Abstract
Enterotoxigenic Escherichia coli (ETEC) remains a formidable cause of diarrheal illness worldwide. At present, there is no vaccine that provides broad-based protection against ETEC. A ′phoA-based self-cloning mutagenesis system, TnphoA.ts, employed to identify novel ETEC surface antigens, led to identification of an ETEC two-partner secretion locus (etpBAC) on the pCS1 virulence plasmid of prototype strain H10407. Cloning and expression of etpBAC in recombinant E. coli LMG194(pJY019) resulted in secretion of a high-molecular-weight (HMW) glycosylated exoprotein. This glycoprotein, EtpA, exhibits linear peptide sequence and predicted structural homologies with known HMW adhesins produced by other two-partner secretion loci. Antibodies directed against recombinant EtpA (anti-rEtpA.6H) recognized an HMW protein in culture supernatants of ETEC strains H10407 and LMG194(pJY019) but not in culture supernatant of strain H10407-P, which lacks the 92-kb pCS1 plasmid, or an isogenic etpA mutant. etpA mutants were deficient in adherence to intestinal epithelial cells in vitro, and anti-rEtpA.6H antibodies inhibited association of H10407 with target epithelial cells. Cloning and expression of etpB in recombinant E. coli were sufficient to confer adherence. Screening of multiple ETEC isolates for the etpBAC locus by colony hybridization and by EtpA immunoblotting suggested that EtpA is one of the most common antigens secreted by these pathogens. Together, these results indicate that the newly identified ETEC two-partner secretion locus directs the secretion of a high-molecular-weight glycosylated protein, EtpA, that in concert with the putative EtpB transporter participates in adherence of H10407 to epithelial cells, thereby expanding the repertoire of potential ETEC virulence proteins and vaccine candidates.
Enterotoxigenic Escherichia coli (ETEC) remains one of the principal causes of infectious diarrhea in travelers and in children in developing countries (72). ETEC is also an emerging cause of diarrheal illness in industrialized countries, including the United States (3, 6, 75). The organisms belonging to this taxon have genetically diverse E. coli pathotypes characterized by the production of heat-labile enterotoxin and/or heat-stable enterotoxin. In the classic paradigm of ETEC infection, these organisms adhere to the small intestinal mucosa via fimbrial colonization factor (CF) molecules, where they elaborate heat-labile enterotoxin and/or heat-stable enterotoxin. While early studies indicated that immunization with CFs provides protection against ETEC infection, development of a broadly protective vaccine has been hampered by the antigenic heterogeneity of these molecules. However, the recent identification of additional ETEC surface molecules (32, 56, 68) suggests that the present pathogenesis paradigm is incomplete and consequently that there may be additional antigens that can be exploited in vaccine development.
Surface expression of virulence molecules by ETEC and other gram-negative pathogens requires export through the inner and outer membranes. In general, gram-negative export of virulence proteins occurs via a number of different types of secretion systems (types I to V). These systems range from very complex multimeric structures that require coordinated assembly of multiple proteins to simple systems consisting of a single protein, such as the autotransporters (type V) (42). Autotransporters consist of three essential domains: an amino-terminal signal peptide, a passenger domain, and a carboxy-terminal transporter. In classical autotransporter molecules (type Va) the carboxy-terminal transporter is thought to form a hydrophilic channel composed of β strands in the outer membrane. This pore in the outer membrane is required to export the amino-terminal passenger domain to the bacterial surface. To date, two autotransporters, TibA and EatA, have been described in ETEC (27, 68).
A second set of type V secretion molecules (type Vb) are also involved in the secretion of large virulence proteins from gram-negative pathogens. In these systems the secreted exoproteins (generically referred to as TpsA proteins) and their requisite transporters (TpsB) are encoded on separate genes. Therefore, they have been referred to as “unlinked autotransporters” (41) or, perhaps more descriptively, two-partner secretion (TPS) pathways (42, 46). A TPS pathway has been described in a variety of pathogens, including Bordetella pertussis (24, 57, 89), Serratia marcescens (69, 79), Proteus mirabilis (87), and Haemophilus influenzae (4, 5). Like the autotransporter passenger domains, the exoproteins secreted via the TPS systems play different roles, and a number of the proteins characterized to date function as adhesins (21, 35, 57, 73, 83, 88) or cytolysins (12, 69, 87).
Some TPS proteins, such as filamentous hemagglutinin (FHA) of B. pertussis, and the high-molecular-weight (HMW) adhesins of H. influenzae, have been the subjects of considerable structural and functional characterization. However, there appear to be many potential TPS systems in other pathogens which have yet to be characterized (45, 46). Indeed, genes for many potential two-partner secretion pathways have been identified in the sequences of the genomes of a variety of pathogens (13, 46). Here we describe identification of a two-partner secretion system in ETEC using a ′phoA-based plasposon (23), TnphoA.ts (Fig. 1), which was developed to facilitate identification of secreted proteins.
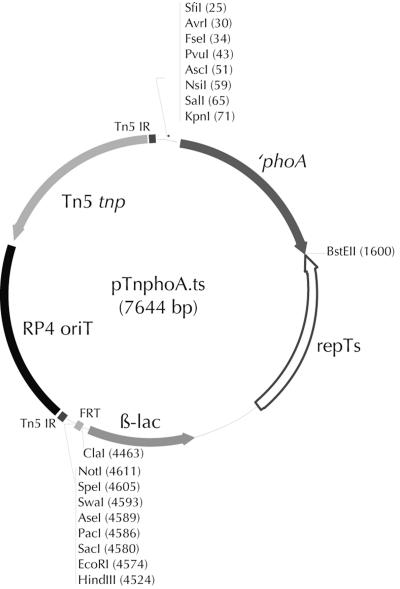
FIG. 1.
pTnphoA.ts plasposon. Between the Tn5 inverted repeat elements (Tn5 IR) the insertion element contains a target site for FLP recombinase (FRT) flanked by multiple restriction sites, a beta-lactamase cassette (β-lac), and a temperature-sensitive origin of replication (repTs) from pST76-A (70), combined with a truncated ′phoA gene and additional restriction sites from pMini-O-phoA (10). The Tn5 transposase (Tn5 tnp) and origin of transfer (RP4 oriT) are located outside the repeat elements.
MATERIALS AND METHODS
Media and strains.
A complete list of the plasmids and bacterial strains employed in this study is shown in Table 1. 5-Bromo-4-chloro-3-indolyl-phosphate (p-toluidine salt) (XP) was prepared as a 20-mg/ml stock solution in N,N-dimethylformamide. Plates for detection of gene fusions with phoA contained XP at a final concentration of 40 μg/ml and 100 μg/ml ampicillin in a base medium composed of tryptone (1%), yeast extract (0.5%), and agar (1.5%) (58).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotype or descriptiona | Reference or source |
|---|---|---|
| Bacterial strains | ||
| H10407 | ETEC serotype 078:H11, LT+LST+ | 29 |
| H10407-P | Derivative of H10407 cured of 92-kb pCS1 plasmid | |
| H10407-S | Spontaneous Smr derivative of H10407 | |
| jf1181 | A10407 (pTnphoA.Ts) Ampr | This study |
| jf1289 | Isogenic etpA deletion mutant | This study |
| jf1358 | etpC deletion mutant generated with cassette from pKD13, Kmr | This study |
| jf1370 | etpC deletion following curing of Kmr cassette using pCP20 helper plasmid | This study |
| NovaBlue | endA1 HsdR17 (rk12− mk12−) supE44 thi-1 recA1 gyrA96 re1A1 lac F′ [proA+B+lacI ZΔM15::Tn10 (Tcr)] | Novagen |
| DH10BT1 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1: endA1 araD139 Δ(ara leu)7697 galU galkλrpsL nupG tonA | 48 |
| LMG194 | F−ΔlacZX74 galE thi rpsL ΔphoA (PvuII) Δara-714 leu::Tn10 | Invitrogen |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu | 51 |
| BL21(DE3) | F−ompT hsdSp (rp−mp−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pSTBlue-1 | 3,851-bp PCR cloning plasmid, Ampr Kmr | Novagen |
| pT7Blue-3 | 3,821-bp PCR cloning plasmid, Ampr Kmr | Novagen |
| pFLAG-CTC | 5,348-bp expression plasmid for construction of C-terminal FLAG epitope fusions, Ampr | Sigma |
| pGPS1.1 | Tn7 transprimer donor plasmid, Kmr | NEBb |
| pGPS2.1 | Tn7 transprimer donor plasmid, Cmr | NEB |
| pBAD/Myc-His A | Arabinose-inducible expression plasmid | Invitrogen |
| pGEM-5zf (+) | Cloning vector, ampr | Promega |
| pST76-A | Temperature-sensitive suicide plasmid, Ampr | 70 |
| pmini-OphoA | 3,362-bp self-cloning phoA minitransposon plasmid, Gmr | 10 |
| pET30a (+) | 5,422-bp His tag expression plasmid, Kmr | Novagen |
| pJY005 | 3,257-bp etpA fragment cloned in frame with polyhistidine tag on pET30a (+) | This study |
| pJFF025 | 3,077-bp amplicon from pST76-A cloned into pSTBlue-1 | This study |
| pTnphoA.ts | 2,983-bp BstEII/SacI fragment from pJFF025 cloned into corresponding sites on pmini-OphoA, yielding a Tn5/′phoA/Ampr-based plasposon with temperature-sensitive origin of replication | This study |
| pCVD442 | pir-dependent SacB+ suicide vector, Ampr | 25 |
| pJML001 | 12-kb plasmid obtained by MluI digestion religation of genomic DNA from TnphoA.ts mutant 220 | This study |
| pJY001 | 2,400-bp PCR amplicon containing etpA in-frame deletion cloned into pT7Blue-3 | This study |
| pJMF1001 | 1,819-bp PCR amplicon containing etpB cloned into EcoRV site of pT7Blue-3 | This study |
| pJMF1002 | 1,811-bp HindIII-EcoRI etpB fragment from pJMF1001 cloned into pFLAG-CTC | This study |
| pJY017 | 5,317-bp amplicon containing etpA cloned into pT7Blue-3 | This study |
| pJY019 | etpBAC locus cloned onto pBAD/Myc-His A | This study |
| pJY028 | 2,112-bp SphI fragement from pJY001 cloned into corresponding site of pCVD442 | This study |
| pJY035 | 5,307-bp EcoRI-BglII fragment from pJY017 cloned into pFLAG-CTC, yielding etpA-FLAG fusion | This study |
| pJY039 | 1,913-bp etpC HindIII-BglII fragment cloned into pFLAG-CTC | This study |
| pJY040 | etpC cloned in frame with FLAG epitope on pFLAG-CTC | This study |
| pKD46 | Temperature-sensitive λ-Red recombinase expression helper plasmid, Ampr | 22 |
| pKD13 | Template plasmid for construction of gene disruption cassette | 22 |
| pCP20 | FLP recombinase helper plasmid | 22 |
Ampr, Kmr, Cmr, Gmr, and Smr, ampicillin, kanamycin, chloramphenicol, gentamicin, and streptomycin resistance, respectively; LT, heat-labile enterotoxin; ST, heat-stable enterotoxin.
NEB, New England Biolabs.
Construction of a TnphoA-based plasposon.
To construct a plasmid that would expedite isolation of TnphoA fusions to potential virulence genes in ETEC, we started with two existing plasmids, pmini-OphoA, which contains a Tn5-based transposon with the cognate transposase outside the inverted repeats (10), and pST76-A, a temperature-sensitive suicide plasmid (70). First, primers jf020904.1 (5′-GATCAGCCAGTGCCAAGC-3′) and jf020904.2 (5′-AAACCGGTTACCGATACAAGAGCCATAAGA-3′) were used to produce a 3,077-bp PCR product from pST76-A. The resulting amplicon was cloned into the EcoRV site of pSTBlue-1. The resulting plasmid, pJFF025, was then digested with BstEII and SacI, yielding a 2,983-bp fragment which was cloned into the corresponding sites of pmini-OphoA to construct the final plasposon. In this plasposon, pTnphoA.ts, the gentamicin resistance cassette and the pMB1/ColE1 origin of replication located between the inverted repeat elements of pmini-OphoA were replaced by the beta-lactamase gene and the conditional temperature-sensitive origin of replication from pST76-A.
Generation of TnphoA.ts mutants in ETEC.
ETEC strain H10407 was transformed with pTnphoA.ts by electroporation. Ampicillin-resistant colonies obtained after overnight growth on Luria agar plates containing 100 μg/ml of ampicillin at 30°C were pooled and used to inoculate Luria broth (LB) containing ampicillin (100 μg/ml), which was incubated overnight at 30°C. After 1:2 dilution in 50% glycerol, this stock (jf1181) was frozen at −80°C, and it was subsequently used as the starting material for generation of mutants.
Briefly, 2-ml aliquots of LB containing ampicillin (100 μg/ml) were inoculated with jf1181 and incubated overnight at 30°C and 250 rpm. The following morning cultures were diluted 1:100 in fresh medium and grown for 90 min at 30°C, after which the cultures were again diluted 1:100 and 100-μl aliquots were plated onto multiple XP plates. Plates inoculated in this way were then incubated at 37°C overnight. Blue colonies selected from these plates were streak purified on fresh XP plates, and isolated colonies were subsequently used to inoculate 96-well plates containing LB with ampicillin. Total genomic DNA isolated from these colonies was digested with a restriction endonuclease (e.g., MluI), ligated, and used to transform E. coli DH10BT1 to ampicillin resistance after growth at 30°C. Plasmid DNA isolated from the resulting Ampr transformants was digested with the corresponding enzyme and subjected to agarose gel electrophoresis. Plasmids exhibiting a single band at ≥5 kb were then used as templates in DNA sequencing reactions. The DNA sequence upstream from the ′phoA region of the transposon was then screened for unique sequences not present in the E. coli K-12 strain MG1655 genome (9) by BLASTN searches.
DNA sequencing of the etpBAC locus.
The focus of these studies, TnphoA.ts mutant 220, was 1 of approximately 250 colonies generated in an initial screening analysis. Plasmid DNA from this mutant was generated as described above following initial digestion of the genomic DNA with MluI. Digestion of the resulting plasmid, pJML001, yielded a single HMW band at approximately 12 kb. To obtain DNA sequences immediately upstream and downstream of the transposon insertion cloned in pJML001, we employed primers TnphoA.179 (5′-CCATCCCATCGCCAATCA-3′) and TnphoA.ts1 (5′-CGAAATTAATACGACTCA-3′), respectively. Additional DNA sequence from this locus was obtained by a combination of primer walking and generation of additional templates using in vitro Tn7 transposition of the GPS1.1 and GPS2.1 transprimers (New England Biolabs) into pJML001. Templates generated in this way were then sequenced using primers N (5′-ACTTTATTGTCATAGTTTAGATCTATTTTG-3′) and S (5′-ATAATCCTTAAAAACTCCATTTCCACCCCT-3′). To sequence through the repeat regions of etpA, this gene was first amplified using primers jf011005.1 (5′-CGGAATTCAATGAACCGTATATATAAA-3′) and jf011005.2 (5′-GAAGATCTTTGCCAGTACACCTCACT-3′) and cloned into the EcoRV site of pT7Blue-3 to create pJY017. This plasmid contained unique vector BamHI and SphI sites that were located 18 and 39 bp downstream from the 3′ end of etpA, respectively. Following restriction endonuclease digestion with these enzymes, approximately 2.5 μg of the linearized DNA was digested with exonuclease III at 37°C, which removed aliquots at 30-s intervals. Each aliquot was then treated with S1 nuclease, repaired with the Klenow fragment, and ligated with T4 DNA ligase. The resulting plasmids having progressive deletions from the 3′ end of etpA were then sequenced using vector primer R-20 (5′-CAGCTATGACCATGATTACG-3′). All sequencing was performed with an Applied Biosystems 373 automated sequencer and were edited and assembled with Sequencher 3.0 (GeneCodes, Ann Arbor, MI).
DNA and protein sequence comparisons: Clustal alignment.
Protein sequences were aligned by using ClustalW (http://www.ch.embnet.org/software/ClustalW.html), and resulting alignments were shaded for similarity using Boxshade 3.2.1 (http://www.ch.embnet.org/software/BOX_form.html).
Molecular cloning.
To generate a recombinant expression plasmid containing orf1 (etpB), primers jf102604.1 (5′-CCCAAGCTTGTGGTGAAATTCATGTCA-3′) and jf102604.3 (5′-GCGAATTCGAACGTTTTCAGGGCTGA-3′) were used to amplify an 1,819-bp fragment which was cloned into the EcoRV site of pT7Blue-3 to create pJMF1001. The corresponding HindIII/EcoRI insert fragment from pJMF1001 was then directionally cloned into the corresponding sites in pFLAG-CTC (Sigma). The resulting plasmid, pJMF1002, contained etpB minus its native stop codon to form an in-frame gene fusion with the region of pFLAG-CTC encoding the FLAG epitope tag. pJMF1002 was sequenced with primers N-26 (5′-CATCATAACGGTTCTGGCAAATATTC-3′) and C-24 (5′-CTGTATCAGGCTGAAAATCTTCTCTC-3′) to verify the construct.
To construct an etpA-polyhistidine tag gene fusion, the etpA gene minus its native stop codon was amplified using primers jf092804.1 (5′-CGGGATCCATGAACCGTATATATAAA-3′) and jf092804.2 (5′-CCGCTCGAGTTGCCAGTACACCTCACT-3′). Following digestion with BamHI and XhoI, the resulting 3,269-bp amplicon was cloned into the corresponding sites on pET30a(+) in frame with the polyhistidine tag coding region to create pJY005. DNA sequencing of pJY005 was performed using the T7 and SP6 vector primers to ensure that the insert encoding the etpA fragment was in frame with the polyhistidine tag.
To construct an EtpA expression plasmid, a 5,308-bp EcoRI-BglII etpA fragment from pJY017 was directionally cloned into the corresponding sites of pFLAG-CTC, placing etpA in frame with a FLAG epitope coding sequence to create pJY035.
Finally, primers jf031505.1 (5′-AATAATCTCGAGAATGGTGGTGAAATTCATG-3′; XhoI site underlined) and jf031505.2 (5′-AATAATAAGCTTTCATTCGGCACCCTCCTC-3′; HindIII site underlined) were used to amplify the entire etpBAC locus for cloning into the corresponding restriction sites on pBAD/Myc-His A to create pJY019.
Construction of deletion mutants.
To construct an isogenic etpA mutant, an amplicon with an in-frame etpA deletion was first constructed by PCR. Primers jf092504.1 (5′-GCATGCCGCGTACCGGCAGACAGT-3′) and jf092704.4 (5′-ATGAACCGTATATATAAAATTACTCTTACCGGGAAC-3′) were used to generate a 1,167-bp amplicon which included the first 6 codons and last 12 codons of the predicted etpA open reading frame (ORF) and 1,065 bp of DNA sequence downstream from the stop codon of this gene. Next, the reverse complement of jf092704.4, primer jf092704.5 (5′-GTTCCCGGTAAGAGTAATTTTATATATACGGTTCAT-3′), and primer jf100104.5 (5′-GCCTCTCCCTGCTGCT GA-3′) were used to generate a 1,308-bp amplicon with the reverse complement of the etpA deletion and 1,272 bp of upstream flanking sequence. The products of these two reactions were combined as the template DNA in a third PCR using the jf100104.5 and jf092504.1 flanking primers, which resulted in a 2,400-bp amplicon that was cloned into pT7Blue-3, yielding pJY001. pJY001 was then sequenced with primers U-19 (5′-GTTTTCCCAGTCACGACG T-3′) and R-20 (5′-CAGCTATGACCATGATTACG-3′) to verify the deletion constructed. Next, a 2,113-bp SphI insert fragment from pJY001was cloned into the corresponding site of pCVD442 to obtain the suicide plasmid pJY028. SM10λpir(pJY028) was then used to introduce the suicide plasmid into H10407-S to construct the in-frame deletion by double homologous recombination, as previously described (25). Resulting Amps Smr colonies were then examined by PCR and Southern blotting to confirm construction of the in-frame deletion.
To construct an etpC deletion mutant, a λ-Red recombinase-based method was employed, as previously described by Datsenko and Wanner (22). Briefly, the λ-Red recombinase expression plasmid pKD46 was introduced into wild-type strain H10407 by electroporation, followed by selection on ampicillin (100 μg/ml) at 30°C. Primers jf093005.1 (5′-GCTGCAGGGCGCACGCTACCGGTTAATTTTTCGTATTCAGGAGTGTAGTGTAGGCTGGAGCTGCTTC-3′) and jf093005.2 (5′-TTCGGCACCCTCCTCCTGACGATGGCTACGCGCCTCCCACAACTGCCAATT CCGGGGATCCGTCGACC-3′) were used to amplify a 1,398-bp PCR product containing etpC flanking sequences (underlined) and a cassette containing kanamycin resistance and FLP recombinase target sites from pKD13. The resulting amplicon was electroporated into H10407(pKD46), and this was followed by selection on kanamycin (25 μg/ml) at 37°C. Kmr Amps colonies were then tested by performing a three-primer PCR with etpC flanking primers jf091504.1 (5′-TGATGAAGAGGGCAACTG-3′) and jf031505.2 (5′-TCATTCGGCACCCTCCTC-3′) and primer K2 (5′-CGGTGCCCTGAATGAACTGC-3′) (22).
Southern hybridization.
An etpA gene probe was amplified from H10407 using primers jf011005.1 (5′-ATGAACCGTATATATAAA-3′) and jf011005.2 (5′-TTGCCAGTACACCTCACT-3′), and the resulting amplicon was then labeled by random primer labeling with digoxigenin (DIG)-UTP (44). Hybridization with plasmid DNA digested with SphI or genomic DNA digested with MluI was carried out under stringent conditions without formamide.
Colony hybridization.
In colony hybridization studies to screen for the presence of etpBAC, DNA probes were amplified using the following primer sets: for etpA (3,257 bp), jf011005.1 (5′-ATGAACCGTATATATAAA-3′) and jf011005.2 (5′-TTGCCAGTACACCTCACT-3′); for etpB (1,812 bp), jf102604.1 (5′-CCCAAGGTGGTGAAATTCATGTCA-3′) and jf102604.2 (5′-GGAATTCTCAGAACGTTTTCAGGGCTGA-3′); and for etpC (1,495 bp), jf092104.1 (5′-CCTGAACGCGGATAACCT-3′) and jf031505.2 (5′-AATAATAAGCTTTCATTCGGCACCCTCCTC-3′). Products were labeled by random primer labeling with digoxigenin and used in hybridization studies carried out as previously described (68, 77) under stringent conditions without formamide.
Adhesion assays.
The Caco-2 (33) and HCT-8 (86) intestinal cell lines used in the adhesion assays were obtained from the American Type Culture Collection. Each cell line was propagated by using American Type Culture Collection protocols. On the day prior to assays, cells were split at a ratio of approximately 1:2, and the resulting cell suspensions were used to inoculate individual wells of 96-well tissue culture plates. Bacteria were grown overnight in 2 ml of LB with appropriate antibiotics and then diluted 1:100 on the morning of the experiment and grown for 90 min to the mid-logarithmic growth phase. Organisms were then added to wells of the tissue culture plates in triplicate at a multiplicity of infection (MOI) of approximately 10:1 (26). After 1 h the monolayers were washed with tissue culture medium and then exposed to 0.1% Triton X-100 in phosphate-buffered saline for 5 min. Triton X-100 lysates containing total cell-associated bacteria were then diluted 1:10 in phosphate-buffered saline and plated onto Luria agar. To determine the number of intracellular bacteria, monolayers were washed three times with RPMI and then incubated with medium containing gentamicin (100 μg/ml) for 2 h. After washing to remove gentamicin, bacteria released by Triton X-100 lysis were plated onto Luria agar. The number of bacteria that remained cell associated relative to the inoculum was then determined by determining the number of CFU. The data were expressed as the percentage of organisms recovered per CFU/inoculum. Alternatively, monolayers were fixed with methanol and stained with Giemsa stain, and the number of adherent bacteria/cell was determined by microscopic examination. In antibody inhibition experiments, antibodies were added to wells at the appropriate dilution immediately prior to introduction of the bacteria.
Antibody production.
Following expression of E. coli BL21(DE3)(pJY005) and purification of recombinant polyhistidine-tagged EtpA (rEtpA.6H) by Ni affinity chromatography, the resulting 110-kDa protein was used to immunize two New Zealand White rabbits, as previously described (31). Polyclonal rabbit antisera were absorbed on an E. coli lysate column (Pierce Biotechnology) and subsequently incubated with lyophilized E. coli AAEC191-A to remove cross-reacting antibodies. Additional preparative purification of antibodies from both preimmune and immune sera was performed using protein G column chromatography (HiTrap Protein G HP; Amersham Biosciences).
For affinity purification of anti-EtpA antibodies we employed rEtpA.6H protein immobilized on nitrocellulose membranes. Following separation by sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE), recombinant protein was transferred to nitrocellulose, and the membrane was blocked with 1% bovine serum albumin in Tris-buffered saline for 1 h at room temperature. Nitrocellulose strips with the immobilized antigen were then incubated with a 1:10 dilution of anti-EtpA rabbit polyclonal antibody overnight at 4°C. The antibody was eluted with 1 ml of 0.1 M glycine (pH 2.7) for 15 min at room temperature. The eluate was sterile filtered and concentrated 50-fold using a 10,000-molecular-weight cutoff centrifugal concentration filter.
Immunoblotting and detection of glycoproteins.
To identify FLAG epitope-tagged EtpA molecules in culture supernatants, proteins were first concentrated by precipitation with trichloroacetic acid (TCA). Briefly, 750 μl of cold TCA (20%) was added to an equal volume of supernatant from an overnight culture, incubated on ice for 30 min, and centrifuged at 4°C and 15,000 × g for 15 min, and the supernatant was discarded. The resulting pellet was washed twice with 500 μl of ice-cold acetone, air dried, and resuspended in 10 μl of 1 M Tris (pH 7.4). Concentrated proteins were separated by SDS-PAGE (54) and transferred to nitrocellulose. Mouse monoclonal antibody M2 (Sigma) directed against the FLAG octapeptide (N-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-C) was used at a dilution of 1:1,000, followed by horseradish peroxidase-labeled goat anti-mouse antibodies (Pierce) at a dilution of 1:80,000. To detect native EtpA secreted into culture supernatants, immunoblotting was performed using preabsorbed rabbit polyclonal anti-rEtpA.6H sera (1:2,000) and goat anti-rabbit immunoglobulin G(Fc)-horseradish peroxidase (1:60,000; Pierce). All blocking and incubation steps were performed at room temperature in Tris-buffered saline (pH 7.4) containing 0.05% Tween 20 and 5% milk. Detection was carried out with a luminal-based chemiluminescent substrate (SuperSignal; Pierce).
After separation by SDS-PAGE and transfer to nitrocellulose, glycoproteins in culture supernatants were detected using DIG-glycan reagents (39, 50, 66) (Roche). Briefly, carbohydrate residues were first oxidized with 10 mM sodium metaperiodate in sodium acetate buffer (pH 5.5) and then labeled with DIG- 0-3-succinyl-ɛ-aminocaproic acid hydrazide. DIG-labeled proteins were then detected with alkaline phosphatase conjugated to anti-digoxigenin antibodies using a 4-nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl-phosphate solution as the substrate.
Fractionation of bacterial cells.
To obtain whole-cell lysates of bacteria, overnight cultures were centrifuged to pellet bacteria, and the pellets were solubilized in nonionic detergent in 20 mM Tris-HCl (pH 7.5) (B-PER; Pierce). The bacterial membranes were fractionated by solubilization of inner membranes in Triton X-100, followed by isolation of outer membrane proteins by ultracentrifugation as previously described (81).
Nucleotide sequence accession number.
Nucleotide data for the etpBAC locus have been deposited in the GenBank database under accession number AY920525.
RESULTS
Identification of a two-partner secretion locus by TnphoA.ts mutagenesis.
Initial DNA sequencing of the region interrupted in TnphoA.ts mutant 220 using primers TnphoA.179 and TnphoA.ts1 revealed no homology to E. coli K-12. Further sequencing of this locus revealed three candidate ORFs in the same transcriptional orientation, as shown in Fig. 2. BLASTP analysis (2) of the predicted proteins suggested that this locus may encode a two-partner secretion pathway (46) (Table 2).
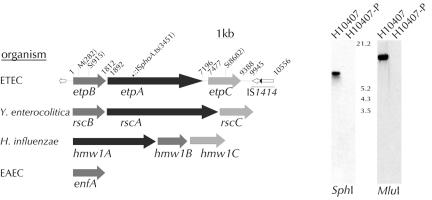
FIG. 2.
Organization of the ETEC two-partner secretion locus. The candidate genes in the etpBAC locus were designated based on the nomenclature used for homologues, including the Y. enterocolitica rscBAC locus (64) and the prototype hmwABC locus, as previously described (5). The numbers (bp) indicate the beginning of each start codon and the end of each stop codon, beginning with etpB. The original TnphoA.ts insertion is in etpA 3,451 nucleotides from the start of etpB. Immediately downstream from the etpBAC locus is a copy of IS1414 (61). The open arrow indicates the direction of transcription of the putative IS1414 transposase gene that is flanked by direct repeats beginning at nucleotide positions 9945 and 10556. In the transposase (tnp) coding sequence is a putative astA gene (solid arrowhead, in +1 reading frame relative to tnp). The Yersinia rscBAC, Haemophilus hmwABC, and enteroaggregative E. coli (EAEC) enfA loci are shown below the ETEC locus for comparison of the organizations and relative sizes of the genes. Genes are shaded to reflect homology in the encoded proteins. The locations of MluI and SphI restriction sites in the etpBAC locus are indicated on the etpBAC map by M(282), S(915), and S(8682). The blots show the results for Southern blot hybridization (using an etpA probe) of plasmid DNA from H10407 and H10407-P digested with SphI and total genomic DNA from the same strains digested with MluI. The numbers indicate the relative locations of digoxigenin-labeled molecular weight markers (in kb).
TABLE 2.
Homologues of predicted proteins encoded by the ETEC two-partner secretion locus
| Candidate ETEC protein (accession no.) | Mol wt (103) | Conserved domain(s)a | Homologue (GenBank accession no.)b | % Indentity (% similarity) | Putative function of homologue | Reference(s) |
|---|---|---|---|---|---|---|
| EtpB (AY920525) | 61.7 | HlyB (8792), | Enf (AAK58509) | 50 (65) | Nonfimbrial adhesin | 63 |
| FhaC (12184) | RscB (AAK77859) | 30 (51) | 57,64 | |||
| Hmw1B (AAA20528.1) | 19 (35) | TPS transporter | 4,5 | |||
| EtpA (AY920526) | 177 | Secretion | RscA (AAK77860) | 29 (44) | 64 | |
| (24084) | HmwA (AAD56660) | 31 (48) | Nonfimbrial adhesin | 88 | ||
| Hmw1A (AAX88733) | 31 (48) | Nonfimbrial adhesin | 4,5 | |||
| EtpC (AY920527) | 72.0 | Spy (13225) | RscC (AAK77861) | 50 (63) | O-linked N-acetylglucosamine | 64 |
| HmwC (AAF00477) | 42 (58) | Transferase? | 88 | |||
| Hmw1C (AAA20529.1) | 42 (58) | Glycotransferase | 4,35 |
Numbers in parentheses refer to conserved domain database reference (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cdd) (59, 60).
BlastP comparisons.
The middle ORF, etpA, interrupted by the transposon insertion in mutant 220, encodes a predicted protein having a molecular mass of approximately 177 kDa which exhibits homology with a family of secreted high-molecular-weight proteins exemplified by the HMW adhesins previously identified in nontypeable H. influenzae strains (83) (Table 2). EtpA exhibits significant homology with HmwA-like putative adhesin proteins from Yersinia pestis (67) and Yersinia pseudotuberculosis (16) and the RscA protein of Yersinia enterocolitica (64). EtpA has a putative secretion domain in its amino terminus (Table 2). This domain is found at the amino-terminal end of a variety of known adhesins, including HMW 1A (4, 5), and filamentous hemagglutinin of B. pertussis (74). In this conserved amino-terminal domain is an Asn-Pro-Asn-Gly-Val sequence (Fig. 3) similar to the Asn-Pro-Asn-Gly-Ile motif found in many exoproteins secreted via a two-partner secretion pathway (46). Mutagenesis of this similar motif in other TpsA proteins interferes with their secretion (82) and/or release (84) from the bacterial cell.
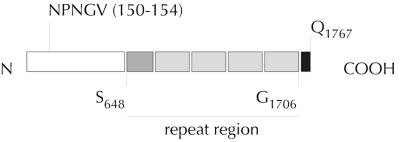
FIG. 3.
Molecular features of EtpA. The predicted EtpA sequence lacks a traditional signal sequence at the amino terminus. The NPNGV sequence at residues 150 to 154 is nearly identical to the NPNGI sequence found at the same location in the homologues RscA (64) and HmwA (14) and is similar to Asn-Pro-Asn motifs found to be involved in secretion of less closely related proteins, such as ShlA (46, 82). A search of the peptide sequence using the RADAR (40) (http://www.ebi.ac.uk/cgi-bin/radar/radar) and REPRO (34) (http://ibivu.cs.vu.nl/programs/reprowww/) algorithms revealed the presence of several repeat regions in the carboxy-terminal region of the molecule. Four major repeat units (∼226 residues) are preceded by a 173-amino-acid partial repeat beginning at residue S648.
The carboxy-terminal region of EtpA (from S648 to G1706) consists of a series of repeated domains (Fig. 3). Four repeats that are approximately 226 amino acids long are preceded by a single 173-residue partial repeat beginning at S684.
Additional in silico analysis of EtpA suggested that there is structural similarity to a number of bacterial virulence proteins and predicted that EtpA assumes a β-helical conformation. Examination of the predicted EtpA peptide with BetaWrap (http://betawrap.lcs.mit.edu/), a program which scores primary amino acid sequences for compatibility with right-handed beta-helix supersecondary structure, yielded highly significant results (P = 7.8 × 10−05). Interestingly, this predicted structure appears to be common among bacterial virulence proteins, including the passenger domain of the B. pertussis autotransporter pertactin (28), but it is found only infrequently in most other protein sequences (11). Similarly, analysis of the EtpA peptide with the 3D-PSSM algorithm (http://www.sbg.bio.ic.ac.uk/servers/3dpssm/) (47) yielded similar structural predictions, again suggesting that there is a β-helical structure similar to that of pertactin.
The etpB gene, immediately upstream from etpA, encodes a predicted 61.7-kDa protein that exhibits considerable homology with Enf (Table 2), a putative afimbrial adhesin of enteroaggregative E. coli serotype O111:H12 (63). In addition, the predicted EtpB protein exhibits similarity to RscB from Y. enterocolitica and to the prototypical TpsB transporter proteins Hmw1B from H. influenzae and FhaC from B. pertussis, as demonstrated in the ClustalW alignment shown in Fig. S1 in the supplemental material. A SignalP (7) search of the amino terminus of EtpB predicted a signal peptide cleavage site between amino acid residues 24 and 25, suggesting that the molecular mass of the mature protein in the outer membrane is approximately 59.2 kDa. This is similar to the size of the mature 58-kDa Enf protein. Further analysis of the EtpB peptide using the 3D-PSSM algorithm supported the possibility that this structure forms a β-barrel in the outer membrane.
The final 1,913-bp ORF in this locus, etpC, encodes a putative 72-kDa protein with homology to RscC of Y. enterocolitica (64) and Hmw1C (4, 5). The predicted EtpC protein contains a domain that is highly conserved in a family of proteins involved in posttranslational modification of other proteins, which work as O-linked N-acetylglucosamine transferases to catalyze the transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to target Ser/Thr residues (38).
Flanking the locus described above are potential mobility elements, including an upstream ORF with homology to ORF 121 from IS91 (62). Downstream from this locus is a copy of IS1414 (61), which contains a copy of astA, the gene for EAST1 (78) in the putative transposase. Because of the close proximity of these potential mobility elements to the etpBAC locus and because other organisms may have more than one closely related TPS locus, such as HMW1 and HMW2 of H. influenzae (5), we sought to determine whether a single copy of the etpBAC locus was present. As shown in Fig. 2, Southern blotting of both plasmid and total genomic DNA from H10407 yielded single bands when the DNA was hybridized with an internal etpA gene probe. These bands were absent after hybridization with DNA obtained from H10407-P, which lacks the large 92-kb pCS1 virulence plasmid, suggesting that a single copy of etpBAC is in this extrachromosomal element.
Taken together, these data suggested that the genes in this locus are dedicated to the formation of a TPS pathway. Therefore, additional studies were undertaken to explore the validity of this suggestion.
Secretion of EtpA.
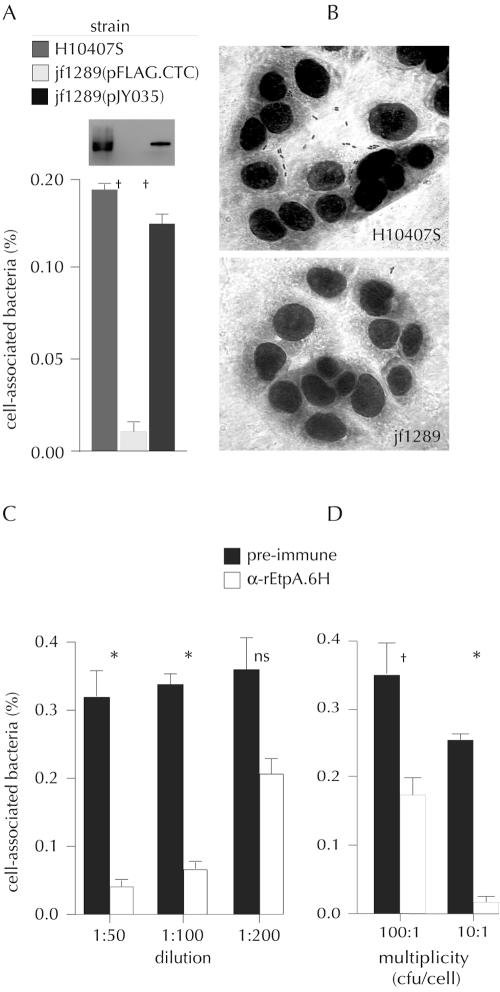
Because the predicted peptide encoded by etpA exhibits significant homology with a number of secreted proteins, we performed experiments to examine the secretion of EtpA. As shown in Fig. 4, antisera raised against a recombinant polyhistidine-tagged EtpA fragment (rEtpA.6H) reacted with an HMW specimen in immunoblots of TCA-precipitated culture supernatants from the H10407 wild-type strain but not in immunoblots of TCA-precipitated culture supernatants from jf1289, an isogenic etpA deletion mutant, suggesting that EtpA is in fact secreted. Restoration of the etpA gene in trans as a FLAG epitope fusion on expression plasmid pJY035 restored secretion of a protein that could be detected either with anti-EtpA antisera (Fig. 5A) or with monoclonal antibodies directed against the FLAG epitope (not shown). Supernatants from H10407-P did not contain a similar reactive protein. Furthermore, cloning and expression of the etpBAC locus in a recombinant E. coli background, LMG194(pJY019), were sufficient to produce the HWM protein in culture supernatant that was recognized by anti-EtpA antisera on Western blots (Fig. 4A).
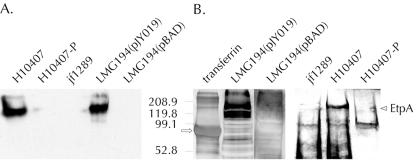
FIG. 4.
Identification of secreted, glycosylated forms of EtpA in TCA-precipitated culture supernatants. (A) Immunoblot of supernatants from H10407, H10407-P, the isogenic ΔetpA strain jf1289, recombinant E. coli LMG194(pJY019) expressing the etpBAC locus, and the vector control LMG194(pBAD) using polyclonal rabbit primary antibodies directed against rEtpA.6H. (B) Detection of glycoproteins in concentrated supernatants using digoxigenin-glycan reagents. The strains examined were recombinant E. coli LMG194 carrying the etpBAC expression plasmid pJY019, the same strain with a control plasmid (pBAD), the isogenic etpA deletion mutant jf1289, wild-type strain H10407, and H10407-P. Transferrin (arrow) was included as a positive control for detection of glycoprotein. The arrowhead on the right indicates the position of EtpA.
FIG. 5.
Involvement of EtpA in bacterial interaction with host epithelial cells. (A) Comparison of the relative abilities of the wild type and the isogenic etpA deletion mutant to adhere to target HCT-8 intestinal epithelial cells. The strains used were wild-type strain H10407S, the isogenic ΔetpA mutant carrying the vector plasmid [jf1289(pFLAG.CTC)], and the ΔetpA deletion mutant complemented in trans with the rEtpA expression plasmid [jf1289(pJY035)]. The values are the numbers of organisms recovered by lysis of monolayers compared to the inoculum. The Western blot above the graph shows identification of EtpA in culture supernatants from the strains following TCA precipitation and immunoblotting with anti-EtpA antibodies. (B) Giemsa-stained HCT-8 monolayers following infection with wild-type strain H10407S or jf1289. (C) Antibodies directed against recombinant EtpA.6H inhibited association of ETEC strain H10407 with target HCT-8 cultured intestinal epithelial cells. (D) Anti-EtpA antibody effect at different multiplicities of infection, expressed as CFU/cell. An asterisk indicates that the P value is <0.01, a dagger indicates that the P value is <0.05, and ns indicates not significant, as determined by a two-tailed Student t test for paired samples.
Although EtpA could be identified easily in concentrated supernatant from H10407, only a small amount of this protein was detected on immunoblots of whole-cell lysates, and we were not able to detect EtpA in outer membrane preparations of this strain (not shown). These findings suggest that the secretion of EtpA is quite efficient and that this exoprotein may be only transiently associated with the bacterial cell surface.
etpBAC locus directs secretion of a high-molecular-weight glycosylated exoprotein.
Based on their sequence homology to proteins involved in secretion of high-molecular-weight glycoproteins by H. influenzae, we predicted that the proteins encoded by the etpBAC locus would also result in the export of a large glycosylated molecule (35). Indeed, as shown in Fig. 4B, we identified HMW glycoprotein in culture supernatants from LMG194(pJY019) and the H10407 parent strain. This glycoprotein was absent in TCA-precipitated supernatants obtained from H10407-P and the isogenic etpA deletion mutant, jf1289.
Role of the etpBAC locus in adherence of ETEC to cultured intestinal epithelial cells.
Because EtpA exhibits homology with a number of previously identified TpsA exoproteins that have been shown to function as adhesins, we tested the ability of etpA mutants to adhere to cultured epithelial cells. As shown in Fig. 5A, jf1289 did not adhere as efficiently to cultured HCT-8 cells as the H10407 wild-type strain adhered (P = 0.02), while complementation of the etpA deletion in trans with pJY035 encoding a FLAG epitope-tagged version of EtpA restored this phenotype to wild-type levels (P = 0.015 for a comparison of the etpA deletion strain and the complemented mutant). To ensure that bacteria were actually cell associated and not adherent to the wells, the experiments included controls in which the same number of organisms was used to inoculate wells containing no target epithelial cells. We routinely failed to recover any bacteria from these wells (data not shown). In addition, Giemsa staining of monolayers (Fig. 5B) following infection with either the wild type or the etpA mutant at an MOI of 10 resulted in identification of approximately 0.8 ± 0.02 wild-type bacterium per epithelial cell, compared to 0.12 ± 0.04 organism/cell for the mutant (P = 0.002). An increase in the MOI to 100:1 resulted in 3.3 ± 1.1 wild-type organisms/cell, compared to 0.29 ± 0.06 mutant bacterium/cell (P = 0.046).
To further determine whether EtpA might play a role in mediating adherence to epithelial cells, we tested the ability of antibodies directed against recombinant EtpA (rEtpA) to inhibit adherence by the ETEC strain H10407 prototype. As shown in Fig. 5C, anti-rEtpA.6H antisera dramatically inhibited association of the wild-type strain with target HCT-8 cells compared to the results obtained with identical concentrations of preimmune sera from the same rabbit. Similar results were obtained using Caco-2 cells or using affinity-purified anti-EtpA antibodies (data not shown). These data suggest that EtpA, like its molecular TpsA homologues, promotes pathogen-host cell interaction. Expression of the entire etpBAC locus (on pJY019) in recombinant E. coli strain LMG194 was sufficient to promote adherence to HCT-8 cells. After induction of LMG194(pJY019) with 0.0002 mM arabinose, 0.44% ± 0.04% of the organisms added to HCT-8 monolayers were recovered in adherence assays, which was significantly more than the value for the recombinant vector control strain, LMG194(pBAD/Myc-His A), for which no bacteria were recovered (P = 0.001), but was less than the value for wild-type ETEC strain H10407 (0.89% ± 0.08) (P = 0.01).
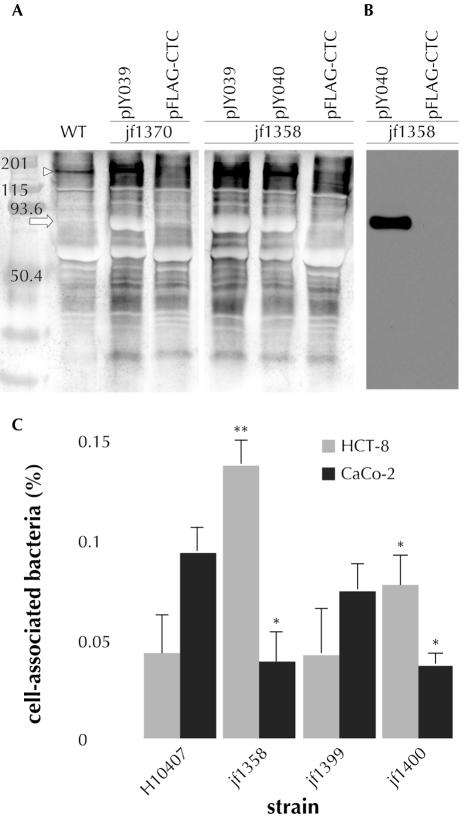
Each of the bacterial glycoprotein adhesins described to date appears to require glycosylation to effectively promote adherence in vitro (8, 35, 56). To investigate the potential role of posttranslational modification of EtpA by glycosylation in the adherence phenotype, we constructed a deletion in etpC. As predicted by the homology of EtpC to HmwC, a protein involved in glycosylation of the HmwA adhesin of H. influenzae, etpC deletion mutants produced nonglycosylated EtpA (Fig. 6A and B). Interestingly, we found that the adherence phenotype of etpC glycosylation mutants was strikingly dependent on the target intestinal cell line employed. Consistent with the observation that glycosylation of HmwA is required for adherence in vitro, we found that etpC deletion mutants were less likely to associate with Caco-2 cells than the wild type was (Fig. 6C). However, the same mutants were actually hyperadherent when HCT-8 cells were used in the assays. This hyperadherent phenotype was due entirely to an increase in adherence as the numbers of organisms that survived gentamicin treatment were quite small and were not significantly different for the wild type and the mutant (6.31E-02% ± 1.80E-02% and 4.85E-02% ± 8.10E-03%, respectively). These data suggest that EtpA may mediate adherence to different receptors in a process that is differentially modulated by glycosylation.
FIG. 6.
Role of EtpC in posttranslational modification of EtpA and interaction with target epithelial cells. (A) Digoxigenin-glycan blot of TCA-precipitated supernatants, demonstrating the absence of EtpA glycoprotein in EtpC mutants jf1358 and jf1370 containing only the pFLAG-CTC vector plasmid. Production of EtpA glycoprotein was restored in etpC mutants containing plasmids expressing EtpC (pJY039) or EtpC-FLAG (pJY040) following induction with 0.1 mM IPTG. EtpC overexpression at this IPTG concentration affected cell viability and led to appearance of EtpC in culture supernatants. (B) Anti-FLAG immunoblot demonstrating expression of epitope-tagged EtpC in the complemented strain jf1358(pJY040). (C) Association of the wild type and etpC mutants with target HCT-8 or Caco-2 intestinal epithelial cell lines. jf1399 was jf1358 complemented with etpC expression plasmid pJY039, and jf1400 was jf1358 complemented with pFLAG-CTC vector plasmid. Assays were performed following induction of bacteria with 0.00001 mM IPTG. Asterisks indicate the results for comparisons with H10407 data for the same cell line, as determined by Student's t test (one asterisk, P < 0.05; two asterisks, P < 0.01).
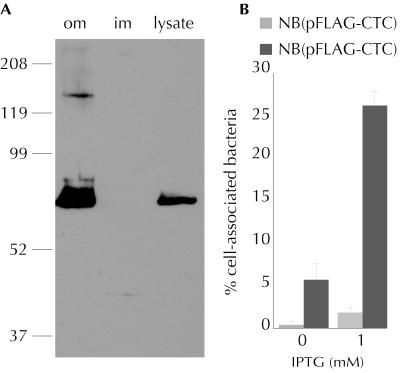
As noted above, the predicted protein encoded by etpB exhibits considerable homology with Enf, a recently identified outer membrane protein adhesin of enteroaggregative E. coli (63). Indeed, when etpB was cloned and expressed in a recombinant E. coli background, rEtpB was largely localized to the outer membrane (Fig. 7A) and conferred the ability to adhere to cultured epithelial cells of gastrointestinal origin. A laboratory strain of E. coli (NovaBlue) bearing pJMF1002 exhibited marked increases in adherence to Caco-2 cells compared to the control strain bearing the expression plasmid vector pFLAG-CTC (Fig. 7B), suggesting that EtpB, independent of its possible function as a TpsB transporter for EtpA, might promote adherence similar to that promoted by the highly homologous Enf afimbrial adhesin. Interestingly, a BLAST-X search of the 192 bp of DNA sequence downstream from the enfA gene available in GenBank accession no. AF196977 indicated the beginning of an ORF that, when translated, exhibited homology to TpsA exoproteins, including the HMW adhesins of H. influenzae (data not shown), raising the possibility that Enf might also be part of a TPS pathway.
FIG. 7.
EtpB is localized to the outer membrane and promotes adherence. (A) Localization of recombinant EtpB-FLAG to the outer membrane of recombinant E. coli in strain Novablue(pJMF1002). om, outer membrane; im, inner membrane and cytosol; lysate, whole-cell lysate fractions probed with anti-FLAG antibody. The cloned etpB gene promoted association of recombinant E. coli with cultured Caco-2 intestinal epithelial cells. Recombinant E. coli NovaBlue containing either the vector plasmid [NB(pFLAG-CTC)] or the etpB expression plasmid [NB(pJMF1002)] was grown to the mid-logarithmic phase with or without IPTG induction and was then used to infect semiconfluent Caco-2 cell monolayers at an MOI of ∼10. After incubation for 1 h at 37°C in the presence of 5% CO2, monolayers were extensively washed and lysed with Triton X-100. The data are the total number of remaining cell-associated bacteria expressed as a percentage of the inoculum, as determined by colony counting.
Multiple ETEC strains secrete the EtpA glycoprotein.
The identification of genes encoding potential mobility elements adjacent to etpBAC led us to examine the prevalence of this locus in other E. coli strains. In screening for the etpBAC locus by DNA hybridization, we examined a variety of diarrheagenic clinical E. coli isolates from different pathotypes, including enterotoxigenic E. coli, enteropathogenic E. coli, enteroaggregative E. coli, enteroinvasive E. coli, and enterohemorrhagic E. coli. We detected hybridization signals from many of the ETEC strains in our initial collection with all three probes (etpB, etpA, and etpC) (Table 3), while no signal was detected in colony hybridizations of enteroaggregative E. coli, enteroinvasive E. coli, enteropathogenic E. coli, or enterohemorrhagic E. coli strains (Table 4). These findings suggested that the locus is unique to ETEC.
TABLE 3.
Screening of ETEC isolates for the etpBAC locus and secretion of EtpA
| Strain | CFA (CS) | Toxin(s) | Serotype | DNA probe
|
EtpA secretion | Origina | ||
|---|---|---|---|---|---|---|---|---|
| etpA | etpB | etpC | ||||||
| M415C1 | I | STb | O2:nm | −c | − | − | − | a |
| M633C1 | I | LT/ST | O78:H11 | − | − | − | − | a |
| M109C2 | I | Rough:H12 | + | + | + | + | a | |
| DS244-1 | I | O6:H16 | + | + | + | + | a | |
| 15758a | I | O78:H10 | + | + | + | + | a | |
| TX1 | I | ST | O78:H12 | + | + | + | + | a |
| DS 229-1 | I | LT/ST | O128:H12 | + | + | + | + | a |
| H410C1 | I | Rough:nm | − | − | − | − | a | |
| H10407 | I | LT/ST | O78:H11 | + | + | + | + | a |
| DS67-1 | I | ST | O153:nm | + | b | |||
| DS97-2 | I | ST | O153:nm | − | b | |||
| DS229-1 | I | LT/ST | O128:H12 | + | b | |||
| DS321-1 | I | LT/ST | O128:H12 | + | b | |||
| ETEC-10 | I | ST | O153:H45 | − | b | |||
| ETEC-13 | I | ST | O153:H45 | − | b | |||
| ETEC-15 | I | O153:H45 | − | b | ||||
| H10407 | I | LT/ST | O78:H11 | + | b | |||
| DS220-4 | II (CS2, CS3) | O11:H33 | − | − | + | − | a | |
| DS373-2 | II (CS2, CS3) | O18:nm | + | + | + | + | a | |
| DS89-1 | II (CS3) | LT/ST | O6:H16 | − | b | |||
| DS61-1 | II (CS2, CS3) | LT/ST | O6:H16 | + | b | |||
| M424C1 | II (CS1, CS3) | LT/ST | O6:H16 | + | b | |||
| DS15-1 | II (CS2, CS3) | LT/ST | O6:H16 | − | b | |||
| DS18-1 | II (CS1, CS3) | LT/ST | O6:H16 | + | b | |||
| DS96-1 | II (CS2, CS3) | LT/ST | O6:H16 | + | b | |||
| DS124-1 | II (CS1, CS3) | LT/ST | O6:H16 | + | b | |||
| DS180-1 | II (CS3) | LT/ST | O6:H16 | + | b | |||
| DS198-1 | II (CS3) | LT/ST | O6:H16 | + | b | |||
| E9199 | II (CS1, CS3) | LT/ST | O6:H16 | − | b | |||
| E5470 | II (CS2, CS3) | LT/ST | O6:H16 | + | b | |||
| DS303-2 | II (CS1, CS3) | LT/ST | O6:H16 | + | b | |||
| ETEC-23 | II | LT/ST | O6:H16 | − | b | |||
| ETEC-26 | II | O6:H16 | − | b | ||||
| ETEC-43 | II | LT/ST | O6:H16 | − | b | |||
| DS7-3 | II (CS3) | LT/ST | O8:H9 | − | b | |||
| DS300-1 | IV (CS4, CS6) | O8:nm | + | − | − | − | a | |
| B4106-1 | IV (CS6) | ST | O27:H7 | − | − | − | − | a |
| B7A | IV (CS6) | LT/ST | O148:H28 | + | − | − | − | a |
| E11881E | IV (CS4, CS6) | O25:H42 | − | b | ||||
| PE423 | IV (CS5) | ST | O115:H40 | − | b | |||
| E17018A | (CS5, CS6) | LT?/ST | O167:H5 | − | b | |||
| E10703 | (CS5, CS6) | ST | O167:H5 | − | b | |||
| B7A | IV (CS6) | LT/ST | O148:H28 | − | b | |||
| E11881/9 | IV (CS4, CS6) | LT | O25:H42 | − | b | |||
| E34420A | CFAIII (CS8) | LT/ST | O167:nm | − | b | |||
| E2539-C1 | NDd | LT | O25:nm | − | − | − | − | a |
| 10614C | ND | O78:nm | − | − | − | − | a | |
| Scott | ND | O78:K80 | + | − | − | − | a | |
| EDL903 | ND | LT | O88:H25 | − | − | − | − | a |
| ThroopDe | ND | LT | O63:nm | + | + | + | + | |
| E7476A | (CS14) | ST | O166:H27 | + | b | |||
| E20738A | (CS17) | LT | O114:H25 | + | b | |||
| DS37-4 | (CS17) | LT | O114:H21 | + | b | |||
| Z16-4 | (CS17) | LT | O8:H29 | − | b | |||
Only the original collection of strains from WRAIR (a) was tested in the DNA hybrization studies. b, Berna Biotech.
ST, heat-stable enterotoxin; LT, heat-labile enterotoxin.
−, negative; +, positive.
ND, not determined.
See reference 30.
TABLE 4.
Screening of other E. coli pathotypes for etpBAC
| Pathotypea | Strain | Serotype | etpA | etpB | etpC | EtpA secretion | Originb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Porcine | F41 | −c | − | − | − | a | |||||||
| ETEC |
|
||||||||||||
| EPEC | B170 | O111:nm | − | − | − | − | a | ||||||
| EPEC | 833-60 | O119:H6 | − | − | − | − | a | ||||||
| EPEC | 3336-54 | O127:nm | − | − | − | − | a | ||||||
| EPEC | E2348 | O127:H6 | − | − | − | − | a | ||||||
| EPECd | RDEC-1 | O15 | − | − | − | − | a | ||||||
| EIEC | SD-61 | NDe | − | − | − | − | a | ||||||
| EIEC | SD-193-1 | ND | − | − | − | − | a | ||||||
| EHEC | EDL880 | O157:H7 | − | − | − | − | a | ||||||
| EHEC | B6914 | O157:H7 | − | − | − | − | a | ||||||
| EHEC | B1409 | O157:H7 | − | − | − | − | a | ||||||
| EHEC | C8632 | O157:H7 | − | − | − | − | a | ||||||
| EHEC | A7785 | O157:H7 | − | − | − | − | a | ||||||
| EHEC | EDL931 | O157:H7 | − | − | − | − | a | ||||||
| EHEC | C984 | O157:H7 | − | − | − | − | a | ||||||
| EAEC | 17-2 | O3:H2 | − | − | − | − | c | ||||||
| EAEC | JM221 | O93:H33 | − | − | − | − | a | ||||||
| EAEC | 309-1-1 | O130:H27 | − | − | − | − | a | ||||||
| EAEC | 103-1-1 | O148:H28 | − | − | − | − | a | ||||||
EPEC, enteropathogenic E. coli; EIEC, enteroinvasive E. coli; EHEC, enterohemorrhagic E. coli; EAEC, enteroaggregative E. coli.
a, WRAIR; b, Berna Biotech; c, James P. Nataro, CVD, University of Maryland School of Medicine.
−, negative.
See reference 15.
ND, not determined.
Immunoblotting studies to detect EtpA in concentrated supernatants from these strains corroborated the results of our hybridization analysis, since only strains that hybridized with all three probes secreted EtpA (Fig. 8). Although some ETEC strains, such as B7A and DS300-1, yielded a positive signal with the etpA gene probe, there was no detectable hybridization with either the etpB or etpC probes, and these strains did not secrete EtpA. Production of HMW glycoprotein by ETEC paralleled the results of immunoblotting experiments since all of the strains exhibiting immunoreactivity with anti-EtpA antibodies also produced a glycoconjugate having the same molecular weight (Fig. 9). Interestingly, several EtpA-negative strains, such as ETEC-10, ETEC-13, ETEC-15, and ETEC-26, yielded a weakly reactive species as determined by DIG-glycan analysis, raising the possibility that these strains may produce other exported HMW glycoproteins.
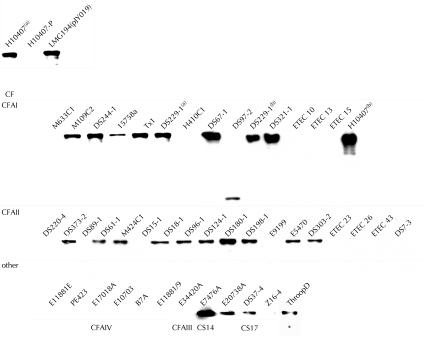
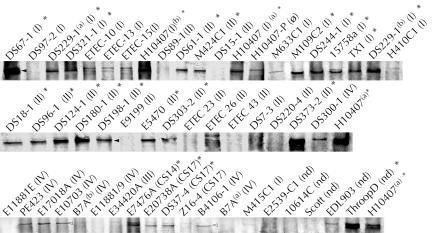
FIG. 8.
Secretion of EtpA by multiple ETEC strains: anti-EtpA immunoblots of concentrated supernatants from ETEC isolates. The results for the positive controls H10407 and LMG194(pJY019) and the negative control H10407-P for EtpA secretion are shown at the upper left. Isolates are clustered according to CFA group. An asterisks indicates an undetermined CF. Superscript letters indicate the origins of duplicate isolates obtained from two different sources, WRAIR (superscript a) and Berna Biotech (superscript b).
FIG. 9.
Secretion of HMW glycoprotein by ETEC: digoxigenin-glycan detection of HMW glycoprotein in concentrated supernatants from ETEC isolates. The position of the EtpA protein is indicated by a solid arrowhead. The open arrowheads indicate the positions of HMW non-EtpA proteins. The CF groups are indicated in parentheses. EtpA immunoblot-positive strains are indicated by asterisks. Superscript letters indicate the origins of duplicate isolates obtained from two different sources, WRAIR (superscript a) and Berna Biotech (superscript b).
Altogether, 59% (10/17) of CFAI-expressing strains, 56% (10/18) of CFAII-expressing strains, and 75% (3/4) of strains expressing CS14 or CS17 expressed EtpA or reactive proteins with a molecular weight identical to that of EtpA. However, strains expressing CS4, CS5, or CS6 (CFAIV strains) were negative as determined by Western blotting. In supernatant from a single ETEC strain, DS97-2 (CFAI, heat-stable enterotoxin, serotype O153:nm), anti-EtpA antibodies identified a protein whose molecular weight was significantly different than that of EtpA.
DISCUSSION
Although a large number genes encoding potential TPS systems have been identified through DNA sequencing of microbial genomes, only a limited number of TPS molecules have been fully characterized so far. Of the few TPS systems that have been characterized, most have been shown to play a role in adhesion (5, 74, 76) or cytolysis (43, 69, 87), while the hxuCBA locus of H. influenzae appears to be involved in iron acquisition (18-20).
Here, we describe identification and initial molecular characterization of a TPS system in a prototypical ETEC strain. Similar to the other TPS systems described to date, the etpBAC locus supports the secretion of a high-molecular-weight exoprotein. The homology of the proteins encoded at this locus to known TpsA and TpsB proteins predicts that EtpB is an outer membrane transporter (TpsB) required for the secretion of EtpA exoprotein (TpsA). This prediction is supported by our findings that both native EtpA and rEtpA proteins were found predominately in culture supernatants and that cloning and expression of the etpBAC locus were sufficient to export EtpA from a recombinant E. coli strain.
Furthermore, based on the homology of EtpC with Hwm1C, a protein previously shown to be involved in the glycosylation of the H. influenzae adhesin HMW1 (35), we predicted that EtpA is also glycosylated. Indeed, in culture supernatants of H10407 and LMG194(pJY019) we were able to identify native EtpA and rEtpA, respectively, using either antibodies which recognize EtpA or DIG-glycan reagents. These findings appear to contradict previous studies which described the first report of glycoprotein production in E. coli, as they suggested that the TibA outer membrane protein is the only glycosylated protein produced by H10407 (56). However, in the previous studies the workers focused on analysis of whole-cell lysates of H10407 rather than on secreted proteins, and the results may simply indicate that very little of the EtpA glycoprotein remains cell associated when cells are grown under some laboratory conditions. Indeed, we were not able to identify EtpA in outer membrane preparations from the wild-type strain, suggesting that it does not remain tightly associated with the outer membrane.
Interestingly, while production of glycoproteins by eukaryotic cells is essential for a number of important biological processes, glycosylation is now increasingly recognized in prokaryotes, particularly bacterial mucosal pathogens (80, 85). Several of the bacterial glycoproteins described to date, including TibA (55, 56), the related E. coli autotransporter AIDA-I (8, 53), HMW1 (35), and a 40-kDa Chlamydia trachomatis major outer membrane protein (52), promote adherence to epithelial cells, and in each instance modification of the protein by glycosylation is required to demonstrate the adherence phenotype. Our studies confirmed that EtpC is required for glycosylation of EtpA, similar to the previously described system in H. influenzae (35). Interestingly, the effect of glycosylation on EtpA-mediated adherence appears to be quite complex. Similar to previous reports of the importance of glycosylation in the adherence phenotype, etpC was required for adherence when Caco-2 intestinal monolayers were used as the target. In contrast, the etpC mutant was shown be significantly hyperadherent compared to the wild-type strain when HCT-8 intestinal cells were employed, suggesting that glycosylation may modulate binding to different molecules on the surfaces of different cell lines. Therefore, while the etpBAC locus may have many similarities to previously described TPS operons, the studies described here revealed important differences in the biology of the different exoprotein adhesin molecules.
The predicted EtpA peptide sequence contains at its amino-terminal end a putative secretion module associated with other TpsA exoproteins, including FHA, a 230-kDa adhesin of B. pertussis. The recent determination of the crystal structure of the filamentous hemagglutinin secretion domain (17) provides insight into the possible mechanism of export via TPS, suggesting that this region is probably involved in formation of the ultimate β-helical structure of the TpsA proteins. The highly conserved Asn-Pro-Asn-Gly motif found in this region of EtpA, which is essential for secretion of other TpsA proteins (36, 82), may be involved in stabilization of the β-helix through formation of type I β-turns (17).
The carboxy-terminal portion of EtpA is comprised of multiple repeat regions. While a combination of intragenic duplication and recombination is thought to underlie the formation of repeats (2), the molecular significance of the repeat regions in EtpA is not clear yet. Repeat regions of proteins are often involved in protein-protein interactions. Indeed, the passenger domain of the ShdA autotransporter of Salmonella enterica serovar Typhimurium has repeat regions in the carboxy-terminal region of the molecule which are involved in its interaction with fibronectin (49).
The homology of EtpB to the afimbrial adhesin Enf from enteroaggregative E. coli serotype O111:H12 led us to investigate whether EtpB had similar properties. Like Enf, the cloned recombinant EtpB outer membrane protein was able to promote adherence to epithelial cells. Together, these findings indicate that EtpB (and perhaps Enf) may have two functions, transporting cognate TpsA exoproteins and functioning independently to promote adherence.
Adherence to epithelial cells plays an important role in the pathogenesis of many mucosal pathogens, and many organisms possess multiple adhesins (65). Here we found that a newly identified TPS system of a prototypical ETEC strain contributes to the interaction of ETEC with target epithelial cells. Our findings that an isogenic etpA mutant strain was demonstrably less adherent than the parent organism and that antibodies raised against recombinant EtpA prevented adherence to cultured epithelial cells strongly suggest that this exoprotein contributes to the adherence phenotype observed in vitro. However, the precise role of the etpBAC locus and the role of the EtpA exoprotein remain undefined. Indeed, the present studies were complicated by the fact that the expression of known fimbrial adhesins, such as CFAI, may not be optimal under the conditions which we employed.
The mechanism by which EtpA might facilitate adherence is not obvious from the present data. Indeed, the role played by FHA, an homologous TpsA adhesin from B. pertussis, is actually quite complex. FHA, which can be found on the bacterial surface and in the external milieu (21), appears to play a multifunctional role in adherence of B. pertussis to host cells, binding to multiple ligands via different functional domains, including binding to lactosylceramide (71) via the carbohydrate binding domain, to sulfated glycoconjugates via the heparin binding domain (37), or to the integrin CR3 via its RGD sequence (73).
Like the recently described eatA autotransporter gene, the etpBAC locus also resides on the pCS1 virulence plasmid of ETEC strain H10407. Several lines of evidence support this conclusion. First, DNA sequencing of the etpBAC locus showed that immediately downstream of etpC is an IS1414 copy bearing an EAST1 gene which was previously found on pCS1 in H10407 (90). In addition, H10407-P, which lacks pCS1, was negative for etpA as determined by PCR, Southern blotting, and colony hybridization studies. Finally, both blotting and detection of concentrated supernatant proteins from H10407-P with either anti-rEtpA.6H antibodies or DIG-glycan reagents failed to detect the high-molecular-weight EtpA protein. Conjugal transfer of pCS1 (91), which contains the etpBAC locus as well as CFAI, heat-stable enterotoxin, and EatA genes (68), may account for the relatively high prevalence of the etpBAC locus in CFAI-expressing strains. The finding that there are potential mobility elements adjacent to this locus may account for its identification in ETEC strains having multiple CF types.
The studies described here, along with other recently described studies of new potential virulence loci of ETEC, highlight the previously unappreciated complexity of the ETEC genome and suggest that there is much to be learned about the pathogenesis of this organism. While the precise role of the etpBAC locus in the pathogenesis of ETEC remains to be completely defined, this locus is an enticing target for further investigation given the apparent structural and functional homology of its encoded proteins to established virulence molecules that have recently proven to be valuable in development of vaccines for other pathogens.
Supplementary Material
Acknowledgments
This work was supported by funds from the Department of Veteran's Affairs (to J.M.F.), by Public Health Service grant RR16190-04 (to J.M.F), and in part by NIH Medical Student Research Program grant T35 DK07405.
We thank Erik Dhondt and Didier Favre of the Division of Live Bacterial Vaccines at Berna Biotech for supplying their collection of ETEC strains, Donald E. Woods for supplying the pmini-OphoA plasmid, and Jing Yu and Bradley Postlethwaite for their contributions. We also thank Harry Courtney and David Hasty for their thoughtful reviews of the manuscript.
Editor: A. D. O'Brien
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Andrade, M. A., C. Perez-Iratxeta, and C. P. Ponting. 2001. Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134:117-131. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 1994. Foodborne outbreaks of enterotoxigenic Escherichia coli-Rhode Island and New Hampshire, 1993. Morb. Mortal. Wkly. Rep. 43:81, 87-89. [PubMed] [Google Scholar]
- 4.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., and J. W. St. Geme, 3rd. 1994. Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 62:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatty, M. E., C. A. Bopp, J. G. Wells, K. D. Greene, N. D. Puhr, and E. D. Mintz. 2004. Enterotoxin-producing Escherichia coli O169:H41, United States. Emerg. Infect. Dis. 10:518-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: Signal P 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 8.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 9.Blattner, F. R., G. Plunkett, 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 10.Bolton, A. J., and D. E. Woods. 2000. Self-cloning minitransposon phoA gene-fusion system promotes the rapid genetic analysis of secreted proteins in gram-negative bacteria. BioTechniques 29:470-472, 474. [DOI] [PubMed] [Google Scholar]
- 11.Bradley, P., L. Cowen, M. Menke, J. King, and B. Berger. 2001. BETAWRAP: successful prediction of parallel beta-helices from primary sequence reveals an association with many microbial pathogens. Proc. Natl. Acad. Sci. USA 98:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun, V., S. Hobbie, and R. Ondraczek. 1992. Serratia marcescens forms a new type of cytolysin. FEMS Microbiol. Lett. 79:299-305. [DOI] [PubMed] [Google Scholar]
- 13.Brown, N. F., C. A. Logue, J. A. Boddey, R. Scott, R. G. Hirst, and I. R. Beacham. 2004. Identification of a novel two-partner secretion system from Burkholderia pseudomallei. Mol. Genet. Genomics 272:204-215. [DOI] [PubMed] [Google Scholar]
- 14.Buscher, A. Z., K. Burmeister, S. J. Barenkamp, and J. W. St. Geme, 3rd. 2004. Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J. Bacteriol. 186:4209-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantey, J. R., and R. K. Blake. 1977. Diarrhea due to Escherichia coli in the rabbit: a novel mechanism. J. Infect. Dis. 135:454-462. [DOI] [PubMed] [Google Scholar]
- 16.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clantin, B., H. Hodak, E. Willery, C. Locht, F. Jacob-Dubuisson, and V. Villeret. 2004. The crystal structure of filamentous hemagglutinin secretion domain and its implications for the two-partner secretion pathway. Proc. Natl. Acad. Sci. USA 101:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cope, L. D., S. E. Thomas, Z. Hrkal, and E. J. Hansen. 1998. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect. Immun. 66:4511-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cope, L. D., S. E. Thomas, J. L. Latimer, C. A. Slaughter, U. Muller-Eberhard, and E. J. Hansen. 1994. The 100 kDa haem:haemopexin-binding protein of Haemophilus influenzae: structure and localization. Mol. Microbiol. 13:863-873. [DOI] [PubMed] [Google Scholar]
- 20.Cope, L. D., R. Yogev, U. Muller-Eberhard, and E. J. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coutte, L., S. Alonso, N. Reveneau, E. Willery, B. Quatannens, C. Locht, and F. Jacob-Dubuisson. 2003. Role of adhesin release for mucosal colonization by a bacterial pathogen. J. Exp. Med. 197:735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domenighini, M., D. Relman, C. Capiau, S. Falkow, A. Prugnola, V. Scarlato, and R. Rappuoli. 1990. Genetic characterization of Bordetella pertussis filamentous haemagglutinin: a protein processed from an unusually large precursor. Mol. Microbiol. 4:787-800. [DOI] [PubMed] [Google Scholar]
- 25.Donnenberg, M., and J. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 27.Elsinghorst, E. A., and J. A. Weitz. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley, P., I. G. Charles, N. F. Fairweather, and N. W. Isaacs. 1996. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381:90-92. [DOI] [PubMed] [Google Scholar]
- 29.Evans, D. G., R. P. Silver, D. J. Evans, Jr., D. G. Chase, and S. L. Gorbach. 1975. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect. Immun. 12:656-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkelstein, R. A., M. L. Vasil, J. R. Jones, R. A. Anderson, and T. Barnard. 1976. Clinical cholera caused by enterotoxigenic Escherichia coli. J. Clin. Microbiol. 3:382-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleckenstein, J. M., J. T. Holland, and D. L. Hasty. 2002. Interaction of an outer membrane protein of enterotoxigenic Escherichia coli with cell surface heparan sulfate proteoglycans. Infect. Immun. 70:1530-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleckenstein, J. M., D. J. Kopecko, R. L. Warren, and E. A. Elsinghorst. 1996. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect. Immun. 64:2256-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fogh, J., J. M. Fogh, and T. Orfeo. 1977. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J. Natl. Cancer Inst. 59:221-226. [DOI] [PubMed] [Google Scholar]
- 34.George, R. A., and J. Heringa. 2000. The REPRO server: finding protein internal sequence repeats through the Web. Trends Biochem. Sci. 25:515-517. [DOI] [PubMed] [Google Scholar]
- 35.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St. Geme, 3rd. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 36.Grass, S., and J. W. St. Geme, 3rd. 2000. Maturation and secretion of the non-typable Haemophilus influenzae HMW1 adhesin: roles of the N-terminal and C-terminal domains. Mol. Microbiol. 36:55-67. [DOI] [PubMed] [Google Scholar]
- 37.Hannah, J. H., F. D. Menozzi, G. Renauld, C. Locht, and M. J. Brennan. 1994. Sulfated glycoconjugate receptors for the Bordetella pertussis adhesin filamentous hemagglutinin (FHA) and mapping of the heparin-binding domain on FHA. Infect. Immun. 62:5010-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartweck, L. M., C. L. Scott, and N. E. Olszewski. 2002. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161:1279-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haselbeck, A., E. Schickaneder, H. von der Eltz, and W. Hosel. 1990. Structural characterization of glycoprotein carbohydrate chains by using diagoxigenin-labeled lectins on blots. Anal. Biochem. 191:25-30. [DOI] [PubMed] [Google Scholar]
- 40.Heger, A., and L. Holm. 2000. Rapid automatic detection and alignment of repeats in protein sequences. Proteins 41:224-237. [DOI] [PubMed] [Google Scholar]
- 41.Henderson, I. R., R. Cappello, and J. P. Nataro. 2000. Autotransporter proteins, evolution and redefining protein secretion. Trends Microbiol. 8:529-532. [DOI] [PubMed] [Google Scholar]
- 42.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirono, I., N. Tange, and T. Aoki. 1997. Iron-regulated haemolysin gene from Edwardsiella tarda. Mol. Microbiol. 24:851-856. [DOI] [PubMed] [Google Scholar]
- 44.Holtke, H. J., W. Ankenbauer, K. Muhlegger, R. Rein, G. Sagner, R. Seibl, and T. Walter. 1995. The digoxigenin (DIG) system for non-radioactive labelling and detection of nucleic acids—an overview. Cell Mol. Biol. (Noisy-Le-Grand) 41:883-905. [PubMed] [Google Scholar]
- 45.Jacob-Dubuisson, F., R. Fernandez, and L. Coutte. 2004. Protein secretion through autotransporter and two-partner pathways. Biochim. Biophys. Acta 1694:235-257. [DOI] [PubMed] [Google Scholar]
- 46.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 47.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 48.Killmann, H., R. Benz, and V. Braun. 1996. Properties of the FhuA channel in the Escherichia coli outer membrane after deletion of FhuA portions within and outside the predicted gating loop. J. Bacteriol. 178:6913-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kingsley, R. A., D. Abi Ghanem, N. Puebla-Osorio, A. M. Keestra, L. Berghman, and A. J. Baumler. 2004. Fibronectin binding to the Salmonella enterica serotype Typhimurium ShdA autotransporter protein is inhibited by a monoclonal antibody recognizing the A3 repeat. J. Bacteriol. 186:4931-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kniep, B., and P. F. Muhlradt. 1990. Immunochemical detection of glycosphingolipids on thin-layer chromatograms. Anal. Biochem. 188:5-8. [DOI] [PubMed] [Google Scholar]
- 51.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 52.Kuo, C., N. Takahashi, A. F. Swanson, Y. Ozeki, and S. Hakomori. 1996. An N-linked high-mannose type oligosaccharide, expressed at the major outer membrane protein of Chlamydia trachomatis, mediates attachment and infectivity of the microorganism to HeLa cells. J. Clin. Investig. 98:2813-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laarmann, S., and M. A. Schmidt. 2003. The Escherichia coli AIDA autotransporter adhesin recognizes an integral membrane glycoprotein as receptor. Microbiology 149:1871-1882. [DOI] [PubMed] [Google Scholar]
- 54.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 55.Lindenthal, C., and E. A. Elsinghorst. 2001. Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect. Immun. 69:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Locht, C., P. Bertin, F. D. Menozzi, and G. Renauld. 1993. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetella spp. Mol. Microbiol. 9:653-660. [DOI] [PubMed] [Google Scholar]
- 58.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Operon and gene fusions, p. 177-184. In Genetic analysis of pathogenic bacteria: a laboratory manual, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 59.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33(Database Issue):D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McVeigh, A., A. Fasano, D. A. Scott, S. Jelacic, S. L. Moseley, D. C. Robertson, and S. J. Savarino. 2000. IS1414, an Escherichia coli insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect. Immun. 68:5710-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mendiola, M. V., Y. Jubete, and F. de la Cruz. 1992. DNA sequence of IS91 and identification of the transposase gene. J. Bacteriol. 174:1345-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monteiro-Neto, V., S. Y. Bando, C. A. Moreira-Filho, and J. A. Giron. 2003. Characterization of an outer membrane protein associated with haemagglutination and adhesive properties of enteroaggregative Escherichia coli O111:H12. Cell. Microbiol. 5:533-547. [DOI] [PubMed] [Google Scholar]
- 64.Nelson, K. M., G. M. Young, and V. L. Miller. 2001. Identification of a locus involved in systemic dissemination of Yersinia enterocolitica. Infect. Immun. 69:6201-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ofek, I., D. L. Hasty, R. J. Doyle, and I. Ofek. 2003. Bacterial adhesion to animal cells and tissues. ASM Press, Washington, D.C.
- 66.O'Shannessy, D. J., and W. L. Hoffman. 1987. Site-directed immobilization of glycoproteins on hydrazide-containing solid supports. Biotechnol. Appl. Biochem. 9:488-496. [DOI] [PubMed] [Google Scholar]
- 67.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 68.Patel, S. K., J. Dotson, K. P. Allen, and J. M. Fleckenstein. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect. Immun. 72:1786-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poole, K., E. Schiebel, and V. Braun. 1988. Molecular characterization of the hemolysin determinant of Serratia marcescens. J. Bacteriol. 170:3177-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Posfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prasad, S. M., Y. Yin, E. Rodzinski, E. I. Tuomanen, and H. R. Masure. 1993. Identification of a carbohydrate recognition domain in filamentous hemagglutinin from Bordetella pertussis. Infect. Immun. 61:2780-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qadri, F., A. M. Svennerholm, A. S. Faruque, and R. B. Sack. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Relman, D., E. Tuomanen, S. Falkow, D. T. Golenbock, K. Saukkonen, and S. D. Wright. 1990. Recognition of a bacterial adhesin by an integrin: macrophage CR3 (alpha M beta 2, CD11b/CD18) binds filamentous hemagglutinin of Bordetella pertussis. Cell 61:1375-1382. [DOI] [PubMed] [Google Scholar]
- 74.Relman, D. A., M. Domenighini, E. Tuomanen, R. Rappuoli, and S. Falkow. 1989. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc. Natl. Acad. Sci. USA 86:2637-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roels, T. H., M. E. Proctor, L. C. Robinson, K. Hulbert, C. A. Bopp, and J. P. Davis. 1998. Clinical features of infections due to Escherichia coli producing heat-stable toxin during an outbreak in Wisconsin: a rarely suspected cause of diarrhea in the United States. Clin. Infect. Dis. 26:898-902. [DOI] [PubMed] [Google Scholar]
- 76.Rojas, C. M., J. H. Ham, W. L. Deng, J. J. Doyle, and A. Collmer. 2002. HecA, a member of a class of adhesins produced by diverse pathogenic bacteria, contributes to the attachment, aggregation, epidermal cell killing, and virulence phenotypes of Erwinia chrysanthemi EC16 on Nicotiana clevelandii seedlings. Proc. Natl. Acad. Sci. USA 99:13142-13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sambrook, J., and D. W. Russell. 2001. Lysing colonies and binding of DNA on filters, p. 1.135-131.137. In N. Irwin (ed.), Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 78.Savarino, S. J., A. Fasano, J. Watson, B. M. Martin, M. M. Levine, S. Guandalini, and P. Guerry. 1993. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc. Natl. Acad. Sci. USA 90:3093-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schiebel, E., H. Schwarz, and V. Braun. 1989. Subcellular location and unique secretion of the hemolysin of Serratia marcescens. J. Biol. Chem. 264:16311-16320. [PubMed] [Google Scholar]
- 80.Schmidt, M. A., L. W. Riley, and I. Benz. 2003. Sweet new world: glycoproteins in bacterial pathogens. Trends Microbiol. 11:554-561. [DOI] [PubMed] [Google Scholar]
- 81.Schnaitman, C. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schonherr, R., R. Tsolis, T. Focareta, and V. Braun. 1993. Amino acid replacements in the Serratia marcescens haemolysin ShIA define sites involved in activation and secretion. Mol. Microbiol. 9:1229-1237. [DOI] [PubMed] [Google Scholar]
- 83.St. Geme, J. W., 3rd, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.St. Geme, J. W., 3rd, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 85.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 86.Tompkins, W. A., A. M. Watrach, J. D. Schmale, R. M. Schultz, and J. A. Harris. 1974. Cultural and antigenic properties of newly established cell strains derived from adenocarcinomas of the human colon and rectum. J. Natl. Cancer Inst. 52:1101-1110. [DOI] [PubMed] [Google Scholar]
- 87.Uphoff, T. S., and R. A. Welch. 1990. Nucleotide sequencing of the Proteus mirabilis calcium-independent hemolysin genes (hpmA and hpmB) reveals sequence similarity with the Serratia marcescens hemolysin genes (shlA and shlB). J. Bacteriol. 172:1206-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Schilfgaarde, M., P. van Ulsen, P. Eijk, M. Brand, M. Stam, J. Kouame, L. van Alphen, and J. Dankert. 2000. Characterization of adherence of nontypeable Haemophilus influenzae to human epithelial cells. Infect. Immun. 68:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weiss, A. A., E. L. Hewlett, G. A. Myers, and S. Falkow. 1983. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect. Immun. 42:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamamoto, T., and P. Echeverria. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamamoto, T., and T. Yokota. 1983. Plasmids of enterotoxigenic Escherichia coli H10407: evidence for two heat-stable enterotoxin genes and a conjugal transfer system. J. Bacteriol. 153:1352-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.