Abstract
Toll-like receptors (TLRs) are a group of highly conserved molecules that initiate the innate immune response to pathogens by recognizing structural motifs expressed by microbes. We have identified a novel TLR, TLR15, by bioinformatic analysis of the chicken genome, which is distinct from any known vertebrate TLR and thus appears to be avian specific. The gene for TLR15 was sequenced and is found on chromosome 3, and it has archetypal TIR and transmembrane domains and a distinctive arrangement of extracellular leucine-rich regions. mRNA for TLR15 was detected in the spleen, bursa, and bone marrow of healthy chickens, suggesting a role for this novel receptor in constitutive host defense. Following in vivo Salmonella enterica serovar Typhimurium infection, quantitative real-time PCR demonstrated significant upregulation of TLR15 in the cecum of infected chickens. Interestingly, similar induction of TLR2 expression following infection was also observed. In vitro studies revealed TLR15 upregulation in chicken embryonic fibroblasts stimulated with heat-killed S. enterica serovar Typhimurium. Collectively, these results suggest a role for the TLR in avian defense against bacterial infection. We hypothesize that TLR15 may represent an avian-specific TLR that has been either retained in chicken and lost in other taxa or gained in the chicken.
Salmonella enterica serovar Typhimurium infections in humans cause typhoid fever and enteritis (24, 27, 35), with the latter constituting the second most common cause of food-borne disease in the United States (19). Animal models for typhoid fever and enteritis have largely been conducted in mice and calves, respectively (24). Although the host immune response to Salmonella infection in chickens is not well characterized, recent work examining the production of proinflammatory cytokines and chemokines following serovar Typhimurium infection in young chickens revealed that many elements of the avian host response are similar to those in mammalian models (29).
Toll-like receptors (TLRs) are pathogen recognition receptors, which initiate the pathways controlling expression of cytokines and chemokines and represent a link between innate and acquired immunity (28). TLRs were initially identified in vertebrates by homology to the transmembrane Toll protein in Drosophila melanogaster which regulates early embryonic development as well as mediating innate immune mechanisms. TLRs are widely expressed by many cell types, including leukocytes and epithelial cells, and function through recognition and interaction with conserved motifs expressed on the surface of invading pathogens, known as pathogen-associated molecular patterns (PAMPs). PAMPs, such as lipopolysaccharides (LPS) and peptidoglycan, are essential structural components of the bacterial cell wall, and mutations within them are deleterious to microbes, hence PAMPs are relatively resistant to mutation and are ideal pathogen recognition receptor ligands.
The carboxy-terminal, cytoplasmic tail of the Toll receptors and all TLRs shares striking homology with the type 1 mammalian interleukin-1 (IL-1) receptor, and this motif is known as the Toll/IL-1 receptor (TIR) domain (21). Activation of each TLR by a relevant PAMP results in the TIR domain initiating a signaling cascade that shares many similarities with the Toll signaling pathway in Drosophila (8). This ultimately results in the translocation of NF-κB to the nucleus and the initiation of appropriate gene transcription leading to the production of many proinflammatory cytokines, gamma/beta interferon, and antimicrobial peptides.
In the chicken, prior to the genome release, in silico clustering of expressed sequence tags revealed homologues of the TLR pathway including two TLRs, one homologous to human TLR3 and the other similar to human and mouse TLR1, TLR6, and TLR10 (16). Other chicken TLR pathway genes identified include those encoding Toll interacting protein (TOLLIP), interleukin-1 receptor-associated kinase 4 (IRAK4), myeloid differentiation factor 88 (MyD88), MyD88-adapter-like protein (Mal or TIRAP), Tak1-binding proteins 1 and 2 (TAB1 and TAB2), tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6), transforming growth factor beta (TGFβ)-activated kinase (TAK1), and IκB kinases α and β (IKKα and IKKβ) (16). Chicken TLR1 (types 1 and 2), TLR2 (types 1 and 2), TLR3, TLR4, TLR5, and TLR7 have previously been described (2, 7, 10, 14, 16, 22, 33) and recently shown to be expressed in chicken heterophils (11). Both forms of chicken TLR2, with identical TIR domains, have been mapped to the same region of chromosome 4 and are thought to have arisen from recent gene duplication (2, 7). Furthermore, TLR2 type 2 was shown to act as a receptor for lipoprotein and, similar to studies of human TLR2 (5), was demonstrated to recognize LPS in the presence of MD-2 (7). Chicken TLR4 has been shown to play a role in resistance to Salmonella infection, with susceptible chickens bearing certain allelic variations in their TLR4 sequence (14). Exposure of cells expressing chicken TLR5 to flagellin induced chicken IL-1β upregulation, and the receptor is thought to be involved in restricting the entry of flagellated Salmonella into systemic sites (10). Exposure of the TLR7-expressing chicken HD11 cell line and splenocytes to TLR7 agonists R848 and loxoribine resulted in increased production of IL-1β, which could be abrogated by addition of 100 μM chloroquine, indicating that chicken TLR7 may be endosomal, as with mammalian TLR7 (22).
During the course of an extensive bioinformatic and functional analysis of immune response genes in the chicken genome, we characterized a new TLR, which we designate chicken TLR15. Here we show the phylogenetic relationship of TLR15 with known TLRs and the tissue distribution of TLR15 mRNA expression in comparison with established chicken TLRs. Furthermore, we report the upregulation of the novel TLR15 mRNA in the cecum of chickens following Salmonella enterica serovar Typhimurium infection and in chicken embryonic fibroblasts following stimulation with heat-killed S. enterica serovar Typhimurium.
MATERIALS AND METHODS
Bioinformatics.
Publicly available protein sequences corresponding to the 10 known human TLRs were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/GenBank). Following its March 2004 release, the DNA sequences of the initial assembled chromosomes of the sequenced chicken genome were searched using the human TLR peptide sequences with the TBLASTN program (1). The subsequent release of the Ensembl dataset of 28,416 predicted chicken genes was also searched for sequences displaying a high degree of similarity to human TLRs. Chromosomal location and strand orientation of the TLRs were determined using the BLAST Like Alignment Tool (BLAT) at the University of California—Santa Cruz genome browser (http://genome.ucsc.edu). Genomic DNA corresponding to putative TLRs was retrieved using BLAT and used for prediction of intron-exon boundaries using the GenScan program (http://genes.mit.edu/GENSCAN.html) (3). Domain predictions for the novel TLR15 were carried out using the SMART programs (25). The proposed chicken homologues of known mammalian TLRs were further analyzed by aligning them with homologous sequences from other vertebrate species using the T-COFFEE multiple sequence alignment program (20), while phylogenetic analysis of the proteins was carried out using Mega v.2.1 (12).
Salmonella infection: 5-week-old challenge model.
Broiler chickens were obtained from a commercial hatchery in Saskatchewan, Canada, at 1 day of age. They were reared in an isolation room until 5 weeks of age. The challenged birds had 0.5 ml of Salmonella enterica serovar Typhimurium strain SL1344 suspended in 0.85% NaCl administered orally. S. enterica serovar Typhimurium was grown in modified N-minimal medium containing 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 100 mM Tris-HCl (pH 7.0), 38 mM glycerol, 0.1% Casamino Acids, 24 mM MgCl2, 337 μM 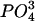 at 37°C with shaking at 200 rpm until an optical density at 600 nm of 0.7 was reached. The cells were harvested by centrifugation and resuspended in 0.85% NaCl to one-tenth the original volume. The number of viable bacteria present in the challenge was determined by viable cell counts on Luria-Bertani (LB) agar plates and calculated to be 3.5 × 109/ml. At 48 h after challenge, birds were euthanized by cervical dislocation. To determine if colonization of the gut by Salmonella had occurred, swabs of the cecal contents were cultured on Brilliant Green Agar. Tissue samples from the tongue, liver, spleen, small intestine, cecum, bursa, and bone marrow were quickly dissected, rinsed in saline, and stored in RNAlater (QIAGEN Ltd., West Sussex, United Kingdom). All animal experiments were conducted according to the guidelines provided by the Canadian Council on Animal Care.
at 37°C with shaking at 200 rpm until an optical density at 600 nm of 0.7 was reached. The cells were harvested by centrifugation and resuspended in 0.85% NaCl to one-tenth the original volume. The number of viable bacteria present in the challenge was determined by viable cell counts on Luria-Bertani (LB) agar plates and calculated to be 3.5 × 109/ml. At 48 h after challenge, birds were euthanized by cervical dislocation. To determine if colonization of the gut by Salmonella had occurred, swabs of the cecal contents were cultured on Brilliant Green Agar. Tissue samples from the tongue, liver, spleen, small intestine, cecum, bursa, and bone marrow were quickly dissected, rinsed in saline, and stored in RNAlater (QIAGEN Ltd., West Sussex, United Kingdom). All animal experiments were conducted according to the guidelines provided by the Canadian Council on Animal Care.
Expression of TLRs in chicken tissues.
Following pulverization of the tissues using a PRO200 Homogenizer (PRO Scientific Inc.), total cellular RNA was purified using the RNeasy Kit (QIAGEN Ltd.) according to the manufacturer's recommendations. The quantity and quality of total RNA was assessed using the Agilent Bioanalyser and UV spectrophotometric analysis. Single-stranded cDNA was synthesized from 1 μg of total RNA using oligo(dT) primer (Promega, Madison, WI) and Omniscript (QIAGEN Ltd.) in a 20-μl reaction mixture following recommendations of the manufacturers. The gene-specific cDNAs were PCR amplified using Taq polymerase (QIAGEN Ltd.) and primers designed internally from the coding sequence of TLR2, TLR4, TLR15, IL-1β, and 18S RNA (intron spanning wherever possible). An initial 94°C step for 8 min was followed by 30 cycles (94°C, 55°C, and 72°C, each for 30 s for denaturation, annealing, and extension, respectively) for all PCR amplifications with the exception of TLR4, which required an annealing temperature of 65°C for 30 s. PCR products were separated by electrophoresis on ethidium bromide containing 2% agarose gels and visualized using an AutoChemi System (Ultra Violet Products Ltd.). Relevant PCR products were sequenced to ensure their correct identity. A list of the PCR primer sequences and their PCR product lengths are shown in Table 1. The TLR15 coding sequence was fully sequenced using a combination of specific forward and reverse primers (F1, 5′-ATGAGGATCCTTATTGGG AG-3′; F2, 5′-TGACTTGTGTGGAGCACCGAT-3′; F3, 5′-TACACCCATCGA AAGCCT-3′; F4, 5′-ATCAGGGAATAAGATCTC-3′; R1, 5′-GCTGTCAGC TCTTCATTAGA-3′; R2, 5′-TGGAGCAGTTGGACACTT-3′; R3, 5′-GAT GGCGTTGTCGCTAATGT-3′; and R4, 5′-TACAGTTCATACTGACACCA-3′) and submitted to GenBank (accession number DQ267901).
TABLE 1.
PCR primer sequences and predicted product lengths
| Target mRNA | 5′ Primer | 3′ Primer | Product size (bp) |
|---|---|---|---|
| TLR2 | 5′-GATTGTGGACAACATCATTGACTC-3′ | 5′-AGAGCTGCTTTCAAGTTTTCCC-3′ | 294 |
| TLR4 | 5′-AGTCTGAAATTGCTGAGCTCAAAT-3′ | 5′-GCGACGTTAAGCCATGGAAG-3′ | 190 |
| TLR15 | 5′-GTTCTCTCTCCCAGTTTTGTAAATAGC-3′ | 5′-GTGGTTCATTGGTTGTTTTTAGGAC-3′ | 262 |
| IL-1β | 5′-CGCTCACAGTCCTTCGACATC-3′ | 5′-CCGCTCATCACACACGACATGT-3′ | 230 |
| 18S RNA | 5′-GCTGAAACTTAAAGGAATTGACG-3′ | 5′-CTGTTATTGCTCAATCTCGGG-3′ | 304 |
Cell culture.
Chicken embryonic fibroblasts were grown in Dulbecco's modified Eagle medium supplemented with 10% (wt/vol) heat-inactivated fetal calf serum (Gibco BRL) and maintained in a CO2-free environment at 37°C. The day before stimulation, cells were seeded at a density of 5 × 105 cells/ml in 25-cm3 cell culture flasks. Cells were treated with 107 heat-killed Salmonella enterica serovar Typhimurium cells (KPL Europe)/ml and incubated for 6, 24, and 48 h in antibiotic-free medium. Total cellular RNA was purified using the RNeasy kit (QIAGEN Ltd.) according to the manufacturer's recommendations.
Real-time PCR.
In order to quantify TLR and IL-1β mRNA expression, quantitative real-time PCR was performed using SYBR Green Taq ReadyMix (Sigma Chemical Co., Poole, United Kingdom) according to the manufacturer's recommendations. Briefly, 10 μl of ReadyMix was added to 2.5 mM MgCl2, 0.2 μM forward and reverse primer, 0.1-μg equivalent of reverse-transcribed RNA in a cDNA reaction mix and made to 20 μl with H2O. Thirty-five cycles of amplification were performed in duplicate on a LightCycler real-time PCR machine (Roche Molecular Biochemicals) at annealing temperatures of 56°C for IL-1β and TLR15 and 58°C for TLR2 and TLR4.
Real-time PCR data were analyzed using the 2−ΔΔCt method (15) to calculate the relative level of each mRNA in each sample and expressed as a ratio relative to 18S rRNA housekeeper genes. The relative abundance of the mRNAs was calculated by dividing the values for Salmonella-infected samples by those values from uninfected samples after the necessary housekeeper gene corrections to give a fold change value (ΔΔCt) (for ΔΔCt values <1, −1/ΔΔCt = fold change). An average of duplicate values gave a more accurate indication of difference. Values from both experimental groups were compared using a Student's t test on logged data to account for the possibility of nonnormal distributions in the variance of the different treatment groups. The data for Salmonella-infected samples are expressed relative to the average value of the control samples ± standard errors of the means (see Fig. 4). The correlation between fold change expression values for each gene was carried out using the CORREL function in Microsoft Excel.
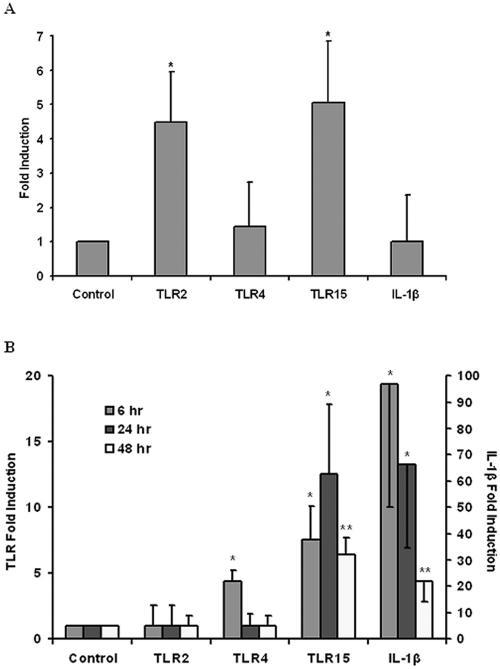
FIG. 4.
(A) Differential expression of TLR2, TLR4, TLR15, and IL-1β mRNAs in the cecum of five 2-day-old Salmonella enterica serovar Typhimurium-infected chickens expressed relative to those of five uninfected control chickens. Quantitative real-time PCR analysis was performed with 18S RNA as an internal positive control. TLR2 and TLR15 mRNAs were significantly upregulated, with P values of less than 0.05 (*). Error bars indicate the standard error of the means. (B) Differential expression of TLR2, TLR4, TLR15, and IL-1β mRNAs in chicken embryonic fibroblasts stimulated with heat-killed Salmonella enterica serovar Typhimurium for 6, 24, and 48 h expressed relative to those of unstimulated control cells. The axis on the left indicates the fold induction of the TLRs, while the axis on the right indicates the fold induction of IL-1β. Quantitative real-time PCR analysis was performed with 18S RNA as an internal positive control. Asterisks indicate P values of less than 0.05 (*) and less than 0.01 (**). The data are representative of four independent experiments. Error bars indicate the standard errors of the means.
Nucleotide sequence accession number.
The TLR15 coding sequence was submitted to GenBank under accession number DQ267901.
RESULTS AND DISCUSSION
To date, 13 TLR family members have been identified in mammals, although not all members of the family are present in all species; humans have only 10 functional TLRs, for example (34). Sequence information for nonmammalian TLRs has only begun to accumulate in recent years. The identification of chicken TLR1 (types 1 and 2), TLR3, TLR5, and TLR7 (2, 7, 10, 16, 22, 33) as well as the previously characterized chicken TLR2 (types 1 and 2) and chicken TLR4 have helped generate a more complete picture of the chicken TLR catalog. As well as confirming the presence of all the above chicken TLRs, initial homology searches identified a further putative TLR coding sequence, which was named chicken TLR15. The entire TLR15 coding sequence was sequenced and submitted to GenBank (accession number DQ267901). The sequence was in agreement with the bioinformatic prediction, except for a synonymous change at base positions 168 (A to G) and 450 (C to T) and a nonsynonymous change at base positions 926 (A to C) and 1363 (T to G). The nonsynonymous changes result in amino acid changes from glutamic acid to alanine and from leucine to valine, respectively. However, both amino acids are located in the variable extracellular region of the protein and not in the conserved TIR domain, and thus they do not compromise the status as a novel TLR. It should be noted that TLR15 was named sequentially, as it appears to have an extracellular domain structure distinctive from that of any of the known vertebrate TLRs.
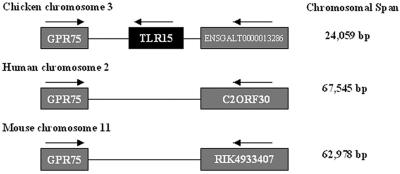
TLR15 was bioinformatically mapped to chromosome 3 (chr3:2925041-2927644) in the February 2004 assembly of the chicken genome. The TLR15 locus and surrounding chromosomal portion display a high degree of synteny with conserved areas on the human 2 and mouse 11 chromosomes (Fig. 1). The syntenic regions in human and mouse were searched for the presence of genes corresponding to the chicken TLR15. No orthologous genes were identified using both the Genscan gene prediction program (3) and a Hidden Markov Model search strategy (6) based on the TIR domain.
FIG. 1.
Comparative genomic synteny in chicken, human, and mouse for flanking genes of chicken TLR15. GPR75, G-protein coupled receptor 75. ENSGALT0000013286, C2ORF30, and RIK4933407 are homologous sequences in chicken, human, and mouse, respectively, for which functionality has not yet been assigned. The figure is not to scale. Syntenic region span lengths are indicated.
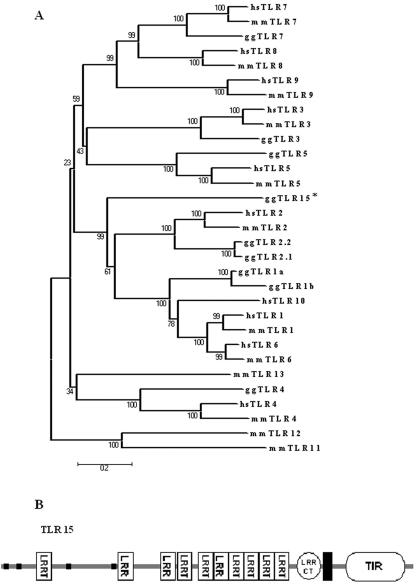
Phylogenetic analysis was carried out using known human and mouse TLRs to construct a multiple-sequence alignment (data not shown) and a phylogenetic tree (Fig. 2A). TLR15 groups with high bootstrap support with the recognized TLR1/TLR2/TLR6/TLR10 clade but displays no close relationship to any particular clade member, the best identity being 30.1% with chicken TLR2. TLR15 is also 29.2% and 29.7% similar to fugu and zebrafish TLR14, respectively. These identities, and the pattern in the phylogenetic tree, are not sufficient to assign a clear 1:1 orthology, thus TLR15 is molecularly distinct from all known TLRs (23). This contrasts with the majority of previously described chicken TLRs (TLR2, TLR3, TLR4, TLR5, and TLR7), which are well conserved as orthologues in amniotes (a taxon that includes birds and mammals). Other parts of the TLR tree show taxon-specific variation, which is in a process of active evolutionary development; TLR11 in mouse recognizes profilin from Toxoplasma gondii (32) and uropathogenic Escherichia coli but is a pseudogene in humans (34). One implication of the phylogenetic tree (Fig. 2A) is that chicken TLR1 may cover the functional specificity of mammalian TLR1, TLR6, and TLR10. It is possible that TLR15 may provide avian-specific functionality to compensate for the lack of variability in TLR1, TLR6, and TLR10, or it may have evolved to counter avian pathogens for which the preferential host is the chicken. Homologues of TLR15 were searched for but not found in any other available avian sequences, but as chicken is the only completely sequenced species in this taxon, absence of evidence is not evidence of absence. TLR evolution is dynamic, because the pathological challenges to vertebrate species do not stand still. Functional characterization studies to determine the antigenic specificity of the proposed TLR15 could allow for a more definitive classification.
FIG. 2.
(A) Neighbor-joining tree generated from amino acid sequences of human (hs), mouse (mm), and chicken (gg) TLR homologues reconstructed using Mega v.2.1. The results of 1,000 bootstrap replicates are shown at each node. Bootstrap analysis is based on multiple resampling of the original data and is the most common method of estimating the degree of confidence in the topology of phylogenetic trees. The asterisk indicates chicken TLR15. (B) Organization of secondary structural domains of TLR15. A horizontal black bar represents the transmembrane domain, while small black squares represent sequences of low compositional complexity. TIR, Toll/IL-1 receptor domain; LRR, leucine-rich repeat; LRRT, leucine-rich repeat typical subfamily; LRRCT, leucine-rich repeat C-terminal domain.
Analysis of the domain structure of TLR15 revealed an archetypal TLR structure comprising a cytoplasmic Toll/IL-1 receptor (TIR) region, a short transmembrane domain, and an extracellular domain with multiple leucine-rich regions of varying length and sequence compositions (Fig. 2B). This evidence supports the proposal of this sequence as a novel chicken TLR. Furthermore, the predicted peptide for chicken TLR15 is coded for by a single exon, a feature common to all the mammalian members of the TLR1/TLR2/TLR6/TLR10 clade.
A panel of seven tissues from broiler chickens (n = 5 to 7) was examined for mRNA expression of TLR2, TLR4, and TLR15 (Fig. 3). In these healthy chickens, expression of TLR2 and TLR15 mRNAs was similar. Interestingly, the transcripts of these TLRs were abundant in the bone marrow and bursa of Fabricius, which are not associated with a primary defense function but with lymphoid development. Slightly lower mRNA expression levels of these TLRs were observed in the spleen, which has primary immunological function. However, the transcript levels of these TLRs were least abundant in the liver, the small intestine, the tongue, and the cecum. Other investigators have also shown that TLR2 mRNA is highly expressed in the bursa and spleen of a White Leghorn chicken, while it is present at lower levels in the small intestine, cecum, and liver (9, 22). However, a study using tissues from a White Leghorn hen detected high expression of TLR2 mRNA in the liver, small intestine, large intestine, and spleen (33). The evidence from our study and the related studies discussed suggests that regulation of TLR2 mRNA expression is complex and variable in healthy chickens.
FIG. 3.
Gene-specific PCR products showing levels of expression of TLR2, TLR4, and TLR15 mRNAs in a panel of tissues from healthy chickens. Total RNA was extracted from 5 to 7 chickens, and cDNAs were made and amplified as described in the text. 18S RNA was used as an internal positive control for normalization. Negative controls (NC) contain no cDNA template.
TLR4 mRNA was expressed primarily in the bone marrow, with very little or no expression in other tissues. This result suggests that TLR4 expression may be strain specific, as White Leghorn chickens showed a broader pattern of tissue expression of TLR4 mRNA through Northern blot analysis (14) and also when primers identical to (9) or different from (33) those used in this study are used to PCR amplify the transcript.
A Salmonella enterica serovar Typhimurium infection model examined the levels of certain key TLR mRNAs following bacterial colonization in the chicken cecum. Cecal swabs were plated to confirm bacterial colonization 48 h postbacterial challenge (two infected chickens had <10 serovar Typhimurium colonies per plate, two had 51 to 100 colonies per plate, and one had >100 colonies per plate; the five uninfected chickens had 0 colonies per plate). Quantitative real-time PCR was performed to determine mRNA fold induction following Salmonella enterica serovar Typhimurium infection. TLR15 was significantly induced fivefold (P = 0.02) at 48 h postinfection compared to controls (Fig. 4A). Interestingly, TLR2 mRNA was also upregulated 4.5-fold (P = 0.01) in the cecum compared to control birds (Fig. 4A). In mammals, TLR2 and TLR4 are known to mediate the response to LPS in vitro (4, 18, 31), and chicken TLR2 (type 2) has been shown to recognize both lipoprotein and LPS (7), similar to human TLR2 (5). However, it has previously been shown that TLR2, and not TLR4, is significantly induced in mouse macrophages following stimulation with LPS and various cytokines (17). We also observed no upregulation of chicken TLR4 transcript levels following Salmonella infection. TLR4 is thought to be involved in a biphasic regulation of the expression of TLR2, initially upregulating it followed by a self-inhibitory effect (26). It has been shown that mouse TLR2 upregulation in response to Salmonella infection is reduced in C3H/HeJ mice, which carry mutated TLR4 (26), supporting a regulatory role for TLR2 of TLR4. Thus, the observed upregulation of TLR2 may be a direct consequence of initial TLR4 activation followed by self downregulation. TLR15, whose transcript level also increases following Salmonella infection, might also be regulated by activation of other TLRs or the production of proinflammatory cytokines. Furthermore, both TLR2 and TLR15 might be responding to similar or identical ligands. If this is the case, it supports the hypothesis that there is redundancy in LPS signaling and that TLR15 can act as a surrogate for TLR2 (18). The lack of IL-1β upregulation was unexpected, as the cecal swabs confirmed bacterial colonization. However, it may be that at 48 h postinfection IL-1β levels have decreased from an initial induction. A recent study in which chickens were similarly infected with Salmonella enterica serovar Typhimurium also reported no increase in IL-1β in the gastrointestinal tract at 1, 3, or 7 days postinfection (30).
As TLRs are known to form both homodimers and heterodimers in order to activate intracellular signaling pathways, TLR2 and TLR15 fold inductions were compared. A high degree of similarity in gene expression patterns was observed (correlation coefficient of 0.99) between individual chickens in the infected group for TLR2 and TLR15 (data not shown). PCR products were sequenced to confirm that distinct transcripts were being amplified (data not shown). This induction of highly correlated fold changes for both genes in response to infection is interesting. Further functional analysis would be necessary to reveal if the novel TLR15 forms a heterodimer with TLR2 following Salmonella enterica serovar Typhimurium infection.
Embryonic fibroblasts have been shown to be suitable model cell lines to study TLR responses in the mouse (13) and chicken (10). In addition to the in vivo infection model, chicken embryonic fibroblasts were stimulated with heat-killed Salmonella enterica serovar Typhimurium cells for 6, 24, and 48 h. TLR15 was upregulated 7.5-fold (P = 0.04) compared to uninfected controls at 6 h poststimulation, 12.5-fold (P = 0.04) at 24 h poststimulation, and 6.5-fold (P = 0.003) at 48 h poststimulation (Fig. 4B). This level of TLR15 upregulation corresponds to that seen in the in vivo model. The proinflammatory cytokine IL-1β was upregulated 97-fold (P = 0.02) at 6 h poststimulation, which is indicative of an early vigorous innate immune response, including activation of TLRs. At subsequent time points, IL-1β upregulation was reduced in a time-dependent manner to a 66-fold increase (P = 0.04) at 24 h poststimulation and a 22-fold increase (P = 0.003) at 48 h poststimulation (Fig. 4B). This time-dependent reduction reflects that observed in the in vivo model, where IL-1β levels ininfected chickens were similar to those of controls at 48 h postinfection. TLR4 was upregulated 4.5-fold (P = 0.01) at 6 h poststimulation, corresponding with the large increase in IL-1β, but was not induced at the later time points; TLR2 remained unchanged at all time points. It may be that TLR2 was not induced in the in vitro model due to the absence of more physiologically relevant TLR-expressing cells, or perhaps the use of heat-killed bacteria, in contrast to live bacteria, may alter the PAMP profile of the pathogen resulting in the lack of TLR2 stimulation.
In conclusion, we report the discovery of the novel chicken TLR15 and initial work on the tissue expression and induction of this and other TLRs. TLR15 was found to be significantly upregulated in the cecum of Salmonella enterica serovar Typhimurium-infected chickens. Interestingly, induction of TLR15 correlated significantly with induction of TLR2. TLR15 was also induced in chicken embryonic fibroblasts following stimulation with heat-killed Salmonella enterica serovar Typhimurium. Further characterization of the chicken TLR repertoire is under way and will represent a significant step in tracing the evolutionary history and divergence pattern of this immunologically important gene family. Furthermore, it is becoming increasingly clear that analysis of the evolutionary relationships among genes can offer significant insights into the different functions of the cognate proteins. TLR15 may represent an avian-specific TLR that has been either retained in chicken and lost in other taxa or gained in the chicken. It is likely that similarly unique TLR genes are present in other genomes that contribute to species- or class-specific immune defense mechanisms tailored to combat particular pathogens.
Acknowledgments
We acknowledge the Vaccine Development Group technical staff and the Animal Care team at the Vaccine and Infectious Disease Organization, Saskatoon, Saskatchewan, Canada, for provision and care of the chickens and sample collection.
This research was supported by the Food Institutional Research Measure (F.I.R.M.), grant 01/R&D/D/135, from the Irish Department of Agriculture, Food, and Rural Development and Genome Prairie, Genome BC, and Inimex Pharmaceuticals through the “Functional Pathogenomics of Mucosal Immunity” project.
All experiments described in this study comply with the current laws of the Republic of Ireland and Canada.
Editor: A. D. O'Brien
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, Y., M. Goodchild, S. Morroll, and N. Bumstead. 2001. Mapping of the chicken and mouse genes for toll-like receptor 2 (TLR2) to an evolutionarily conserved chromosomal segment. Immunogenetics 52:294-298. [DOI] [PubMed] [Google Scholar]
- 3.Burge, C., and S. Karlin. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268:78-94. [DOI] [PubMed] [Google Scholar]
- 4.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 5.Dziarski, R., Q. Wang, K. Miyake, C. J. Kirschning, and D. Gupta. 2001. MD-2 enables Toll-like receptor 2 (TLR2)-mediated responses to lipopolysaccharide and enhances TLR2-mediated responses to gram-positive and gram-negative bacteria and their cell wall components. J. Immunol. 166:1938-1944. [DOI] [PubMed] [Google Scholar]
- 6.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 7.Fukui, A., N. Inoue, M. Matsumoto, M. Nomura, K. Yamada, Y. Matsuda, K. Toyoshima, and T. Seya. 2001. Molecular cloning and functional characterization of chicken toll-like receptors. A single chicken toll covers multiple molecular patterns. J. Biol. Chem. 276:47143-47149. [DOI] [PubMed] [Google Scholar]
- 8.Hallman, M., M. Ramet, and R. A. Ezekowitz. 2001. Toll-like receptors as sensors of pathogens. Pediatr. Res. 50:315-321. [DOI] [PubMed] [Google Scholar]
- 9.Iqbal, M., V. J. Philbin, and A. L. Smith. 2005. Expression patterns of chicken Toll-like receptor mRNA in tissues, immune cell subsets and cell lines. Vet. Immunol. Immunopathol. 104:117-127. [DOI] [PubMed] [Google Scholar]
- 10.Iqbal, M., V. J. Philbin, G. S. Withanage, P. Wigley, R. K. Beal, M. J. Goodchild, P. Barrow, I. McConnell, D. J. Maskell, J. Young, N. Bumstead, Y. Boyd, and A. L. Smith. 2005. Identification and functional characterization of chicken toll-like receptor 5 reveals a fundamental role in the biology of infection with Salmonella enterica serovar Typhimurium. Infect. Immun. 73:2344-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kogut, M. H., M. Iqbal, H. He, V. Philbin, P. Kaiser, and A. Smith. 2005. Expression and function of Toll-like receptors in chicken heterophils. Dev. Comp. Immunol. 29:791-807. [DOI] [PubMed] [Google Scholar]
- 12.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 13.Kurt-Jones, E. A., F. Sandor, Y. Ortiz, G. N. Bowen, S. L. Counter, T. C. Wang, and R. W. Finberg. 2004. Use of murine embryonic fibroblasts to define Toll-like receptor activation and specificity. J. Endotoxin Res. 10:419-424. [DOI] [PubMed] [Google Scholar]
- 14.Leveque, G., V. Forgetta, S. Morroll, A. L. Smith, N. Bumstead, P. Barrow, J. C. Loredo-Osti, K. Morgan, and D. Malo. 2003. Allelic variation in TLR4 is linked to susceptibility to Salmonella enterica serovar Typhimurium infection in chickens. Infect. Immun. 71:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC[T]) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 16.Lynn, D. J., A. T. Lloyd, and C. O'Farrelly. 2003. In silico identification of components of the Toll-like receptor (TLR) signaling pathway in clustered chicken expressed sequence tags (ESTs). Vet. Immunol. Immunopathol. 93:177-184. [DOI] [PubMed] [Google Scholar]
- 17.Matsuguchi, T., T. Musikacharoen, T. Ogawa, and Y. Yoshikai. 2000. Gene expressions of Toll-like receptor 2, but not Toll-like receptor 4, is induced by LPS and inflammatory cytokines in mouse macrophages. J. Immunol. 165:5767-5772. [DOI] [PubMed] [Google Scholar]
- 18.Matsuguchi, T., K. Takagi, T. Musikacharoen, and Y. Yoshikai. 2000. Gene expressions of lipopolysaccharide receptors, toll-like receptors 2 and 4, are differently regulated in mouse T lymphocytes. Blood 95:1378-1385. [PubMed] [Google Scholar]
- 19.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 21.O'Neill, L. 2000. The Toll/interleukin-1 receptor domain: a molecular switch for inflammation and host defence. Biochem. Soc. Trans. 28:557-563. [DOI] [PubMed] [Google Scholar]
- 22.Philbin, V. J., M. Iqbal, Y. Boyd, M. J. Goodchild, R. K. Beal, N. Bumstead, J. Young, and A. L. Smith. 2005. Identification and characterization of a functional, alternatively spliced Toll-like receptor 7 (TLR7) and genomic disruption of TLR8 in chickens. Immunology 114:507-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roach, J. C., G. Glusman, L. Rowen, A. Kaur, M. K. Purcell, K. D. Smith, L. E. Hood, and A. Aderem. 2005. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. USA 102:9577-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335-1344. [DOI] [PubMed] [Google Scholar]
- 25.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Totemeyer, S., N. Foster, P. Kaiser, D. J. Maskell, and C. E. Bryant. 2003. Toll-like receptor expression in C3H/HeN and C3H/HeJ mice during Salmonella enterica serovar Typhimurium infection. Infect. Immun. 71:6653-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 28.Werling, D., and T. W. Jungi. 2003. TOLL-like receptors linking innate and adaptive immune response. Vet. Immunol. Immunopathol. 91:1-12. [DOI] [PubMed] [Google Scholar]
- 29.Withanage, G. S., P. Kaiser, P. Wigley, C. Powers, P. Mastroeni, H. Brooks, P. Barrow, A. Smith, D. Maskell, and I. McConnell. 2004. Rapid expression of chemokines and proinflammatory cytokines in newly hatched chickens infected with Salmonella enterica serovar Typhimurium. Infect. Immun. 72:2152-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Withanage, G. S., P. Wigley, P. Kaiser, P. Mastroeni, H. Brooks, C. Powers, R. Beal, P. Barrow, D. Maskell, and I. McConnell. 2005. Cytokine and chemokine responses associated with clearance of a primary Salmonella enterica serovar Typhimurium infection in the chicken and in protective immunity to rechallenge. Infect. Immun. 73:5173-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, R. B., M. R. Mark, A. Gray, A. Huang, M. H. Xie, M. Zhang, A. Goddard, W. I. Wood, A. L. Gurney, and P. J. Godowski. 1998. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature 395:284-288. [DOI] [PubMed] [Google Scholar]
- 32.Yarovinsky, F., D. Zhang, J. F. Andersen, G. L. Bannenberg, C. N. Serhan, M. S. Hayden, S. Hieny, F. S. Sutterwala, R. A. Flavell, S. Ghosh, and A. Sher. 2005. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 308:1626-1629. [DOI] [PubMed] [Google Scholar]
- 33.Yilmaz, A., S. Shen, D. L. Adelson, S. Xavier, and J. J. Zhu. 2005. Identification and sequence analysis of chicken Toll-like receptors. Immunogenetics 56:743-753. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, D., G. Zhang, M. S. Hayden, M. B. Greenblatt, C. Bussey, R. A. Flavell, and S. Ghosh. 2004. A toll-like receptor that prevents infection by uropathogenic bacteria. Science 303:1522-1526. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, S., R. A. Kingsley, R. L. Santos, H. Andrews-Polymenis, M. Raffatellu, J. Figueiredo, J. Nunes, R. M. Tsolis, L. G. Adams, and A. J. Baumler. 2003. Molecular pathogenesis of Salmonella enterica serotype Typhimurium-induced diarrhea. Infect. Immun. 71:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]