Abstract
Bordetella pertussis, the causative agent of whooping cough or pertussis, is an obligate human pathogen with multiple high-affinity iron transport systems. Maximal expression of the dedicated heme utilization functions encoded by the hurIR bhuRSTUV genes requires an iron starvation signal to relieve Fur repression at the hurIR promoter-operator and an inducing signal supplied by heme for HurI-mediated transcriptional activation at the bhuRSTUV promoter. The BhuR outer membrane receptor protein is required for heme uptake and for heme sensing for induction of bhuRSTUV transcription. It was hypothesized that heme utilization contributed to the success of B. pertussis as a pathogen. In this study, virulence attenuation resulting from inactivation of the B. pertussis heme system was assessed using mixed infection competition experiments in a mouse model. As a measure of in vivo fitness, the ability of a B. pertussis heme utilization mutant to colonize and persist was determined relative to that of an isogenic coinfecting wild-type strain. Relative fitness of the mutant strain declined significantly after 7 days postinfection and continued to decline throughout the remainder of the 28-day infection time course. In parallel infections using inocula supplemented with an inducing 2 μM concentration of hemin chloride, hemin coadministration augmented the competitive advantage of the wild-type strain over the mutant. The results confirm that heme utilization contributes to the pathogenesis of B. pertussis in the mouse infection model and indicate that heme utilization may be most important for adaptation to host conditions existing during the later stages of infection.
Bordetella pertussis is a versatile pathogen with diverse high-affinity transport systems for the assimilation of nutritional iron required for multiplication (4). It is hypothesized that specific iron transport systems serve distinct roles in responding to changes in iron source availability in the host environment during the course of infection. The heme utilization systems encoded by the hurIR bhuRSTUV genes of B. pertussis and Bordetella bronchiseptica (11, 12, 13) are novel hybrid systems combining the heme transport machinery of gram-negative pathogens with extracytoplasmic function (ECF) regulators of the iron starvation subfamily (12). Transcriptional regulation of Bordetella heme utilization genes is complex. BhuR, the TonB-dependent outer membrane heme receptor that is required for heme uptake and utilization, participates directly in regulation of heme system gene expression. BhuR has a predicted molecular mass of 90 kDa and has conserved characteristics of microbial heme receptors, including a TonB box C motif and the FRAP/NPNL amino acid sequence motif common to many hemin/hemoglobin receptors (11). Unique among heme receptor proteins, however, BhuR also has the N-terminal extension domain characteristic of TonB-dependent receptors that communicate with ECF regulators (11). Expression of the regulatory hurIR genes is Fur and iron repressible, and expression of the transport and utilization genes bhuRSTUV is heme inducible by a mechanism involving transmembrane transduction of the BhuR heme receptor occupancy signal via the anti-sigma factor membrane regulator HurR to the ECF sigma factor HurI for transcriptional activation of the bhuRSTUV promoter (12, 13).
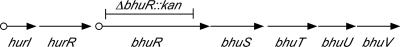
The global response of a microbial pathogen to iron starvation in the host contributes fundamentally to in vivo bacterial fitness (6). Mixed infection competition experiments (1, 5, 10) use the mouse or other appropriate host organism as a selective environment to assess the relative fitness of coinfecting strains for survival and multiplication in the host environment. The goal of this study was to determine the importance of B. pertussis heme utilization for multiplication and survival in mice using mixed infection competition experiments with strain UT25Sm1 (BhuR+ Strr) (3) and the isogenic ΔbhuR::kan mutant strain PM5 (BhuR− Strr Kanr) (11). In previous studies, PM5 was found to have no measurable in vitro growth defect, except in heme utilization during iron starvation (11). The ΔbhuR::kan mutation of PM5 exerts polar effects on the downstream, cotranscribed heme utilization genes bhuSTUV (11) (Fig. 1); thus, PM5 is defective in expression of all functions required for utilization of heme as an iron source. Receptor mutants are well suited to analysis by mixed infection competition since they cannot be trans-complemented by a receptor-proficient coinfecting strain; the receptor does not function as a diffusible factor and can only transport substrates into the receptor-producing cell. In this study, the ability of a bhuR heme utilization mutant to colonize and persist was determined relative to that of an isogenic coinfecting wild-type strain. The results demonstrate that heme utilization functions contribute fundamentally to the fitness of B. pertussis in the experimental murine host system.
FIG. 1.
Genetic organization of the 8-kb heme utilization gene cluster of B. pertussis. Arrows indicate the transcriptional orientations and spatial limits of the ECF regulator pair genes hurIR and the bhuRSTUV heme utilization operon. Circles represent the known Fur and iron-repressible promoter-operator region upstream of hurIR and the HurI-activated, heme-inducible promoter upstream of bhuRSTUV. Genes and products: hurI, transcriptional activator (ECF sigma factor); hurR, sensor/regulator protein; bhuR, outer membrane heme receptor protein; bhuS, heme chaperone protein; bhuT, periplasmic binding protein; bhuU, membrane permease; bhuV, ATP-binding protein. The deleted bhuR genetic region that was replaced by a kanamycin resistance cassette in B. pertussis mutant strain PM5 is indicated by the bar labeled ΔbhuR::kan.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. pertussis strain UT25Sm1 (Strr) (3) and the construction and phenotypic characterization of the isogenic ΔbhuR::kan mutant strain PM5 (Strr Kanr) (11) have been described previously. B. pertussis strains were maintained on Bordet-Gengou agar plates (BG) (2) with appropriate selective antibiotics. Liquid cultures used modified Stainer-Scholte defined medium (SS) (9). Streptomycin was used at 30 μg/ml, and kanamycin was used at 50 μg/ml.
In vitro growth competition.
B. pertussis strains UT25Sm1 and PM5 were subcultured from fresh BG plate growth to SS with the appropriate antibiotics at an initial cell density corresponding to an A600 of 0.1. SS cultures were grown at 37°C with shaking (300 rpm) for 36 h. Bacterial cells were recovered by centrifugation and washed with sterile saline, and each strain suspension was diluted to an estimated concentration of 2 × 108 CFU/ml in sterile saline. Equal volumes of wild-type and mutant strain cell suspensions were combined and used to inoculate 10 ml iron-replete SS (36 μM iron), iron-depleted SS (no added iron), and iron-depleted SS supplemented with 10 μM bovine hemin chloride (initial density of 0.1 A600) (Sigma Chemical Co.). These three liquid cultures lacking selective antibiotics were grown at 37°C with shaking (300 rpm) and were sampled at 0, 6, 12, 24, and 48 h for differential enumeration of wild-type and mutant CFU/ml by standard plate counting performed in triplicate using BG with appropriate selective antibiotics. For each culture condition, the competitive index (CI) at each time point sampled was calculated as the mutant/wild-type ratio in the output divided by the initial mutant/wild-type ratio.
Preparation of inoculum for mouse infections.
B. pertussis strains were subcultured from BG to SS with the appropriate antibiotics and grown at 37°C with shaking at 300 rpm for 36 h. Bacteria were subcultured to SS (18 μM iron) without antibiotics and grown at 37°C with shaking for 36 h. Cells were harvested by centrifugation, washed with phosphate-buffered saline (PBS), and diluted to an estimated concentration of 2 × 108 CFU/ml in PBS. Equal volumes of UT25Sm1 and PM5 suspensions were combined to use as inoculum. The CFU in the inoculum were enumerated for determination of the mutant/wild-type input ratio by standard plate counting. For infection experiments involving hemin coadministration, bacterial suspensions were prepared from the same bacterial cultures using PBS containing 2 μM bovine hemin chloride.
In vivo mixed infection competition.
All experimental animals were handled in accordance with institutional guidelines. Twenty-five female HSD:ICR (CD-1) mice (8 to 20 g) (Harlan Sprague-Dawley, Inc.) were mildly sedated by isoflurane inhalation and infected with ∼2 × 106 total CFU of a 1:1 mixture of wild-type B. pertussis strain UT25Sm1 and the isogenic ΔbhuR::kan heme receptor mutant PM5 (10-μl volume instilled intranasally). A parallel (“with hemin”) subject group received the same inoculum, except that the bacterial suspension was supplemented with 2 μM hemin chloride. At 3 days, 7 days, 14 days, 21 days, and 28 days postinfection, five mice from each group were euthanized and respiratory tissue (lung and trachea) homogenates were plated in the presence of appropriate antibiotics for differential enumeration of bacterial strains.
Statistical methods.
The CI was calculated as the mutant/wild-type CFU ratio recovered at each time point divided by the mutant/wild-type CFU ratio in the input inoculum. The CI values for the in vitro competition experiments were derived from the mean CFU/ml (determined in triplicate) of each strain in the inoculum and from the output recovered from the cultures at the specified time points. For the in vivo studies, each CI value for both subject groups (with or without hemin coadministration) is the mean of five independent mouse infections. Student's t test was used to determine whether the mean CI at each time point differed significantly from the hypothesized mean value of 1.00 (the predicted mean CI if there were no difference in fitness between the two strains used in mixed infections or in vitro cultures) and whether hemin coadministration resulted in a significant change in the mean CI compared with infections without hemin coadministration (hypothesized mean CI difference of 0.00 between subject groups). Probabilities (P) of ≤0.05 were considered significant.
RESULTS AND DISCUSSION
Heme utilization mutant PM5 and wild-type parent strain UT25Sm1 exhibit comparable growth in vitro unless supplied with heme as the sole iron source.
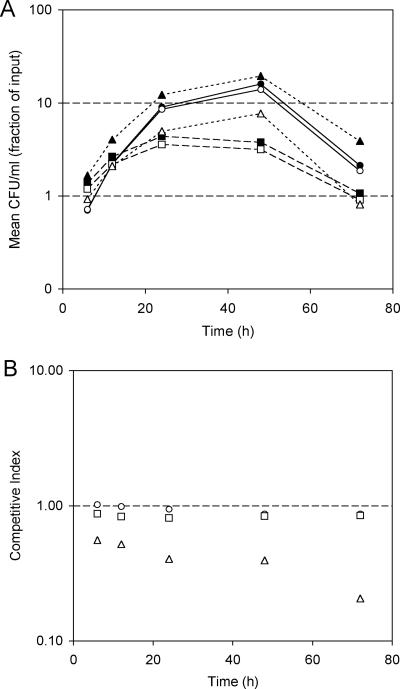
PM5 does not produce the TonB-dependent outer membrane heme receptor protein BhuR and cannot transport and utilize heme as an iron source (11). Previous studies established that PM5 had no demonstrable growth defects when cultured in vitro in SS (11; T. J. Brickman, C. K. Vanderpool, and S. K. Armstrong, unpublished observations). In this study, in vitro growth competition experiments verified that PM5 displays a duration of lag phase, exponential growth rate, total biomass accumulation, and rate of decline similar to those of its wild-type parent strain, UT25Sm1, during batch coculture in iron-replete or iron-depleted SS (Fig. 2A). Accordingly, CI values were similar to the hypothesized value of 1.00 (Fig. 2B). In contrast, when supplied with heme as the sole iron source in iron-depleted SS, heme utilization mutant PM5 was defective in growth and hence was outcompeted by its heme utilization-proficient parent, UT25Sm1 (Fig. 2B). In iron-depleted SS with hemin supplement, the CI decreased from the hypothesized CI of 1.00 to 0.56 by 6 h (Fig. 2B) and to 0.21 by 72 h. That is, after 72 h of culture in vitro with hemin chloride as the sole iron source, the mutant-to-wild-type CFU ratio was reduced to one-fifth of its initial value.
FIG. 2.
Growth of B. pertussis strains cocultured in vitro under various nutritional iron conditions. (A) Growth of strains UT25Sm1 (filled symbols) and heme utilization mutant PM5 (open symbols) cocultured over a 72-h period in iron-replete SS (circles and solid lines), iron-depleted SS (squares and dashed lines), and iron-depleted SS supplemented with 10 μM bovine hemin chloride (triangles and dotted lines). Mean CFU/ml (n = 3) of each strain, represented as the fraction of input CFU, is plotted as a function of time (h). (B) Competitive index values calculated from mean CFU/ml of cocultured strains sampled at various times. Circles, iron-replete SS; squares, iron-depleted SS; triangles, iron-depleted SS with hemin supplementation. The dashed line at a y value of 1.00 corresponds to the hypothesized CI if there were no difference in fitness between the strains cocultured under the specified growth conditions.
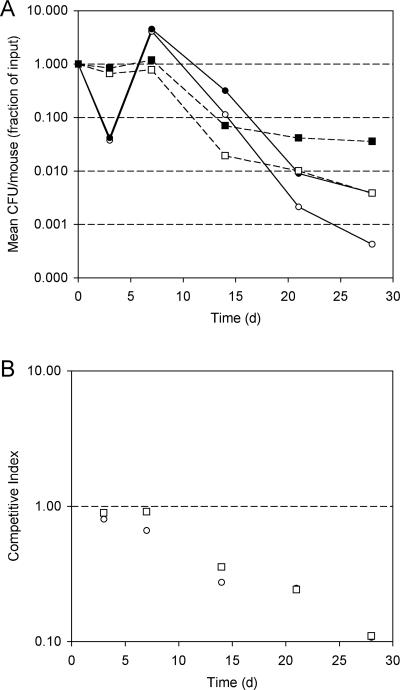
Heme utilization is important for virulence of B. pertussis during primary infection in mice.
Mice were coinfected with a mixture of isogenic wild-type and heme utilization mutant strains, with or without hemin coadministration, and CFU of the two strains in mouse lung and tracheal homogenates were determined at five different time points over a 28-day period. The CI values for the “no hemin” infection group (Table 1; Fig. 3B) indicate that heme utilization mutant strain PM5 had a significantly reduced ability to survive and multiply in mice compared to the wild-type coinfecting strain. These results confirm that the heme utilization mutant exhibits a marked reduction in relative fitness, supporting the hypothesis that heme utilization contributes to the virulence of B. pertussis in the mouse infection model. The mean CI declined significantly after 7 days postinfection (day 14 mean CI = 0.36, P < 0.0001) and continued to decline until the mean CI at 28 days was 0.11. The fact that the CI reduction was observed only after 7 days indicates that heme utilization contributed little to initial survival and multiplication and thus may be more important for adaptation to host conditions that exist later in infection.
TABLE 1.
Relative in vivo fitness of a B. pertussis heme utilization mutant
| Time postinfection (days) | Mean CIa
|
Mean CI differencec (CIno hemin − CIwith hemin) | |
|---|---|---|---|
| No hemin coadministration | With heminb coadministration | ||
| 3 | 0.90 ± 0.15 (0.1990) | 0.80 ± 0.12 (0.0223) | 0.102 (0.0003) |
| 7 | 0.91 ± 0.14 (0.2396) | 0.66 ± 0.11 (0.0021) | 0.252 (<0.0001) |
| 14 | 0.36 ± 0.05 (<0.0001) | 0.27 ± 0.03 (<0.0001) | 0.082 (0.0001) |
| 21 | 0.24 ± 0.03 (<0.0001) | 0.25 ± 0.03 (<0.0001) | −0.002 (0.3739) |
| 28 | 0.11 ± 0.01 (<0.0001) | 0.11 ± 0.01 (<0.0001) | 0.000 (—d) |
Mean CI value ± standard deviation, n = 5 mice (P value). P value is the probability that the mean CI value at each time point postinfection is the same as the reference value of 1.00 (the predicted mean CI if there were no difference in multiplication and survival between the two strains used in mixed infections). The test used was the one-sample t test with a hypothesized mean of 1.00. Probabilities (P) of ≤0.05 were considered significant.
Bovine hemin chloride (2 μM) coadministered with B. pertussis.
Mean CI difference between “no hemin” and “with hemin” infection groups (P value). P value is the probability that the mean CI for infections with 2 μM hemin coadministration is the same as the mean CI for infections without hemin coadministration at each time point postinfection. The test used was the paired t test with a hypothesized difference of 0.00. Probabilities (P) of ≤0.05 were considered significant.
No difference in mean CI.
FIG. 3.
Recovery of B. pertussis CFU from infected mice. (A) Mean CFU/mouse (n = 5) in output recovered from lung and tracheal homogenates, represented as the fraction of the input CFU (∼2 × 106 total CFU of a 1:1 mixture of wild-type strain UT25Sm1 [filled symbols] and heme utilization mutant strain PM5 [open symbols]), as a function of time postinfection (days). Circles, infections without hemin coadministration; squares, infections with coadministration of 2 μM bovine hemin chloride. Standard deviations did not exceed 15% of any mean CFU value determined. (B) Competitive index values calculated from mean CFU/ml of coinfecting strains at various times postinfection. Squares, infections without hemin coadministration; circles, infections with coadministration of 2 μM bovine hemin chloride. The dashed line at a y value of 1.00 corresponds to the hypothesized CI if there were no difference in fitness between the strains in vivo.
Comparing the “no hemin” and “with hemin” subject groups, mean CI values were significantly lower for infections with hemin than for those without hemin coadministration at 3 days, 7 days, and 14 days postinfection (Table 1; Fig. 3B), indicating that hemin coadministration augments the competitive advantage of the wild-type strain over the heme utilization mutant. Furthermore, in contrast with the “no hemin” group, which showed no reduction in CI until later in infection (after 7 days), hemin coadministration was associated with a significant decline in CI as early as 3 days (Table 1; Fig. 3B).
Hemin coadministration alters B. pertussis multiplication and survival in vivo.
Heme compounds promote the growth of B. pertussis in laboratory cultures, even when inorganic iron is supplied in abundance (8; C. K. Vanderpool and S. K. Armstrong, unpublished observations). In the present study, hemin coadministration increased the numbers of CFU of both strains recovered at 3 days and reduced the rate of clearance after 14 days compared with the “no hemin” group (Fig. 3A). Interestingly, although hemin coadministration enhanced the competitive advantage of the wild-type strain over the mutant (Table 1; Fig. 3B), it caused a reduction in the maximal level of bacterial multiplication normally observed at 7 days for infections without added hemin (Fig. 3A). Bacteria that were administered without hemin decreased in number more than fourfold over the initial infecting dose by 3 days postinfection but then multiplied approximately 100-fold between 3 days and 7 days, reaching maximal numbers that were nearly fivefold greater than the “with hemin” infection at 7 days. In contrast, with hemin coadministration, numbers of CFU at 3 days and 7 days were sustained at levels similar to the infecting dose but then began to decline and never surpassed the infecting dose. Although the mechanism by which hemin coadministration limited the maximal numbers of CFU compared with the peak CFU of the “no hemin” infections at 7 days is unknown, it is hypothesized that activation of the heme system at the initiation of infection may provide the inducing signal at an inappropriate stage of infection, thus interfering with the ability of B. pertussis to optimally multiply in the host. It has been proposed that, early during natural infection, host heme sources available on respiratory mucosa may exist at lower concentrations compared to later stages of infection when pathological effects of B. pertussis toxins would be predicted to increase host heme availability (11). A different iron-scavenging mechanism, perhaps the alcaligin siderophore system, may be more effectively implemented to scavenge iron at the initial stages of colonization (4).
Surprisingly, even the heme receptor mutant seemed to benefit from hemin coadministration as evidenced by its sustained numbers at 3 days (Fig. 3A) and its reduced rate of clearance after 14 days. This suggests that this hemin-associated effect may not be related to iron supply per se but could instead reflect a more general protective mechanism afforded by heme. In laboratory-grown liquid cultures, heme compounds promote B. pertussis aggregation (C. K. Vanderpool and S. K. Armstrong, unpublished observations); perhaps aggregate formation improves initial survival of B. pertussis in a mammalian host. Alternatively, it may be relevant that certain porphyrin-containing bacterial hemoglobin-like proteins have been implicated in protection of pathogenic microbes from damage by reactive nitrogen species such as might be encountered in the host defensive environment (7). Heme may provide a measure of protection by a similar or related mechanism.
Success of B. pertussis as an obligate human pathogen depends on its ability to colonize the appropriate host site, multiply, and disseminate using the adaptive ecological strategies that it has refined through selection and coevolution with its host. The pathogenic potential of B. pertussis is defined by functions that either improve its in vivo fitness or decrease host fitness. Therefore, any function that contributes to B. pertussis fitness within a host may be considered to be a virulence determinant. By that criterion, these studies establish heme utilization as an important virulence determinant for B. pertussis in the murine respiratory infection model. Of key importance was the finding that other iron transport systems of B. pertussis were unable to compensate for the mutant's inability to utilize heme, confirming that the heme system has an independent role in virulence and is not redundant with other iron transport systems despite their common fundamental biological objective.
Acknowledgments
This work was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases and by University of Minnesota grant-in-aid 19473.
Editor: D. L. Burns
REFERENCES
- 1.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 2.Bordet, J., and O. Gengou. 1906. Le microbe de la coqueluche. Ann. Inst. Pasteur (Paris) 20:731-741. [Google Scholar]
- 3.Brickman, T. J., and S. K. Armstrong. 1996. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J. Bacteriol. 178:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brickman, T. J., C. K. Vanderpool, and S. K. Armstrong. 2004. Bordetella, p. 311-328. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, D.C.
- 5.Freter, R., B. Allweiss, P. C. O'Brien, S. A. Halstead, and M. S. Macsai. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vitro studies. Infect. Immun. 34:241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahan, M. J., D. M. Heithoff, R. L. Sinsheimer, and D. A. Low. 2000. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu. Rev. Genet. 34:139-164. [DOI] [PubMed] [Google Scholar]
- 7.Poole, R. K. 2005. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 33:176-180. [DOI] [PubMed] [Google Scholar]
- 8.Rowatt, E. 1957. Some factors affecting the growth of Bordetella pertussis. J. Gen. Microbiol. 17:279-296. [DOI] [PubMed] [Google Scholar]
- 9.Schneider, D. R., and C. D. Parker. 1982. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect. Immun. 38:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanderpool, C. K., and S. K. Armstrong. 2003. Heme-responsive transcriptional activation of Bordetella bhu genes. J. Bacteriol. 185:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanderpool, C. K., and S. K. Armstrong. 2004. Integration of environmental signals controls expression of Bordetella heme utilization genes. J. Bacteriol. 186:938-948. [DOI] [PMC free article] [PubMed] [Google Scholar]