be resistant to attachment by C. pneumoniae (data not shown). We can conclude that the phenotypes of these mutagenized cell lines reflect at least one cellular component required in common for attachment of C. trachomatis, C. psittaci, and C. pneumoniae that is separate from postadherence processes ultimately leading to productive infection.
FIG. 2.
Immunofluorescence staining of C. pneumoniae (CWL029) elementary bodies attached to CHO-K1 and CHO-6. Elementary bodies were added to monolayers, incubated for 1 h at 4°C, fixed with methanol, and stained with a C. pneumoniae-specific monoclonal antibody conjugated with fluorescein isothiocyanate.
Continued on following page