Abstract
Brucellosis is a costly disease of water buffaloes (Bubalus bubalis). Latent infections and prolonged incubation of the pathogen limit the efficacy of programs based on the eradication of infected animals. We exploited genetic selection for disease resistance as an approach to the control of water buffalo brucellosis. We tested 231 water buffalo cows for the presence of anti-Brucella abortus antibodies (by the agglutination and complement fixation tests) and the Nramp1 genotype (by PCR-denaturing gradient gel electrophoresis). When the 231 animals (58 cases and 173 controls) were divided into infected (seropositive) and noninfected (seronegative) groups and the Nramp1 genotypes were compared, the seropositive subjects were 52 out of 167 (31%) in the Nramp1A+ (Nramp1AA or Nramp1AB) group and 6 out of 64 (9.4%) in the Nramp1A− (Nramp1BB) group (odds ratio, 4.37; 95% confidence limits, 1.87 to 10.19; χ2, 11.65 for 1 degree of freedom). Monocytes from Nramp1BB subjects displayed significantly (P < 0.01) higher levels of Nramp1 mRNA than Nramp1AA subjects and also a significantly (P < 0.01) higher ability in controlling the intracellular replication of several Brucella species in vitro. Thus, selection for the Nramp1BB genotype can become a valuable tool for the control of water buffalo brucellosis in the areas where the disease is endemic.
The water buffalo (Bubalus bubalis) occupies an economically important place in the livestock industry in many parts of the world. One of these is the south of Italy. Brucellosis causes serious economic losses and is relevant also as a zoonosis (8). The causative agent is Brucella abortus, a facultative intracellular pathogen which infects host macrophages. Only a few water buffalo cows that become infected develop clinical signs of the disease (spontaneous abortion). However, many infected cows shed B. abortus in the milk. Eradication programs involving the slaughter of infected animals have been carried out for more than 20 to 30 years. However, latent infections, prolonged incubation of the pathogen, incomplete protection provided by vaccines, and difficulties in distinguishing serologically between vaccinated and naturally infected animals have limited the efficacy of eradication programs. This paper exploits selective breeding for disease-resistant genotypes as a new approach to the control of water buffalo brucellosis.
Remarkably, even in water buffalo herds heavily infected with B. abortus, about 20% of the subjects remain negative by the serological tests and presumably noninfected all the time. This observation suggests that genetic variation within the host may play a part in the resistance to brucellosis. In cattle, it is known that the resistance to brucellosis is genetically determined (15, 38, 40). These circumstances, and the widespread presence of genes protecting against bacterial infections in livestock (15, 23, 30), humans (6, 25, 34, 43), mice (27, 36), and invertebrates (26, 29) prompted the search for polymorphisms conferring resistance to brucellosis in the water buffalo.
The Nramp1 gene, first identified in the mouse (49), is a member of a large family of genes coding for metal ion-transporting proteins. Homologues of this gene are present in genetically distant organisms, such as mammals, insects, worms, plants, yeasts, and bacteria (11). The presence of Nramp1 in bacteria and mammals has suggested that intracellular pathogens and host cells compete for the same nutrient, each competitor attempting to steal essential cations for its own benefit (18). The mouse Nramp1 gene confers resistance to several unrelated intracellular pathogens, including Salmonella enterica serovar Typhimurium, Leishmania donovani, and Mycobacterium bovis BCG (37, 44). The human Nramp1 gene confers resistance to Mycobacterium tuberculosis (6), and the cattle Nramp1 gene confers resistance to Brucella abortus (15). As to the mechanism by which Nramp1 confers innate resistance to intracellular pathogens, it has been proposed that the product of Nramp1 may limit microbial replication in the phagosome by subtracting critical nutrients to invading microbes (18).
Here it is shown that in the water buffalo, as in cattle, the resistance to B. abortus infection is associated with the gene Nramp1. The two alleles Nramp1A and Nramp1B (for brevity referred to as allele A and allele B) differ in the number of guanine and thymine (GT) microsatellites, the presence in the A allele of an insertion at position 17 and a point mutation at position 98. Animals homozygous for the B allele can control the replication of B. abortus inside the macrophages.
MATERIALS AND METHODS
Study design.
The inheritance of the A and B alleles was studied in 166 water buffaloes (the totality of the animals from an experimental herd with accurate paternity records and located in the province of Salerno, Italy). This herd, free from brucellosis, was not included in the association study. For this purpose, the interest focused on two herds characterized by an exceptionally high incidence of brucellosis (up to 20% of the subjects were positive in the serological tests for brucellosis). The two herds are about 30 km distant, and both are located in the province of Caserta, Italy. The 231 water buffalo cows included in the study (age, 2 to 8 years) were chosen randomly among a total of about 500 present in the two herds. The 231 animals were all equally exposed to infection since birth and then were grown in one of the two infected herds. The animals that were positive by the agglutination and complement fixation tests at least twice within a 3-month period were classified as cases; the animals negative by the tests were classified as controls. Genotype analysis was carried out blindly (without knowing in advance the results from the serological tests). To avoid stratification (39), cases and controls contain equal proportions of animals (29 cases and 86 controls) from each herd.
PCR-DGGE analysis.
The Nramp1 genotype of the subjects included in the present study was determined by PCR-denaturing gradient gel electrophoresis (DGGE). DNA was phenol-chloroform extracted from venous blood as previously described (41). The 3′ untranslated region (3′UTR) (nucleotide positions 1745 to 1955) of the water buffalo Nramp1 gene was amplified using the forward primer 5′ GTGGAATGAGTGGGCACAGT 3′ and the reverse primer 5′ CTCTCCGTCTTGCTGTGCAT 3′ (22). A guanine-cytosine clamp was added to the forward primer (35). PCR was carried out in a Progene thermocycler (Techne, Cambridge, United Kingdom). The 50-μl total reaction mixture contained 50 ng DNA, 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, pH 9, 0.1% Triton X-100), 0.2 mM deoxynucleoside triphosphates, 1.5 mM MgCl2, 0.4 mM of each primer, and 2 U of Taq polymerase (Promega). The thermal profile included one cycle at 94°C for 2 min and 35 cycles at 94°C for 30 s, 53°C for 30 s, and 72°C for 30 s. The extension step was carried out at 72°C for 5 min. PCR products were electrophoresed through 8% polyacrylamide gels containing a 25 to 50% urea-formamide denaturing gradient using the Bio-Rad Dcode apparatus (Hercules, CA). After electrophoresis, the gels were stained in ethidium bromide solution for 5 min and then washed in distilled water for 20 min. Bands were visualized with the Gel Doc 2000 apparatus (Bio-Rad).
Sequencing of the Nramp1 alleles.
PCR products from three AA and three BB animals were sequenced in both directions. The nucleotide sequence was determined using version 2.0 of the Big Dye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) and the ABI 310 PRISM genetic analyzer (Applied Biosystems). The length of the capillary was 47 cm, and the section was 50 μm. The separation medium was the POP-4 polymer (Applied Biosystems). The sequence data were analyzed using GeneScan and Sequencing software (Applied Biosystems).
DNA typing.
The DNAs from the 231 subjects included in the association study were genotyped with 11 microsatellite markers from the commercially available ABI “StockMarks” kit for cattle DNA typing (Applied Biosystems) using the procedure described by the supplier. The results were analyzed on an ABI model 310 automated DNA fragment analyzer. Of the 11 markers used, 2 (SPS115 and TGLA227) provided unambiguous results with water buffalo samples.
Nramp1 messenger level measurement.
Peripheral blood mononuclear cells were separated by gradient centrifugation (Lympholyte-Mammal; Cederlane, Hornby, Ontario, Canada). Total RNA was isolated by the Trizol reagent (Invitrogen, Milan, Italy). Synthesis of cDNA was carried out with the ImProm-II reverse transcriptase (Promega, Madison, WI). Amplification of the internal standard (the CD64 [FcγRI] gene) and of the target gene (Nramp1) was carried out under the following conditions: 3 min at 95°C, 40 cycles each of 15 s at 95°C, and then 45 s at 60°C. The primers were 5′ GAGTCACAATGGCATCTATCACTG 3′ (sense) and 5′ AGAAGGATGTTCT CA GCACTGG 3′ (antisense) for CD64 and 5′ ACATTGAGTCGGATCTTCAGG 3′ (sense) and 5′ GGGCACCTTAGGGTAGTAGAG 3′ (antisense) for Nramp1. The sizes of the CD64 and Nramp1 amplicons were 116 and 170 bp, respectively. The amplification mixture contained 12.5 μl iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) and 1 μM primers in a final volume of 25 μl. Each experiment included as negative control a nontemplate reaction tube. Monocytes were infected in vitro with B. abortus 2308 (104 bacteria and 104 monocytes/well for 4 h). Relative expression levels were calculated by the comparative cycle threshold method (16). Each genotypic class included three animals. Each animal was tested in triplicate. Preliminary experiments showed that the internal standard (the CD64 gene) is not induced by the experimental conditions and is expressed at a level comparable with that of Nramp1.
In vitro antibacterial activity of the Nramp1 alleles.
Intracellular bacteria were counted as described previously (38). Briefly, peripheral blood mononuclear cells from seronegative AA, AB, or BB cows were separated by gradient centrifugation and infected with B. abortus 2308, B. melitensis biovar 1, biovar 2, biovar 3, or B. suis biovar 1 (approximately 104 cells and 104 bacteria/well), centrifuged at 750 × g for 5 min, and incubated at 37°C (5% CO2) for 30 min. Extracellular bacteria were then killed by gentamicin (4 μg/well). Cells were washed three times with Dulbecco's modified Eagle medium (to remove the antibiotic), fed on fresh medium, incubated for 18 h, and finally lysed with 0.5% Tween 20 (15 μl/well). The content of each well was properly diluted with phosphate-buffered saline and plated on tryptose soy agar plates. The percentage of bacteria surviving at 18 h postchallenge was determined as described previously (38). Control wells were set up as described previously (38).
Other procedures.
The serological tests for brucellosis were carried out by agglutination and complement fixation tests (3). B. abortus was isolated from vaginal swabs on blood agar (Oxoid, England, United Kingdom) and identified by PCR (14). The P value as predicted by the Fisher's exact test, and the odds ratio (OR) with 95% confidence intervals (CI) were calculated by using SPSS software (Chicago, IL).
Nucleotide sequence accession numbers.
The nucleotide sequences of A and B alleles are available in the GenBank/DDBJ/EMBL databases under the accession numbers DQ095780 and DQ095781, respectively.
RESULTS
Identification of the A and B alleles in the 3′ untranslated region of the Nramp1 gene.
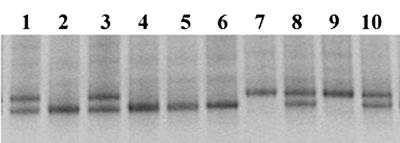
The genomic DNA samples from 166 water buffaloes were analyzed for the presence of a dinucleotide repeat polymorphism in the 3′ untranslated region (3′UTR) of the Nramp1 gene. The analysis, carried out by DGGE, identified the A and the B variants. All tested animals displayed either the A or B variant or both; the phenotype characterized by the lack of both variants was absent (Fig. 1). This pattern immediately suggested that the presence of the variants was regulated by two codominant alleles. Family data confirmed the proposed model of inheritance (data not shown). It was found that heterozygous animals could belong to either sex, thereby indicating that the Nramp1 locus is not sex linked. The frequencies of the alleles A and B, calculated on the genotype of 81 offspring, were 0.47 and 0.53, respectively. Sequence analysis displayed that the two alleles differ in the number of GT microsatellites (33 in the A allele and 36 in B) and the presence in the A allele of a GG insertion at position 17 and a point mutation (A versus G) at position 98. The A and B alleles are distinct from the alleles located in introns 4 and 5 and in exon V of the Nramp1 gene described in different cattle and buffalo breeds (1).
FIG. 1.
Representative DGGE profiles of Nramp1AA (lanes 7 and 9), Nramp1AB (lanes 1, 3, 8, and 10), and Nramp1BB (lanes 2, 4, 5, and 6) water buffalo genotypes.
The B allele confers resistance to brucellosis.
We next examined the influence of the A and B alleles on the presence of antibodies against B. abortus, a strong indicator of infection following contact with the microbe. For this purpose, 231 animals were tested by the agglutination and complement fixation tests to detect the presence of anti-B. abortus antibodies and by DGGE to establish the Nramp1 genotype. When the 231 animals were divided into infected (seropositive) and noninfected (seronegative) groups and the Nramp1 genotypes were compared, the seropositive subjects were 52 out of 167 (31%) in the A+ (AA or AB) group and 6 out of 64 (9.4%) in the A− (BB) group. Homozygosity for the B allele is thus significantly associated with resistance to B. abortus infection (OR, 4.37; 95% confidence limits, 1.87 to 10.19; χ2, 11.65 for 1 degree of freedom; P < 0.001). Records of serological tests for brucellosis relative to 21 water buffaloes demonstrate that BB animals remain seronegative in spite of prolonged exposure (up to 10 years) to B. abortus. The DNA from the 231 subjects included in the association study was analyzed for deviation from Hardy-Weinberg equilibrium of allele frequencies between B. abortus infected and noinfected animals. The analysis included the candidate gene (Nramp1) and the two polymorphic loci SPS115 and TGLA227. The latter represent the only polymorphic loci that were reproducibly detected using the cattle “StockMarks” kit. The SPS115 and TGLA227 loci did not display significant differences in genotype frequencies between infected (SPS115, χ2 = 0.78; TGLA227, χ2 = 2.34) and noninfected (SPS115, χ2 = 1.82; TGLA227, χ2 = 1.78) animals. The genotypic distribution of the A and B alleles of the Nramp1 gene displayed a highly significant (P = 0.012) departure from Hardy-Weinberg equilibrium when frequencies between infected and noninfected animals were compared (Table 1).
TABLE 1.
Evaluation of Hardy-Weinberg equilibrium in the frequencies of Nramp1A and Nramp1B alleles in water buffaloes infected with B. abortus and left noninfecteda
| Subject | No. of buffaloes for genotype:
|
Gene frequency
|
χ2 | P | ||||
|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | Total | A | B | |||
| Infected | 13 (18.2) | 39 (28.6) | 6 (11.2) | 58 | 0.56 | 0.44 | 7.68 | 0.021 |
| Noninfected | 28 (29.08) | 87 (83.7) | 58 (60.22) | 173 | 0.41 | 0.59 | 0.25 | 0.882 |
Figures in parentheses refer to the expected number of subjects in each class. The frequency of the B allele in the infected and noninfected subjects is significantly different (P = 0.012).
Milk and vaginal mucus samples were collected at weekly intervals for 10 weeks from 10 BB-seronegative cows. One sample of milk and vaginal mucus also was collected from 10 seropositive AA or AB cows. Brucella abortus was isolated on blood agar and identified by PCR. Milk and vaginal mucus samples from the BB-seronegative cows were all negative. Milk and/or vaginal mucus samples from seven of the seropositive AA or AB cows were instead positive. PCR tests included as a control a milk and vaginal mucus sample from one of the AA or AB cows positive by the PCR test. The absence of B. abortus in the body fluids and of the corresponding antibodies in the blood of the BB-seronegative animals, if confirmed by more extensive experiments, will indicate that these animals do not carry the pathogen.
Level of the Nramp1 mRNA in genetically resistant or susceptible animals.
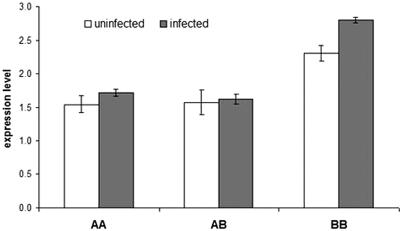
Real-time reverse transcription-PCR did not display any significant difference in Nramp1 expression between the AA, AB, and BB genotypes when the internal standard was the glyceraldehydes-3-phosphate dehydrogenase (GAPDH) gene (data not shown). The expression of housekeeping genes traditionally considered suitable internal standards (such as GAPDH) can vary considerably between cell types and can obscure differences in the expression level of the target gene (16, 48). When GAPDH was replaced with the CD64 (FcγRI) gene as an internal control, the BB animals displayed a significantly higher level of the Nramp1 messenger (Fig. 2). No significant difference in the expression of Nramp1 was observed between the AA and AB individuals, which are both susceptible to brucellosis.
FIG. 2.
Nramp1 relative expression levels in monocytes from AA, AB, and BB monocytes, noninfected and infected (4 h) with Brucella abortus 2308. The CD64 gene was used as an internal standard. Relative expression levels were calculated by the comparative cyclic threshold method (16). Each genotypic class includes three animals, each animal tested in triplicate. Nramp1 expression level in BB-induced monocytes is significantly different (P < 0.01) from the level in BB noninduced monocytes and from the levels in AA and AB monocytes, induced and noninduced. Nramp1 expression level in BB noninduced monocytes is significantly different (P < 0.05) from the levels in AA and AB monocytes, induced and noninduced.
Nramp1BB animals limit the growth in vitro of several Brucella species.
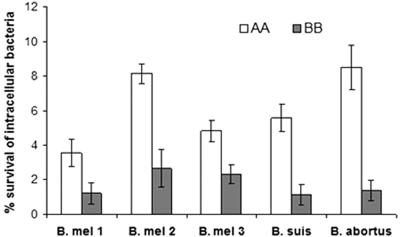
Water buffaloes can be infected by several Brucella species in addition to B. abortus. The BB subjects were therefore tested for their capacity to control the replication in vitro of B. abortus, B. melitensis, and B. suis. Monocytes from the BB animals displayed the ability to control the intracellular growth of the three Brucella species tested (Fig. 3). These results do not necessarily contradict the report in which Nramp1 was found to be of limited efficacy in controlling Brucella melitensis in infected mice (19). The discrepancy in the results may reflect differences in the host (mice versus water buffalo) or between in vivo (19) and in vitro (this study) replication of Brucella. Infection of the BB animals with Brucella species was not carried forward, having been vetoed by the sanitary authority. The importance of the BB genotype for the natural resistance of water buffaloes to B. melitensis and B. suis therefore remains to be assessed.
FIG. 3.
Ability of AA and BB subjects to control the intracellular replication of Brucella species. Each genotypic class includes three animals, each animal tested in triplicate. Intracellular bacteria were counted 18 h postinfection. Differences in intracellular survival between AA and BB subjects are all significantly different (P < 0.01). B. mel 1, B. melitensis serovar 1; B. mel 2, B. melitensis serovar 2; B. mel 3, B. melitensis serovar 3.
DISCUSSION
The search for alleles influencing susceptibility to pathogens is not new. One of the earliest examples (and certainly the best known) of these studies describes the protection of the human hemoglobin S allele from Plasmodium falciparum (2). In recent years, high-throughput searches for polymorphisms yielded a large number of reports on the association of allelic variants with diseases. Unfortunately, many of these associations proved not to be reproducible (10, 24, 32).
Criteria to attenuate the pitfalls afflicting these studies have been proposed (24, 28, 31). When carrying out genetic case-control studies, the biological plausibility of the candidate gene assumes primary importance. The association is likely to be meaningful (and reproducible) if there is evidence, from an animal model or a homologue gene, of a biological relation between trait and candidate gene (28). The association reported here certainly makes biological sense. The Nramp1 gene in cattle is associated with resistance to B. abortus (15) and in the mouse with resistance to several intracellular bacteria that share with B. abortus the preference for an intracellular life (37, 44). More importantly, the resistance-associated allele of the bovine Nramp1 gene, when expressed in stably transfected RAW264.7 macrophages, controls the in vitro replication of B. abortus (4).
In case-control studies, the proper selection of controls also assumes much value. A genetic heterogeneity between cases and controls, due to selection, recent admixture, or any other cause, may show up as disease association (10, 28); obviously, in this case the association would be artifactual. Analogously, if some of the control subjects have not been exposed to the pathogen, a true association may be missed, because unexposed subjects capable of becoming infected are included as controls (47). More intuitively, including in the control group subjects not exposed to infection is like trying to find malaria resistance genes in a sample of people living in an area free of mosquitoes. Thus, to prevent spurious associations, cases and controls need to be homogeneous for genetic composition and exposure to the pathogen. Cases and controls included in the present study fulfill the above requisites. The SPS115 and TGLA227 loci, each including four alleles, did not display a significant difference in genotype frequencies between the two groups. Second, both cases and controls are from farms where up to 20% of the subjects were seropositive by the test for brucellosis. Thus, exposure to B. abortus was sufficiently long for susceptible animals to become infected.
The majority of the BB subjects (58 out of 64) remain B. abortus antibody negative. Thus, the role of the water buffalo B allele in B. abortus infection reminds us of the human CKR5 allele in human immunodeficiency virus (HIV) infection, where the individuals homozygous for CKR5 are resistant to HIV infection and are HIV antibody negative (13). The absence of B. abortus antibodies in the majority of the BB animals is compatible with in vivo and in vitro studies showing that the NRAMP1 protein mediates resistance in the initial phase of infection (17).
The role of the BB genotype in the containment of B. abortus infection is strengthened by the following observations. First, the frequency of the B allele is significantly higher in the control population (n = 173) than in the case population (n = 58) (0.44 versus 0.59; Fisher's exact test; P = 0.012). Second, the BB animals remain seronegative even after prolonged exposure to the pathogen. Third, B. abortus was absent in the vaginal swabs collected weekly from 10 BB-seronegative cows over a 10-week period of time. Fourth, BB animals displayed higher levels of Nramp1 mRNA and higher antibacterial activity than AA animals (Fig. 2 and 3).
A minority (6 out of 64) of the BB animals were found to be weakly positive by the serological test of brucellosis (antibody titer, 20 to 40 IU). These animals were culled immediately. We therefore can only speculate on what might have caused this result. One explanation is that the six subjects, following a recent infection with B. abortus, mounted a successful immune response and killed the pathogen. Accordingly, the antibodies detected by the serological test were only transient. Two lines of evidence support this interpretation. The first is that the same subjects in the course of previous screens for brucellosis were found repeatedly seronegative. The second is that cattle resistant to the brucellosis, when challenged with live B. abortus 2308, develop low transient antibody titers (38). Alternative explanations are that a new strain of B. abortus able to infect the BB animals is emerging in the population; stressful circumstances or a particular route of infection increased the vulnerability of these animals to brucellosis.
The possibility that the association described in this study is due to a gene in linkage disequilibrium with Nramp1 cannot be formally dismissed. However, the biological congruence between candidate gene and trait (discussed above) strongly suggests that Nramp1 is the gene or, more likely, one of the genes (21) conferring resistance to B. abortus infection. Several studies (4, 7) have described the molecular basis of the association between the Nramp1 gene and diseases. These studies provide support to speculate how this gene might influence the resistance to brucellosis in water buffaloes as well. The microsatellite polymorphism in the promoter region of the human Nramp1 gene (42) controls the resistance to tuberculosis by regulating the level of the NRAMP1 protein: the allele promoting a high level of the protein (allele 3) confers resistance to tuberculosis, and the allele promoting a low level of the same protein (allele 2) confers susceptibility (7). The microsatellite polymorphisms in the 3′UTR region of human (9) and cattle (4) Nramp1 genes influence disease predisposition by the same mechanism. Based upon the above evidence and the results reported in Fig. 2, we suggest that the microsatellite polymorphism identified in the 3′UTR region of the water buffalo Nramp1 gene shapes susceptibility or resistance to B. abortus by determining a low (in the presence of one or two copies of the A allele) or high (in the presence of two copies of the B allele) level of the NRAMP1 protein.
The persistence in water buffaloes of the A allele causing susceptibility to a diffuse pathogen (B. abortus) is enigmatic. Why has it not been eliminated by natural selection? The alleles 2 and 3 of the human Nramp1 gene are maintained by balanced polymorphism. Allele 3 protects against infectious diseases but predisposes to autoimmune diseases; allele 2 protects against autoimmune diseases but predisposes to the infectious ones (7). This result has been replicated in numerous independent studies (7). On the basis of this evidence, we suggest balanced polymorphism as the mechanism probably maintaining the A and B alleles in the water buffalo. It is interesting to note that the examples of balanced polymorphism so far known are all associated with resistance to infectious diseases: the HbS variant of hemoglobin (12), the loci controlling the leukocyte antigens (HLA) in humans (20), glucose-6-phosphate dehydrogenase (46), NRAMP1 (6), and the prion protein (33).
The present study was undertaken to test the validity of a selection program for resistance to brucellosis in the water buffalo. We would like therefore to comment on what might be the results of this program. We cannot exclude that water buffaloes selected for resistance to the existing strains of B. abortus in the future might test susceptible to new strains of the same pathogen. A warning comes from sheep that are genetically resistant to the known strains of scrapie but that are susceptible to new strains (5). While the possibility that the good gene of today may become the bad gene of tomorrow cannot be excluded, selection for resistance still remains the most suitable strategy to temper the host-pathogen interaction. The rapid elimination of the subjects susceptible to the newly emerging B. abortus strains would delay bacterial evolution by continually imposing on the pathogen new attack strategies (50). Disease resistance is a trait shaped by tradeoffs with other fitness components (50). Host resistance to brucellosis may therefore carry a fitness cost in the form of, for example, reduced fertility, reduced milk production, or increased susceptibility to other diseases. This outcome, while undesirable, would not compromise irreparably plans for a selection program. It is possible, in fact, to combine selection for traits displaying a negative genetic correlation (45). Finally, the minority of BB hosts displaying anti-B. abortus antibodies, if they really are susceptible, would not represent a limit to the project, since they would be protected by the herd immunity.
In conclusion, we have detected in water buffalo a correlation between the BB genotype and resistance to B. abortus infection. Genetic selection as a means to increase the resistance to brucellosis therefore seems a reasonable approach. The selective breeding of BB males will spread rapidly the allele conferring resistance while conserving the gene pool of the herds.
Editor: V. J. DiRita
REFERENCES
- 1.Ables, G. P., M. Nishibori, M. Kanemaki, and T. Watanabe. 2002. Sequence analysis of the NRAMP1 genes from different bovine and buffalo breeds. J. Vet. Med. Sci. 64:1081-1083. [DOI] [PubMed] [Google Scholar]
- 2.Allison, A. C. 1954. Protection afforded by the sickle-cell trait against subtertian malarial infection. Br. Med. J. 1:290-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alton, G. G., W. M. Jones, and D. E. Pietz. 1975. Laboratory techniques in brucellosis, p. 64-124. WHO monograph series 55. World Health Organization, Geneva, Switzerland. [PubMed]
- 4.Barthel, R., J. Feng, J. A. Piedrahita, D. N. McMurray, J. W. Templeton, and L. G. Adams. 2001. Stable transfection of the bovine NRAMP1 gene into murine RAW264.7 cells: effects on Brucella abortus survival. Infect. Immun. 69:3110-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baylis, M., and K. M. McIntyre. 2004. Scrapie control under new strain. Nature 432:810-811. [DOI] [PubMed] [Google Scholar]
- 6.Bellamy, R., C. Ruwende, T. Corrah, K. P. McAdam, H. Whittle, and A. V. Hill. 1998. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africa. N. Engl. J. Med. 338:640-644. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell, J. M., S. Searle, H. Mohamed, and J. K. White. 2003. Divalent cation transport and susceptibility to infectious and autoimmune disease: continuation of the Ity/Lsh/Nramp1/Slc11a1 gene story. Immunol. Lett. 85:197-203. [DOI] [PubMed] [Google Scholar]
- 8.Boschiroli, M., V. Foulongne, and D. O'Callighan. 2001. Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4:58-64. [DOI] [PubMed] [Google Scholar]
- 9.Buu, N. T., M. Cellier, P. Gros, and E. Schurr. 1995. Identification of a highly polymorphic length variant in the 3′UTR of NRAMP1. Immunogenetics 42:428-429. [DOI] [PubMed] [Google Scholar]
- 10.Cardon, L. R., and J. I. Bell. 2001. Association study designs for complex diseases. Nat. Rev. Genet. 2:91-99. [DOI] [PubMed] [Google Scholar]
- 11.Cellier, M., A. Belouchi, and P. Gros. 1996. Resistance to intracellular infections: comparative genomic analysis of Nramp. Trends Genet. 12:201-205. [DOI] [PubMed] [Google Scholar]
- 12.Cooke, G. S., and A. V. Hill. 2001. Genetics of the susceptibility to human infectious disease. Nat. Rev. Genet. 2:967-977. [DOI] [PubMed] [Google Scholar]
- 13.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. Smith, R. Allikmets, J. J. Goedet, S. P. Buchbinder, E. Vittinghoff, E. E. Gompert, S. Donfiels, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, and R. Detels. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 14.Ewald, D. R., and B. J. Bricker. 2000. Validation of the abbreviated Brucella AMOS PCR as a rapid screening method for differentiation of Brucella abortus field strain isolates and vaccine strains, 19 and RB51. J. Clin. Microbiol. 38:3085-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, J., Y. Li, M. Hashad, M. Schurr, P. Gros, L. G. Adams, and J. W. Templeton. 1996. Bovine natural resistance associated macrophage protein 1 (Nramp1) gene. Genome Res. 6:956-964. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Crespo, D., R. A. Juste, and A. Hurtado. 2005. Selection of ovine housekeeping genes for normalization by real-time RT_PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Vet. Res. doi: 10.1186/1746-6148-1-3. [DOI] [PMC free article] [PubMed]
- 17.Gros, P., E. Skamene, and A. Forget. 1983. Cellular mechanisms of genetically controlled host resistance to Mycobacterium bovis (BCG). J. Immunol. 131:1966-1972. [PubMed] [Google Scholar]
- 18.Gruenheid, S., and P. Gros. 2000. Genetic susceptibility to intracellular infections: Nramp1, macrophage function and divalent cations transport. Curr. Opin. Microbiol. 3:43-48. [DOI] [PubMed] [Google Scholar]
- 19.Guilloteau, L. A., J. Dornand, A. Gross, M. Olivier, F. Cortade, Y. Le Vern, and D. Kerboeuf. 2003. Nramp1 is not a major determinant in the gene control of Brucella melitensis infection in mice. Infect. Immun. 71:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, A. V., J. Elvin, A. C. Willis, M. Aidoo, C. E. Allsopp, F. M. Gotch, X. M. Gao, M. Takiguchis, B. M. Greewood, A. R. Townsend, A. J. McMichael, and H. C. Whittle. 1992. Molecular analysis of the association of HLA-B53 and resistance to severe malaria. Nature 360:434-439. [DOI] [PubMed] [Google Scholar]
- 21.Ho, M., and C. Cheers. 1982. Resistance and susceptibility of mice to bacterial infection. IV. Genetic and cellular basis of resistance to chronic infection with Brucella abortus. J. Infect. Dis. 146:381-387. [DOI] [PubMed] [Google Scholar]
- 22.Horîn, P., I. Rychlik, J. W. Templeton, and L. G. Adams. 1999. A complex pattern of microsatellite polymorphism within the bovine NRAMP1 gene. Eur. J. Immunogenet. 26:311-313. [DOI] [PubMed] [Google Scholar]
- 23.Hu, J., N. Bumstead, P. Barrow, G. Sebastiani, L. Olien, K. Morgan, and D. Malo. 1997. Resistance to salmonellosis in the chicken is linked to NRAMP1 and TNC. Genome Res. 7:693-704. [DOI] [PubMed] [Google Scholar]
- 24.Ioannidis, J. P., E. E. Ntzani, T. A. Trikalinos, and D. G. Contopoulos-Ioannidis. 2001. Replication validity of genetic association studies. Nat. Genet. 29:306-309. [DOI] [PubMed] [Google Scholar]
- 25.Jang, Z. D., P. C. Okhuysen, D. C. Guo, R. He, T. M. King, H. DuPont, and M. Milewicz. 2003. Genetic susceptibility to enteroaggressive Escherichia coli diarrhoea: polymorphism in the interleukin-8 promoter region. J. Infect. Dis. 188:506-511. [DOI] [PubMed] [Google Scholar]
- 26.Khush, R. S., F. Leulier, and B. Lemaitre. 2002. Pathogen surveillance - the flies have it. Science 296:273-275. [DOI] [PubMed] [Google Scholar]
- 27.Kramnick, I., W. F. Dietrich, P. Dermant, and B. R. Bloom. 2000. Genetic control of resistance to experimental infection with virulent Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 97:8560-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lander, E. S., and N. Schork. 1994. Genetic dissection of complex traits. Science 265:2037-2048. [DOI] [PubMed] [Google Scholar]
- 29.Lazzaro, B. P., B. K. Sceurman, and A. G. Clark. 2004. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303:1873-1876. [DOI] [PubMed] [Google Scholar]
- 30.Lessard, M., D. L. Hutchings, and J. L. Spencer. 1995. Cell-mediated and humoral immune responses in chicken infected with Salmonella typhimurium. Avian Dis. 39:230-238. [PubMed] [Google Scholar]
- 31.Lohmueller, K. E., C. L. Pearce, M. Pike, E. S. Lander, and J. N. Hirschhorn. 2003. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common diseases. Nat. Genet. 33:177-182. [DOI] [PubMed] [Google Scholar]
- 32.Lucentini, J. 2004. Gene association studies typically wrong. Scientist 18:20. [Google Scholar]
- 33.Mead, S., M. P. Stumpf, J. Whitfield, J. A. Beck, M. Poulter, T. Campbell, J. B. Uphill, D. Goldstein, M. Alpers, E. M. Fisher, and J. Collinge. 2003. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science 300:640-643. [DOI] [PubMed] [Google Scholar]
- 34.Mira, M. T., A. Alcais, V. T. Nguyen, M. O. Moraes, C. Di Flumeri, H. T. Vu, C. P. Mai, T. H. Nguyen, N. B. Nguyen, P. Khoa, E. Sarno, A. Alter, A. Montpetit, A. M. Moraes, J. Moraes, C. Dorè, C. Gallant, P. Lepage, A. Verner, E. van de Vesse, T. Hudson, L. Abet, and E. Schurr. 2004. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 427:636-640. [DOI] [PubMed] [Google Scholar]
- 35.Muyzer, G. E., C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Brien, A. D., D. L. Rosenstreich, I. Scher, G. H. Campbell, and R. P. MacDermott. 1980. Genetic control of susceptibility to Salmonella typhimurium infection in mice: role of the Lps gene. J. Immunol. 124:20-22. [PubMed] [Google Scholar]
- 37.Plant, J. E., J. M. Blackwell, A. D. O'Brien, D. J. Bradley, and A. A. Glynn. 1982. Are the Lsh and Ity disease resistance genes at one locus on mouse chromosome 1? Nature 297:510-511. [DOI] [PubMed] [Google Scholar]
- 38.Price, R. E., J. W. Templeton, R. Smith III, and L. G. Adams. 1990. Ability of mononuclear phagocytes from cattle naturally resistant or susceptible to brucellosis to control in vitro intracellular survival of Brucella abortus. Infect. Immun. 58:879-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pritchard, J. K., and N. A. Rosenberg. 1999. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 65:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qureshi, T., J. W. Templeton, and L. G. Adams. 1996. Intracellular survival of Brucella abortus, Mycobacterium bovis BCG, Salmonella serovar Dublin, and Salmonella thyphimurium in macrophages from cattle genetically resistant to Brucella abortus. Vet. Immunol. Immunopathol. 50:55-65. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1998. Appendix E: commonly used techniques in molecular cloning, p. E3-E4. In N. Irwin, N. Ford, and C. Nolan (ed.), Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Searle, S., and J. M. Blackwell. 1999. Evidence for a functional repeat polymorphism in the promoter of the human NRAMP1 gene that correlates with autoimmune versus infectious disease susceptibility. J. Med. Genet. 36:295-299. [PMC free article] [PubMed] [Google Scholar]
- 43.Segal, S., and A. V. Hill. 2003. Genetic susceptibility to infectious disease. Trends Microbiol. 11:445-448. [DOI] [PubMed] [Google Scholar]
- 44.Skamene, E., P. Gros, A. Forget, P. A. Kongshavn, C. St-Charles, and B. A. Taylor. 1982. Genetic regulation of resistance to intracellular pathogens. Nature 297:506-509. [DOI] [PubMed] [Google Scholar]
- 45.Stear, M. J., S. C. Bishop, B. A. Mallard, and H. Raadsma. 2001. The sustainability, feasibility and desirability of breeding livestock for disease control. Res. Vet. Sci. 71:1-7. [DOI] [PubMed] [Google Scholar]
- 46.Tishkoff, S., R. Varkonyi, N. Cahinhinan, S. A. Argyropoulos, G. Destro-Bisol, A. Drousiotou, B. Dangerfield, G. Lefranc, J. Loiselet, A. Piro, M. Stoneking, A. Tagarelli, G. Tagarelli, E. H. Touma, S. M. Williams, and A. G. Clark. 2001. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293:455-462. [DOI] [PubMed] [Google Scholar]
- 47.Thio, C. L., D. L. Thomas, and M. Carrington. 2000. Chronic viral hepatitis and the human genome. Hepatology 31:819-827. [DOI] [PubMed] [Google Scholar]
- 48.Warrington, J. A., A. Nair, M. Mahadevappa, and M. Tsyganskaya. 2000. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genomics 2:143-147. [DOI] [PubMed] [Google Scholar]
- 49.Vidal, S. M., D. Malo, K. Vogan, E. Skamene, and P. Gros. 1993. Natural resistance to infection with intracellular parasites: isolation of a candidate gene for Bcg. Cell 73:469-485. [DOI] [PubMed] [Google Scholar]
- 50.Woolhouse, M. E., J. P. Webster, E. Domingo, B. Charlesworth, and B. R. Lewin. 2002. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat. Genet. 32:569-577. [DOI] [PubMed] [Google Scholar]