Abstract
The binding of Borrelia burgdorferi OspE, OspF, and family 163 (Elp) proteins to factor H/factor H-like protein 1 (FHL-1) and other serum proteins from different animals was assessed. OspE paralogs bound factor H and unidentified serum proteins from a subset of animals, while OspF and Elp proteins did not. These data advance our understanding of factor H binding, the host range of the Lyme spirochetes, and the expanding role of OspE in pathogenesis.
Borrelia burgdorferi, a causative agent of Lyme disease, binds the complement regulatory proteins factor H and factor H-like protein 1 (FHL-1) (4, 19, 22, 31, 32). Factor H/FHL-1 function as cofactors in the factor I-mediated cleavage of C3b (3, 6, 19, 21, 29). C3b cleavage at the cell surface inhibits opsonophagocytosis and facilitates immune evasion. FHL-1 binding may also promote adherence and invasion, as demonstrated for group A streptococci (35) and postulated for Treponema denticola (27). B. burgdorferi B31MI produces several factor H-binding proteins, including BBA68 (also referred to as BbCRASP-1) and the OspE proteins (3, 6, 19, 21, 29). BBA68 binds both factor H and FHL-1, while OspE binds only factor H. In B31MI, the OspE protein family consists of three members, designated by TIGR as BBL39, BBN38, and BBP38. BB stands for Borrelia burgdorferi, the next letter designates the plasmid of origin (plasmid L, N, or P), and the number indicates its numerical open reading frame (ORF) designation and relative location on its specific plasmid (www.tigr.org) (15). The genetic elements that carry the ospE genes are circular molecules of 32 kb that have been referred to as cp32s, for circular plasmids of 32 kb (39). However, Eggers and Samuels had originally hypothesized that the cp32s are actually prophages (13) and Zhang and Marconi recently provided evidence to support this hypothesis (44). Some B. burgdorferi strains carry as many as nine copies of these prophages (10, 24, 44, 47), but only three carry ospE genes, and of these ospE genes, BBL39 and BBP38 are identical in sequence. Hence, in this report we do not differentiate between BBL39 and BBL38 and refer to them collectively as BBL39. One or more OspE paralogs have been demonstrated to be expressed during infection in ticks and mammals, indicating that they may carry out important functions in these environments (16-18, 20, 32-34, 38). Interestingly, while BBA68 has clear factor H/FHL-1 binding ability, it is not expressed during infection; thus, its role in pathogenesis is unclear (6, 26, 42, 43).
The ospE, ospF, and elp genes have been collectively referred to by some as erp (OspE-related protein) genes (36). B. burgdorferi B31MI carries 12 highly divergent genes that have been included in the Erp group (1, 23, 24, 39, 40) (www.tigr.org). However, it is now evident that this diverse group of genes actually constitutes three distinct gene families (10, 18, 24, 28, 41) that are transcriptionally responsive to different environmental signals and differentially expressed (1, 5, 8, 9, 14, 17, 18, 28). One published study suggested that these diverse proteins may still share a common functional role and be key players in factor H-mediated complement evasion (37). Furthermore, it was hypothesized that some OspE, OspF, and Elp proteins may differentially and preferentially bind to factor H from different animals and that this would serve to increase the potential host range of the Lyme disease spirochetes (37). While the literature is in strong agreement regarding the ability of OspE to bind factor H, there are conflicting reports regarding factor H binding to the OspF and Elp proteins (2, 4, 19, 26, 32, 37). Accurate identification of the Borrelia proteins involved in factor H binding and their binding specificity is an essential step in defining the contribution of this virulence mechanism to Lyme disease pathogenesis and in determining if differential factor H binding influences host range. The goals of this study were to address remaining questions regarding the factor H binding ability and specificity of the B. burgdorferi B31MI OspE (BBL39 and BBN38), OspF (BBM38, BBO39, and BBR42), and Elp (BBN39, BBO40, and BBP39) proteins.
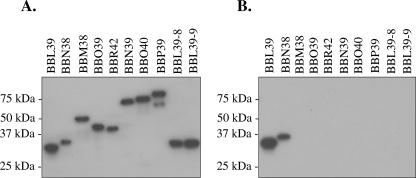
To examine factor H/FHL-1 binding, recombinant proteins were generated using a PCR-based approach with primers (Table 1) that carry tails to allow for ligase-independent cloning into the pET32 Ek/LIC vector (Novagen) (45). The resulting recombinant proteins, which carry N-terminal S and His tags, were purified as previously described (32). Size, integrity, and purity of the recombinant proteins were assessed by Coomassie staining (data not shown) and by screening an immunoblot with horseradish peroxidase (HRP)-conjugated S protein (1:40,000; Novagen). The HRP-conjugated S protein interacts with the N-terminal tag in an antibody-independent manner, a point which is of significance in assessing the “reverse” affinity ligand binding immunoblot (ALBI) analyses presented below. All recombinant proteins were found to be of high purity, with little or no breakdown products (Fig. 1A). Binding of human factor H to the recombinant proteins was determined using the ALBI assay approach as previously described (32), with some modifications. Briefly, the recombinant proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 15% Criterion precast gels (Bio-Rad), immunoblotted, and incubated with purified human factor H (5 ng μl−1; Calbiochem), and bound factor H was detected using goat anti (α)-human factor H antiserum (1:8,000; Calbiochem), followed by HRP-conjugated rabbit α-goat immunoglobulin G (1:40,000). Consistent with earlier studies, the OspE paralogs BBL39 and BBN38 readily bound human factor H while each individual OspF and Elp protein did not exhibit any detectable binding (2, 19, 21, 30, 32). One earlier study had reported that OspF and Elp protein family members bind human factor H (37). However, analysis of the immunoblots presented in that study revealed that factor H binding to the OspF and Elp proteins, if occurring at all, is at a very low level. In addition, in some cases the weak binding that was observed was to proteins with masses not consistent with those of known OspE, Elp, or OspF proteins. Hence, in contrast to the suggestion that factor H binding is a property common to all members of the OspE, OspF, and Elp protein families (37), the data presented here demonstrate that this activity is unique to the OspE proteins.
TABLE 1.
Primer sequences (5′ to 3′)
| Primer | Sequence |
|---|---|
| BBL39-LIC-F | GACGACGACAAGATGCTTATAGGTGCTTGCAAG |
| BBL39-LIC-R | GAGGAGAAGCCCGGTTTATTTTAAATTTCTTTT AAGCTC |
| BBN38-LIC-F | GACGACGACAAGATGCTTATAGGTGCTTGCAAA ATTC |
| BBN38-LIC-R | GAGGAGAAGCCCGGTTTATTTTAAATTTTTTTA AGCAC |
| BBN39-LIC-F | GACGACGACAAGATCAAGAATTTTGCAACTGGT AAAG |
| BBN39-LIC-R | GAGGAGAAGCCCGGTTTACTGACTGTCACTGAT GTATCC |
| BBO40-LIC-F | GACGACGACAAGATTAAAAATTATGCAACTGGT AAAG |
| BBO40-LIC-R | GAGGAGAAGCCCGGTTCAATATGAATTACTATC CTCAATG |
| BBP39-LIC-F | GACGACGACAAGATTAAGAATTATGCAATTAAA GAT |
| BBP39-LIC-R | GAGGAGAAGCCCGGTTTAATCTTCTTCATCATA ATTATCC |
| BBM38-LIC-F | GACGACGACAAGATTTACGCAAGTGGTGAAGATG |
| BBM38-LIC-R | GAGGAGAAGCCCGGTTTATTCTTTTTTATTAGA ATC |
| BBO39-LIC-F | GACGACGACAAGATTTATGCAAGTGGTGAA |
| BBO39-LIC-R | GAGGAGAAGCCCGGTTTATTCTTTTTTATCTTC TTC |
| BBR42-LIC-F | GACGACGACAAGATTGATGTAACTAGTAAAGAT TTA |
| BBR42-LIC-R | GAGGAGAAGCCCGGTCTTTATTCTTTTTTACCT TCTACA |
FIG. 1.
Analysis of factor H/FHL-1 binding by the B. burgdorferi B31MI OspE, OspF, and Elp proteins. Recombinant S-tagged fusion proteins (the TIGR-assigned ORF designations are indicated above each lane) were generated, purified, separated by SDS-PAGE (15% Criterion precast gels), and immunoblotted. To assess expression and integrity of the recombinant proteins, a blot was screened with HRP-conjugated S protein (A). The proteins were then tested for factor H/FHL-1 binding by use of the ALBI assay (B), as described in the text. Molecular mass standards are indicated.
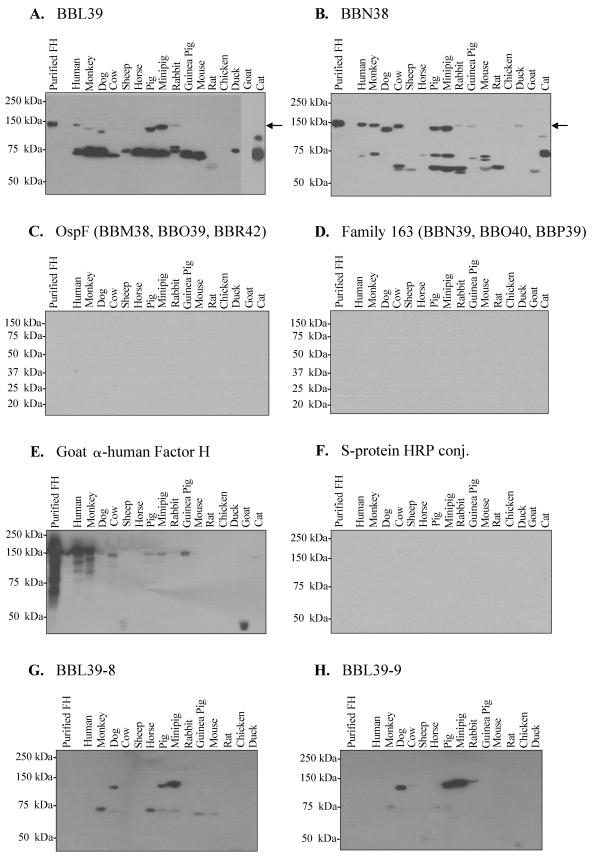
The formation and presentation of the factor H-binding pocket of BBA68 and the OspE proteins have been demonstrated to be dependent on conformational or structural determinants that are defined by highly stable coiled-coil domains (25, 30). For both OspE and BBA68, these structural determinants either are resistant to denaturation during SDS-PAGE or rapidly reform following immunoblotting, as demonstrated for herpes simplex virus glycoprotein D (11). However, it is possible that OspF and Elp proteins did not bind factor H because they are more sensitive than OspE to structural perturbations introduced by the assay conditions employed. To test the abilities of the OspE, OspF, and Elp proteins to bind to human factor H and factor H from other animals under different experimental conditions, a “reverse” ALBI was developed and employed. In this assay, sera from 16 different animals were diluted 1:4 with phosphate-buffered saline and each sample was separated by SDS-PAGE under nonreducing conditions with 7.5% or 12.5% Criterion precast Tris-HCl gels (Bio-Rad). The conditions used for nonreducing electrophoresis have been previously described (32). Nonreducing conditions were employed to maintain disulfide bonds that may be important in factor H/FHL-1 functional activity (46). As in several other studies, serum served as the source of factor H in these assays because only purified factor H from humans is commercially available. The serum proteins were immunoblotted, and a series of identical blots were incubated with individual recombinant S-tagged proteins, as indicated above each panel of Fig. 2 (room temperature, 3 h; 100 μg ml−1 in blocking buffer). Unbound protein was removed by washing, and bound protein was detected using HRP-conjugated S protein (1:40,000 dilution; Novagen). As a negative control, a blot was screened with HRP-conjugated S protein alone, and no signal was observed (Fig. 2F). The OspE proteins were found to bind factor H and to several additional serum proteins (Fig. 2A and B), while the OspF and Elp proteins did not (Fig. 2C and D).
FIG.2.
“Reverse” ALBI analyses: analysis of the abilities of recombinant OspE, OspF, and Elp proteins to bind to immobilized serum proteins. Serum proteins from different animals (indicated in each panel) were separated by SDS-PAGE with either 7.5% or 12.5% Criterion precast gels under nonreducing conditions. The proteins were immunoblotted and incubated with individual recombinant S-tagged proteins (100 μg ml−1), as indicated in each panel. Note that the blots shown in panels C and D were screened with BBO39 and BBN39, respectively. Identical results were obtained with BBM38, BBR42, BBO40, and BBP39 (data not shown). In all assays, bound recombinant protein was detected using HRP-conjugated (conj.) S protein. All methods were as described in the text. Purified human factor H (FH) was included as a positive control with all immunoblots. The location of factor H on the immunoblots presented in panels A and B is indicated by an arrow for reference. Note that in panel E the serum proteins were separated using reducing conditions and were screened with goat α-human factor H antiserum. With longer exposure, factor H was detected in all animals except duck, horse, rat, and chicken.
Focusing first on a discussion of factor H binding, while in several serum samples the OspE paralogs bound to a 150-kDa protein that is presumed to be factor H, no binding of OspF or Elp proteins to a similarly sized protein in any of the samples tested was observed (Fig. 2C and D). To determine the migration position of factor H separated under nonreducing SDS-PAGE conditions, purified human factor H was included in all experiments. In addition, the presence of factor H in the serum samples was confirmed by screening an immunoblot with goat α-human factor H antiserum (Fig. 2E). The goat α-human factor H antiserum recognized factor H from human, monkey, dog, cow, pig, mini-pig, rabbit, guinea pig, and cat. With longer exposure, factor H was detected in all animals except duck, rat, chicken, and horse (data not shown). The quantitative differences in detection level are likely due to factor H sequence divergence among animals. For example, while both mouse and human express factor H at a high level, mouse factor H was not detected by the antiserum. This is not surprising since these proteins exhibit only 61% identity. It is important to note that the immunoblot used in this specific control was generated using serum samples separated by SDS-PAGE under reducing conditions. The data presented here do not support the suggestion put forth by Stevenson et al. that factor H binding is a universal feature of the OspE, OspF, and Elp proteins (37).
The overall factor H and serum protein binding patterns for the two different OspE paralogs were found to exhibit significant differences. BBN38 bound to factor H from a broader group of species than did BBL39. This observation suggests that the contribution of individual OspE paralogs in factor H-mediated complement evasion may differ within different animals. The fact that factor H binding patterns differed for BBL39 and BBN38 raises the possibility that OspE sequence variation as it develops in nature could serve as an adaptive or selective response that facilitates persistence in different mammalian reservoirs. The inability of OspE to bind to mouse factor H is particularly noteworthy. Mice serve as a major reservoir for B. burgdorferi and are the primary animal model for the study of Lyme disease pathogenesis. It was previously reported that the OspE paralogs can bind mouse factor H (37). This discrepancy may be due to the absence of a critical control in that study. Stevenson et al. incubated mouse serum with membrane-immobilized recombinant OspE, and then factor H binding was detected by screening the blots with goat α-human factor H antiserum. However, a control blot of the recombinant proteins screened with α-human factor H antiserum alone was not presented. Metts demonstrated that when immunoblots of recombinant OspE are incubated with goat α-human factor H antiserum with no exogenous factor H added, the antiserum reacts with OspE (32). It was postulated that either the antiserum cross-immunoreacts with OspE or goat factor H present in the goat α-human factor H antiserum binds to OspE, thus allowing for the subsequent detection of bound factor H by the antiserum. However, as demonstrated here, BBL39 and BBN38 do not bind to goat factor H, so the latter explanation seems unlikely. In any event, the binding of mouse factor H by OspE reported by Stevenson et al. appears to represent an experimental artifact. Alitalo et al. also reported that an OspE variant, designated P21 (12), binds to mouse factor H (2). However, the interaction of P21 with mouse factor H, as measured by surface plasmon resonance, was approximately 50% of that observed with human factor H (2). P21 exhibits 85% and 71% amino acid sequence identity with BBN38 and BBL39, respectively. The differing abilities of OspE paralogs to bind mouse factor H are most likely due to specific sequence differences. This suggestion is supported by the paralog-specific serum protein binding patterns that we observed in this report for BBN38 and BBL39. The inability of the OspE paralogs BBN38 and BBL39 to bind to mouse factor H also suggests that the mouse model may not be an appropriate system for examining the effects of ospE gene inactivation on factor H-mediated immune evasion by the Lyme spirochetes.
Regarding the binding of OspE to other non-factor H serum proteins, the binding patterns differed significantly among paralogs. BBL39 bound one or more proteins in the 70- to 75-kDa-size range in 13 of the 16 serum samples tested. In contrast, BBN38 bound only weakly to these proteins (Fig. 2, compare panels A and B) and instead bound to one or more proteins of ∼60 kDa in eight of the species tested. None of these proteins were bound by the OspF or Elp proteins. It is important to note that when the serum samples were separated by SDS-PAGE under reducing conditions the binding of OspE to factor H and other serum proteins was completely abolished. This finding suggests that the OspE binding sites on these proteins require disulfide bond formation for their presentation. As discussed above, the short consensus repeats of factor H/FHL-1 possess highly conserved Cys residues that are important in defining their structure and functional activity (46).
Several observations indicate that the binding of OspE to serum proteins is a specific interaction and not mediated by charge or influenced by the S tag. First, none of the other S-tagged proteins analyzed bound to serum proteins and hence it can be concluded that the interaction is not due to the S tag. The differential binding of BBN38 and BBL39 to different sets of proteins also points to a specific interaction. To further assess the specific nature of protein binding by BBL39, the abilities of two BBL39 variants (BBL39-8 and BBL39-9) to bind to factor H and other serum proteins were determined. These BBL39 variants, which were generated by random mutagenesis, harbor a single (but different) amino acid substitution (relative to the wild type) that was previously demonstrated to abolish human factor H binding ability (30). Specifically, BBL39-8 has a G61-to-R change while BBL39-9 has an S82-to-L change. As demonstrated here, these variants did not bind factor H from most animals and exhibited attenuated binding to other serum proteins (Fig. 2G and H). It is unlikely that two independent, single-amino-acid changes would disrupt binding if the interaction was nonspecific. Of note is the fact that the binding of BBL39-8 and BBL39-9 to factor H from dog, pig, mini-pig, and rabbit was unaffected. This suggests that different determinants of OspE may be involved in binding to factor H from different animals.
The selective and differential binding of BBL39 and BBN38 to factor H and other serum proteins has important implications for understanding the host-pathogen interaction, the role of OspE in pathogenesis, and the enzootic cycle of B. burgdorferi. First, the species selectivity of the OspE-factor H interaction indicates that OspE may play an important role in mediating factor H-based immune evasion in some animals but not in others. It is also evident that different OspE paralogs within a cell may have different functions and contribute to pathogenesis in distinctly different ways. The generation of sequence diversity within OspE as a result of mutation or recombination could influence factor H and serum protein binding characteristics. Recent analyses have demonstrated that the circular plasmids of 32 kb that carry the ospE genes are prophages (13, 44), and a large late operon consisting of 32 ORFs has been defined (44). ospE, which lies outside of this operon, appears to represent a classic “moron” (44). Morons are defined by Brussow et al. as phage-carried genes of bacterial origin that serve to enhance the fitness of the lysogen (7). The natural development of sequence variation within ospE and the lateral transfer of these prophage-carried genes could serve as a fitness-enhancing, adaptive mechanism that ensures survival of the spirochetes at the population level in diverse animals. With regard to the binding of serum proteins by OspE, the biological significance of this interaction in Borrelia pathogenesis is currently unknown. If factor H and serum proteins compete for binding to OspE, one possibility is that competitive binding could influence or modulate OspE-mediated factor H complement evasion. Another possibility is that serum protein binding serves as a form of molecular mimicry that conveys host-like properties to the spirochetes, which shields them from the immune response. Last, OspE binding to factor H and serum proteins could facilitate adherence to endothelium or immune effector cells, that present serum proteins on their surface. It is becoming increasingly apparent that the potential role of OspE in virulence may extend beyond solely factor H-mediated immune evasion. The identity of OspE-binding serum proteins, their relative affinities for OspE, and possible competition among these ligands for binding are important areas for future research.
Acknowledgments
This work was supported in part by grants from NIAID NIH to R.T.M. and an F31 award from NINDS to K.M.H. J.V.M. was supported in part by an NIAID training grant in Molecular Pathogenesis to the Department of Microbiology and Immunology of Virginia Commonwealth University.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alitalo, A., T. Meri, T. Chen, H. Lankinen, Z.-Z. Cheng, T. S. Jokiranta, I. J. T. Seppala, P. Lahdenne, P. S. Hefty, D. R. Akins, and S. Meri. 2004. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 172:6195-6201. [DOI] [PubMed] [Google Scholar]
- 3.Alitalo, A., T. Meri, L. Rämö, T. S. Jokiranta, T. Heikkilä, I. J. T. Seppälä, J. Oksi, M. Viljanen, and S. Meri. 2001. Complement evasion by Borrelia burgdorferi: serum-resistant strains promote C3b inactivation. Infect. Immun. 69:3685-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alitalo, A., T. Meri, H. Lankinen, I. Seppala, P. Lahdenne, P. S. Hefty, D. Akins, and S. Meri. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847-3853. [DOI] [PubMed] [Google Scholar]
- 5.Babb, K., J. D. McAlister, J. C. Miller, and B. Stevenson. 2004. Molecular characterization of Borrelia burgdorferi erp promoter/operator elements. J. Bacteriol. 186:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, C. S., S. R. Vuppala, A. M. Jett, A. Alitalo, S. Meri, and D. R. Akins. 2005. Complement regulator-acquiring surface protein 1 imparts resistance to human serum in Borrelia burgdorferi. J. Immunol. 175:3299-3308. [DOI] [PubMed] [Google Scholar]
- 7.Brussow, H., C. Canchaya, and W.-D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caimano, M. J., C. H. Eggers, C. A. Gonzalez, and J. D. Radolf. 2005. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J. Bacteriol. 187:7845-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caimano, M. J., C. H. Eggers, K. R. O. Hazlett, and J. D. Radolf. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 72:6433-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlyon, J. A., C. LaVoie, S.-Y. Sung, and R. T. Marconi. 1998. Analysis of the organization of multicopy linear- and circular-plasmid-carried open reading frames in Borrelia burgdorferi sensu lato isolates. Infect. Immun. 66:1149-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, G. H., V. J. Isola, J. Kuhns, P. W. Berman, and R. J. Eisenberg. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J. Virol. 60:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das, S., S. W. Barthold, S. S. Giles, R. R. Montgomery, S. R. Telford III, and E. Fikrig. 1997. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and the mammalian host. J. Clin. Investig. 99:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggers, C., and D. S. Samuels. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eggers, C. H., M. J. Caimano, and J. D. Radolf. 2004. Analysis of promoter elements involved in the transcriptional initiation of RpoS-dependent Borrelia burgdorferi genes. J. Bacteriol. 186:7390-7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, C., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.F.Tomb, R. D. Fleischman, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 16.Hefty, P. S., C. S. Brooks, A. M. Jett, G. L. White, S. K. Wikel, R. C. Kennedy, and D. R. Akins. 2002. OspE-related, OspF-related, and Elp lipoproteins are immunogenic in baboons experimentally infected with Borrelia burgdorferi and in human Lyme disease patients. J. Clin. Microbiol. 40:4256-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, and D. R. Akins. 2002. Changes in temporal and spatial patterns of outer surface lipoprotein expression generate population heterogeneity and antigenic diversity in the Lyme disease spirochete Borrelia burgdorferi. Infect. Immun. 70:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of the OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. T. Seppala, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 20.Kraiczy, P., J. Hellwage, C. Skerka, H. Becker, M. Kirschfink, M. M. Simon, V. Brade, P. F. Zipfel, and R. Wallich. 2004. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 279:2421-2429. [DOI] [PubMed] [Google Scholar]
- 21.Kraiczy, P., C. Skerka, M. Kirschfink, P. F. Zipfel, and V. Brade. 2001. Mechanism of complement resistance of pathogenic Borrelia burgdorferi isolates. Int. Immunopharmacol. 1:393-401. [DOI] [PubMed] [Google Scholar]
- 22.Kraiczy, P., C. Skerka, and P. F. Zipfel. 2001. Further characterization of complement regulator-acquiring surface proteins of Borrelia burgdorferi. Infect. Immun. 69:7800-7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam, T. T., T.-P. K. Nguyen, R. R. Montgomery, F. S. Kantor, E. Fikrig, and R. A. Flavell. 1994. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marconi, R. T., S. Y. Sung, C. N. Hughes, and J. A. Carlyon. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 178:5615-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDowell, J. V., M. E. Harlin, E. Rogers, and R. T. Marconi. 2005. Putative coiled-coil structural elements of the BBA68 protein of the Lyme disease spirochetes are required for formation of its factor H binding site. J. Bacteriol. 187:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDowell, J. V., K. M. Hovis, H. Zhang, E. Tran, J. Lankford, and R. T. Marconi. Evidence that the factor H binding BBA68 (BbCRASP-1) protein of the Lyme disease spirochetes does not contribute to factor H mediated immune evasion in humans and other animals. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 27.McDowell, J. V., J. Lankford, L. Stamm, T. Sadlon, D. L. Gordon, and R. T. Marconi. 2005. Demonstration of factor H-like protein 1 binding to Treponema denticola, a pathogen associated with periodontal disease in humans. Infect. Immun. 73:7126-7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDowell, J. V., S.-Y. Sung, G. Price, and R. T. Marconi. 2001. Demonstration of the genetic stability and temporal expression of select members of the Lyme disease spirochete OspF protein family during infection in mice. Infect. Immun. 69:4831-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDowell, J. V., E. Tran, D. Hamilton, J. Wolfgang, K. Miller, and R. T. Marconi. 2003. Analysis of the ability of spirochete species associated with relapsing fever, avian borreliosis, and epizootic bovine abortion to bind factor H and cleave C3b. J. Clin. Microbiol. 41:3905-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDowell, J. V., J. Wolfgang, L. Senty, C. M. Sundy, M. J. Noto, and R. T. Marconi. 2004. Demonstration of the involvement of outer surface protein E coiled-coil structural domains and higher order structural elements in the binding of infection-induced antibody and the complement-regulatory protein, factor H. J. Immunol. 173:7471-7480. [DOI] [PubMed] [Google Scholar]
- 31.McDowell, J. V., J. Wolfgang, E. Tran, M. S. Metts, D. Hamilton, and R. T. Marconi. 2003. Comprehensive analysis of the factor H binding capabilities of Borrelia species associated with Lyme disease: delineation of two distinct classes of factor H binding proteins. Infect. Immun. 71:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metts, S., J. V. McDowell, M. Theisen, P. R. Hansen, and R. T. Marconi. 2003. Analysis of the OspE determinants involved in the binding of factor H and OspE targeting antibodies elicited during infection in mice. Infect. Immun. 71:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. C., K. von Lackum, K. Babb, J. D. McAlister, and B. Stevenson. 2003. Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle. Infect. Immun. 71:6943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen, T.-P. K., T. T. Lam, S. W. Barthold, S. R. Telford III, R. A. Flavell, and E. Fikrig. 1994. Partial destruction of Borrelia burgdorferi within ticks that engorged on OspE- or OspF-immunized mice. Infect. Immun. 62:2079-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandiripally, V., L. Wei, C. Skerka, P. F. Zipfel, and D. Cue. 2003. Recruitment of complement factor H-like protein 1 promotes intracellular invasion by group A streptococci. Infect. Immun. 71:7119-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson, B., J. L. Bono, T. G. Schwan, and P. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson, B., T. G. Schwan, and P. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevenson, B., K. Tilly, and P. A. Rosa. 1996. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J. Bacteriol. 178:3508-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sung, S.-Y., C. P. LaVoie, J. A. Carlyon, and R. T. Marconi. 1998. Genetic divergence and evolutionary instability in ospE-related members of the upstream homology box gene family in Borrelia burgdorferi sensu lato complex isolates. Infect. Immun. 66:4656-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sung, S. Y., J. V. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect. Immun. 68:1319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72:5419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallich, R., O. Jahraus, T. Stehle, T. T. T. Tran, C. Brenner, H. Hofmann, L. Gern, and M. M. Simon. 2003. Artificial-infection protocols allow immunodetection of novel Borrelia burgdorferi antigens suitable as vaccine candidates against Lyme disease. Eur. J. Immunol. 33:708-719. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, H., and R. T. Marconi. 2005. Demonstration of cotranscription and 1-methyl-3-nitroso-nitroguanidine induction of a 30-gene operon of Borrelia burgdorferi: evidence that the 32-kilobase circular plasmids are prophages. J. Bacteriol. 187:7985-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, H., A. Raji, M. Theisen, P. R. Hansen, and R. T. Marconi. 2005. bdrF2 of Lyme disease spirochetes is coexpressed with a series of cytoplasmic proteins and is produced specifically during early infection. J. Bacteriol. 187:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zipfel, P. F., C. Skerka, J. Hellwage, S. T. Jokiranta, S. Meri, V. Brade, P. Kraiczy, M. Noris, and G. Remuzzi. 2002. Structure-function studies of the complement system. Biochem. Soc. Trans. 30:971-978. [DOI] [PubMed] [Google Scholar]
- 47.Zuckert, W. R., and J. Meyer. 1996. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 178:2287-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]