Abstract
Cholera toxin (CT) moves from the plasma membrane to the endoplasmic reticulum (ER) by retrograde vesicular traffic. In the ER, the catalytic CTA1 polypeptide dissociates from the rest of the toxin and enters the cytosol by a process that involves the quality control mechanism of ER-associated degradation (ERAD). The cytosolic CTA1 then ADP ribosylates Gsα, resulting in adenylate cyclase activation and intoxication of the target cell. It is hypothesized that the C-terminal A13 subdomain of CTA1 plays two crucial roles in the intoxication process: (i) it contains a hydrophobic domain that triggers the ERAD mechanism and (ii) it facilitates interaction with the cytosolic ADP-ribosylation factors (ARFs) that serve as allosteric activators of CTA1. In this study, we examined the role(s) of the CTA13 subdomain in CT intoxication. Full-length CTA1 constructs and truncated CTA1 constructs lacking the A13 subdomain were generated and used to conduct two-hybrid studies of interactions with ARF6, in vitro enzyme assays, in vivo toxicity assays, and in vivo processing/degradation assays. Direct, plasmid-mediated expression of CTA1 constructs in the ER or cytosol of transfected CHO cells was used to perform the in vivo assays. With these methods, we found that the A13 subdomain of CTA1 is important both for interaction with ARF6 and for full expression of enzyme activity in vivo. Surprisingly, however, the A13 subdomain was not required for ERAD-mediated passage of CTA1 from the ER to the cytosol. A possible alternative trigger for CTA1 to activate the ERAD mechanism is discussed.
Cholera toxin (CT), produced by Vibrio cholerae, is an AB5-type protein toxin that consists of an A moiety with latent enzymatic activity and a cell-binding B moiety (reviewed in references 2 and 35). The A moiety is initially synthesized as a single CTA polypeptide that undergoes proteolytic processing to yield a disulfide-linked A1/A2 heterodimer. CTA1 is a 22-kDa ADP ribosyltransferase with three distinct subdomains: CTA11 (residues 1 to 132) comprises the catalytic core of the toxin; CTA12 (residues 133 to 161) is a short, flexible subdomain; and CTA13 (residues 162 to 192) is a globular region that interfaces with CTA11 and has many hydrophobic residues and a single cysteine residue that provides the disulfide linkage to CTA2 (52). The 5.5-kDa CTA2 polypeptide maintains numerous noncovalent interactions with the B moiety and thereby acts as a linker between the catalytic and cell-binding components of CT. The homopentameric B moiety of CT is assembled from 11.6-kDa monomers, binds to GM1 gangliosides on the eukaryotic plasma membrane, and serves as a vehicle for CTA1 delivery to the endoplasmic reticulum (ER) (3, 15, 33).
CT is internalized from the eukaryotic plasma membrane by an endocytic mechanism that utilizes lipid rafts (27, 51). The active pool of internalized toxin subsequently moves to the endoplasmic reticulum by retrograde vesicular transport (13, 14, 19, 23, 26, 29). In the ER, the disulfide bond linking CTA1 to CTA2/CTB5 is reduced (18, 25, 45). Protein disulfide isomerase mediates the dissociation of CTA1 from CTA2/CTB5, and CTA1 is then translocated to the cytosol in a process involving the quality control system of ER-associated degradation (ERAD) (30, 39-41, 44-46, 50). The cytosolic pool of CTA1 ADP ribosylates and irreversibly activates Gsα. Activated Gsα stimulates the action of adenylate cyclase, which in turn leads to elevated levels of intracellular cyclic AMP (cAMP). Chloride efflux into the intestinal lumen is triggered by cAMP-dependent signaling events, and the resulting osmotic movement of water into the gut generates the massive watery diarrhea characteristic of cholera (reviewed in references 9 and 35).
The CTA13 subdomain is thought to be involved with two important events in the process of cholera intoxication: (i) it may act as a trigger for ERAD-mediated export to the cytosol, and (ii) it may promote an interaction with the cytosolic ADP-ribosylation factors (ARFs) that serve as allosteric activators of CTA1. Both of these events are discussed in more detail below.
ERAD recognizes misfolded or misassembled proteins in the ER and exports them to the cytosol for ubiquitination and proteasomal degradation (11, 20, 46). A hydrophobic region in the CTA13 subdomain, exposed when CTA1 dissociates from holotoxin, was postulated to act as a signal for protein misfolding and would therefore trigger the ERAD-mediated export of CTA1 into the cytosol (4). CTA1 is believed to avoid ubiquitin-dependent proteasomal degradation in the cytosol, because it has only two lysine residues to serve as potential sites for ubiquitination (4, 28). Other plant and bacterial toxins are also thought to masquerade as misfolded proteins in order to exploit ERAD for entry into the cytosol (4, 17). Hydrophobic regions in the A moieties of two of these toxins, ricin and Shiga toxin, have been implicated in toxin transfer from the ER to the cytosol (32, 36). However, no studies have yet directly addressed the role of the hydrophobic CTA13 subdomain in CTA1 passage from the ER to the cytosol.
ARFs are low-molecular-weight GTPases involved with membrane trafficking in the biosynthetic and endocytic transport pathways (48). In vitro, ARFs also act as allosteric activators of the catalytic activity of CTA1 (48). Several residues in the CTA13 subdomain are important for ARF interaction with CTA1 in a bacterial two-hybrid system, and holotoxins containing mutations in these residues exhibit little toxicity (7). Although the reduced toxicity of these CT mutants could have resulted from the loss of in vivo ARF interaction, mutations in the A13 subdomain could also inhibit toxicity through alterations to the enzymatic activity of CT, the trafficking of CT to the ER, CTA1 entry into the cytosol, or the in vivo stability of either the CT holotoxin or the CTA1 fragment. The intracellular trafficking of CT thus makes it difficult to correlate in vitro assays of CTA1 activity with in vivo toxicity studies.
In this work, we assessed the toxicities of wild-type and mutant forms of CTA1 by using a plasmid-based system to directly express various CTA1 constructs in either the ER or the cytosol of transfected cells. This system bypasses the retrograde trafficking itinerary of CT and the need for intact holotoxin. CTA1 activity (monitored by cAMP accumulation in transfected cells) and CTA1 processing can therefore be examined without the confounding problems related to CT trafficking. This in vivo expression system was combined with two-hybrid studies and in vitro enzyme assays to assess the role(s) of the CTA13 subdomain in ARF-stimulated toxin activity and toxin entry into the cytosol. An intact CTA13 subdomain was shown to be required for efficient interaction with ARF6 and for full enzyme activity, but toxin entry into the cytosol was not inhibited in an ER-localized CTA1 construct lacking the A13 subdomain. Toxin entry into the cytosol may instead depend upon an inherent physical property of the CTA1 polypeptide.
MATERIALS AND METHODS
Materials.
Lipofectamine and the pcDNA3.1 expression plasmid were purchased from Invitrogen (Carlsbad, CA), the purified CTA1/CTA2 heterodimer was from Calbiochem (La Jolla, CA), cell culture reagents were purchased from Gibco BRL (Grand Island, NY), bacterial-culture reagents were from Fisher Scientific (Pittsburg, PA), chemicals were from Sigma-Aldrich (St. Louis, MO), [35S]methionine was from NEN Life Sciences (Boston, MA), and the [125I]cAMP Biotrak kit was from Amersham-Pharmacia (Arlington Heights, IL). Immobilized anti-CTA antibodies (5) were generated by a 4°C overnight incubation with 50 mg protein G-agarose (Sigma-Aldrich) in 1 ml of phosphate-buffered saline (PBS) supplemented with 0.1% bovine serum albumin. Talon metal affinity resin was from BD Biosciences (Palo Alto, CA).
Generation of CTA1 constructs.
A T7-based clone expressing mature CTA1 with a C-terminal His6 tag was made in multiple steps by first replacing the mature CTB gene from a derivative of pT7CTBR with XbaI deleted (16) with the mature CTA1 gene from pNPCT (47) to make pT7CTA1. Then, the carboxyl end of a CTA1 derivative encoded by pMGJ6701 (6) and a His6 tag derived from pT7SH6 (43) were cloned together into pT7CTA1 in several steps to create pT7CTA1h6, encoding a mature CTA1 polypeptide ending in CGNSHHHHHHXX (single-amino-acid notation, where N is Asn-189 of mature CTA1 and XX denotes two termination codons). A deletion of the A13 domain was first made in a holotoxin backbone by cloning an XbaI-SmaI fragment of a PCR product, made by using a vector primer and CTA13SmaR (CTCCCCGGGAAACCTGCCAATCC), into a derivative of a holotoxin-encoding clone, pMGJ142 (8), which has a silent mutation introducing an SmaI site encoding Pro-185/Gly-186 of CTA1 to create pMGJ197. The XbaI-SmaI fragment was then subcloned into an XbaI-EcoRI (filled-in) digest of pT7CTA1h6, creating pT7CTA1d3h6, encoding CTA1 ending in GFPSHHHHHH (where P is Pro-168 of mature CTA1).
For eukaryotic cytoplasmic expression of CTA1 derivatives, the wild-type CTA1h6 and CTA1d3h6 were moved into the backbone of pcDNA3.1/mCTA1 (42) to create pcDNAmatCTA1h6 (encoding mCTA1·His6) and pcDNAmatCTA1d3h6 (encoding mCTA11-168·His6), respectively. A native, non-His-tagged variant of the A13 deletion clone was made by replacing a HindIII-EcoRI fragment of pcDNA3.1/mCTA1 with a similarly digested PCR fragment made using a T7 universal primer and CTA1d3ER (GGGAATTCATTACGGAGGGAAACCTGCC), which represents a mature CTA1 gene terminating at Pro-169. Inactivating Glu-to-Asp mutations at residues 110 and 112 of mature CTA1 were moved into pcDNA3.1/mCTA1 to create pcDNAmatCTA1dd (encoding mCTA1dd). Construction of the pcDNA3.1 clones producing ssCTA1 and ssCTA1dd has been described previously (42). The pcDNA3.1 clone encoding an ER-localized form of the CTA13 deletion derivative was made by replacing a BglII-PflMI fragment of pcDNAmatCTA1d3h6 with the same-cut fragment from pcDNA3.1/ssCTA1, encoding the signal peptide of CTA1.
mCTA1·His6 and mCTA11-168·His6 protein purification.
Escherichia coli strain BL21(pLysS) was transformed with an inducible mCTA1·His6 or mCTA11-168·His6 expression plasmid and grown at 37°C in 500 ml Luria-Bertani broth (LB) (21) to an A600 of 0.6. CTA1 expression was then induced by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to the growth medium. At 4 h postinduction, the cells were pelleted, resuspended in Tris-buffered saline (TBS) (20 mM Tris-HCl, pH 7.6, 137 mM NaCl) containing 8 M urea, and lysed with three freeze-thaw cycles. The insoluble lysate fraction was removed after a 45-min spin at 10,000 × g. The soluble fraction was then incubated with Talon resin at room temperature for 30 min. The resin was subsequently poured into a plastic column and washed with 10 column volumes of TBS containing 8 M urea. The CTA1 construct was then eluted from the column with 8 to 10 column volumes of elution buffer (TBS, 8 M urea, and 100 mM imidazole). One-milliliter fractions were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for the presence of CTA1. Before use in the ADP-ribosylation assay, the fractions containing CTA1 were dialyzed against three changes of 4 liters of TBS containing 2 M urea. With the use of a buffer exchange column (Pierce, Rockford, IL), the dialyzed protein was transferred to buffer containing 20 mM dithiothreitol (DTT), 0.1 mg bovine serum albumin/ml, and 0.2 M potassium phosphate, pH 7.5.
ARF6 protein purification.
E. coli strain BL21(pLysS) was transformed with an inducible ARF6 expression plasmid (pT7Arf6; kindly provided by Joel Moss, National Heart, Lung, and Blood Institute, Bethesda, MD) and grown at 37°C in 500 ml LB to an A600 of 0.6. ARF6 production was then induced by the addition of 1 mM IPTG to the growth medium. At 4 h postinduction, the cells were pelleted, resuspended in TBS, and lysed with three freeze-thaw cycles. The insoluble lysate fraction was removed after a 45-min spin at 10,000 × g. The remaining soluble lysate was dialyzed against two changes of 4 liters low-salt buffer (20 mM Tris, pH 7.6, 50 mM NaCl, and 1 mM EDTA). The sample was then put over a DEAE column, and the flowthrough was collected. As assessed by SDS-PAGE, the flowthrough contained ARF6 and very few other proteins. The ARF6-enriched flowthrough was dialyzed against three changes of 4 liters no-salt buffer (200 mM Tris, pH 7.6, 1 mM MgCl, and 1 mM DTT) and placed over a Q-Sepharose column. The fraction containing ARF6 was eluted with buffer containing 15 mM NaCl, 200 mM Tris, pH 7.6, 1 mM MgCl, and 1 mM DTT. The purity of the eluted ARF6 was confirmed by SDS-PAGE and Coomassie staining.
Nondenaturing polyacrylamide gel electrophoresis.
All samples, gels, and electrophoresis buffers were prepared without SDS or DTT. The samples were then run on a nondenaturing 12% polyacrylamide minigel in a Tris-glycine buffer. The gels were run at 25 milliamps until the dye front reached the bottom of the gel. The samples were then visualized with Coomassie stain.
Two-hybrid assay.
Interactions between CTA1 derivatives and ARF6 were determined using modifications of a bacterial two-hybrid assay (7). The A13 domain deletion from pMGJ197 was subcloned as an XbaI-ClaI fragment in place of the native CTA1 domain of the pCTA1R7KT18 plasmid to create pCTA1d3T18. Wild-type and deletion derivatives were coexpressed with pCT25ARF6 in a new lac+ mal+ reporter strain, DC8, a ΔcyaA1403::Km mutant of E. coli MM294 (CGSC 6315 from New England Biolabs, Beverly, MA). Interaction was assessed by assaying the cAMP-induced production of β-galactosidase activity in log-phase cultures as described by Miller (21). The background signal (32 Miller units) from cells harboring pCT25arf6 and pAT18-9 vectors was subtracted from the presented experimental data.
ADP-ribosylation assay.
Diethylamino(benzylidine-amino)guanidine (DEA-BAG), an artificial substrate for ADP ribosylation that absorbs light at ∼355-nm wavelength and loses its ability to bind to Bio-Rad AG (Hercules, CA). Ion-exchange resin after ADP ribosylation, was synthesized as described previously (34). Dilutions of the His6-tagged CTA1 protein were mixed with 2 mM DEA-BAG in 0.2 ml of assay buffer (20 mM DTT, 0.1 mg bovine serum albumin/ml, 10 mM NAD, and 0.2 M potassium phosphate, pH 7.5). Where indicated, 10 μg ARF6 was also added to the assay buffer. After 2 h at 30°C, the reaction was stopped by binding unreacted DEA-BAG with 1 ml of a 40% slurry of Bio-Rad AG-50W-X4 resin. The DEA-BAG-Bio-Rad AG-50W-X4 suspension was vortexed for 30 s and spun for 10 min at full speed in a microcentrifuge; 0.4 ml of the resulting supernatant was removed and analyzed for fluorescence (excitation wavelength, 361 nm; emission wavelength, 440 nm) with a VersaFluor fluorometer (Bio-Rad, Hercules, CA). Three reactions were performed for each data point.
Cell culture and transfection.
The Chinese hamster ovary (CHO) fibroblasts used in this study are auxotrophic for proline and were provided by the late Henry Wu (Uniformed Services University of the Health Sciences, Bethesda, MD). The cells were maintained in Ham's F-12 medium supplemented with 10% fetal bovine serum. Growth conditions were set at 37°C and 5% CO2 in a humidified incubator. For transient transfections, 1 μg of plasmid DNA was mixed with 5 μl Lipofectamine for 20 min at room temperature. The mixture was then added for 3 h to cells that had been seeded at 75% confluence in six-well plates. The transfected cells were analyzed at either 4 or 16 h posttransfection (cAMP assay) or at 16 to 24 h posttransfection (metabolic labeling).
Transfection-cAMP assay.
Transfected cells were solubilized in 0.75 ml acidic ethanol (1 N HCl-ethanol at a 1:100 ratio) for 15 min at 4°C. The cell extracts were transferred to microcentrifuge tubes for a 10-min 4°C spin. The supernatant was collected, and the pellet was reextracted with 0.75 ml of an ice-cold ethanol-H2O solution at a 2:11 ratio. Supernatants from both extractions were combined and lyophilized overnight. The lyophilized cell extracts were reconstituted in 1.5 ml of buffer, and cAMP levels in the reconstituted extracts were quantitated with the use of a 125I-labeled cAMP Biotrak competition assay. Background cAMP levels from cells transfected with the empty pcDNA3.1 vector (typically 1,000 fmol cAMP/well) were subtracted from the values obtained for our experimental conditions.
Metabolic labeling and immunoprecipitation of native and truncated CTA1.
Transfected cells were washed twice with PBS, incubated in 0.5 ml methionine-free medium for 1 h, and exposed to 0.5 ml of 150 μCi [35S]methionine/ml for 1 h. After two additional PBS washes, the cells were either solubilized in 1 ml lysis buffer (25 mM Tris, pH 7.4, 20 mM NaCl, 1% deoxycholic acid, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 μg pepstatin/ml, and 1 μg leupeptin/ml) for 20 min at 4°C or returned to serum-free medium containing 0.5 mg methionine/ml. Additional cell extracts were generated immediately after the specified chase intervals. Triton-insoluble material was removed from the cell extracts by centrifugation, and immobilized anti-CTA antibodies were added to the cleared supernatants for overnight incubation at 4°C. The immunoprecipitated material was washed twice with NDET (1% Nonidet P-40, 0.4% deoxycholic acid, 5 mM EDTA, 10 mM Tris, pH 7.4, 150 mM NaCl) and once with water before resuspension in sample buffer. SDS-PAGE (15% polyacrylamide gels) with phosphorimager analysis (Bio-Rad) was used to visualize and quantitate the immunoisolated samples. Results for the chase intervals were expressed as percentages of the values obtained from the pulse-labeled cells.
RESULTS
ARF interaction with CTA1 and CTA11-168.
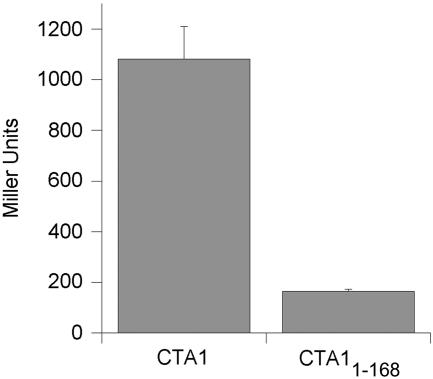
An established bacterial two-hybrid assay (7) was used to confirm that an intact A13 subdomain was required for CTA1 to interact with ARF. With this assay, the catalytic domain of Bordetella pertussis adenylate cyclase was separated into two complementary fragments (T25 and T18) that lacked adenylate cyclase activity. The T25 fragment was fused to ARF6, and the T18 fragment was fused to CTA1 or to a truncated CTA1 variant lacking the A13 subdomain (CTA11-168). Both CTA1 constructs were detoxified with arginine 7-to-lysine substitutions in order to facilitate their expression in the cytoplasm of E. coli DC8 (7). The T25-ARF6 and CTA1-T18 fusion proteins interacted to produce functional adenylate cyclase, resulting in production of intracellular cAMP and expression of the β-galactosidase reporter gene. In contrast, coexpression of T25-ARF6 and CTA11-168-T18 produced only a minimal increase in the level of genes encoding β-galactosidase (Fig. 1), corresponding to about 15% of the level observed with ARF6 and the full-length CTA1 construct. Thus, the presence of the CTA13 subdomain appeared to be important for the interaction of ARF6 with CTA1.
FIG. 1.
ARF6 interaction with CTA1 and CTA11-168 in a bacterial two-hybrid system. The cAMP-dependent expression of β-galactosidase activity was measured from triplicate cultures of E. coli coexpressing T25/ARF6 and CTA1/T18 or CTA11-168/T18. β-Galactosidase activity is expressed in Miller units and reflects the amount of cAMP produced as a consequence of the interaction between ARF6 and CTA1 or CTA11-168. The error bars represent standard deviations of the mean.
In vitro activity of CTA1·His6 and CTA11-168·His6.
To corroborate the lack of interaction of CTA11-168 with ARF6, we purified ARF6, CTA1, and CTA11-168 for use in an in vitro ADP-ribosylation assay. Members of the ARF family of proteins serve as allosteric activators of CTA1 (48), so the inability of CTA11-168 to interact with ARF6 would be expected to prevent any stimulatory effect of ARF6 on the enzymatic activity of CTA11-168. Recombinant ARF6 was purified from lysates of E. coli transformed with an inducible ARF6 expression plasmid. CTA1 and CTA11-168 were modified with a C-terminal His6 tag for Talon column purification from lysates of E. coli transformed with inducible CTA1 expression plasmids. The recombinant CTA1 proteins were called mCTA1·His6 and mCTA11-168·His6 to denote the mature CTA1 polypeptide (m) and the His6 tag.
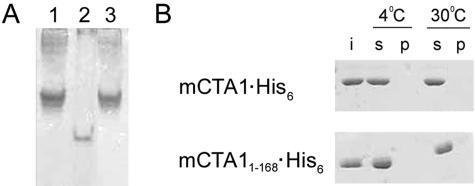
Two control experiments were performed to verify the in vitro stabilities of mCTA1·His6 and mCTA11-168·His6 (Fig. 2). First, nondenaturing PAGE resolved both mCTA1·His6 and mCTA11-168·His6 as monomeric species with the expected mobilities (Fig. 2A). Identical mobilities were observed for mCTA1·His6 and the CTA1 polypeptide from a reduced CTA1-CTA2 heterodimer. Next, an established protocol for CTA unfolding and aggregation (50) failed to detect aggregated (i.e., pelleted) forms of our His6-tagged constructs (Fig. 2B). These experiments demonstrated that mCTA1·His6 and mCTA11-168·His6 were purified as soluble, monomeric proteins.
FIG. 2.
In vitro stabilities of mCTA1·His6 and mCTA11-168·His6. (A) One-microgram samples of the reduced CTA1/CTA2 heterodimer (lane 1), mCTA11-168·His6 (lane 2), and mCTA1·His6 (lane 3) were resolved by nondenaturing PAGE and visualized by Coomassie staining. (B) Three hundred nanograms of mCTA1·His6 and mCTA11-168·His6 were incubated at 4°C or 30°C for 20 min before a 15-min, 4°C spin at 16,000 × g. The input (i) starting material, as well as the resulting supernatants (s, reflecting soluble protein) and pellets (p, reflecting aggregated protein) were visualized by SDS-PAGE with Coomassie staining.
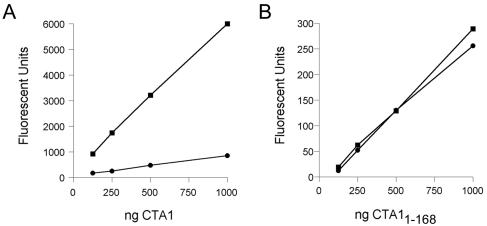
The enzymatic activities of mCTA1·His6 and mCTA11-168· His6 were assessed by ADP ribosylation of the substrate DEA-BAG (Fig. 3). The activity of mCTA1·His6 was stimulated ∼7-fold by the addition of ARF6 (Fig. 3A). In contrast, no stimulation of mCTA11-168·His6 activity was observed in the presence of ARF6 (Fig. 3B). mCTA11-168·His6 was also less active than mCTA1·His6, exhibiting only 14% of the basal catalytic activity of an equimolar amount of full-length CTA1. Removal of the CTA13 subdomain thus decreased the catalytic activity of mCTA11-168·His6 and abolished the stimulation of its catalytic activity by ARF6.
FIG. 3.
ARF6 stimulation of mCTA1·His6 and mCTA11-168·His6 enzyme activity. Twofold dilutions of mCTA1·His6 (A) or mCTA11-168·His6 (B) were mixed with 2 mM DEA-BAG in 0.2 ml of ADP-ribosylation assay buffer. Toxins were incubated in the absence (circles) or presence (squares) of ARF6. After 2 h at 30°C, the reaction was stopped. The ADP-ribosylation of DEA-BAG was then assessed by fluorometry; increasing fluorescent units correspond to increasing levels of ADP ribosylation. Assays with mCTA1·His6 and mCTA11-168·His6 were performed in parallel but are shown on separate graphs because of the difference in scale. In the absence of ARF, mCTA11-168·His6 exhibited only 14% of the activity recorded for an equimolar amount of mCTA1·His6 (n = 2), and ARF6 failed to stimulate the catalytic activity of mCTA11-168·His6. Three reactions were performed for each data point; the error bars represent the standard deviations.
In vivo activities of CTA1 and CTA11-168/9.
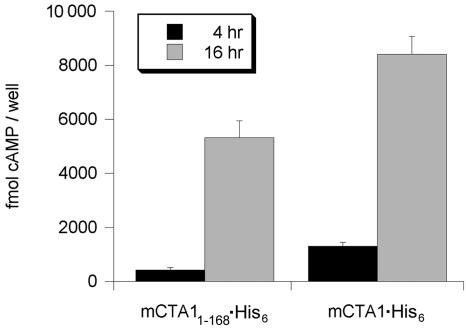
To examine the in vivo activities of our CTA1 constructs, the coding sequences of mCTA1·His6 and mCTA11-168·His6 were introduced into the pcDNA3.1 eukaryotic expression vector for use in a transfection-cAMP assay. With this assay, a plasmid-based system was used to produce mature CTA1 constructs directly in the cytosol of transfected CHO cells. cAMP levels in transfected cells were then used as a measure of CTA1 activity against Gsα. Previous work showed that transfected, CTA1-producing CHO cells produce a cAMP response within 4 hours of posttransfection chase (42). If the in vivo activity of CTA1 required ARF interaction, this cAMP response would be expected to be markedly decreased or absent from cells producing CTA11-168·His6.
Transfected cells producing either mCTA1·His6 or mCTA11-168· His6 generated detectable cAMP responses at both 4 and 16 h of posttransfection chase (Fig. 4). Cells producing mCTA11-168· His6 produced a cAMP signal that was 1.4 times above background at 4 h of chase. This signal rose to 7.3 times above background by 16 h of chase. However, cells producing the full-length mCTA1·His6 construct produced substantially more cAMP than cells expressing mCTA11-168·His6 at both 4 and 16 h of chase. mCTA11-168·His6 thus exhibited a basal level of ADP-ribosyltransferase activity that was sufficient to cause an easily detectable increase in production of cAMP in vivo, but it was significantly less active than the full-length mCTA1·His6 construct.
FIG. 4.
In vivo activity of mCTA1·His6 and mCTA11-168·His6. cAMP levels in transfected CHO cells expressing mCTA1·His6 or mCTA11-168·His6 were determined at 4 and 16 h of posttransfection chase. Background levels of cAMP (typically 1,000 fmol cAMP/well) from cells transfected with an empty vector were subtracted from the experimental results. Duplicate samples were used for each experiment; the graph presents the means ± standard deviations of the means of four independent experiments.
Transfected cells synthesized 2.3-fold more mCTA11-168·His6 than mCTA1·His6 (data not shown; n = 4). When the raw cAMP responses presented in Fig. 4 were corrected for this difference in the CTA1 production level, we found that mCTA11-168·His6 produced 15% (4-h chase) or 29% (16-h chase) of the cAMP signal obtained with mCTA1·His6.
The C-terminal His6 tag could potentially alter the catalytic activities of mCTA1·His6 and mCTA11-168·His6. To control for this possibility, we repeated the transfection-cAMP assay with untagged variants of mCTA1 and mCTA11-168 (Fig. 5). An untagged mCTA1 variant with glutamate-to-aspartate substitutions at residues 110 and 112 in the CTA1 active site was also examined in these studies. This construct is called mCTA1dd to denote the mature CTA1 polypeptide (m) and the two active-site mutations (dd). CT holotoxin variants containing glutamate-to-aspartate substitutions at either residue 110 or 112 in CTA1 exhibited less than 1% of wild-type holotoxin enzyme activity in vitro (5).
FIG. 5.
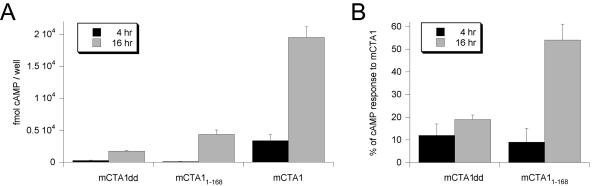
In vivo activity of mCTA1dd, mCTA11-168, and mCTA1. (A) cAMP levels in transfected CHO cells expressing mCTA1dd, mCTA11-168, or mCTA1 were determined at 4 and 16 h of posttransfection chase. Background levels of cAMP (typically 1,000 fmol cAMP/well) from cells transfected with an empty vector were subtracted from the experimental results. Duplicate samples were used for each experiment; the graph presents the means ± standard errors of the means of four independent experiments. (B) The cAMP responses of cells expressing mCTA1dd or mCTA11-168 were calculated as percentages of the cAMP response from cells expressing mCTA1. SDS-PAGE and phosphorimager analysis were used to quantitate the pools of mCTA1dd, mCTA11-168, and mCTA1 immunoprecipitated from metabolically labeled cells at 16 h posttransfection chase. Transfected cells synthesized 1.8-fold more mCTA1 than mCTA1dd and 2.4-fold more mCTA1 than mCTA11-168. The data presented in panel A were standardized to these differences in CTA1 expression levels before the relative activities of mCTA1dd and mCTA11-168 were determined. The means plus standard deviations of the means of four independent experiments are presented in the graph.
CHO cells expressing the genes for mCTA1dd, mCTA11-168, or mCTA1 were assayed for cAMP content at 4 and 16 h of posttransfection chase (Fig. 5A). mCTA1-producing cells generated a robust cAMP response at 4 h of chase, but only a minimal cAMP response was recorded at 4 h of chase for cells expressing mCTA1dd or mCTA11-168. However, by 16 h of chase, all three mCTA1 constructs elicited easily detectable increases in the cAMP responses from transfected cells. These results were consistent with the data obtained for the His6-tagged constructs and demonstrated that mCTA11-168 was a weak but functional toxin that exhibited significantly more activity than mCTA1dd. The data also demonstrated the high sensitivity of this system for detecting low residual levels of catalytic activity in the mutant forms of CTA1.
Transfected cells synthesized 1.8-fold more mCTA1 than mCTA1dd and 2.4-fold more mCTA1 than mCTA11-168. After standardizing the data in Fig. 4A to these differences in expression levels, the activities of mCTA1dd and mCTA11-168 were expressed as percentages of mCTA1 activity (Fig. 5B). mCTA1dd exhibited only 12 to 19% of the activity recorded for mCTA1 over the course of the experiment. In contrast, the relative activity of mCTA11-168 increased dramatically over the course of the experiment: mCTA11-168 exhibited only 9% of the activity recorded for mCTA1 at 4 h of chase, but at 16 h of chase, mCTA11-168 exhibited 54% of mCTA1 activity.
CTA1 constructs that were expressed directly in the eukaryotic cytosol were used to generate the data presented in Fig. 5. Essentially identical results were obtained when CTA1dd, CTA11-169, and CTA1 were initially directed into the ER lumen (Fig. 6). Cotranslational insertion of CTA1 variants into the ER was accomplished by placing the coding region for the CTA signal sequence directly upstream of the mCTA1dd, mCTA11-169, or mCTA1 coding sequence. The resulting variants are referred to as ssCTA1dd, ssCTA11-169, and ssCTA1 to denote the presence of the CTA signal sequence (ss). In eukaryotes, the CTA signal sequence acts as a targeting determinant for cotranslational delivery of CTA1 into the ER lumen and is proteolytically removed from CTA1 in the ER. Protease protection assays, complete cleavage of the CTA signal sequence, and the addition of oligosaccharides to recombinant CTA1 constructs with N-linked glycosylation sites have demonstrated that the entire pool of newly synthesized CTA1 is inserted into the ER lumen when the CTA signal sequence is present (30, 39). The ER-localized pool of CTA1 is then transferred to the cytosol and degraded by the proteasome in a process involving the ERAD mechanism (39).
FIG. 6.
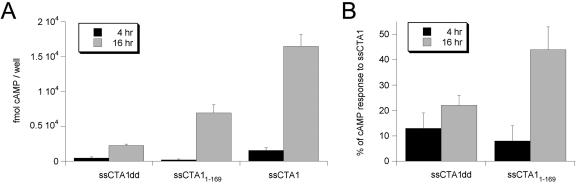
In vivo activity of ssCTA1dd, ssCTA11-169, and ssCTA1. (A) cAMP levels in transfected CHO cells expressing ssCTA1dd, ssCTA11-169, or ssCTA1 were determined at 4 and 16 h of posttransfection chase. Background levels of cAMP (typically 1,000 fmol cAMP/well) from cells transfected with an empty vector were subtracted from the experimental results. Duplicate samples were used for each experiment; the graph presents the means plus standard errors of the means of four independent experiments. (B) The cAMP responses of cells expressing ssCTA1dd or ssCTA11-169 were calculated as percentages of the cAMP response from cells expressing ssCTA1. The data presented in panel A were standardized to differences in CTA1 expression levels before the relative activities of ssCTA1dd and ssCTA11-169 were determined. The means plus standard deviations of the means of four independent experiments are presented in the graph.
The transfection-cAMP assay was performed with constructs producing ssCTA1dd, ssCTA11-169, and ssCTA1 in order to examine the role of the A13 subdomain in CTA1 export from the ER to the cytosol. If a hydrophobic region in the A13 subdomain served as a trigger to promote ERAD-mediated passage of CTA1 into the cytosol, then truncation of the A13 subdomain would be expected to eliminate the signal for export and prevent CTA1 from entering the cytosol. As such, ssCTA11-169 would fail to elicit a cAMP response from transfected cells because it would not exit the endomembrane system to reach its Gsα target in the cytosol. However, in contrast to this model, we found that ssCTA11-169 was as toxic as mCTA11-168: the ER-localized and cytosolic variants of CTA11-168/9 both generated ∼80 fmol cAMP/well at 4 h of chase and 4,000 to 7,000 fmol cAMP/well at 16 h of chase (Fig. 5A and 6A). Thus, it appeared that the A13 subdomain was not required for CTA1 passage into the cytosol.
As with the mCTA1 variants, the raw data presented for the ssCTA1 constructs in Fig. 6A were corrected for differences in CTA1 expression levels and expressed as percentages of wild-type ssCTA1 activity (Fig. 6B). ssCTA1dd exhibited only 13 to 22% of the activity recorded for ssCTA1 over the course of the experiment. In contrast, the relative activity for ssCTA11-169 increased substantially from 4 h to 16 h of posttransfection chase, from 8% of the activity recorded for ssCTA1 at 4 h of chase to 44% of the activity recorded for ssCTA1 at 16 h of chase. The nearly equivalent relative activities of mCTA11-168 and ssCTA11-169 at both 4 h (∼10% of wild-type activity) and 16 h (∼50% of wild-type activity) of posttransfection chase again demonstrated that the removal of the A13 subdomain did not prevent ER-localized CTA1 from reaching its cytosolic target. The similar results obtained with mCTA11-168 and ssCTA11-169 also indicated that, over the course of a 4- or 16-h experiment, the ER-to-cytosol export of ssCTA1 was not a rate-limiting event for CTA1 interaction with Gsα. This interpretation is supported by our previous work, which demonstrated a relatively rapid rate of CTA1 export from the ER to the cytosol (39). Collectively, our data suggest that the CTA13 subdomain is required for efficient expression of enzyme activity but not for toxin entry into the cytosol.
Intracellular degradation of CTA1 and CTA11-168/9.
To further examine the role of the A13 subdomain in CTA1 export from the ER to the cytosol, we monitored the intracellular processing of ssCTA1 and ssCTA11-169 (Fig. 7A). The ER-localized pool of CTA1 is transferred to the cytosol and degraded in a proteasome-dependent manner (36, 37). If the A13 subdomain contains the trigger for export from the ER, then export of ssCTA11-169 to the cytosol would be expected to be slower than that of ssCTA1, resulting in decreased proteolysis and an increase in the half-life for ssCTA11-169 relative to ssCTA1.
FIG. 7.
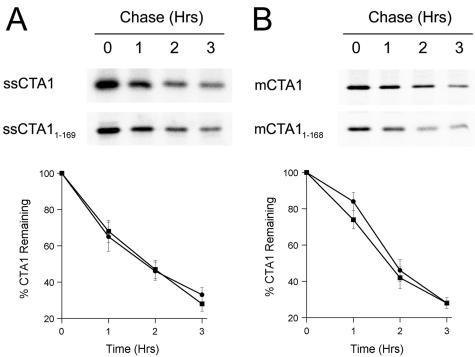
Degradation of CTA1 and CTA11-168/9. (A) Transfected CHO cells expressing ssCTA1 (circles) or ssCTA11-169 (squares) were incubated for 1 h with 150 μCi [35S]methionine/ml and chased in serum-free medium containing an excess of cold methionine. Anti-CTA immunoprecipitates from cell extracts generated at the stated intervals were visualized and quantitated by SDS-PAGE with phosphorimager analysis. Results for the chase intervals were expressed as percentages of the values obtained from the pulse-labeled cells. One of four independent experiments is presented in the gel. The graph presents the means ± standard errors of the means of all four experiments. (B) Experiments identical to the ones described above were performed with transfected CHO cells expressing mCTA1 (circles) or mCTA11-168 (squares). One of four independent experiments is presented in the gel. The graph presents the means ± standard deviations of the means of all four experiments.
To test this hypothesis, CHO cells producing ssCTA1 or ssCTA11-169 were preincubated in methionine-free medium for 1 h, labeled with 150 μCi [35S]methionine/ml for 1 h, and chased for various intervals in serum-free medium containing an excess of unlabeled methionine. Cell extracts generated after the stated intervals were subjected to immunoprecipitation with immobilized anti-CTA antibodies. The immunoisolated samples were then resolved by SDS-PAGE. Phosphorimager quantitation of the immunoisolated samples demonstrated that ssCTA1 and ssCTA11-169 were degraded at identical rates, with 2-h half-lives (Fig. 7A). We found that mCTA1 and mCTA11-168, produced as cytoplasmic proteins, also exhibited ∼2-h half-lives (Fig. 7B). These results were consistent with the data obtained from our transfection-cAMP assays and demonstrated that the A13 subdomain does not play an essential role in CTA1 export from the ER to the cytosol.
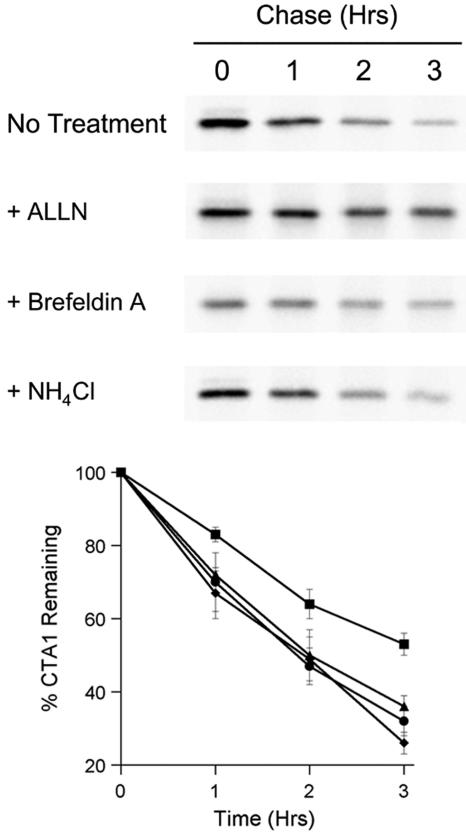
ERAD substrates are efficiently retained in the proximal compartments of the secretory pathway before passage into the cytosol for proteasome-mediated degradation (11, 20, 46). The processing of ER-localized CTA1 occurs in a similar manner (39). To ensure that ssCTA11-169 was also processed as an ERAD substrate, we monitored the turnover of ssCTA11-169 in cells exposed to N-acetyl-Leu-Leu-Norleu-Al (ALLN), brefeldin A (BfA), or NH4Cl (Fig. 8). ALLN inhibits the proteasome (12), BfA inhibits trafficking through the secretory pathway (10), and NH4Cl inhibits the activity of acid-dependent lysosomal proteases (31). The 2-h half-life of ssCTA11-169 was extended to just over 3 h in ALLN-treated cells, thus demonstrating a role for the proteasome in ssCTA11-169 degradation. However, neither BfA nor NH4Cl had a significant impact on the turnover of ssCTA11-169. Furthermore, no secreted pools of ssCTA11-169 were detected in the extracellular medium of our transfected cells (data not shown). These results collectively demonstrated that ssCTA11-169 was processed as an ERAD substrate despite the absence of the A13 subdomain and its putative hydrophobic trigger for ERAD recognition.
FIG. 8.
ERAD processing of ssCTA11-169. Transfected CHO cells incubated with 150 μCi [35S]methionine/ml for 1 h were chased in serum-free medium containing excess cold methionine for the indicated times. Exposure to 100 μM ALLN (squares), 1 μg BfA/ml (triangles), or 20 mM NH4Cl (diamonds) was begun 1 h prior to labeling and was maintained throughout the experiment. Untreated cells are represented by circles. Anti-CTA immunoprecipitates from cell extracts generated at the stated intervals were visualized and quantitated by SDS-PAGE with phosphorimager analysis. Results for the chase intervals were expressed as percentages of the values obtained from the pulse-labeled cells. One of four independent experiments is presented in the gel; the graph presents the means ± standard deviations of the means of all four experiments.
DISCUSSION
Previous work has shown that certain hydrophobic residues in the CTA13 subdomain are important for ARF association with CTA1 (7). A C-terminal hydrophobic region in the A1 polypeptide of E. coli heat-labile toxin, which is 80% homologous to CTA1, is also required for interaction with the ARF proteins (53). In this work, we found that an intact CTA13 subdomain was essential for a functional interaction between ARF6 and CTA1. Our two-hybrid assay demonstrated that the level of interaction between ARF6 and CTA11-168 was only 15% of that observed for ARF6 and full-length CTA1. Additional experiments with purified proteins showed that ARF6 failed to stimulate the catalytic activity of mCTA11-168 · His6. Our data thus provide physical and functional evidence for the role of the CTA13 subdomain in ARF stimulation of CTA1 activity.
The recently published CT/ARF6 cocrystal suggests that ARF6 binding to CTA1 does not require amino acids 168 to 192 of CTA1 (24). However, our results here show that ARF6 does not physically or functionally interact with CTA11-168. This simply indicates that the inhibition of ARF6 binding to CTA1 can result from structural effects other than loss of the specific ARF6 binding site. For example, we have previously suggested that the surface exposure of certain aromatic amino acids in the A13 subdomain may create an initial ARF interaction motif (7). Alternatively, deletion of the A13 subdomain may induce a conformational change in CTA11-168 that inhibits ARF6 binding. Either possibility is consistent with the collective results of the current study and the CT/ARF6 cocrystal study.
CTA11-168/9 variants were able to produce attenuated and time-dependent cAMP responses in transfected CHO cells, despite their inability to interact with ARF: mCTA11-168 and ssCTA11-169 elicited 8 to 9% of the cAMP responses obtained from cells producing the corresponding full-length CTA1 constructs at 4 h of chase, but by 16 h of chase the CTA11-168/9 variants generated 44 to 54% of the cAMP responses from full-length CTA1-producing cells. The time required for substantial cAMP accumulation in cells producing mCTA11-168 or ssCTA11-169 was a consequence of the low basal activity of CTA11-168/9 and is consistent with a time-dependent increase in the cellular levels of ADP-ribosylated Gsα and activated adenylate cyclase. These findings indicate that ARF stimulation of CTA1 activity may facilitate, but is not required for, activation of adenylate cyclase in target cells.
The cAMP responses of CTA1dd-producing cells were much less dramatic than the responses from CTA11-168/9-producing cells. For cells expressing mCTA1dd or ssCTA1dd, cAMP levels rose by a factor of only 1.7 from 4 h to 16 h of posttransfection chase. In contrast, for cells expressing mCTA11-168 or ssCTA11-169 there was a sixfold increase in cAMP levels from 4 to 16 h of chase. The differential cAMP responses of CTA1dd-producing cells versus CTA11-168/9-producing cells could not be attributed to differences in protein turnover, as ssCTA1dd had the same 2-h half-life as ssCTA11-169 (data not shown). These findings suggest that the balance between toxin activity and toxin degradation is less favorable for CTA1dd than for CTA11-168/9 and that the basal enzymatic activity of CTA1dd was not sufficient to produce a substantial increase in the pool of ADP-ribosylated Gsα in vivo during the interval from 5 to 16 h posttransfection. Previous studies have also suggested that the intoxication of target cells involves a balance between toxin activity and toxin degradation (1, 41, 49).
Both the CTA1dd and CTA11-168/9 variants exhibited substantially more activity in vivo than in vitro. Differences between in vivo and in vitro activities have also been reported for the plasmid-encoded toxin of enteroaggregative E. coli: a variant of this toxin with no in vitro activity was found to have substantial in vivo activity when synthesized directly in the eukaryotic cytosol with a plasmid-based expression system (37). Collectively, these results simply indicate that in vivo and in vitro assay conditions are not always equivalent.
The hydrophobic CTA13 subdomain was necessary for a functional interaction with ARF6 but was dispensable for toxin entry into the cytosol. Results from the transfection-cAMP assay established that the ER-localized ssCTA11-169 variant was as toxic as the cytosolic mCTA11-168 construct. Since elimination of the CTA1 export signal would be expected to prevent ER-localized CTA1 from reaching its Gsα target in the cytosol, the similar in vivo activities of ssCTA11-169 and mCTA11-168 demonstrated that residues in the A13 subdomain were not required to trigger ERAD-mediated passage of CTA1 into the cytosol.
Data from our transfection-cAMP assay could potentially be affected by a small, undetectable pool of ssCTA11-169 that was mistargeted and synthesized directly in the cytosol. However, the processing of metabolically labeled ssCTA11-169 by transfected CHO cells also indicated that the A13 subdomain was not required for toxin entry into the cytosol. An inhibition of export from the ER to the cytosol prevents the proteasome-mediated degradation of CTA1 (40), yet ssCTA11-169 was degraded at the same rate as the full-length ssCTA1 construct. No secreted pools of ssCTA11-169 were detected, and the proteasome-dependent turnover of ssCTA11-169 was unaffected by treatments that inhibit lysosomal proteolysis or trafficking in the secretory pathway. Thus, although ssCTA11-169 lacked the A13 subdomain and the putative trigger for entry into the cytosol, it was efficiently retained in the ER and exported to the cytosol for proteasomal degradation by the ERAD system. These experiments, which directly monitored CTA1 protein levels and would not be affected by a small pool of mistargeted ssCTA11-169 in the cytosol, provided independent support for our interpretation of Fig. 6—namely, that the A13 subdomain does not contain the sole signal for ERAD recognition and processing of CTA1.
It was possible that the normal signal for CTA1 entry into the cytosol was located in the A13 subdomain, but removal of this region generated a misfolded CTA1 variant that was still recognized by the ERAD system. Export of ssCTA11-169 to the cytosol might therefore result from a general defect in protein folding rather than from a specific trigger residing in the ssCTA11-169 polypeptide. Based on the crystal structure of CTA1, truncation of the A13 subdomain would not generate obvious difficulties for protein folding. A flexible, 28-amino-acid molecular tether (the A12 subdomain) links the A13 subdomain to the compact, catalytic A11 subdomain (52). The structures of the globular A11 subdomain and the flexible A12 linker would thus likely remain intact in CTA1 constructs lacking the A13 subdomain. Three experimental findings also suggested that the CTA11-168/9 variants were not dramatically misfolded: (i) all truncated CTA1 constructs retained some enzymatic and biological activity; (ii) the CTA11-168/9 variants were degraded with the same kinetics, but not faster than, the corresponding full-length CTA1 variants; and (iii) all truncated CTA1 variants retained the capacity to bind anti-CTA antibodies and to be recovered by immunoprecipitation. A grossly misfolded protein would not be expected to retain biological activity, yet all three CTA11-168/9 variants exhibited readily detectable in vivo enzyme activity after 16 h of posttransfection chase. Furthermore, a misfolded protein would likely be degraded at a higher rate than a properly folded variant of the same protein. The identical half-lives of ssCTA11-169 and ssCTA1 (as well as mCTA11-168 and mCTA1) thus indicated that the structural organization of CTA1 was not dramatically altered by deletion of the A13 subdomain. Finally, the ability to immunoprecipitate CTA11-168/9 with anti-CTA antibodies indicated that the CTA1 epitopes required for anti-CTA antibody interaction were preserved in the truncated variants. As the CTA11-168/9 constructs appeared to fold properly, our findings do not support the hypothesis that the A13 subdomain contains the sole signal for ERAD recognition and translocation of CTA1 into the cytosol.
The signal for ERAD-mediated export to the cytosol may reflect an inherent physical property of CTA1 rather than a specific region or motif within the polypeptide. Fourier-transform infrared spectroscopy has shown that the structural stability of CTA1 is temperature dependent, with a loss of CTA1 secondary structure in the disulfide-bridged CTA1/CTA2 heterodimer occurring between 40°C and 46°C (38). Furthermore, circular dichroism and fluorescence techniques have recently shown that the reduced CTA1/CTA2 heterodimer is in a fully folded conformation at 25°C and a partially unfolded state at 37°C (K. Teter, D. Moe, A. H. Pande, M. G. Jobling, J. K. Tinker, S. A. Tatulian, and R. K. Holmes, submitted for publication). It has also been reported that, in the absence of ARFs or phospholipids, the reduced CTA1/CTA2 heterodimer has greater enzymatic activity in vitro at 25°C than at 37°C (22). Thermal instability in the reduced CTA1 polypeptide thus generates a partially unfolded conformation at 37°C, and this unfolded state could act as the trigger for ERAD-mediated export of CTA1 to the cytosol. In vivo, ARF and/or phospholipid activation of CTA1 might stabilize the active conformation of CTA1 or compensate for a decreased basal level of enzymatic activity resulting from a partially unfolded CTA1 conformation (22).
In conclusion, CTA1 utilizes the A13 subdomain for functional interaction with ARF6 and for efficient expression of in vivo enzyme activity. Surprisingly, however, the A13 subdomain is not required for ERAD-mediated toxin passage into the cytosol. Thermal instability at 37°C may instead generate a partially unfolded CTA1 polypeptide that is recognized by the ERAD system as a signal for export to the cytosol. Work is under way to examine this possibility.
Acknowledgments
This work was initiated at the University of Colorado Health Sciences Center (NIH grant support F32 AI10394 to K. Teter and R01 AI31940 to R. K. Holmes) and completed at the University of Central Florida (NIH grant support K22 AI054568 to K. Teter).
We thank Joel Moss for the gift of the pT7ARF6 expression plasmid.
Editor: J. D. Clements
REFERENCES
- 1.Deeks, E. D., J. P. Cook, P. J. Day, D. C. Smith, L. M. Roberts, and J. M. Lord. 2002. The low lysine content of ricin A chain reduces the risk of proteolytic degradation after translocation from the endoplasmic reticulum to the cytosol. Biochemistry 41:3405-3413. [DOI] [PubMed] [Google Scholar]
- 2.De Haan, L., and T. R. Hirst. 2004. Cholera toxin: a paradigm for multi-functional engagement of cellular mechanisms. Mol. Membr. Biol. 21:77-92. [DOI] [PubMed] [Google Scholar]
- 3.Fujinaga, Y., A. A. Wolf, C. Rodighiero, H. Wheeler, B. Tsai, L. Allen, M. G. Jobling, T. Rapoport, R. K. Holmes, and W. I. Lencer. 2003. Gangliosides that associate with lipid rafts mediate transport of cholera and related toxins from the plasma membrane to endoplasmic reticulm. Mol. Biol. Cell 14:4783-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazes, B., and R. J. Read. 1997. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry 36:11051-11054. [DOI] [PubMed] [Google Scholar]
- 5.Jobling, M. G., and R. K. Holmes. 2001. Biological and biochemical characterization of variant A subunits of cholera toxin constructed by site-directed mutagenesis. J. Bacteriol. 183:4024-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jobling, M. G., and R. K. Holmes. 1992. Fusion proteins containing the A2 domain of cholera toxin assemble with B polypeptides of cholera toxin to form immunoreactive and functional holotoxin-like chimeras. Infect. Immun. 60:4915-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jobling, M. G., and R. K. Holmes. 2000. Identification of motifs in cholera toxin A1 polypeptide that are required for its interaction with human ADP-ribosylation factor 6 in a bacterial two-hybrid system. Proc. Natl. Acad. Sci. USA 97:14662-14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jobling, M. G., L. M. Palmer, J. L. Erbe, and R. K. Holmes. 1997. Construction and characterization of versatile cloning vectors for efficient delivery of native foreign proteins to the periplasm of Escherichia coli. Plasmid 38:158-173. [DOI] [PubMed] [Google Scholar]
- 9.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klausner, R. D., J. G. Donaldson, and J. Lippincott-Schwartz. 1992. Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116:1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostova, Z., and D. H. Wolf. 2003. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 13.Lencer, W. I., C. Constable, S. Moe, M. G. Jobling, H. M. Webb, S. Ruston, J. L. Madara, T. R. Hirst, and R. K. Holmes. 1995. Targeting of cholera toxin and Escherichia coli heat labile toxin in polarized epithelia: role of COOH-terminal KDEL. J. Cell Biol. 131:951-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lencer, W. I., J. B. de Almeida, S. Moe, J. L. Stow, D. A. Ausiello, and J. L. Madara. 1993. Entry of cholera toxin into polarized human intestinal epithelial cells. Identification of an early brefeldin A sensitive event required for A1-peptide generation. J. Clin. Investig. 92:2941-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lencer, W. I., and B. Tsai. 2003. The intracellular voyage of cholera toxin: going retro. Trends Biochem. Sci. 28:639-645. [DOI] [PubMed] [Google Scholar]
- 16.L'Hoir, C., A. Renard, and J. A. Martial. 1990. Expression in Escherichia coli of two mutated genes encoding the cholera toxin B subunit. Gene 89:47-52. [DOI] [PubMed] [Google Scholar]
- 17.Lord, J. M., and L. M. Roberts. 1998. Toxin entry: retrograde transport through the secretory pathway. J. Cell Biol. 140:733-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majoul, I., D. Ferrari, and H. D. Soling. 1997. Reduction of protein disulfide bonds in an oxidizing environment. The disulfide bridge of cholera toxin A-subunit is reduced in the endoplasmic reticulum. FEBS Lett. 401:104-108. [DOI] [PubMed] [Google Scholar]
- 19.Majoul, I. V., P. I. Bastiaens, and H. D. Soling. 1996. Transport of an external Lys-Asp-Glu-Leu (KDEL) protein from the plasma membrane to the endoplasmic reticulum: studies with cholera toxin in Vero cells. J. Cell Biol. 133:777-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCracken, A. A., and J. L. Brodsky. 2003. Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD). Bioessays 25:868-877. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. E. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 22.Murayama, T., S. C. Tsai, R. Adamik, J. Moss, and M. Vaughan. 1993. Effects of temperature on ADP-ribosylation factor stimulation of cholera toxin activity. Biochemistry 32:561-566. [DOI] [PubMed] [Google Scholar]
- 23.Nambiar, M. P., T. Oda, C. Chen, Y. Kuwazuru, and H. C. Wu. 1993. Involvement of the Golgi region in the intracellular trafficking of cholera toxin. J. Cell. Physiol. 154:222-228. [DOI] [PubMed] [Google Scholar]
- 24.O'Neal, C. J., M. G. Jobling, R. K. Holmes, and W. G. Hol. 2005. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science 309:1093-1096. [DOI] [PubMed] [Google Scholar]
- 25.Orlandi, P. A. 1997. Protein-disulfide isomerase-mediated reduction of the A subunit of cholera toxin in a human intestinal cell line. J. Biol. Chem. 272:4591-4599. [PubMed] [Google Scholar]
- 26.Orlandi, P. A., P. K. Curran, and P. H. Fishman. 1993. Brefeldin A blocks the response of cultured cells to cholera toxin. Implications for intracellular trafficking in toxin action. J. Biol. Chem. 268:12010-12016. [PubMed] [Google Scholar]
- 27.Orlandi, P. A., and P. H. Fishman. 1998. Filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J. Cell Biol. 141:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodighiero, C., B. Tsai, T. A. Rapoport, and W. I. Lencer. 2002. Role of ubiquitination in retro-translocation of cholera toxin and escape of cytosolic degradation. EMBO Rep. 3:1222-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandvig, K., O. Garred, and B. van Deurs. 1996. Thapsigargin-induced transport of cholera toxin to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 93:12339-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz, A., H. Herrgen, A. Winkeler, and V. Herzog. 2000. Cholera toxin is exported from microsomes by the Sec61p complex. J. Cell Biol. 148:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seglen, P. O. 1983. Inhibitors of lysosomal function. Methods Enzymol. 96:737-764. [DOI] [PubMed] [Google Scholar]
- 32.Simpson, J. C., J. M. Lord, and L. M. Roberts. 1995. Point mutations in the hydrophobic C-terminal region of ricin A chain indicate that Pro250 plays a key role in membrane translocation. Eur. J. Biochem. 232:458-463. [DOI] [PubMed] [Google Scholar]
- 33.Smith, D. C., J. M. Lord, L. M. Roberts, and L. Johannes. 2004. Glycosphingolipids as toxin receptors. Semin. Cell Dev. Biol. 15:397-408. [DOI] [PubMed] [Google Scholar]
- 34.Soman, G., J. Narayanan, B. L. Martin, and D. J. Graves. 1986. Use of substituted (benzylidineamino)guanidines in the study of guanidino group specific ADP-ribosyltransferase. Biochemistry 25:4113-4119. [DOI] [PubMed] [Google Scholar]
- 35.Spangler, B. D. 1992. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol. Rev. 56:622-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suhan, M. L., and C. J. Hovde. 1998. Disruption of an internal membrane-spanning region in Shiga toxin 1 reduces cytotoxicity. Infect. Immun. 66:5252-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sui, B. Q., P. R. Dutta, and J. P. Nataro. 2003. Intracellular expression of the plasmid-encoded toxin from enteroaggregative Escherichia coli. Infect. Immun. 71:5364-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surewicz, W. K., J. J. Leddy, and H. H. Mantsch. 1990. Structure, stability, and receptor interaction of cholera toxin as studied by Fourier-transform infrared spectroscopy. Biochemistry 29:8106-8111. [DOI] [PubMed] [Google Scholar]
- 39.Teter, K., R. L. Allyn, M. G. Jobling, and R. K. Holmes. 2002. Transfer of the cholera toxin A1 polypeptide from the endoplasmic reticulum to the cytosol is a rapid process facilitated by the endoplasmic reticulum-associated degradation pathway. Infect. Immun. 70:6166-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teter, K., and R. K. Holmes. 2002. Inhibition of endoplasmic reticulum-associated degradation in CHO cells resistant to cholera toxin, Pseudomonas aeruginosa exotoxin A, and ricin. Infect. Immun. 70:6172-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teter, K., M. G. Jobling, and R. K. Holmes. 2003. A class of mutant CHO cells resistant to cholera toxin rapidly degrades the catalytic polypeptide of cholera toxin and exhibits increased endoplasmic reticulum-associated degradation. Traffic 4:232-242. [DOI] [PubMed] [Google Scholar]
- 42.Teter, K., M. G. Jobling, and R. K. Holmes. 2004. Vesicular transport is not required for the cytoplasmic pool of cholera toxin to interact with the stimulatory alpha subunit of the heterotrimeric G protein. Infect. Immun. 72:6826-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripet, B., M. W. Howard, M. Jobling, R. K. Holmes, K. V. Holmes, and R. S. Hodges. 2004. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 279:20836-20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai, B., and T. A. Rapoport. 2002. Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J. Cell Biol. 159:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai, B., C. Rodighiero, W. I. Lencer, and T. A. Rapoport. 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104:937-948. [DOI] [PubMed] [Google Scholar]
- 46.Tsai, B., Y. Ye, and T. A. Rapoport. 2002. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell. Biol. 3:246-255. [DOI] [PubMed] [Google Scholar]
- 47.Vadheim, K. L., Y. Singh, and J. M. Keith. 1994. Expression and mutagenesis of recombinant cholera toxin A subunit. Microb. Pathog. 17:339-346. [DOI] [PubMed] [Google Scholar]
- 48.Welsh, C. F., J. Moss, and M. Vaughan. 1994. ADP-ribosylation factors: a family of approximately 20-kDa guanine nucleotide-binding proteins that activate cholera toxin. Mol. Cell Biochem. 138:157-166. [DOI] [PubMed] [Google Scholar]
- 49.Wesche, J., A. Rapak, and S. Olsnes. 1999. Dependence of ricin toxicity on translocation of the toxin A-chain from the endoplasmic reticulum to the cytosol. J. Biol. Chem. 274:34443-34449. [DOI] [PubMed] [Google Scholar]
- 50.Winkeler, A., D. Godderz, V. Herzog, and A. Schmitz. 2003. BiP-dependent export of cholera toxin from endoplasmic reticulum-derived microsomes. FEBS Lett. 554:439-442. [DOI] [PubMed] [Google Scholar]
- 51.Wolf, A. A., M. G. Jobling, S. Wimer-Mackin, M. Ferguson-Maltzman, J. L. Madara, R. K. Holmes, and W. I. Lencer. 1998. Ganglioside structure dictates signal transduction by cholera toxin and association with caveolae-like membrane domains in polarized epithelia. J. Cell Biol. 141:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, R. G., D. L. Scott, M. L. Westbrook, S. Nance, B. D. Spangler, G. G. Shipley, and E. M. Westbrook. 1995. The three-dimensional crystal structure of cholera toxin. J. Mol. Biol. 251:563-573. [DOI] [PubMed] [Google Scholar]
- 53.Zhu, X., E. Kim, A. L. Boman, A. Hodel, W. Cieplak, and R. A. Kahn. 2001. ARF binds the C-terminal region of the Escherichia coli heat-labile toxin (LTA1) and competes for the binding of LTA2. Biochemistry 40:4560-4568. [DOI] [PubMed] [Google Scholar]