Abstract
4-Coumarate:coenzyme A ligase (4CL) plays a key role in phenylpropanoid metabolism, providing precursors for a large variety of important plant secondary metabolites, such as lignin, flavonoids, and phytoalexins. Although 4CLs have been believed to be specific to plants, a gene encoding a 4CL-like enzyme which shows more than 40% identity in amino acid sequence to plant 4CLs was found in the genome of the gram-positive, filamentous bacterium Streptomyces coelicolor A3(2). The recombinant enzyme, produced in Escherichia coli with a histidine tag at its N-terminal end, showed distinct 4CL activity. The optimum pH and temperature of the reaction were pH 8.0 and 30°C, respectively. The Km value for 4-coumarate and kcat were determined as 131 ± 4 μM and 0.202 ± 0.007 s−1, respectively. The Km value was comparable to those of plant 4CLs. The substrate specificity of this enzyme was, however, distinctly different from those of plant 4CLs. The enzyme efficiently converted cinnamate (Km, 190 ± 2 μM; kcat, 0.475 ± 0.012 s−1), which is a very poor substrate for plant 4CLs. Furthermore, the enzyme showed only low activity toward caffeate and no activity toward ferulate, both of which are generally good substrates for plant 4CLs. The enzyme was therefore named ScCCL for S. coelicolor A3(2) cinnamate CoA ligase. To determine the amino acid residues providing the unique substrate specificity of ScCCL, eight ScCCL mutant enzymes having a mutation(s) at amino acid residues that probably line up along the substrate-binding pocket were generated. Mutant A294G used caffeate as a substrate more efficiently than ScCCL, and mutant A294G/A318G used ferulate, which ScCCL could not use as a substrate, suggesting that Ala294 and Ala318 are involved in substrate recognition. Furthermore, the catalytic activities of A294G and A294G/A318G toward cinnamate and 4-coumarate were greatly enhanced compared with those of the wild-type enzyme.
The enzyme 4-coumarate:coenzyme A (CoA) ligase (4CL; EC 6.2.1.12) catalyzes the last reaction of the general phenylpropanoid pathway in plants. This pathway channels carbon flow from primary metabolism to different branch pathways of secondary phenolic metabolism via the sequential action of the enzymes phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase, and 4-coumarate:CoA ligase (4CL). PAL catalyzes the conversion of phenylalanine to cinnamate, and cinnamate 4-hydroxylase catalyzes the subsequent 4-hydroxylation of cinnamate to 4-coumarate. Finally, 4CL catalyzes the conversion of 4-coumarate (4-hydroxycinnamate) and other substituted cinnamates, such as caffeate (3,4-dihydroxycinnamate) and ferulate (3-methoxy-4-hydroxycinnamate), into the corresponding CoA thiol esters, which are used for the biosynthesis of numerous phenylpropanoid-derived compounds, such as lignins, lignans, suberins, flavonoids, isoflavonoids, and various small phenolic compounds. These compounds serve diverse functions in plants, including mechanical support and rigidity in cell walls, attractants for insect pollinators, protection against biotic and abiotic stresses, and roles in signaling (4, 8, 9, 14, 29). Because of the importance of phenylpropanoid-derived products in plants, 4CL has been the subject of extensive study for 25 years.
4CL-catalyzed CoA ester formation takes place via a two-step reaction. During the first step, 4-coumarate and ATP form a coumaroyl-adenylate intermediate with the simultaneous release of pyrophosphate. In the second step, the coumaroyl group is transferred to the sulfhydryl group of CoA, and AMP is released (2, 19). The mechanism of formation of an adenylate intermediate is common among a number of enzymes with divergent functions, including luciferases, fatty acyl-CoA ligases, acetyl-CoA ligases, and specialized domains within peptide synthetase multienzymes. Despite their low overall amino acid sequence identity, similar reaction mechanisms and the presence of conserved peptide motifs were used as criteria to classify them in a superfamily of adenylate-forming enzymes (13). The relationship of 4CL to other adenylate-forming enzymes was substantiated recently by functional analysis of key 4CL amino acid residues that are conserved in other adenylate-forming enzymes (25). Phylogenetic analyses within the superfamily of adenylate-forming enzymes show that 4CL forms a monophyletic plant-specific group that is most closely related to luciferases rather than long-chain acyl-CoA ligases and acetyl-CoA ligases (7).
The filamentous, soil-living, gram-positive bacterial genus Streptomyces is characterized by the ability to produce a wide variety of secondary metabolites, including antibiotics, and by complex morphological differentiation culminating in sporulation (17). Streptomyces coelicolor A3(2) is the best genetically characterized strain within the Streptomyces genus, and recently the whole genome has been sequenced (http://www.sanger.ac.uk/Projects/S_coelicolor). In the database, a gene (SCD10.15) encoding a 522-amino-acid protein has been annotated as a 4CL gene. The protein shows higher similarity in amino acid sequence to plant 4CLs than to bacterial acyl-CoA ligases; it shows 44% identity and 58% similarity to Arabidopsis thaliana 4CL2. This is the first bacterial protein which shows end-to-end sequence similarity to plant 4CLs over 40% identity.
The purpose of this study was to reveal the enzymatic properties of the 4CL homologue. We cloned and expressed the SCD10.15 gene in Escherichia coli. The recombinant protein showed distinct 4CL activity, but its substrate specificity turned out to be unique; the enzyme efficiently converted cinnamate, which is a very poor substrate for plant 4CL. The enzyme was therefore named ScCCL for S. coelicolor A3(2) cinnamate:CoA ligase. Site-directed mutagenesis of ScCCL was also carried out to elucidate the amino acid residues providing the unique substrate specificity of ScCCL.
MATERIALS AND METHODS
Bacterial strains, plasmids, and general recombinant DNA studies.
S. coelicolor A3(2) M130 was obtained from D. A. Hopwood, John Innes Research Centre, Norwich, United Kingdom (16). Plasmid pUC19 and E. coli JM109 for DNA manipulation were purchased from Takara Shuzo. Plasmid pET16b and E. coli BL21(DE3) for producing histidine-tagged ScCCL were purchased from Novagen. Restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were purchased from Takara Shuzo. DNA was manipulated in E. coli as described earlier (23).
Plasmid construction.
On the basis of the nucleotide sequence of SCD10.15 in the DNA database, the ScCCL gene (AL359988) was amplified by PCR with S. coelicolor A3(2) chromosomal DNA as a template and the following two primers: N-Hind-Nde (5′-GCGaagcttCATATGTTCCGCAGCGAGTACGCAGAC-3′; the italic letters indicate the start codon of the ScCCL gene, and the lowercase letters and underlined letters indicate HindIII and NdeI sites, respectively) and C-Bam (5′-CGCGGATCCTCATCGCGGCTCCCTGAGCTGTC-3′; the italic letters indicate the termination codon of the ScCCL gene, and the underlined letters indicate a BamHI site). The amplified fragment was digested with HindIII plus BamHI and cloned into pUC19. The absence of undesired alterations was checked by nucleotide sequencing. The ScCCL gene was then excised with NdeI and BamHI and cloned into pET16b, generating pET-ScCCL. The expression plasmid pET-ScCCL directs the synthesis of a protein (calculated Mr, 58,000) with the structure Met-Gly-His10-Ser2-Gly-His-Ile-Glu-Gly-Arg-His-ScCCL.
Generation of mutant ScCCL.
The ScCCL sequence in pUC19-ScCCL was divided into three fragments, and restriction sites were introduced by PCR amplification with the following primers: fragment I-F, 5′-GCGaagcttCATATGTTCCGCAGCGAGTACGCAGAC-3′ (the lowercase letters and underlined letters indicate HindIII and NdeI sites, respectively); fragment I-R, 5′-ACGGCATGCTCGGTTCGAGCTGGGCGAGGT-3′ (the underlined letters indicate an SphI site); fragment II-F, 5′-CGTGCATGCCGTCCGCGCCCGGCGA-3′ (the underlined letters indicate an SphI site); fragment II-R, 5′-CTCCTGCAGGGAGGTCCGTGCCCGGGTC-3′ (the underlined letters indicate a PstI site); fragment III-F, 5′-TCCCTGCAGGAGAGTCCGGGGAGATCCT-3′ (the underlined letters indicate a PstI site); and fragment III-R, 5′-CGCGGATCCTCATCGCGGCTCCCTGAGCTGTC-3′ (the underlined letters indicate a BamHI site). The resulting three fragments were cloned into pUC19.
Fragment III was excised and ligated into pUC19-fragment I to generate pUC19-fragment I-fragment III. Fragment II was mutagenized with the QuikChange multisite-directed mutagenesis kit (Stratagene) with the following primers: T228N, 5′-ATCTACGGCCTGAACGCCCTGATGAACGCCCCG-3′ (the altered nucleotides are shown in italics); Y265A, 5′-CGCATCACCAGCCTGGCCGTCGCCCCGCCG-3′; A294G, 5′-TACATCGTCAGCGGCGCCGCCCCGCTCGACGCG-3′; A318G, 5′-CCGCCCGTCGGCCAGGGCTACGGCATGACC-3′; and G327V, 5′-GAACTGTCCCCGGTCACCCACGTCGTCCCC-3′. Mutagenized fragment II was excised and ligated between fragment I and fragment III in pUC19-fragment I-fragment III. The mutated ScCCL sequences were recombined with pET16b and expressed as described below.
Expression and purification of histidine-tagged ScCCL.
We examined the culture conditions of E. coli BL21(DE3) harboring pET16b-ScCCL to produce the histidine-tagged ScCCL in a soluble form as much as possible, because the protein was found in both soluble and insoluble fractions under the usual culture conditions. After cultivation temperature, inoculum size, and induction of the T7 promoter via the lac operator with isopropyl-β-d-thiogalactopyranoside (IPTG) had been examined, we established the following cultivation conditions. E. coli BL21(DE3) harboring pET-ScCCL was cultured at 37°C for 8 h in Luria-Bertani broth (1% Bacto-tryptone, 0.5% Bacto yeast extract, and 1% NaCl) containing 50 μg of ampicillin per ml, and 60 μl of the seed culture was transferred to 30 ml of the same medium containing 100 μg of ampicillin per ml and cultured at 26.5°C for 18 h without induction with IPTG. The cells were harvested from 30 ml of culture by centrifugation and suspended in 600 μl of buffer A (40 mM NaH2PO4, 240 mM NaCl, 8 mM imidazole, and 20% ethylene glycol, pH 8.0). After the cells had been disrupted by sonication, a soluble fraction obtained by centrifugation at 20,000 × g of the sonicate was applied to the Ni-nitrilotriacetic acid spin column (Qiagen), and the histidine-tagged ScCCL was eluted according to the manual of the supplier. Purification was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Enzyme assays.
4CL activity was determined by the spectrophotometric assay, as previously described (20), with various cinnamate derivatives as phenolic substrates. Cinnamate and its derivatives were purchased from Sigma-Aldrich. CoA was purchased from Oriental Yeast. The changes in absorbance during CoA ester formation were monitored at wavelengths of 333, 311, 369, 345, and 352 nm, depending on the absorption maxima for 4-coumaroyl-CoA, cinnamoyl-CoA, caffeoyl-CoA, feruloyl-CoA, and sinapoyl-CoA, respectively (24). The standard reaction mix, in a total volume of 1 ml, contained 400 mM Tris-HCl (pH 7.8), 5 mM ATP, 5 mM MgCl2, 0.3 mM CoA, 0.5 mM phenolic substrate, and 40 to 50 μg of ScCCL. After the reaction mixture had been incubated at 30°C for 5 min, the reaction was initiated by adding CoA and the enzyme. The reaction was carried out at 30°C. The change in absorbance was measured by successive scanning of the wavelength with a Beckman DU7400 spectrophotometer, with the first measurement taken for 2 min. Protein concentrations were determined by the method of Bradford (5) with bovine serum albumin as a standard.
Determination of kinetic parameters.
The kinetic constants Km and kcat for cinnamate derivatives were estimated by linear regression of 1/v against 1/[S] (Lineweaver-Burk plot) at fixed concentrations of ATP (5 mM) and CoA (0.3 mM). The concentration of the phenolic substrates was varied from 0.05 to 0.5 mM.
Phylogenetic reconstruction.
Proteins similar to adenylate-forming enzymes in a total of 29 sequences were aligned with Clustal W (27). The phylogenetic tree was constructed by the neighbor-joining method based on multiple alignment with MEGA version 2.1 (S. Kumar, K. Tamura, I. B. Jakobsen, and M. Nei, 2001, Arizona State University). A total of 490 amino acid sites were considered without gap regions in the alignment. The following sequences were used (the EMBL or GenBank accession numbers are given in parentheses): Arabidopsis thaliana 4CL1 (U18675), A. thaliana 4CL2 (AF106086), A. thaliana 4CL3 (AF106088), aspen 4CL1 (AF041049), aspen 4CL2 (AF041050), Lithospermum 4CL1 (D49366), Lithospermum 4CL2 (D49367), parsley 4CL1 (X13324), parsley 4CL2 (X13325), pine 4CL1 (U12012), pine 4CL2 (U12013), poplar 4CL1 (AF008184), poplar 4CL2 (AF008183), potato 4CL1 (M62755), potato 4CL2a (AAD40664), rice 4CL1 (X52623), rice 4CL2 (L43362), soybean 4CL16 (X69955), tobacco 4CL0 (D43773), tobacco 4CL1 (U50845), tobacco 4CL2 (U50846), vanilla 4CL (X75542), Arabidopsis 4CL-like (chromosome 1) (AC011000), Arabidopsis 4CL-like (chromosome 4-14) (AL161502), Arabidopsis 4CL-like (chromosome 4-49) (AL161549), Hotaria luciferase (AAC37253), Photinus luciferase (CAA59281), Pyrocoelia luciferase (AAC37254), and S. coelicolor A3(2) ScCCL (AL359988).
RESULTS
Comparison of 4CL-like protein from S. coelicolor A3(2) with three 4CL isoforms from A. thaliana.
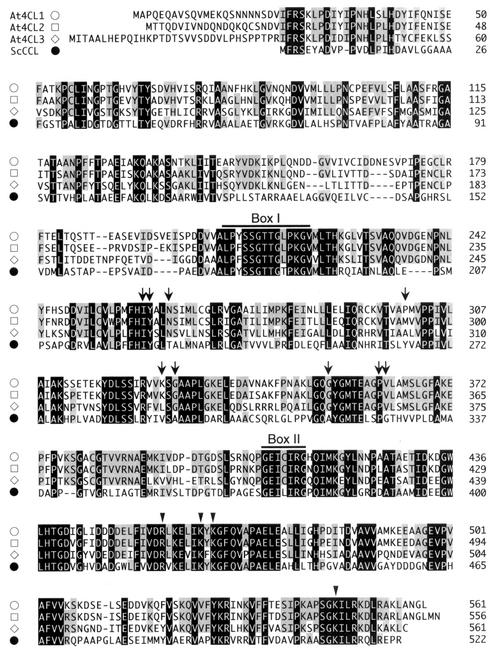
The deduced amino acid sequence of SCD10.15, a 4CL gene of S. coelicolor A3(2), was multiple-aligned with three 4CL isoforms from A. thaliana (Fig. 1). The 4CL homologue (ScCCL) from S. coelicolor A3(2) showed end-to-end similarity to the Arabidopsis 4CLs. ScCCL had 44% identity and 58% similarity to A. thaliana 4CL2. Among the 4CL enzymes, two highly conserved peptide motifs, box I (SSGTTGLPKGV) and box II (GEICIRG), have been identified (1, 2, 10, 11, 18, 26, 28). In the ScCCL sequence, both motifs were also highly conserved. Although a conserved Cys in box II was replaced with Leu in ScCCL, Cys403 of A. thaliana 4CL2 was reported to be nonessential for the enzymatic activity (25). Furthermore, several residues important for the activity of A. thaliana 4CL2 (25), such as Arg449, Lys455, Lys457, and Lys540, were all conserved in ScCCL.
FIG. 1.
Alignment of deduced amino acid sequences of the 4CL homologue (ScCCL) from S. coelicolor A3(2) and three 4CL isoforms from A. thaliana (At4CL1, At4CL2, and At4CL3). The amino acids identical in all four sequences and in three of the four sequences are indicated by solid and gray boxes, respectively. Two highly conserved peptide motifs (box I and box II) within the 4CL enzymes are indicated. Nine amino acid residues which are suggested to form the substrate-binding pocket of the A. thaliana 4CLs (see Fig. 4) are indicated by arrows. Four amino acids which are important for the activity of A. thaliana 4CL2, as revealed by site-directed mutagenesis (26), are indicated by arrowheads.
The information obtained from the crystal structure of the phenylalanine-activating domain of gramicidin S synthase (6) predicted that nine residues formed a substrate-binding pocket in A. thaliana 4CLs (26) (Fig. 1; see also Fig. 4). Of the nine residues, seven are highly conserved in plant 4CLs. Three (Ile252, Tyr253, and Pro354) of the seven residues are conserved in ScCCL, but others (Asn256, Gly322, Gly346, and Val355) are not. The involvement of these residues in the substrate selectivity of ScCCL is discussed below.
FIG. 4.
Comparison of amino acid residues that probably line up along the substrate-binding pocket in plant 4CLs, ScCCL, and its mutants. The amino acid positions of A. thaliana 4CL2 (At4CL2) and ScCCL are represented. Residues that are highly conserved in the sequences are shaded. The substituted amino acids in mutant enzymes are indicated with underlining.
4CL activity of 4CL-like protein from S. coelicolor A3(2).
We cloned the ScCCL gene from the S. coelicolor A3(2) genome by PCR and constructed an expression plasmid, pET-ScCCL, to produce histidine-tagged ScCCL. Improvement of the culture conditions of E. coli BL21(DE3) harboring pET-ScCCL led to production of a considerable amount of the recombinant protein in the soluble fraction (Fig. 2). The recombinant protein was purified to homogeneity from the soluble fraction with an Ni-nitrilotriacetic acid spin column, giving a single protein band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 2). From 30 ml of culture, about 150 μg of histidine-tagged ScCCL was obtained.
FIG. 2.
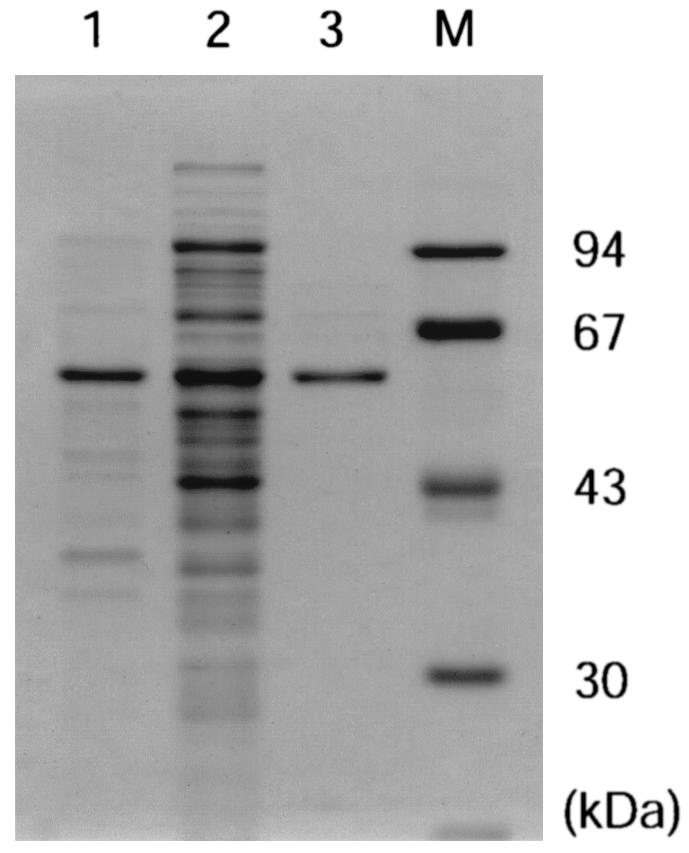
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of His-tagged ScCCL. Lane 1, insoluble fraction; lane 2, soluble fraction; lane 3, His-tagged ScCCL after affinity chromatography with His-binding resin; and lane M, size markers. Each lane contains appropriate amounts of protein.
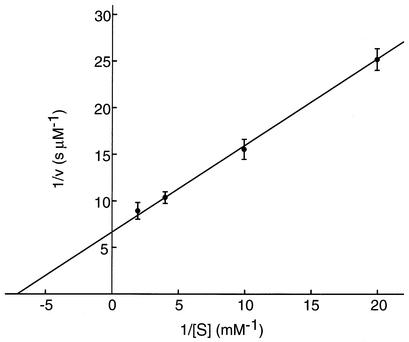
With this recombinant protein, we examined the 4CL activity of ScCCL from S. coelicolor A3(2) by the spectrophotometric assay. Under standard conditions used for plant 4CLs, it showed distinct 4CL activity. We first determined the optimum pH and temperature of the 4-coumaroyl-CoA formation catalyzed by ScCCL. The optimum pH was determined to be pH 8.0 when assayed with Tris-HCl buffer. The optimum temperature was determined to be 30°C. These values were similar to those reported for plant 4CLs (19, 20). We then determined the kinetic constants, Km for 4-coumarate and kcat, under the optimum conditions. The Km for 4-coumarate and kcat were calculated to be 131 ± 4 μM and 0.202 ± 0.007 s−1, respectively, on the basis of the Lineweaver-Burk plot (Fig. 3). The Km of ScCCL was comparable to those of plant 4CLs; the Km values for 4-coumarate were 38 μM (A. thaliana 4CL1), 252 μM (A. thaliana 4CL2), and 23 μM (A. thaliana 4CL3) when assayed with recombinant proteins produced in E. coli (10). Because Vmax values per microgram of protein, instead of kcat values, were described in most cases for plant 4CL-catalyzed reactions and the values were quite different among the individual 4CLs, the kcat value of ScCCL could not be compared directly with those of plant 4CLs. To our knowledge, this is the first bacterial protein showing distinct 4CL activity.
FIG. 3.
Determination of Km and kcat values for 4-coumaroyl-CoA formation by ScCCL. The initial reaction velocities (v) of 4-coumaroyl-CoA formation by ScCCL were determined from five independent experiments. The kinetic constants Km for 4-coumarate and kcat were estimated by linear regression of 1/v against 1/[S] (Lineweaver-Burk plot) at fixed concentrations of ATP (5 mM) and CoA (0.3 mM). The plot gave a line with the x intercept equal to −1/Km and with the y intercept equal to 1/kcat [ScCCL]0.
Substrate specificity of ScCCL from S. coelicolor A3(2).
The range of substrates used by plant 4CL isoforms varies within and between plant species, but generally 4-coumarate (4-hydroxycinnamate) and caffeate (3,4-dihydroxycinnamate) are the best substrates, followed by ferulate (3-methoxy-4-hydroxycinnamate), with cinnamate itself being a very poor substrate (9). 4CL activity toward sinapate (3,5-dimethoxy-4-hydroxycinnamate) has been described in a few cases in angiosperms (19, 21), but no such activity of heterologously expressed 4CL isoforms has so far been described.
ScCCL was found to have low activity toward caffeate and no activity toward ferulate (Table 1), both of which are generally good substrates for plant 4CLs. ScCCL also showed no activity toward sinapate. On the other hand, ScCCL efficiently converted cinnamate to the corresponding thiol ester (Table 1), while cinnamate is a very poor substrate for plant 4CLs. The Km for cinnamate and kcat values were estimated to be 190 ± 2 μM and 0.475 ± 0.012 s−1, respectively. The Km value for cinnamate is 1.5-fold higher than that for 4-coumarate, but the kcat value for cinnamoyl-CoA formation is 2.4-fold higher than that for 4-coumaroyl-CoA formation, resulting in a 1.7-fold-higher efficiency (kcat/Km) for cinnamate than for 4-coumarate. Thus, ScCCL turned out to have a unique substrate specificity.
TABLE 1.
Kinetic properties of S. coelicolor A3(2) ScCCL and its mutantsa
| Enzyme | Cinnamate
|
4-Coumarate
|
Caffeate
|
Ferulate
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | |
| Wild type | 190 | 0.475 | 2,499 | 131 | 0.202 | 1,545 | ND | ND | ND | — | — | — |
| Y265A | 106 | 0.195 | 1,840 | 59 | 0.027 | 460 | — | — | — | — | — | — |
| A294G | 162 | 1.278 | 7,889 | 131 | 0.302 | 2,305 | 976 | 0.077 | 79 | — | — | — |
| A318G | 664 | 0.389 | 586 | 433 | 0.101 | 233 | — | — | — | — | — | — |
| A294G/A318G | 176 | 0.664 | 3,773 | 77 | 0.146 | 1,894 | 715 | 0.089 | 124 | 242 | 0.106 | 438 |
| A294G/G327V | 423 | 0.179 | 423 | — | — | — | — | — | — | — | — | — |
| A294G/A318G/G327V | 170 | 1.048 | 6,165 | 218 | 0.156 | 716 | — | — | — | — | — | — |
| T228N/A294G/A318G/G327V | 861 | 0.315 | 366 | 680 | 0.034 | 50 | — | — | — | — | — | — |
| T228N/Y265A/A294G/A318G/G327V | 157 | 0.011 | 70 | — | — | — | — | — | — | — | — | — |
Values are the means of four independent experiments with standard errors of less than 15%. ND, not determined, because activity was too low; —, no enzymatic activity.
Site-directed mutagenesis of ScCCL and activities of the mutant enzymes.
We assumed that the differences in substrate specificity between ScCCL and plant 4CLs could be ascribed to the differences in the amino acid residues that line up along the catalytic pocket, on the basis of the recent study in which the residues important for the substrate specificity of A. thaliana 4CL2 were identified by site-directed mutagenesis (26). Nine residues probably forming the catalytic pocket have been predicted from the crystal structure of the phenylalanine-activating domain of gramicidin S synthase (6) and the results of the study on A. thaliana 4CLs (26) (Fig. 4). Two of these residues (Ile252 and Tyr253; the numbers of the A. thaliana 4CL2 sequence are used) are conserved among all plant 4CLs and ScCCL. Pro354 is also conserved among almost all plant 4CLs (except for potato 4CL2a) and ScCCL. The amino acid residues at positions 293 and 320 varied among plant 4CLs, although the differences are restricted to a few amino acids. The remaining four amino acid residues (Asn256, Gly322, Gly346, and Val355) are completely conserved in plant 4CLs, whereas the corresponding residues of ScCCL are Thr228, Ala294, Ala318, and Gly327, respectively.
To determine whether amino acid replacement at these positions changes the substrate specificity of ScCCL, we generated eight ScCCL mutant enzymes (Y265A, A294G, A318G, A294G/A318G, A294G/G327V, A294G/A318G/G327V, T228N/A294G/A318G/G327V, and T228N/Y265A/A294G/A318G/G327V)by using the amino acid residues of A. thaliana 4CL2 at these positions as references (Fig. 4).
(i) Y265A.
Three 4CL isoforms from A. thaliana have different substrate utilization profiles despite their high sequence identity. All three isoforms are able to activate their typical substrates, 4-coumarate and caffeate. However, only A. thaliana 4CL1 and A. thaliana 4CL3 are able to convert ferulate to the corresponding thiol esters, whereas A. thaliana 4CL2 is not (10). Stuible and Kombrink (26) revealed that the introduction of a small amino acid at either position 293 or 320 promoted ferulate activation by A. thaliana 4CL2. Furthermore, they revealed that introduction of bulky aromatic amino acids at position 293 of A. thaliana 4CL2 resulted in a 10-fold increase in the Km value for caffeate (26).
Because the amino acid residues at both positions 293 and 320 of A. thaliana 4CL2 have been suggested to form the substrate-binding pocket, the bulkiness of the amino acids lining the substrate-binding pocket is an important factor that determines substrate specificity in the 4CL family. In the ScCCL sequence, the amino acid residue corresponding to position 293 of A. thaliana 4CL2 is Tyr265. We speculated that this bulky aromatic amino acid residue at this position would explain the weak and no activity of ScCCL toward caffeate and ferulate, respectively. Therefore, we generated a mutant ScCCL, Y265A. The Km value for cinnamate and 4-coumarate of Y265A was decreased, although the kcat value was also decreased, resulting in a decrease in kcat/Km. Contrary to our expectation, however, Y265A could not use caffeate and ferulate as substrates (Table 1). These results suggested that other amino acid residues would hinder the enzyme's ability to use caffeate and ferulate as substrates.
(ii) A294G, A318G, and A294G/A318G.
Judging from the substrate specificity of ScCCL, the substrate-binding pocket of ScCCL is probably smaller than those of plant 4CLs. Therefore, we first replaced Ala294 and Ala318, because the corresponding residues in plant 4CLs are Gly, which has a shorter side chain than Ala. A decrease in Km and an increase in kcat for cinnamate and 4-coumarate in A294G led to generation of a mutant enzyme with higher catalytic activity than native ScCCL. Probably due to the high catalytic activity, A294G catalyzed the reaction toward caffeate to a detectable extent (Table 1). Furthermore, A294G/A318G was active toward caffeate and ferulate, although its kinetic parameters for cinnamate and 4-coumarate were similar to those of native ScCCL (Table 1). This implies that the combination of changes at positions 294 and 318 reduces the Km value for ferulate. It is noteworthy that A294G and A294G/A318G showed higher catalytic activities toward cinnamate and 4-coumarate than the wild-type enzyme (Table 1).
(iii) A294G/G327V, A294G/A318G/G327V, T228N/A294G/A318G/G327V, and T228N/Y265A/A294G/A318G/G327V.
In contrast to the results with A294G/A318G, the combination of mutations at positions 294 and 327 lowered the catalytic activity even for 4-coumarate. Addition of a mutation at position 318 to A294G/G327V restored the enzyme activity, up to that of A294G, for cinnamate but not for 4-coumarate. These observations suggested that an appropriate combination of amino acid replacements not only of the nine amino acids lining up along the substrate-binding pocket but also of other amino acids forming the pocket and maintaining each of the catalytic residues at correct orientations is necessary to maintain the enzyme activity and change the substrate specificity of ScCCL. Consistent with this idea, the mutant enzymes T228N/A294G/A318G/G327V and T228N/Y265A/A294G/A318G/G327V, which had the same amino acid residues in the substrate-binding pocket as plant-type 4CLs, showed almost no activity toward 4-coumarate.
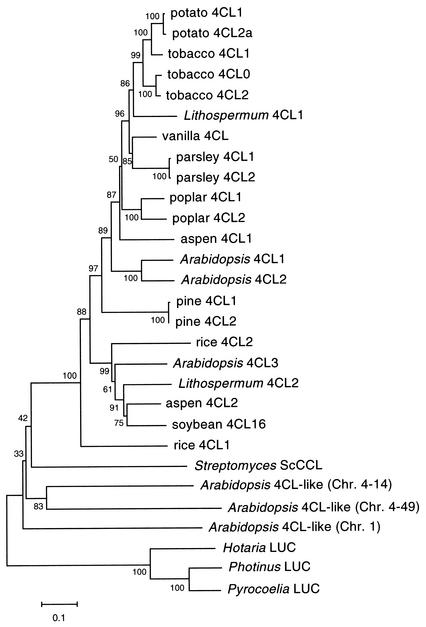
Phylogenetic reconstruction of 4CLs and 4CL-like proteins.
The Arabidopsis genome database (http://www.arabidopsis.org/agi.html) contains at least three 4CL-like genes. The proteins encoded by these genes are similar in size to bona fide 4CLs and are about 40% identical to bona fide 4CLs, with significant similarity over their whole lengths. We generated a phylogenetic tree of 4CLs including these 4CL-like proteins from A. thaliana and the bacterial ScCCL with three luciferases as the outgroup to root the tree (Fig. 5). The result showed that ScCCL as well as three Arabidopsis 4CL-like proteins was out of a distinct clade containing bona fide 4CLs. Nevertheless, ScCCL was shown to have distinct 4CL activity in this study. This would be important for deducing the possible enzymatic functions of Arabidopsis 4CL-like proteins, although Stuible and Kombrinks (26) reported that one of these proteins (AC011000, 4CL-like chromosome 1 in Fig. 5) showed no 4CL activity.
FIG. 5.
Phylogenetic reconstruction of 4CLs and 4CL-like proteins. The phylogenetic tree, constructed on the basis of the neighbor-joining method, was constructed based on multiple alignment with MEGA version 2.1. A total of 490 amino acid sites were considered without gap regions in the alignment. The bar indicates a 10% difference in amino acid sequence. The number at a branch point indicates the percentage of 1,000 bootstrap repetitions. Chr., chromosome; LUC, luciferase.
DISCUSSION
4CLs have been believed to be specific to plants. However, a gene encoding a 4CL-like enzyme which shows more than 40% identity in amino acid sequence to plant 4CLs was found in the genome of the soil-inhabiting, filamentous bacterium S. coelicolor A3(2). Although this 4CL homologue (ScCCL) was not included in a distinct clade containing bona fide 4CLs in the phylogenetic analyses, we revealed that ScCCL had distinct 4CL activity. To our knowledge, this is the first bacterial protein showing 4CL activity. This finding suggests that proteins having 4CL activity are more widespread than expected.
ScCCL was able to convert cinnamate and 4-coumarate efficiently to the corresponding thiol esters in vitro. The physiological substrate(s) of ScCCL in S. coelicolor A3(2) is not clear, because cinnamate and 4-coumarate are not general metabolites in bacteria. PAL is an essential enzyme for cinnamate and 4-coumarate production from phenylalanine. Although PAL activity in Streptomyces verticillatus was reported more than 30 years ago (3, 12), no gene encoding a PAL homologue is present in the genome of S. coelicolor A3(2). S. coelicolor A3(2) has a histidine ammonia lyase (histidase), an enzyme closely related to PAL, but PAL activity of the enzyme has not been reported. Therefore, if cinnamate and 4-coumarate are the physiological substrates, they may be supplied from dead plants in the surrounding environment, although no uptake system for cinnamate or 4-coumarate is known in Streptomyces spp.
Speculating on the in vivo function of ScCCL, we point out that four genes which would be involved in the conversion of CoA ester compounds exist in the neighborhood of the ScCCL gene on the S. coelicolor A3(2) genome. These genes encode probable α and β subunits of acetyl/propionyl-CoA carboxylase, a probable acyl-CoA dehydrogenase, and a probable enoyl-CoA hydratase. The termination codon of ScCCL overlaps the initiation codon of the putative enoyl-CoA hydratase gene, which suggests that these two genes are transcriptionally and translationally coupled. Because functionally related genes are usually clustered in the bacterial genome, ScCCL, together with these enzymes, is perhaps involved in the production of some secondary metabolite.
We point out the following biosynthetic pathway in which a 4CL-like enzyme is responsible for secondary metabolite formation. A biosynthetic pathway for benzoyl-CoA, similar to the plant systems, has recently been found in Streptomyces maritimus (15, 22). The genes involved in this pathway are members in the biosynthesis gene cluster for the enterocin and wailupemycin family of polyketides. Therefore, the pathway would not be ubiquitous in Streptomyces spp. but specific to strains. In this pathway, similar to the plant system, EncH is suggested to be a cinnamoyl-CoA ligase. In contrast to ScCCL, however, EncH shows higher sequence similarity to bacterial acyl-CoA ligases than to plant 4CLs.
ScCCL is quite different from plant 4CLs in its substrate specificity. Three 4CL isoforms from A. thaliana have different substrate utilization profiles despite their high sequence identity. Stuible and Kombrink (26) showed the importance of the bulkiness of the amino acid residues at positions 293 and 320 for the recognition of the 3-methoxy or 3-hydroxy group of the substrate, on the basis of the observation that the introduction of small amino acids at either position 293 or 320 promoted ferulate activation by A. thaliana 4CL2 and that the introduction of bulky aromatic amino acids at position 293 of A. thaliana 4CL2 resulted in a 10-fold increase in the Km value for caffeate. Because the amino acid residues at both positions 293 and 320 of A. thaliana 4CL2 are suggested to be part of the substrate-binding pocket, the bulkiness of the amino acids lining the substrate-binding pocket is an important factor that determines substrate specificity in the 4CL family.
We therefore focused on determining how the nine amino acid residues that perhaps line up along the substrate-binding pocket might affect substrate specificity. Contrary to our expectation, mutant Y265A, having an amino acid replacement from Tyr to Ala at the position corresponding to 293 of A. thaliana 4CL2, showed low catalytic activity toward cinnamate and 4-coumarate and no activity toward caffeate and ferulate. Although point mutations in these amino acid residues failed to change completely the substrate specificity of ScCCL into those of plant 4CLs, some of the mutants showed different substrate specificities than the wild-type enzyme. A294G and A294G/A318G could convert caffeate more efficiently than the wild-type enzyme, and A294G/A318G could utilize ferulate efficiently, suggesting that Ala294 and Ala318 are involved in substrate recognition. Interestingly, the activities of A294G and A294G/A318G for cinnamate and 4-coumarate were increased.
It is surprising that the mutant enzymes with higher activity than the wild-type enzyme were generated, although this may be related to the idea that ScCCL uses certain compounds, different from the compounds used in this study, as a physiological substrate. The discovery of an enzyme having cinnamate/4-coumarate:CoA ligase activity in bacteria is useful for the production of various compounds by combinatorial biosynthesis.
Acknowledgments
We thank H. Nishida for helpful discussion about phylogenetic analyses.
This work was supported by a Research Grant from the Noda Institute for Scientific Research, by a grant from the Industrial Technology Research Grant Program 2000 of the New Energy and Industrial Technology Development Organization of Japan (00A03004), and by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (BDP-02-VI-2-7).
REFERENCES
- 1.Allina, S. M., A. Pri-Hadash, D. A. Theilmann, B. E. Ellis, and C. J. Douglas. 1998. 4-Coumarate:coenzyme A ligase in hybrid poplar. Plant Physiol. 116:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker-André, M., P. Schulze-Lefert, and K. Hahlbrock. 1991. Structural comparison, modes of expression, and putative cis-acting elements of the two 4-coumarate:CoA ligase genes in potato. J. Biol. Chem. 266:8551-8559. [PubMed] [Google Scholar]
- 3.Bezanson, G. S., D. Desaty, A. V. Emes, and L. C. Vining. 1970. Biosynthesis of cinnamide and detection of phenylalanine ammonia-lyase in Streptomyces verticillatus. Can. J. Microbiol. 16:147-151. [DOI] [PubMed] [Google Scholar]
- 4.Boudet, A. M. 2000. Lignins and lignification: selected issues. Plant Physiol. Biochem. 38:81-96. [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Conti, E., T. Stachelhaus, M. A. Marahiel, and P. Brick. 1997. Structural basis for activation of phenylalanine in the nonribosomal biosynthesis of gramicidin S. EMBO J. 14:4174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cukovic, D., J. Ehlting, J. A. VanZiffle, and C. J. Douglas. 2001. Structure and evolution of 4-coumarate:CoA ligase (4CL) gene family. Biol. Chem. 382:645-654. [DOI] [PubMed] [Google Scholar]
- 8.Dixon, R. A., and N. L. Paiva. 1995. Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas, C. J. 1996. Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci. 1:171-178. [Google Scholar]
- 10.Ehlting, J., D. Büttner, Q. Wang, C. J. Douglas, I. E. Somssich, and E. Kombrink. 1999. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19:9-20. [DOI] [PubMed] [Google Scholar]
- 11.Ehlting, J., J. J. K. Shin, and C. J. Douglas. 2001. Identification of 4-coumarate:coenzyme A ligase (4CL) substrate recognition domains. Plant J. 27:455-465. [DOI] [PubMed] [Google Scholar]
- 12.Emes, A. V., and L. C. Vining. 1970. Partial purification and properties of L-phenylalanine ammonia-lyase from Streptomyces verticillatus. Can. J. Microbiol. 48:613-622. [DOI] [PubMed] [Google Scholar]
- 13.Fulda, M., E. Heinz, and F. P. Wolter. 1994. The fadD gene of Escherichia coli K-12 is located close to rnd at 39.6 min of the chromosomal map and is a new member of the AMP-binding protein family. Mol. Gen. Genet. 242:241-249. [DOI] [PubMed] [Google Scholar]
- 14.Hahlbrock, K., and D. Scheel. 1989. Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40:347-369. [Google Scholar]
- 15.Hertweck, C., and B. S. Moore. 2000. A plant-like biosynthesis of benzoyl-CoA in the marine bacterium “Streptomyces maritimus.” Tetrahedron 56:9115-9120. [Google Scholar]
- 16.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 17.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859-864. [DOI] [PubMed] [Google Scholar]
- 18.Hu, W. J., A. Kawaoka, C. J. Tsai, J. H. Lung, K. Osakabe, H. Ebinuma, and V. L. Chiang. 1998. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc. Natl. Acad. Sci. USA 95:5407-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knobloch, K. H., and K. Hahlbrock. 1975. Isoenzymes of p-coumarate:CoA ligase from cell suspension cultures of Glycine max. Eur. J. Biochem. 52:311-320. [DOI] [PubMed] [Google Scholar]
- 20.Knobloch, K. H., and K. Hahlbrock. 1977. 4-Coumarate:CoA ligase from cell suspension cultures of Petroselinum hortense Hoffm. Arch. Biochem. Biophys. 184:237-248. [DOI] [PubMed] [Google Scholar]
- 21.Kutsuki, H., M. Shimada, and T. Higuchi. 1981. Distribution and roles of p-hydroxycinnamate:CoA ligase in lignin biosynthesis. Phytochemistry 21:267-271. [Google Scholar]
- 22.Piel, J., C. Hertweck, P. R. Shipley, D. M. Hunt, M. S. Newman, and B. S. Moore. 2000. Cloning, sequencing and analysis of the enterosin biosynthesis gene cluster from the marine isolate “Streptomyces maritimus”: evidence for the derailment of an aromatic polyketide synthase. Chem. Biol. 7:943-955. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 24.Stöckigt, J., and M. H. Zenk. 1975. Chemical synthesis and properties of hydroxycinnamoyl-coenzyme A derivatives. Z. Naturforsch. 30c:352-358. [DOI] [PubMed] [Google Scholar]
- 25.Stuible, H. P., D. Büttner, J. Ehlting, K. Hahlbrock, and E. Kombrink. 2000. Mutational analysis of 4-coumarate:CoA ligase identifies functionally important amino acids and verifies its close relationship to other adenylate-forming enzymes. FEBS Lett. 467:117-122. [DOI] [PubMed] [Google Scholar]
- 26.Stuible, H. P., and E. Kombrink. 2001. Identification of the substrate specificity-conferring amino acid residues of 4-coumarate:coenzyme A ligase allows the rational design of mutant enzymes with new catalytic properties. J. Biol. Chem. 276:26893-26897. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlmann, A., and J. Ebel. 1993. Molecular cloning and expression of 4-coumarate:coenzyme A ligase, an enzyme involved in the resistance of soybean (Glycine max) against pathogen infection. Plant Physiol. 102:1147-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisshaar, B., and G. I. Jenkins. 1998. Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1:251-257. [DOI] [PubMed] [Google Scholar]