Abstract
It is thought that Trypanosoma cruzi, the protozoan that causes Chagas' disease, modulates the extracellular matrix network to facilitate infection of human cells. However, direct evidence to document this phenomenon is lacking. Here we show that the T. cruzi gp83 ligand, a cell surface trans-sialidase-like molecule that the parasite uses to attach to host cells, increases the level of laminin γ-1 transcript and its expression in mammalian cells, leading to an increase in cellular infection. Stable RNA interference (RNAi) with host cell laminin γ-1 knocks down the levels of laminin γ-1 transcript and protein expression in mammalian cells, causing a dramatic reduction in cellular infection by T. cruzi. Thus, host laminin γ-1, which is regulated by the parasite, plays a crucial role in the early process of infection. This is the first report showing that knocking down the expression of a human gene by RNAi inhibits the infection of an intracellular parasite.
Trypanosoma cruzi, the causative agent of Chagas' disease, is an obligate intracellular parasite that infects several cell types in the body, including those of the cardiac, muscular, nervous, and digestive systems. During infection, invasive trypomastigotes must navigate through the extracellular matrix (ECM) before they can attach to and invade host cells. Chagas' disease causes significant morbidity and mortality in the Americas, yet it remains incurable. Chagas' disease is now viewed as an emerging illness associated with human immunodeficiency virus type 1 infection, where it induces dramatic brain pathology and early death (14).
Oligonucleotide and cDNA microarrays have emerged as indispensable research tools for studying global gene expression profiles in cells (3), including studies of host cell-microbial interactions (5). RNA interference (RNAi) has recently been used to reduce the expression of host genes involved in human immunodeficiency virus type 1 infection (13).
T. cruzi surface trans-sialidases and trans-sialidase-like molecules play roles in the process of infection (6). One of the T. cruzi trans-sialidase-like molecules is surface gp83, which the parasite uses as a ligand to attach to phagocytic and nonphagocytic cells in order to promote entry (19). gp83 binds to cells in a ligand-receptor interaction manner, and neutralizing antibodies to gp83 block both the binding of gp83 to mammalian cells and the infection of these cells in vitro and in vivo (19). gp83 is glycosylphosphatidylinositol anchored to the membrane, is expressed only in infective trypomastigotes, and is expressed more in highly than in weakly infective trypomastigote clones (10). This molecule is released from the invasive trypanosome by cleavage via the trypanosome phospholipase C, whereby it signals human cells to enhance entry by activating protein kinase C (18) and the mitogen-activated protein kinase pathway (17).
Previous studies have shown that parasite molecules bind to immobilized laminin (7) and that human galectin-3 enhances this interaction (12), suggesting that the trypanosome interacts with laminin. Since nearly all of the cells that T. cruzi infects are surrounded by basement membranes, of which several laminin isoforms are the major constituent, the ability of the parasite to effectively regulate and interact with laminin is critically important for its passage through the membrane barrier. Laminin γ-1 is the most abundant isoform of laminin in humans (15). In this study, we tested the hypothesis that T. cruzi gp83 binds to human cells to regulate the expression of laminin γ-1, which is required for T. cruzi infection. Our findings indicate that preexposure of T. cruzi gp83 to human coronary artery smooth cells up-regulates T. cruzi infection and the expression of laminin γ-1 and that knocking down the expression of laminin γ-1 by RNAi dramatically reduces T. cruzi infection of human cells.
MATERIALS AND METHODS
Parasite cultures.
The highly infective trypomastigote clone MMC 20A, derived from the Tulahuen strain of T. cruzi (10), was used. Pure culture trypomastigotes were obtained from the supernatant of heart myoblast monolayers as previously described (10).
Infection assays.
Parasite binding to human cells was evaluated at 2 h by fluorescence microscopy with fluorescein isothiocyanate (FITC)-labeled antibodies to a trypomastigote surface protein and 4′,6′-diamidino-2-phenylindole (DAPI) (9). The number of bound, FITC-fluorescent parasites per 200 cells was determined. Parasite entry was evaluated at the same 2-h time point. The number of internalized parasites was obtained by subtracting the number of bound, FITC-fluorescent parasites from the total number of DAPI-stained kinetoplast DNA parasites per 200 host cells. Parasite multiplication within cells was evaluated at 72 h by standardized procedures (10). Infection assays with primary human coronary artery smooth muscle (HCASM) cells and HeLa cells were done in triplicate, and experiments were repeated three times.
T. cruzi r-gp83 ligand.
The gp83 ligand gene (GenBank accession no. AY513728) was expressed in Escherichia coli as a six-His-tagged protein and purified over a Ni-nitrilotriacetic acid column. The amino acid sequence of gp83 does not have tyrosine 120 and proline 284, which are required for trans-sialidase activity (4). The protein was further purified by fast protein liquid chromatography and passed through a Detoxi-Gel endotoxin-removing gel column (Pierce, Rockford, IL) as previously described (17). A highly purified preparation of endotoxin-free recombinant gp83 (r-gp83) was used in all experiments. Endotoxin levels were assayed with the kinetic-QCL kit for kinetic chromogenic assays (Cambrex, Walkersville, MD) as previously described in detail (17). HCASM cells or HeLa cells were preexposed to the gp83 ligand for 30 min, and then cells were washed with Dulbecco modified Eagle medium and exposed to trypomastigotes to evaluate entry.
cDNA microarrays and gene analysis.
Monolayers of primary HCASM cells (Cambrex, Walkersville, MD) were washed with smooth muscle cell basal medium (SmBM) from Cambrex and incubated with either 0.4-μg/ml r-gp83 in SmBM or with SmBM alone for 30, 60, 120, or 180 min at 37°C. Monolayers were solubilized with TRIzol (Invitrogen, Grand Island, NY). Purified total RNA was treated with RNase-free DNase, and mRNA was purified over an Oligotex column (QIAGEN, Valencia, CA) and analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Hybridizations were carried out with 10K human cDNA microarrays prepared on glass slides by the Vanderbilt Microarray Shared Resource with clones from the Research Genetics Sequence Verified Library (ResGen Division of Invitrogen Corp., Carlsbad, CA). This array contains clones from the IMAGE consortium collection with significant number of ECM cDNAs. mRNA (2 μg) was reverse transcribed and labeled with Cy3 or Cy5 dye in 50 mM Tris (pH 8.3)-75 mM KCl-15 mM MgCl2 with 10 mM dithiothreitol; 200 μM each dATP, dGTP, and dTTP; 60 μM dCTP; and 60 μM Cy-dye dCTP. Labeled RNA extracts were hydrolyzed with 0.5 M NaOH-0.25 M EDTA, neutralized with HCl, and purified over QIAquick PCR purification columns (QIAGEN, Valencia, CA). Glass slides were rinsed in 0.25% sodium dodecyl sulfate (SDS), prehybridized in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS-0.1% bovine serum albumin, and hybridized with the Cy-dye-labeled extracts for 14 to 16 h at 42°C. The slides were then washed in 2× SSC containing 0.1% SDS, in 1× SSC, and lastly in 0.1× SSC. The slides were dried and scanned with an Axon Genepix 4000 scanner. Changes in gene expression profiles after incubation with r-gp83 were analyzed with GeneSpring (Agilent Technologies, Palo Alto, CA). Two independent experiments were performed under the same conditions. Changes were considered significant if the P value was <0.01.
Quantitation of host gene expression by real-time PCR.
First-strand cDNA was synthesized from 5 μg of RNA with SuperScript II as described by the manufacturer (Invitrogen, Grand Island, NY). The cDNAs were used for real-time PCR experiments to determine the transcript level with an iCycler (Bio-Rad, Richmond, CA). The SYBR green reaction mixture (25 μl) was set up as described by the manufacturer (Bio-Rad, Richmond, CA). The cycling conditions were 95°C for 3 min and 40 cycles of 94°C for 30 s and 56°C for 45 s. Melting curve analysis was performed as follows. PCR products were heated to 95°C for 1 min and cooled down to 55°C for 1 min, and then the temperature was steadily increased, at 0.4°C/0.08 s, 100 times with continuous fluorescence readings. The cycling protocol for human 18S RNA was the same but with an annealing temperature of 60°C. The melting curve was similarly determined. The quantity of human 18S RNA transcript in each sample was used to normalize the amount of transcript. The primers used for quantitative real-time PCR were designed with the Beacon Designer 4.0 program (Premier Biosoft, Palo Alto, CA). The primer sequences were 5′-CTCCATCAACCTCACGCTG-3′ (sense) and 5′-CGG CTGGTGTGGAACTTG-3′ (antisense) for laminin γ-1 and 5′-CGGACAGGA TTGACAGATTGATAGC-3′ (sense) and 5′-TGCCAGAGTCTCGTTCGTTAT CG-3′ (antisense) for human 18S RNA.
Generation of short hairpin RNA (shRNA) constructs and transfections.
The Oligonucleotide Retriever program of Cold Spring Harbor Laboratory (http://www.cshl.org/public/SCIENCE/hannon.html/) was used to generate sequences that code for shRNA of laminin γ-1 (GenBank accession no. J03202).
The oligonucleotide ends were modified and synthesized (IDT, Coralville, IA). The synthetic oligonucleotides were annealed and directionally ligated into the XbaI and SalI sites of the IMG-800 vector (Imgenex, San Diego, CA) and used to transform competent DH5α cells (Invitrogen, Grand Island, NY). Positive E. coli colonies were grown, and plasmid DNA was purified with an EndoFree plasmid purification kit (QIAGEN, Valencia, CA). The sequences of the cloned oligonucleotides were verified by automated DNA sequencing. HeLa cells were used for stable transfections because of the lack of RNAi vectors to efficiently transfect primary cell lines. Cells were transfected with 5 μg of plasmid in OPTI-MEM (Invitrogen, Grand Island, NY) with a lipid-based transfection reagent and selected with G418 (600 μg/ml) as previously described (Imgenex, San Diego, CA).
Immunoblotting.
Cell monolayers transfected either with the vector alone or with the vector containing the RNAi oligonucleotides were solubilized in 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) containing protease inhibitors (GE Healthcare, Piscataway, NJ). The same protein concentrations of solubilized samples were separated by SDS-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, probed with goat immunoglobulin G (IgG) to laminin γ-1, and developed with peroxidase-conjugated mouse anti-goat IgG by enhanced-chemiluminescence (GE Healthcare, Piscataway, NJ) as previously described (1). Blots were stripped and reprobed with a monoclonal IgG to β-actin.
Presentation of results and statistical analysis.
The results obtained in this work were from triplicate determinations and represent three independent experiments performed by identical methods. The results are expressed as the mean ± 1 standard deviation. For cDNA microarrays, two independent experiments were performed under the same conditions and the data are presented as the mean ± 1 standard deviation. Differences were considered to be statistically significant if the P value was <0.05 by the Student t test.
RESULTS
T. cruzi trypomastigote gp83 ligand up-regulates trypanosome entry into HCASM cells and HeLa cells.
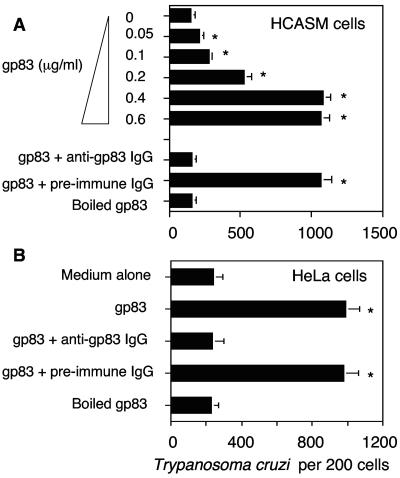
To investigate whether the T. cruzi gp83 ligand regulates the infection of human cells, primary HCASM cells were preincubated with increasing concentrations of soluble recombinant gp83 (0 to 0.6 μg/ml), followed by exposure to invasive trypomastigotes. Our results show that the gp83 ligand induced a significant increase in the number of trypanosomes internalized by HCASM cells in a concentration-dependent and saturable manner (Fig. 1A). The maximum enhancement and saturation were observed at 0.4-μg/ml gp83, where the number of parasites per 200 cells was five times higher than in mock-treated cells. The effect was abolished when the gp83 was denatured or by preincubation with anti-gp83 IgG but not with preimmune IgG. Preincubation of gp83 (0.4 μg/ml) with HeLa cells under similar conditions also up-regulated T. cruzi entry (Fig. 1B). Similarly, the effect was abolished when the gp83 was denatured or preincubated with anti-gp83 IgG but not when it was preincubated with preimmune IgG.
FIG. 1.
T. cruzi trypomastigote gp83 ligand up-regulates trypanosome entry into human cells. (A) gp83 ligand up-regulates T. cruzi entry into HCASM cells. HCASM cells were preincubated with increasing concentrations of gp83, gp83 preincubated with antibodies, or boiled gp83 for 30 min, washed, and exposed to trypomastigotes at a ratio of 10 parasites per cell for 2 h, and the number of internalized trypanosomes was determined. (B) gp83 ligand up-regulates T. cruzi entry into HeLa cells. HeLa cells were preincubated with gp83 (0.4 μg/ml) under the same conditions as in panel A, and entry was evaluated similarly. Bars represent the mean results ± 1 standard deviation obtained with triplicate samples in one representative experiment selected from three experiments with similar results. *, significant difference compared to control values (P < 0.05).
Exposure of T. cruzi gp83 ligand to HCASM cells up-regulates the expression of laminin γ-1 as analyzed by cDNA microarrays and real-time PCR.
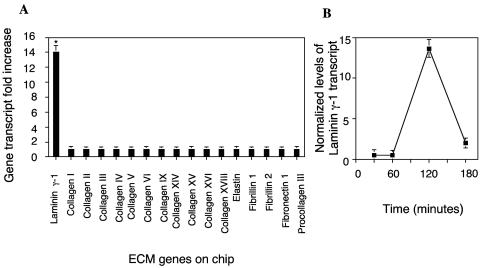
Since extracellular parasites must cross the ECM and interact with ECM proteins in order to attach to and invade host cells, we explored the possibility that T. cruzi gp83 modulates the ECM gene expression profile in HCASM cells. Following incubation of HCASM cells with r-gp83, we used cDNA microarrays to evaluate the changes in the expression profile of ECM genes by GeneSpring software analysis. Figure 2A shows that the laminin γ-1 transcript increased 14-fold at 120 min compared to the transcript levels of other ECM genes on the chip (collagens I, II, III, IV, V, VI, IX, XIV, XV, XVI, and XVIII; elastin; fibrillin 1; fibrillin 2; fibronectin 1; and procollagen III), which did not statistically significantly change. To validate the increase in the laminin γ-1 transcript level on the chips, we evaluated the laminin γ-1 transcript kinetics in HCASM cells exposed to the same concentration of gp83 by real-time PCR. The laminin γ-1 transcript was found to increase to a maximum of 14-fold at 120 min and to return to a minimum increase of 2.5-fold at 180 min of exposure of HCASM cells to r-gp83 (Fig. 2B). Real-time PCR also confirmed the lack of change in other ECM genes on the chips. These results indicate that exposure of HCASM cells to r-gp83 up-regulates the laminin γ-1 transcript. We also found that exposure of HeLa cells to gp83 up-regulates the expression of the laminin γ-1 transcript up to 2.2 times ± 0.1 time (P < 0.05) compared to the level of the laminin γ-1 transcript in mock-treated HeLa cells, as evidenced by real-time PCR. Moreover, exposure of either HCASM cells or HeLa cells to T. cruzi at a ratio of 10 parasites per cell for 2 h increased the levels of the laminin γ-1 transcript up to 5.2 ± 0.2 (P < 0.05) in HCASM cells and up to 2.7 ± 0.4 (P < 0.05) in HeLa cells. These results were obtained from duplicate microarray experiments and confirmed by real-time PCR under the conditions described above.
FIG. 2.
Exposure of T. cruzi gp83 ligand to HCASM cells up-regulates the expression of laminin γ-1. (A) Exposure of gp83 to HCASM cells up-regulates the expression of laminin γ-1 as determined by cDNA microarray analysis. RNA from HCASM cells exposed or not exposed to gp83 was reverse transcribed to labeled cDNA and hybridized onto cDNA microarray slides. Duplicate hybridizations were normalized against their respective controls and analyzed with GeneSpring Software. Bars represent the mean ± 1 standard deviation of ECM gene expression of duplicate experiments with similar results. *, significant difference between laminin γ-1 gene expression and the expression of the other ECM genes (P < 0.01). (B) Kinetics of laminin γ-1 transcript upon exposure of gp83 to HCASM cells. RNA from cells exposed or not exposed to gp83 was reversed transcribed to cDNA, and the laminin γ-1 transcript levels were evaluated by real-time-PCR. Data are the mean ± 1 standard deviation of normalized levels of laminin γ-1 transcript against human 18S RNA in one representative experiment of three independent experiments performed in triplicate with similar results. Each point is the mean of results for triplicate samples in one representative experiment (±1 standard deviation). The P value was <0.05 for points at 120 and 180 min with respect to points at 0 and 60 min.
RNAi with laminin γ-1 knocks down laminin γ-1 expression and blocks T. cruzi infection.
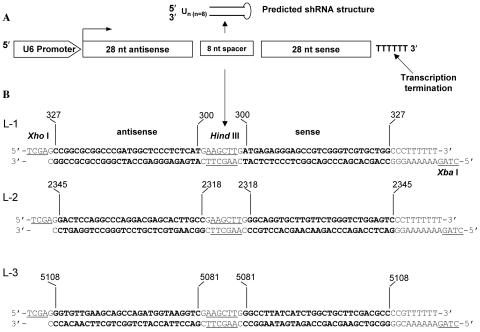
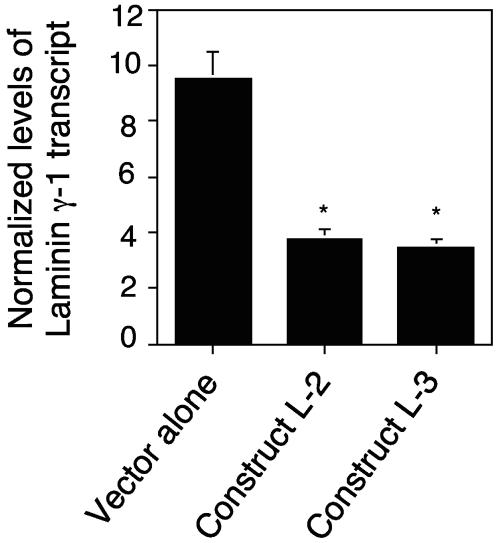
The fact that the T. cruzi ligand up-regulates the expression of the laminin γ-1 transcript prompted us to determine the function of the laminin γ-1 gene in the process of T. cruzi infection of human cells with stable RNAi assays. Accordingly, we investigated whether shRNA targeted to laminin γ-1 could knock down its transcript. Three short nucleotide sequences derived from the laminin γ-1 coding sequence were cloned into the IMG-800 vector (Imgenex, San Diego, CA). The sequences were from nucleotides 300 to 327 (L1), 2318 to 2345 (L2), and 5081 to 5108 (L3). Figure 3A shows a schematic representation of the predicted shRNA structure. We cloned oligonucleotide sequences behind a strong U6 promoter, which constitutively drives the expression of shRNAs. Figure 3B illustrates the constructs designed for the generation of hairpins targeted to the laminin γ-1 gene in HeLa cells. The oligonucleotide ends were modified to facilitate directional cloning into the SalI and XbaI sites of the IMG-800 vector. The HindIII sites separating antisense and sense sequences create the loop in the final shRNA. The morphology and growth rate of human cells stably transfected with the cloned constructs and selected with neomycin remained unchanged following several passages. With the aid of quantitative real-time PCR, we determined the level of the laminin γ-1 transcript in the transfected cells. We observed that two of the three constructs significantly decreased the laminin γ-1 gene transcript level by 60% (L-2) and 65% (L-3), respectively, compared to the vector alone (Fig. 4), indicating that expression of two of the shRNAs was effective in knocking down the laminin γ-1 transcript. The efficiencies of the L-2 and L-3 constructs in knocking down the laminin γ-1 transcript were similar.
FIG. 3.
Laminin γ-1 RNAi constructs used for generating shRNA in HeLa cells. (A) Schematic representation of the predicted shRNA structure. The orientation of the oligonucleotide sequences cloned behind a strong U6 promoter is shown. (B) Design of oligonucleotide constructs from laminin γ-1 cDNA to generate shRNA. The 5′ and 3′ termini underlined show the modification for cloning into the IMG-800 vector. Numbers on oligonucleotides show the locations on the laminin γ-1 cDNA.
FIG. 4.
RNAi with the laminin γ-1 gene reduces laminin γ-1 transcript levels in HeLa cells. RNA from HeLa cells stably transfected with either the IMG-800 vector alone or containing construct L-2 or L-3 was reversed transcribed to cDNA, and the laminin γ-1 transcript level was evaluated by real-time PCR. The results were normalized against human 18S RNA. Bars represent the means of results from triplicate samples in one representative experiment (±1 standard deviation) selected from three independent experiments with similar results. *, significant difference compared to control value, cells transfected with the vector alone (P < 0.05).
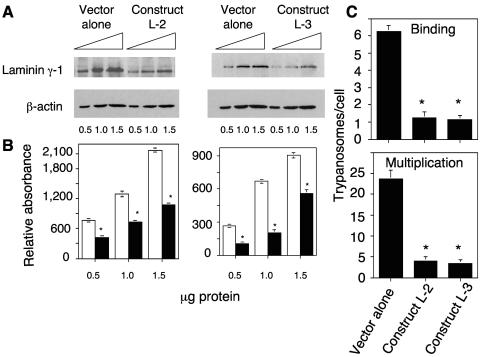
We then determined whether the reduction of the laminin γ-1 gene transcript by RNAi was reflected at the protein level by quantitative immunoblotting analysis. Our results indicate that human cells stably transfected with either the L-2 or the L-3 construct had significantly reduced expression of laminin γ-1 compared to cells transfected with the vector alone (Fig. 5A). Determination of the relative absorbance of laminin γ-1 bands and β-actin bands showed that RNAi with laminin γ-1 significantly decreased the level of protein in the cells by 50% (L-2) and 60% (L-3) (Fig. 5B).
FIG. 5.
RNAi with laminin γ-1 knocks down laminin γ-1 expression and blocks T. cruzi infection of HeLa cells. (A) shRNA constructs targeted to the laminin γ-1 gene reduce the level of laminin γ-1 protein expression. Different protein concentrations of HeLa cells stably transfected with either the IMG-800 vector alone or containing construct L-2 or L-3 were probed with either anti-laminin γ-1 antibodies or β-actin antibodies in immunoblot assays. The results shown are from a representative experiment of three performed with similar results. (B) Densitometric scanning of immunoblots normalized to their corresponding β-actin absorbance as shown in panel A. The mean of three independent experiments ± 1 standard deviation is plotted. Open bars and solid bars represent normalized absorbance of laminin γ-1 bands from cells transfected with the vector alone or the chimeric vector containing constructs, respectively. *, significant difference compared to control values, cells transfected with the vector alone (P < 0.05). (C) RNAi with laminin γ-1 significantly reduces trypanosome infection. HeLa cells stably transfected with the IMG-800 vector alone or with the vector containing construct L-2 or L-3 were exposed to trypomastigotes at a ratio of 15 parasites per cell for 2 h, and trypanosome binding was evaluated (top). T. cruzi multiplication in cells transfected with IMG-800, L-2, or L-3 was evaluated at 72 h (bottom). Bars in panel C represent the means ± 1 standard deviation of results from triplicate samples in one representative experiment (±1 standard deviation) selected from three experiments with similar results. *, significant difference compared to control values, cells transfected with the vector alone (P < 0.05).
To determine the function of the laminin γ-1 gene in the process of T. cruzi infection, human cells with the new phenotype were challenged with invasive trypomastigotes. We found that RNAi with laminin γ-1 dramatically reduced the number of trypomastigotes attached to human cells stably transfected with construct L-2 or L-3 compared to cells transfected with the vector alone. Human cells harboring effective shRNA constructs had, on average, one parasite bound per cell, compared to mock-transfected cells, which showed six parasites bound per cell after 2 h of exposure (Fig. 5C, top). Since fewer trypanosomes bound to cells stably transfected with the L2 and L3 constructs (Fig. 5C) at 2 h, fewer parasites entered and multiplied within these cells at 72 h with respect to cells transfected with the vector alone. Analysis of parasite multiplication at 72 h indicated that cells transfected with the vector alone had a higher number of parasites per cell, whereas cells transfected with constructs L-2 and L-3 had a significantly lower number of parasites per cell (Fig. 5C, bottom).
DISCUSSION
This report describes novel findings that indicate that the T. cruzi gp83 ligand, which the parasite uses to attach to host cells, increases laminin γ-1 transcript and expression levels in mammalian cells, leading to an increase in cellular infection, and that stable RNAi with host cell laminin γ-1 knocks down the laminin γ-1 transcript and protein expression levels in mammalian cells, causing a dramatic reduction in cellular infection by T. cruzi. These results indicate that host laminin γ-1, which is thereby regulated by the parasite, plays a crucial role in the early process of T. cruzi infection. The rationale behind this study was to identify what ECM proteins would be modulated by the released gp83 ligand and the function of its gene in the process of early T. cruzi infection. Whether or not gp83 interacts with the ECM is unknown.
Our findings also indicate that the trypanosome gp83 ligand induces higher T. cruzi infection of human cells. Since gp83 is a membrane glycosylphosphatidylinositol-anchored molecule that is cleaved by the trypanosome phospholipase C, this may represent a mechanism of escape in which the invasive trypomastigotes release gp83 to efficiently gain entry into human cells.
This report also indicates that RNAi with laminin γ-1 substantially reduced the transcript and encoded protein levels, rendering these human cells substantially less susceptible to T. cruzi, indicating that laminin γ-1 is required for the process of T. cruzi infection. The residual T. cruzi infection seen indicates that the parasite can infect cells through other mechanisms that are laminin γ-1 independent. It is known that laminin isoforms signal the cell through integrin and non-integrin receptors which activate multiple signal transduction pathways involving various components such as G proteins, intracellular calcium, phospholipase D, mitogen-activated protein kinases, focal adhesion kinase, small GTPases of the Rho family, and cytoskeleton components (8). It is also known that cellular invasion by T. cruzi requires the activation of some of the same signaling pathways induced by laminin in cells. Therefore, we suggest that down-regulation of laminin would reduce some of the signaling events that T. cruzi requires to invade cells, causing a reduction in cellular infection. RNAi with laminin γ-1 decreases its expression up to 60%, resulting in inhibition of trypanosome binding and consequent inhibition of parasite multiplication of up to 85%. This apparent lack of correlation seen may be due to a possible dual role played by laminin γ-1 in the process of T. cruzi infection. First, it is possible that laminin γ-1 interacts with specific trypanosome surface proteins to facilitate passage through the membrane barrier. Second, it is possible that this interaction triggers host cell signal transduction events mediated by laminin γ-1 that are required for infection, which would enhance infection. This possible dual effect may be responsible for the higher level of inhibition of T. cruzi infection seen.
A previous study by Avalos et al. (2) with partial human cDNA microarrays showed that there is no significant increase in the expression level of genes in human foreskin fibroblasts during the first 6 h after infection with T. cruzi. The microarrays used in that study contained the probe for Lam-2, but no up-regulation of laminin by T. cruzi was seen. However, our results indicate that the T. cruzi gp83 ligand or T. cruzi significantly up-regulated early laminin γ-1 expression in HCASM cells or HeLa cells, as evidenced by human cDNA microarray experiments, confirmed by real-time PCR. The different results found in the two studies may be due to the fact that different cell types were used and to specific experimental conditions in the design of the experiments.
T. cruzi infection causes extensive fibrosis and severe cardiomyopathology, which is in part vasculopathy, leading to cardiac arrest, which is frequently followed by death (16). T. cruzi must navigate through the basal lamina, which contains laminin γ-1, and surrounds individual muscle cells such as HCASM cells before infecting these cells. The fact that a T. cruzi trypomastigote ligand increased the laminin γ-1 transcript level in HCASM cells, as described in this report, correlates with the finding that laminin is deposited in the hearts of patients infected with Chagas' disease (11), suggesting that the results described in this report might explain, in part, the cause of this pathology.
Our observation that the T. cruzi gp83 ligand remodels the ECM by up-regulating the expression of laminin γ-1, together with the report that T. cruzi presents laminin receptors on its surface (7), indicates that the parasite exploits laminin γ-1 to navigate through the ECM to facilitate infection. It was suggested that T. cruzi binds to laminin (7) and that this interaction is enhanced by human galectin-3 (12); however, the involvement of the ECM protein and the exact laminin chain involved are unknown. Here we show that the γ-1 chain of laminin is required for the infection process of T. cruzi, as evidenced by RNAi with that specific chain. Thus, the T. cruzi gp83 ligand is a virulence factor that modifies laminin γ-1 expression in the ECM to contribute to the pathogenesis of T. cruzi infection in human heart cells.
Acknowledgments
This work was supported in part by NIH grants GM 08037, GM 059994, AI 07281, AI 056667, HL 007737, MD 000104, and RR 003032.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Alexander, D. A., F. Villalta, and M. F. Lima. 2003. Transforming growth factor α binds to Trypanosoma cruzi amastigotes to induce signaling and cellular proliferation. Infect. Immun. 71:4201-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avalos, S. V., I. J. Blader, M. Fischer, J. C. Boothroyd, and B. A. Burleigh. 2002. Immediate early response to Trypanosoma cruzi infection involves minimal modulation of host cell transcription. J. Biol. Chem. 277:639-644. [DOI] [PubMed] [Google Scholar]
- 3.Blohm, D. H., and A. Guiseppi-Elie. 2001. New developments in microarray technology. Curr. Opin. Biotechnol. 12:41-47. [DOI] [PubMed] [Google Scholar]
- 4.Buschiazzo, A., G. A. Tavares, O. Campetella, S. Spinelli, M. L. Cremona, G. Paris, M. F. Amaya, A. C. C. Frasch, and P. M. Alzari. 2000. Structural basis of sialyltransferase activity in trypanosomal sialidases. EMBO J. 19:16-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Detweiler, C. S., D. B. Cunanan, and S. Falkow. 2001. Host microarray analysis reveals a role for the Salmonella response regulator phoP in human macrophage cell death. Proc. Natl. Acad. Sci. USA 98:5850-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frasch, A. C. 2000. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol. Today 16:282-286. [DOI] [PubMed] [Google Scholar]
- 7.Giordano, R., D. L. Fouts, D. Tewari, W. Colli, J. E. Manning, and M. J. M. Alves. 1999. Cloning of a surface membrane glycoprotein specific for the infective form of Trypanosoma cruzi having adhesive properties to laminin. J. Biol. Chem. 274:3461-3468. [DOI] [PubMed] [Google Scholar]
- 8.Givant-Horwitz, V., B. Davidson, and R. Reich. 2005. Laminin-induced signaling in tumor cells. Cancer Lett. 223:1-10. [DOI] [PubMed] [Google Scholar]
- 9.Kleshchenko, Y. Y., T. N. Moody, V. A. Furtak, J. Ochieng, M. F. Lima, and F. Villalta. 2004. Human galectin-3 promotes Trypanosoma cruzi adhesion to human coronary artery smooth muscle cells. Infect. Immun. 72:6717-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lima, M. F., and F. Villalta. 1989. Trypanosoma cruzi trypomastigote clones differentially express a parasite cell adhesion molecule. Mol. Biochem. Parasitol. 33:159-170. [DOI] [PubMed] [Google Scholar]
- 11.Milei, J., J. Sanche, R. Storino, Z. X. Yu, B. Denduchis, and V. J. Ferrans. 1993. Antibodies to laminin and immunohistochemical localization of laminin in chronic chagasic cardiomyopathy: a review. Mol. Cell. Biochem. 129:161-170. [DOI] [PubMed] [Google Scholar]
- 12.Moody, T. N., J. Ochieng, and F. Villalta. 2000. Novel mechanism that Trypanosoma cruzi uses to adhere to the extracellular matrix mediated by human galectin-3. FEBS Lett. 470:305-308. [DOI] [PubMed] [Google Scholar]
- 13.Qin, X., D. S. An, I. S. Y. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosemberg, S., C. J. Chaves, M. L. Higuchi, M. B. Lopes, L. H. Castro, and L. R. Machado. 1992. Fatal meningoencephalitis caused by reactivation of Trypanosoma cruzi infection in a patient with AIDS. Neurology 42:640-642. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki, T., R. Fassler, and E. Hohenester. 2004. Laminin: the crux of basement membrane assembly. J. Cell Biol. 164:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanowitz, H. B., L. V. Kirchhoff, D. Simon, S. A. Morris, L. M. Weiss, and M. Wittner. 1992. Chagas' disease. Clin. Microbiol. Rev. 5:400-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villalta, F., Y. Zhang, K. Bibb, J. M. Jr., Burns, and M. F. Lima. 1998. Signal transduction in human macrophages by gp83 ligand of Trypanosoma cruzi: trypomastigote gp83 ligand up-regulates trypanosome entry through the MAP kinase pathway. Biochem. Biophys. Res. Commun. 249:247-252. [DOI] [PubMed] [Google Scholar]
- 18.Villalta, F., Y. Zhang, K. Bibb, S. Pratap, J. M. Burns, Jr., and M. F. Lima. 1999. Signal transduction in human macrophages by gp83 ligand of Trypanosoma cruzi: trypomastigote gp83 ligand up-regulates trypanosome entry through protein kinase C activation. Mol. Cell. Biol. Res. Commun. 2:67-70. [DOI] [PubMed] [Google Scholar]
- 19.Villalta, F., C. Smith, R. Ruiz-Ruano, and M. F. Lima. 2001. A ligand that Trypanosoma cruzi uses to bind to mammalian cells to initiate infection. FEBS Lett. 505:383-388. [DOI] [PubMed] [Google Scholar]