Abstract
Shigella spp. are the causative agent of bacillary dysentery. Infection results in acute colonic injury due to the host inflammatory response. The mediators of the damage, infiltrating polymorphonuclear leukocytes (PMN), also resolve the infection. Shigella flexneri's virulence effectors are encoded on its large virulence plasmid and on pathogenicity islands in the chromosome. The SHI-2 pathogenicity island encodes the virulence factor ShiA, which down-regulates Shigella-induced inflammation. In the rabbit ileal loop model, infection with a shiA null strain (ΔshiA) induces a more severe inflammation than wild-type infection. Conversely, a Shigella strain that overexpresses ShiA (ShiA+) is less inflammatory than the wild-type strain. To determine the host responses modulated by ShiA, we performed infection studies using the mouse lung model, which recapitulates the phenotypes observed in the rabbit ileal loop model. Significantly, ShiA+ strain-infected mice cleared the bacteria and survived infection, while wild-type- and ΔshiA strain-infected mice could not clear the bacteria and ultimately died. Surprisingly, microarray analysis of infected lungs revealed the regulation of genes involved in innate T-cell responses to infection. Immunohistochemistry showed that wild-type- and ΔshiA strain-infected animals have greater numbers of PMN and T cells in their lungs over the course of infection than ShiA+ strain-infected animals. These results suggest that the T-cell innate response is suppressed by ShiA in Shigella infections.
Shigella flexneri is a causative agent of bacillary dysentery, infecting approximately 165 million people per year worldwide. Children under 5 years of age represent 61% of the 1.1 million annual deaths (19, 50). Shigella infection provokes a highly inflammatory response in the colon of the host. Symptoms of shigellosis can range from a mild diarrhea to bloody, mucopurulent stools (36).
Upon ingestion of contaminated food or water, Shigella uses specialized antigen-sampling cells, called M cells, to cross the colonic epithelial barrier (33, 41, 48). The bacteria are then taken up by the underlying resident macrophages (16, 42), where they escape from the phagosome into the cytoplasm (9, 23). Once there, the bacteria secrete the virulence factor IpaB (invasion plasmid antigen B), which activates caspase-1 (5, 43), leading to cytotoxicity. Concomitant to caspase-1-induced cell death, interleukin-1β (IL-1β) and IL-18 are released (12, 13). Subsequently, Shigella invades the basolateral side of intestinal epithelial cells (27), resulting in the release of IL-8 (38). Together, IL-1β, IL-18, and IL-8 initiate a proinflammatory cascade that recruits polymorphonuclear leukocytes (PMN) (35, 37, 38). PMN migrate to the colon and cross the epithelial cell barrier to the lumen. Crossing this barrier disrupts the epithelial cell tight junctions and provides Shigella additional access to the tissue. Thus, the infection is initially exacerbated by the PMN (34). The PMN are, however, crucial for resolution of infection because they kill Shigella (22) and eradicate the infection. Shigella spp. manipulate host innate immunity by inducing inflammation via cytokines to enhance their infectivity; however, it is this host innate response that ultimately clears the bacteria.
The proinflammatory response to Shigella is induced by the delivery of bacterial effector proteins through the type 3 secretion system. Shigella invasion and cytotoxicity genes, such as ipaB, are encoded on a large virulence plasmid. Plasmid-cured strains are not pathogenic (40). However, while the large virulence plasmid is necessary for infection, several chromosomal loci are required for a full inflammatory response (8, 39). One of these chromosomal loci contains the Shigella pathogenicity island SHI-2, a region that encodes several cryptic mobility genes, an aerobactin operon, and seven novel open reading frames (26, 47).
ShiA, a product of one of the novel open reading frames in SHI-2, is a suppressor of inflammation (14). A strain disrupted in shiA (ΔshiA) induces more severe inflammation characterized by an exacerbated PMN influx in the rabbit ileal loop model. Conversely, a strain overexpressing ShiA (ShiA+) provoked very little inflammation and PMN recruitment compared to wild-type infection. Interestingly, ShiA is not involved in Shigella invasion, cytotoxicity, or cytokine regulation in vitro. This suggests that ShiA affects a previously unknown Shigella virulence mechanism.
In this study, we investigate the mechanism by which ShiA regulates inflammation in the mouse lung model of infection (45, 46). This model recapitulates the acute inflammation seen in natural infections and allows us to investigate survival and kinetics of infection using the many tools available for the mouse. We first demonstrate that the phenotypes of wild-type Shigella, ΔshiA, and ShiA+ strain infections in the mouse model are similar to those seen in the rabbit model, and we then use microarray, real-time PCR, and enzyme-linked immunosorbent assay (ELISA) analyses to investigate differences in gene and protein expression during lung infections. We find that many proinflammatory genes are expressed at higher levels in wild-type or ΔshiA strain infection than in ShiA+ strain infection. Interestingly, a subset of these gene products either recruits or is secreted from T cells. Furthermore, we observe that there are significantly more T cells at the site of wild-type and ΔshiA strain infection than at the site of ShiA+ strain infection. A role for T cells in the innate immune response to Shigella infection was recently suggested by Le-Barillec et al. (20). Our results suggest that in S. flexneri infections, ShiA down-regulates an innate inflammatory response that includes the recruitment of T cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type S. flexneri strain M90T (40) contains the empty pUC19 vector. The ΔshiA strain contains a chloramphenicol cassette inserted into shiA (26) and the empty pUC19 vector, and the ShiΑ+ strain is M90T carrying a pUC19 plasmid encoding shiA (14). Shigella strains were grown at 37°C in tryptic soy broth or on Luria broth plates containing ampicillin (100 μg/ml) when appropriate.
Immunoblot.
Cultures of wild-type Shigella, ShiA+, and ΔshiA strains grown overnight were subcultured for 3 h, and bacterial equivalents, as determined by optical density (OD) measurement, were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblot with a polyclonal mouse anti-ShiA antibody and a polyclonal rabbit anti-RecA antibody.
Mouse infections.
All experimental protocols were approved by the Institutional Animal Care and Use Committee. Mice were housed in filter-top cages and provided with sterile water and food ad libitum. C57BL/6 mice were obtained from Charles River Laboratories (Wilmington, MA, or Sulzfeld, Germany), from Elevage Janvier (Le Genest Saint Isle, France), or from the Bundesinstitut für Risikobewertung (Berlin, Germany). Within an experiment, all mice came from the same source. Mouse experiments were performed as previously described (45). Briefly, mice were anesthetized by intramuscular injection of ketamine (12 mg/ml; Ketavet Pharmacia, Erlangen, Germany) and xylazine (4 mg/ml; Rompun Bayer, Leverkusen, Germany) in phosphate-buffered saline (PBS). Bacterial strains were resuspended in PBS and inoculated intranasally in a single application of 20 μl (inoculum of 2 × 107 to 5 × 107 CFU/mouse in each experiment, with all mice receiving the same inoculum within a single experiment, as determined by OD measurement and dilution plating of the inoculum). At different time points, mice were sacrificed, and the lungs were removed and processed for bacterial CFU enumeration, cytokine analysis, histology, or RNA isolation.
Lungs were homogenized in 1 ml of PBS, diluted, and plated onto the appropriate antibiotic agar plates for CFU enumeration. Homogenized lungs were then microcentrifuged at top speed for 5 min, and the supernatants were used for cytokine analysis. Murine IL-17 and IL-6 ELISAs were obtained from R&D Systems (Minneapolis, MN). For histology, lungs were fixed in 4% paraformaldehyde and embedded in paraffin for further processing as described below. For RNA isolation, lungs were homogenized in 1 ml TRIzol (Invitrogen, Carlsbad, CA) solution and frozen for future processing.
Immunohistochemistry of mouse lung sections.
Paraffin sections were processed using standard techniques. For LPS and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling, sections were digested with 20 μg/ml proteinase K for 6 min at room temperature. Blocking buffer included goat normal immune serum, fish gelatin, bovine serum albumin, and Tween 20. Primary antibody was applied overnight at 4°C (for LPS staining, 1:40 dilution of rabbit S. flexneri serotype 5 anti-LPS [Accurate Chemical & Scientific Corp., Westbury NY]; for PMN staining, 1:200 dilution of anti-PMN-specific membrane protein [MCA771G; Serotec]; for T-cell staining, 1:200 dilution of rabbit anti-human anti-CD3 [Dako, Carpinteria, CA]). Secondary antibody was applied for 2 h at 37° (goat anti-rabbit immunoglobulin G [IgG] [Cy3 conjugated], goat anti-rabbit IgG [Cy2 conjugated], and goat anti-rat IgG [Cy2] [Jackson Immunoresearch Europe, Cambridgeshire, United Kingdom]). Hoechst dye was used to visualize nuclei. Sections were analyzed using ImageJ (http://rsb.info.nih.gov/ij/).
Microarrays.
Briefly, RNA was purified from homogenized lung samples with the RNeasy Clean Up kit (QIAGEN, Hilden, Germany) and quality controlled using the Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA). RNA amplification, direct labeling, hybridization, and slide scanning were carried out by Icoria (Research Triangle Park, NC). All samples were hybridized to mouse custom microarrays purchased from Agilent Technologies. The arrays were designed in concert between the Max Planck Institute for Infection Biology and Agilent Technologies. The design includes genes relevant for innate and adaptive immunity as well as unique coding sequences from several organs (e.g., lung, intestine, spleen, liver, and stomach).
Microarray scans were first subjected to Agilent Technologies image analysis software (G2567AA Feature Extraction software [version A6.1.1.1]). All subsequent analyses were carried out using the Rosetta Resolver gene expression data analysis system, version 4.0 (Rosetta Inpharmatics, Seattle, WA). Expression patterns were identified by a stringent analysis using twofold expression change and error-weighted cutoffs using a P value of <0.05.
Real-time PCR.
RNA samples were treated with DNase (Invitrogen, Carlsbad, CA) before cDNA synthesis. Reverse transcription was performed according to the manufacturer's instructions using random primers (Superscript III reverse transcriptase and Random Primers; Invitrogen, Carlsbad, CA). Real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Darmstadt, Germany) in an ABI Prism 7000 sequence detection system PCR machine (Applied Biosystems, Darmstadt, Germany). Data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers used in this study are as follows: GAPDH forward, CATGGCCTTCCGTGTTCCT; GAPDH reverse, GCGGCACGTCAGATCCA; IL-17 forward, CCCTTGGCGCAAAAGTGA; IL-17 reverse, CGTGGAACGGTTGAGGTAGTCT; IL-6 forward, ACCACGGCCTTCCCTACTTC; IL-6 reverse, TTGGGAGTGGTATCCTCTGTGAA; IL-1β forward, CTGGTGTGTGACGTTCCCATT; IL-1β reverse, CGACAGCACGAGGCTTTTTT; Scyb2/macrophage inflammatory protein 2 (MIP-2) forward, CCTCAACGGAAGAACCAAAGAG; Scyb2/MIP-2 reverse, AGGCACATCAGGTACGATCCA; Scya2/monocyte chemoattractant protein 1 (MCP-1) forward, GGCTCAGCCAGATGCAGTTAA; Scya2/MCP-1 reverse, CCTACTCATTGGGATCATCTTGCT; GRO-1/keratinocyte-derived protein chemokine (KC) forward, CCTCAAGAACATCCAGAGCTTGA; growth-regulated oncogene 1 (GRO-1)/KC reverse, AGTGTGGCTATGACTTCGGTTTG.
Relative quantitation of gene expression was calculated using the comparative cycle threshold method. This method is outlined in the ABI User Bulletins (http://dna-9.int-med.uiowa.edu/RealtimePCRdocs/Compar_Anal_Bulletin2.pd).
RESULTS
ShiA+ strain-infected mice survive infection.
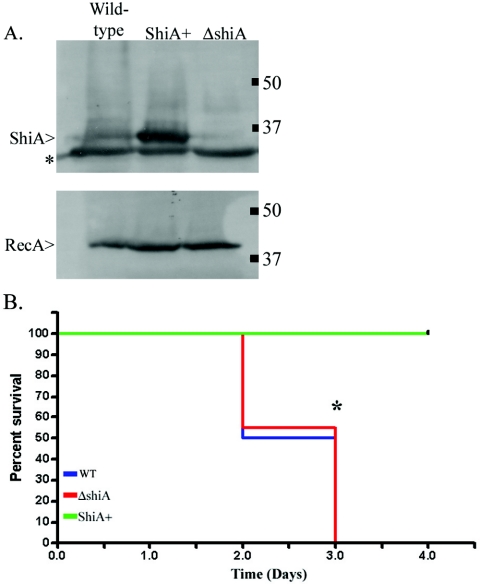
Our previous study established that ShiA down-regulates the host inflammatory response to infection (14). To further characterize the regulation of inflammation mediated by ShiA, we used the mouse lung model of infection. The mouse lung model recapitulates many of the inflammatory characteristics of intestinal Shigella infection (45) while giving us more avenues of study than are possible in the rabbit model. To determine the effect of ShiA on survival of infected animals, we inoculated anesthetized C57BL/6 wild-type mice intranasally with bacteria. We compared three strains (described in Materials and Methods): (i) a wild-type Shigella strain (40), (ii) the hyperinflammatory ΔshiA strain (14), and (iii) the hypoinflammatory ShiA+ strain (14). These three strains are isogenic, with the exception of their level of ShiA expression. Immunoblot experiments confirmed that the ΔshiA strain does not expresses ShiA while the ShiA+ strain overexpresses ShiA in comparison to expression in the wild-type strain (Fig. 1A).
FIG. 1.
ShiA+ strain-infected mice survive infection. (A) Cultures of wild-type Shigella, ShiA+, and ΔshiA strains grown overnight were subcultured for 3 h, and bacterial equivalents, as determined by OD measurement, were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblot with a polyclonal mouse anti-ShiA antibody. In the top panel, the top band is ShiA, while the second band (*) is an unrelated protein that cross-reacts with the antibody. The bottom panel is immunoblotted with a polyclonal rabbit anti-RecA antibody used as a loading control. (B) Mice were infected with the wild type (WT) or the ΔshiA or ShiA+ strain (n = 20 mice/group). Only ShiA+ strain-infected mice survived the infection (green line). Wild-type- and ΔshiA strain-infected animals succumbed to infection with the same kinetics (blue and red lines, respectively). An * denotes a statistical difference in survival between ShiA+ strain infection and wild-type or ΔshiA strain infection (P = 0.007 using the log-rank test). Similar results were obtained from two independent experiments.
Six hours postinfection, all mice had ruffled fur and a stooped posture, indicating sickness. Wild-type- and ΔshiA strain-infected animals appeared moribund until time of death. In contrast, ShiA+ strain-infected animals appeared healthy as early as 10 h postinfection and were very active for the duration of the experiment. There was no difference in the survival of animals infected with wild-type Shigella or ΔshiA strains (Fig. 1B). Both infections resulted in death on or before day 3. In contrast, ShiA+ strain-infected mice survived until the termination of the experiment.
ShiA+ strain-infected mice clear their bacterial load.
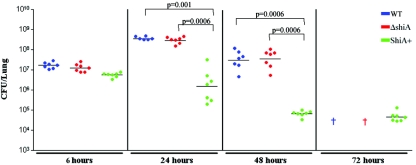
To determine if the differences in survival depended on bacterial clearance, we assessed the number of bacteria in the lung. Mice were infected with wild-type bacteria or ΔshiA or ShiA+ strains and sacrificed at 6, 24, 48, and 72 h. At 6 h, all mice had similar bacterial loads, regardless of the infecting strain, demonstrating that all strains can establish infection equally well (Fig. 2). This result correlated with our observations using the rabbit model (14). Bacterial counts in mice infected with wild-type Shigella or ΔshiA strains increased dramatically (1.5 logs) between 6 h and 24 h and then began to decrease only at 48 h (Fig. 2, blue [wild type] and red [ΔshiA] symbols). Similar to the survival experiments, all wild-type- and ΔshiA strain-infected animals died by 72 h. In contrast, in a ShiA+ strain infection, the number of CFU decreased by more than 2 logs from 6 h to 72 h (Fig. 2, green symbols). At 24 h, there was a further 2 log difference in bacterial counts between ShiA+ strain infected animals and either wild-type- or ΔshiA strain-infected animals. This difference increased to greater than 3 logs at 48 h. Differences between ShiA+ and wild-type strain CFU and ShiA+ and ΔshiA strain CFU are statistically significant at 24 h (wild type, P = 0.001; ΔshiA strain, P = 0.0006) and 48 h (P = 0.0006, both comparisons). The CFU data correlate with the survival data where wild-type- and ΔshiA strain-infected animals have high bacterial counts in their lungs and die while the ShiA+ strain-infected animals clear the infection and survive.
FIG. 2.
Only the ShiA+ strain is cleared from the lungs of infected mice. Mice were infected with the wild type (WT) or the ΔshiA or ShiA+ strain (n = 7 mice/group). To determine bacterial load, infected mouse lungs were homogenized and plated for CFU at the indicated times. A † indicates dead wild-type- and ΔshiA strain-infected mice at 72 h. An * denotes that ShiA+ strain CFU are statistically different from CFU from wild-type infection (P = 0.001) and ΔshiA strain infection at 24 h (P = 0.0006). A # denotes that ShiA+ strain CFU are statistically different from CFU from wild-type infection and ΔshiA strain infection at 48 h (P = 0.0006). Statistics were determined by Mann-Whitney nonparametric test. Similar results were obtained from three independent experiments with 7 to 10 mice per group.
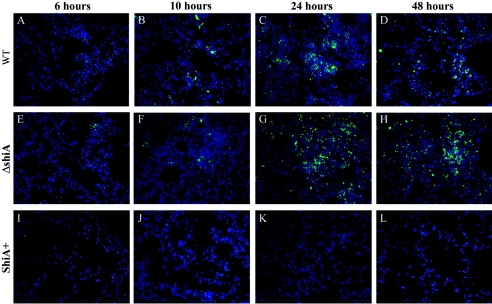
To determine the distribution of the bacterial load in infected animals, mice were infected with wild-type Shigella, ΔshiA, or ShiA+ strains and sacrificed at 6, 10, 24, and 48 h. Mouse lungs were immunostained with S. flexneri serotype 5-specific anti-LPS antibody. In all infections, very few bacteria could be seen at 6 h (Fig. 3A, E, and I). This is due to the low CFU relative to lung volume. Beginning at 10 h, wild-type and ΔshiA strain infections showed bacteria localized as both individual cells and aggregates in the lung (Fig. 3B to D and F to H). The aggregates were most apparent at 24 h (Fig. 3C and G) and decreased slightly at 48 h (Fig. 3D and H) in these two infections. These data correlate with the large increase and subsequent decrease in CFU at 24 and 48 h, respectively, in these infections (Fig. 2). In ShiA+ strain-infected tissue, we never observed bacterial aggregates (Fig. 3I to L), correlating with the lower CFU throughout this infection (Fig. 2).
FIG. 3.
ShiA+ strain-infected mice carry a reduced bacterial load in the lungs. Mice were infected with (A to D) wild-type (WT), (E to H) ΔshiA, and (I to L) ShiA+ strains. To visualize bacteria, infected lungs were removed at specified times and immunostained with an anti-LPS antibody specific for Shigella LPS serotype 5 (green). DNA was counterstained with Hoechst dye (blue). Similar results were obtained with duplicate lungs from two independent experiments.
ShiA decreases inflammation in the lung.
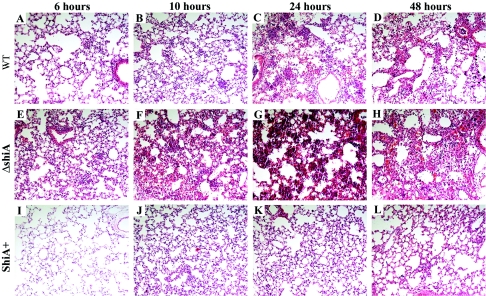
To determine the degree of inflammation in each infection, we infected mice with wild-type Shigella, ΔshiA, and ShiA+ strains. Animals were sacrificed at 6, 10, 24, and 48 h, and lungs were stained with hematoxylin and eosin. As expected, wild-type infection resulted in a strong inflammatory infiltrate and thickening of the alveolar walls at 6 and 10 h postinfection (Fig. 4A and B). At the 24-h time point, lungs presented a severe PMN infiltration with edema (Fig. 4C). Interestingly, inflammation decreased slightly at 48 h (Fig. 4D), and lungs showed signs of resolution. ΔshiA strain infection provoked the strongest inflammatory response (Fig. 4E to H). As early as 6 h, the lungs showed pronounced PMN infiltration and significant edema (Fig. 4E). The inflammatory infiltrate and edema increased over time, and the lung architecture was completely altered (Fig. 4E to H). As in wild-type infection, infection with the ΔshiA strain showed a slight decrease in the severity of inflammation at 48 h (Fig. 4H). The decrease in inflammation seen in the histology at 48 h in wild-type and ΔshiA strain infection correlates with the decrease in bacterial counts in these same infections at 48 h (Fig. 2). In contrast, ShiA+ strain infection induced very mild inflammation in the mouse lung over time (Fig. 4I to L). Lungs remained mostly clear and structurally intact, with only a few infiltrating PMN throughout the duration of the experiment.
FIG. 4.
ShiA attenuates Shigella-induced inflammation in the mouse lung. Mice were infected with (A to D) wild-type (WT), (E to H) ΔshiA, or (I to L) ShiA+ strains. To visualize inflammation, infected lungs were removed at specified times and hematoxylin and eosin stained. Similar results were obtained with duplicate lungs from two independent experiments.
Most significantly, at the 6-h time point, the amount of inflammation was different (Fig. 4A, E, and I) while the number of bacteria was the same in each infection (Fig. 2). Intensity of inflammation increased from minimal in the ShiA+ strain infection to moderate in the wild-type infection to severe in the ΔshiA strain infection. This observation suggests that it is the presence and amount of ShiA, early in infection, that determine the severity of inflammation in infection, rather than the bacterial load.
First, these data demonstrate that the ShiA+ strain is hypoinflammatory and that the ΔshiA strain is hyperinflammatory compared to wild-type infection in the mouse lung model, thereby corroborating our findings in the rabbit ileal loop model of infection that ShiA can down-regulate inflammation during infection. Second, these data demonstrate that ShiA exerts its effects early in infection to repress inflammation pathways, as differences in inflammation are observed as soon as 6 h postinfection.
Microarray analysis suggests that T cells are present in a Shigella infection.
Having determined the ShiA-associated phenotypes in the mouse model, we used microarray analysis of gene transcription to elucidate the host responses that are altered by ShiA during infection. The microarrays consisted of 8,014 sequences of cytokines, receptors, and other components of the immune response; unique coding sequences (expressed sequence tags); and housekeeping controls. We compared wild-type Shigella infection to ShiA+ strain infection using these custom-designed mouse microarrays. We chose to compare infections with these strains because the differences between wild-type and ShiA+ strain infection (survival, CFU, and histology) show greater contrast than those between wild-type and ΔshiA strain infections in the same assays. To exclusively identify genes differently regulated between the two infections, we chose comparative microarrays. With this technique, two samples are simultaneously applied to the same array, eliminating all genes that are regulated in the same manner during the two infections from our analysis.
In two independent experiments, we infected mice with wild-type Shigella or the ShiA+ strain and sacrificed animals at 6 and 10 h. At these time points, wild-type and ShiA+ strain infections showed histological differences, suggesting that ShiA had already acted to down-regulate inflammation mechanisms. We generated samples by pooling three mouse lung homogenates from the same time point, strain infection, and experiment. RNA was purified, amplified, labeled, and hybridized to the arrays as described in Materials and Methods. Each hybridization included equal amounts of differently labeled RNA from ShiA+ strain- and wild-type-infected lungs, resulting in a two-color array. All microarray data presented here are the ratio of wild-type infection to ShiA+ strain infection, where a positive number indicates a greater abundance of signal in the wild-type infection than in the ShiA+ strain infection under the same conditions.
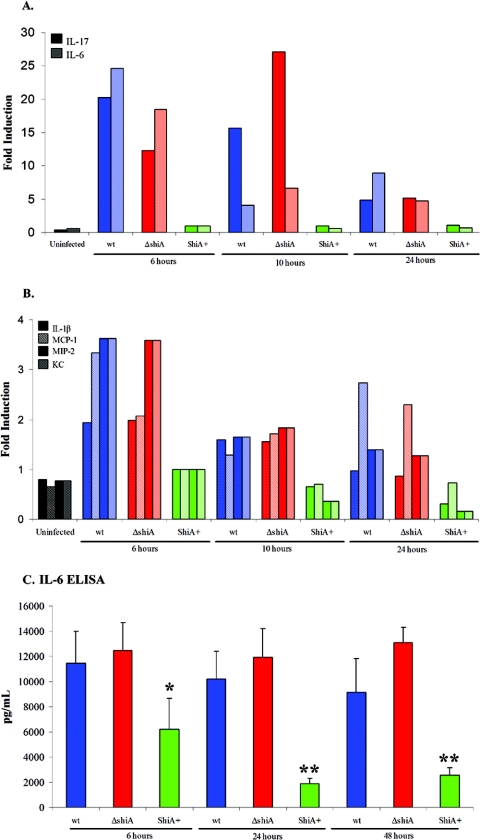
Using stringent analysis criteria (defined in Materials and Methods), we identified 27 genes at 6 h and 28 genes at 10 h that were differently regulated during infection (see Table S1 in the supplemental material). The total number of genes regulated at the two time points is greater than 46 because some genes showed regulation at both time points. We grouped significant genes, or genes that had an absolute change greater than twofold and a P value of less than 0.05, into the following categories: (i) inflammatory genes, with a subset having T-cell-associated function, and (ii) genes with functions that have not yet been implicated in inflammation. The identification of proinflammatory genes in these infections was expected. Shigella induces very strong inflammation in the host (36). However, some genes that we identified suggested that ShiA might down-regulate a T-cell-associated proinflammatory response. We inferred the presence of T cells based on the abundant expression of the T-cell-associated cytokines in wild-type infection compared to ShiA+ strain infection. Microarray analysis of whole organs is not sensitive to changes in small amounts of RNA or to changes in RNA from very small cell populations. Therefore, we interpreted the presence of T-cell cytokines to mean that either there was a great abundance of these cytokines being produced by T cells or there was a large number of T cells at the site of infection. Included in our data set are IL-17, secreted exclusively from T cells; IL-6 and MIP-2, which can be secreted from T cells and recruit PMN (24, 25, 28); and T-cell and PMN recruitment cytokines such as MIP-2, MCP-1, and interferon gamma-inducible 10-kDa protein (IP-10) (2, 4, 6, 24, 25, 29) (more information about this data set, including additional annotation and relevant literature citations, can be found in Table S1 in the supplemental material).
These data suggest that T cells may play an early, innate role in the response to Shigella infection. The fact that all of these genes are less abundantly expressed during ShiA+ strain infection suggests that ShiA may directly or indirectly dampen the T-cell response to infection.
Proinflammatory T-cell-associated effectors are more abundant in wild-type and ΔshiA strain infections.
We validated the microarray data and quantified the levels of expression using real-time PCR for samples from animals infected with wild-type Shigella, ShiA+, and ΔshiA strains. We tested the expression of IL-17, IL-6, MCP-1, IL-1β, MIP-2, and KC. MIP-2 and KC are mouse orthologs of human IL-8. Relative quantitation of gene expression was calculated with the comparative cycle threshold method using GAPDH for normalization.
In two independent experiments, we infected mice with wild-type Shigella or the ΔshiA or ShiA+ strain and sacrificed animals at 6, 10, and 24 h. cDNA was generated from total RNA pooled from three mice per group for each condition. At 6 h, IL-17 was induced 20-fold and 12-fold in wild-type- and ΔshiA strain-infected tissues, respectively, over ShiA+ strain-infected tissue (Fig. 5A). At 10 h, IL-17 increased 16-fold and 27-fold in wild-type- and ΔshiA strain-infected samples, respectively, over ShiA+ strain-infected samples. IL-17 levels dropped at 24 h to five- and sixfold in wild-type and ΔshiA strain infections, respectively, over ShiA+ strain infection levels. In ShiA+ strain infections, IL-17 remained at background, uninfected levels at all time points (Fig. 5A). IL-6 expression displayed a profile similar to that of IL-17 (Fig. 5A). At 6 h, IL-6 showed a substantial increase in induction (25-fold for the wild type and 18-fold for the ΔshiA strain) over ShiA+ strain-infected tissue levels (Fig. 5 A). IL-6 dropped approximately threefold at 10 and 24 h in wild-type and ΔshiA strain infections, compared to the 6-h level, although the expression level remained higher than that with ShiA+ infection at all times (Fig. 5A). In tissues infected with the ShiA+ strain, IL-6 remained at background, uninfected levels at all time points. Other proinflammatory cytokines such as MCP-1, IL-1β, MIP-2, and KC followed a similar trend but at lower levels of induction (Fig. 5 B). Similar to IL-17 and IL-6, the level of transcription of each cytokine tested was greater in tissues infected with wild-type Shigella and ΔshiA strains than in tissues infected with the ShiA+ strain (Fig. 5B). For all cytokines tested, the levels of transcription in ShiA+ strain-infected tissues was near the expression levels of uninfected tissue (Fig. 5A and B).
FIG. 5.
Wild-type- and ΔshiA strain-infected animals have increased levels of IL-17 and IL-6 mRNA. (A and B) Mice were infected with wild-type (wt), ΔshiA, and ShiA+ strains (n = 3 mice/group). To assess gene transcription levels, lungs were removed at 6, 10, and 24 h, homogenized in TRIzol, and pooled, and total RNA was extracted. RNA was reverse transcribed to cDNA, which served as the template for real-time PCR for (A) IL-17 and IL-6 and (B) IL-1β, MCP-1, MIP-2, and KC expression. Uninfected controls represent six mice. Similar results were obtained with triplicate lungs from two independent experiments. (C) Mice were infected with wild-type, ΔshiA, and ShiA+ strains (n = 4 mice/group). To assess IL-6 protein levels, lungs were removed at 6, 24, and 48 h and homogenized in PBS. Supernatants from homogenized lungs served as the sample for IL-6 ELISA. IL-6 levels from uninfected mouse lungs were at the limit of detection and are therefore not displayed on the graph. * denotes a statistically significant difference from wild-type or ΔshiA strain infection (P < 0.05). ** denotes a statistically significant difference from wild-type or ΔshiA strain infection (P < 0.005). Statistics were determined by unpaired t test. Similar results were obtained from three experiments.
To show conclusively that mRNA expression levels were reflective of the amount of protein in infection, we confirmed protein expression in vivo by ELISA of IL-6. We infected mice with wild-type Shigella or the ΔshiA or ShiA+ strain and sacrificed animals at 6, 24, and 48 h. At 6 h, IL-6 concentrations were 12,000 pg in wild-type- and ΔshiA strain-infected lungs (Fig. 5 C). Wild-type- and ΔshiA strain-infected tissue IL-6 levels maintained consistently high protein levels at 24 and 48 h. In contrast, in the ShiA+ strain-infected tissue, the IL-6 protein concentration peaked at 6 h at 6,000 pg, half that of wild-type and ΔshiA strain IL-6 levels. The IL-6 concentration decreased in ShiA+ strain-infected tissue at 24 h (1,900 pg) and 48 h (2,600 pg). In uninfected tissue, IL-6 concentrations were at the limit of detection. IL-6 protein levels in ShiA+ strain infection are statistically different from wild-type and ΔshiA strain infection levels at 6 h (P < 0.05) and at 24 and 48 h (P < 0.005).
Overall, microarray, real-time PCR, and ELISA analyses show a similar pattern of regulation for the cytokines we tested. The cellular sources of these cytokines include macrophages, fibroblasts, and epithelial, endothelial, and T cells, suggesting that ShiA induces a global down-regulation of inflammatory signals early in infection. Of particular note, we observed suppression of T-cell-associated cytokines during ShiA+ strain infection, suggesting both the presence of T cells during the acute phase of wild-type Shigella infection and that ShiA inflammatory down-regulation includes suppression of T-cell-regulated inflammation.
PMN infiltration and T-cell number increase in wild-type and ΔshiA strain infections.
Our microarray and real-time PCR data indicated that cytokines relevant to PMN recruitment were elevated in wild-type and ΔshiA infections and were expressed only at background, uninfected levels in ShiA+ strain infections. IL-17, IL-6, MIP-2, and KC induce PMN maturation and migration (18, 21, 44). In addition, to date, one study has addressed the involvement of T cells in the innate immune response to Shigella infection (20). Our expression data indicated that T-cell-associated cytokines are elevated in wild-type and ΔshiA strain infections. We reasoned that the high expression level of these cytokines observed in wild-type and ΔshiA strain infections would result in greater numbers of PMN and T cells at the site of infection. The converse would also apply, where PMN influx into ShiA+ strain-infected tissues should be less than that in a wild-type infection, and fewer T cells would be present. Therefore, we examined infected tissue to evaluate the impact of the cytokine expression levels on PMN and T-cell numbers at the site of infection.
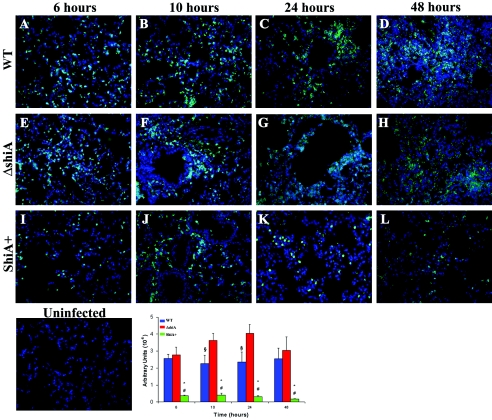
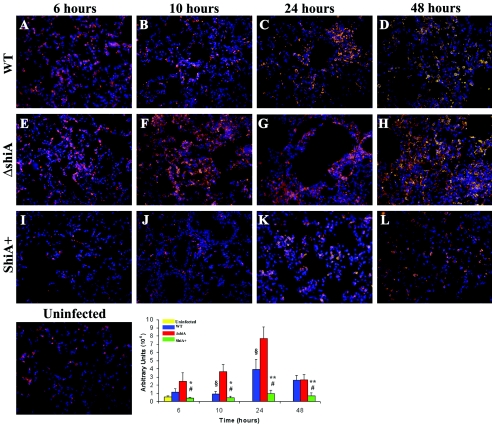
In two independent experiments, we infected mice with wild-type Shigella or the ΔshiA or ShiA+ strain and sacrificed animals at 6, 10, 24, and 48 h. Representative lung sections were stained with an antibody recognizing a PMN surface protein (Fig. 6) or an antibody against CD3, a pan-T-cell marker (Fig. 7). To quantify the amount of PMN migration or the number of T cells in the infected tissue, we digitized randomly selected stained sections from each time point and infection and assessed the overall level of intensity of the staining. Intensity is expressed as an arbitrary number, and the values from six sections from each infection and time point are represented in Fig. 6 (PMN) and Fig. 7 (T cells).
FIG. 6.
ShiA suppresses PMN infiltration into the lungs. Mice were infected with (A to D) wild-type (WT), (E to H) ΔshiA, and (I to L) ShiA+ strains. To visualize PMN, infected lungs were removed at specified times and immunostained with an antibody to a PMN surface marker (green). DNA was counterstained with Hoechst dye (blue). Uninfected controls showed no signal for PMN staining. Tissue section images (6 sections/condition) from mouse lungs infected for the indicated times were digitized and assessed for intensity of staining (arbitrary units) using ImageJ 1.33A. * denotes a statistically significant difference from wild-type infection (P < 0.001); # denotes a statistically significant difference from ΔshiA strain infection (P < 0.001); § denotes a statistically significant difference from ΔshiA strain infection (P < 0.05). Statistics were determined by unpaired t test. Similar results were obtained from two independent experiments.
FIG. 7.
ShiA suppresses the T-cell-number increase in the lungs. Mice were infected with (A to D) wild-type (WT), (E to H) ΔshiA, and (I to L) ShiA+ strains. To visualize T cells, infected lungs were removed at specified times and immunostained with anti-CD3, a pan-T-cell marker (orange). DNA was counterstained with Hoechst dye (blue). Uninfected controls reveal the resident T-cell population. Tissue section images (6 sections/condition) from mouse lungs infected for the indicated times were digitized and assessed for intensity of staining (arbitrary units) using ImageJ 1.33A. * and ** denote a statistically significant difference from wild-type infection (*P < 0.05; **P < 0.005); # denotes a statistically significant difference from ΔshiA strain infection (P < 0.001); § denotes a statistically significant difference from ΔshiA infection (P < 0.05). Statistics were determined by unpaired t test. Similar results were obtained from two independent experiments.
As expected, uninfected mouse lungs did not contain PMN (Fig. 6). In infected lungs, PMN infiltrated shortly after infection (6 h) with all strains (Fig. 6A, E, and I). In both wild-type and ΔshiA strain infections, there was greater PMN infiltration over time than in ShiA+ strain infection (Fig. 6A to D and E to H, respectively). These data confirm that ShiA is a suppressor of inflammation. The number of PMN remained at a constant high level in wild-type-infected tissues (Fig. 6A to D, graph). In ΔshiA strain-infected tissues, PMN infiltration increased steadily over the course of infection until 48 h, at which point it decreased (Fig. 6E to H, graph). The level of infiltration in ΔshiA strain-infected tissues was greater than that for the wild type at all time points, although significantly so only at 10 and 24 h (P < 0.05). Over the course of ShiA+ infection, very few PMN migrated into the tissue (Fig. 6I to L), and the level of PMN migration was statistically different from that of wild-type or ΔshiA strain infection (P < 0.001) at all time points. PMN infiltration correlates with the overall amount of inflammation seen in these three infections (Fig. 4).
In contrast to the PMN staining, but not unexpectedly, uninfected mouse lung sections had resident intraepithelial T cells (Fig. 7) (11). In wild-type and ΔshiA strain infections, the number of T cells was greater than that in ShiA+ infections as early as 6 h and increased in infected tissue over time (Fig. 7A to D and E to H, respectively). The number of T cells was greatest in tissues infected with the ΔshiA strain and is statistically different from that found in wild-type infection at 10 and 24 h (P < 0.05). The number of T cells in ShiA+ strain-infected tissue remained similar to that of uninfected tissue at all time points (Fig. 7I to L) and is statistically different from that found with a wild-type or ΔshiA strain infection (P < 0.05 for the wild type at 6 and 10 h; P < 0.005 for the wild type at 24 and 48 h; P < 0.001 for ΔshiA at 6, 10, 24, and 48 h). These results correlate with the increase in induction of IL-17 mRNA observed in the microarrays (see Table S1 in the supplemental material) and real-time PCR (Fig. 5A and B) in wild-type infection. Conversely, the lack of IL-17 in ShiA+ strain infection can be explained by the near lack of T cells in infected tissues. These results suggest that T cells could play a role in the innate immune response to Shigella infection and that the number of T cells in an infection is dependent on the amount of ShiA present. The number of T cells correlates with the amount of inflammation induced in infection, suggesting that the T cells positively influence inflammation.
DISCUSSION
ShiA induces global repression of inflammation.
ShiA has a profound effect on inflammation in Shigella infections. Using the rabbit ileal loop model, we showed that ShiA down-regulates inflammation. This down-regulation is independent of phenotypes encoded in the virulence plasmid, since invasion and cytotoxicity were not affected in shiA mutants (14). Analogously, in the mouse lung model, infection with the ShiA+ strain causes less tissue damage (Fig. 4), lower levels of proinflammatory cytokine expression (Fig. 5), and less PMN (Fig. 6) and T-cell (Fig. 7) accumulation than in wild-type infections. Accordingly, infection with the ΔshiA strain provokes more tissue damage (Fig. 4) and an increased number of PMN (Fig. 6) and T cells (Fig. 7) at the site of infection. These data indicate that both PMN and T-cell influx are down-regulated by ShiA. Whether ShiA affects the influx of these cells directly cannot be determined by this study. Importantly, though, the amount of ShiA, rather than the number of bacteria, directly regulates this mechanism (Fig. 1A). At 6 h postinfection, tissues infected by all strains carried similar bacterial loads (Fig. 2) but presented differences in inflammation (Fig. 4) as well as PMN (Fig. 6) and T-cell (Fig. 7) accumulation.
We used microarray analysis to determine which host genes were modulated by ShiA. Proinflammatory cytokines such as IL-6, IL-17, MCP-1, IL-1β, MIP-2, and KC were expressed at lower levels in ShiA+ strain infection than in wild-type infection (see Table S1 in the supplemental material). Indeed, real-time PCR showed similar cytokine expression levels in ShiA+ strain-infected tissue and uninfected tissue (Fig. 5A and B). Accordingly, ELISA (Fig. 5C) analysis showed that there was less IL-6 in ShiA+ strain infections than in wild-type or ΔshiA strain infections. Correlating with the lower cytokine expression, there were fewer PMN (Fig. 6) and T cells (Fig. 7) in ShiA+ strain-infected tissues. Finally, we observed that ShiA suppresses inflammation as early as 6 h, suggesting that ShiA acts early in infection.
Different cells, including monocytes, macrophages, fibroblasts, and epithelial, endothelial, and T cells, can make the cytokines we tested. This suggests that the cellular target of ShiA is likely to be upstream of the activation of these cell types. Hence, ShiA probably does not directly affect PMN or T cells, since IL-6, a cytokine expressed by many cell types, is almost absent from ShiA+ strain-infected tissues (Fig. 5A). This suggests that IL-6 is repressed before either PMN or T cells are involved. Furthermore, IL-1β expression, a cytokine released by macrophages (Fig. 5B; see Table S1 in the supplemental material), is regulated by ShiA, suggesting a suppression mechanism upstream of macrophage activation. In vitro infection of epithelial cells, macrophages, and PMN with the ShiA+ strain or wild-type Shigella results in similar cytokine release (data not shown), suggesting that ShiA acts on another cell type or on an unidentified mediator. The direct effect of ShiA on inflammation remains to be elucidated. Our microarray analysis showed the regulation of genes either with unknown function or not thought to be involved in inflammation. One or more of these genes may be a critical player in the early initiation of inflammation by ShiA and are under investigation.
We proposed that ShiA evolved to achieve an optimal level of inflammation. So why is the ShiA+ strain cleared from infected lungs in the absence of a significant PMN infiltrate? Shigella can only invade the basolateral side of the epithelium (27). The amplification of Shigella invasion depends on the disruption of the epithelium by PMN transmigration allowing greater access to the basolateral surface (33). We speculate that in ShiA+ strain infections, the suppression of inflammation and the decrease in PMN influx result in less disruption of the epithelium, and hence, the infection is not amplified. It is possible that resident phagocytes and the few recruited PMN are sufficient to clear the bacteria.
T cells play a role in the immune response to Shigella.
Inflammation in Shigella infections was thought to be initiated by macrophages and epithelial cells through IL-1β, IL-18, and IL-8. In this study, we show that T cells may also be involved in the innate immune response to Shigella infection.
T cells play a key role in the regulation of adaptive immunity. More recently, however, T cells have also been implicated in initiating innate inflammatory responses (7, 10). In patients infected with Shigella, specific T-cell subsets are significantly increased in the lamina propria compared to uninfected individuals (15). It is likely that this response is induced by activated macrophages and dendritic cells, as they can stimulate T cells to produce IL-17 in vitro (1, 32). IL-17 activates many cell types, including fibroblasts and epithelial and endothelial cells (51), to express tumor necrosis factor alpha, IL-1β, IL-8, KC, MIP-2, and other CC and CXC chemokines (3, 17, 18, 31, 52). These cytokines also stimulate PMN migration (49). We observed an increase in the number of T cells at the site of infection (Fig. 7) and detected increased levels of IL-17 and other T-cell cytokines in infected tissues (Fig. 5; see Table S1 in the supplemental material). The role of T cells in Shigella infections is also supported by the study of Le-Barillec et al. (20), who demonstrated that mice lacking T, B, and NK cells (RAG-2 and common γ-chain-deficient RAG2/γc mice) are more susceptible to infection. The phenotype is restored when the mice are reconstituted with αβ T cells (20). Taken together, these data indicate that T cells help regulate the innate immune response in Shigella infections.
What is the role of T cells in Shigella pathogenesis? We hypothesize that resident dendritic cells and macrophages recruit T cells early in infection. In turn, these T cells regulate PMN recruitment to the infection site. Dendritic cells sample antigens directly from the lumen of the intestine (30), and macrophages capture invading Shigella cells as they traverse M cells (33, 41, 48). Therefore, these cells are likely to be the first cells to encounter Shigella. We propose that activated dendritic cells and macrophages then stimulate T cells to produce cytokines such as IL-17 and IL-6. These cytokines would then act synergistically with IL-1β, IL-18, and IL-8 to recruit PMN to the site of infection. Our hypothesis is in agreement with the data described previously by Le-Barillec et al. (20). They demonstrate that in the absence of T cells, mice succumb to Shigella infection at day 4, compared to controls that survive the infection. PMN are abundant, however, at the site of infection, suggesting that T cells are necessary for defense against infection but that PMN recruitment is independent of T-cell function. Le-Barillec and colleagues evaluated PMN recruitment only at day 4, late in infection. It would be of interest to test whether there is a difference in PMN recruitment early in infection in the system described previously by Le-Barillec et al. It is possible that the timely recruitment of PMN, perhaps by T cells, is essential for survival. We observed an increase in T cells and PMN as early as 6 h postinfection. Alternatively, PMN recruitment may not be dependent on T cells, and the PMN deficiency observed during ShiA+ strain infection may be due to the repression of inflammatory signals from epithelial cells and macrophages. However, it is clear from the study described previously by Le-Barillec et al. and our study that T cells are required for an adequate innate immune response to Shigella infections. A comprehensive study to assess the kinetics of the influx of PMN and T cells would determine the role of T cells in Shigella infection as well as the consequences of PMN influx in the absence of T-cell cytokine signaling.
Supplementary Material
Acknowledgments
We thank Kathryn Stockbauer and Björn Albrecht for critical reading of the manuscript; Bärbel Raupach for many helpful suggestions, discussions, and support; Christian Goosman for technical support in staining histological sections; and Hans Mollenkopf for advice concerning microarray analysis.
Editor: J. B. Bliska
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aggarwal, S., N. Ghilardi, M. H. Xie, F. J. de Sauvage, and A. L. Gurney. 2003. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 278:1910-1914. [DOI] [PubMed] [Google Scholar]
- 2.Agostini, C., M. Facco, M. Siviero, D. Carollo, S. Galvan, A. M. Cattelan, R. Zambello, L. Trentin, and G. Semenzato. 2000. CXC chemokines IP-10 and mig expression and direct migration of pulmonary CD8+/CXCR3+ T cells in the lungs of patients with HIV infection and T-cell alveolitis. Am. J. Respir. Crit. Care Med. 162:1466-1473. [DOI] [PubMed] [Google Scholar]
- 3.Awane, M., P. G. Andres, D. J. Li, and H. C. Reinecker. 1999. NF-kappa B-inducing kinase is a common mediator of IL-17-, TNF-alpha-, and IL-1 beta-induced chemokine promoter activation in intestinal epithelial cells. J. Immunol. 162:5337-5344. [PubMed] [Google Scholar]
- 4.Cardona, A. E., P. A. Gonzalez, and J. M. Teale. 2003. CC chemokines mediate leukocyte trafficking into the central nervous system during murine neurocysticercosis: role of γδ T cells in amplification of the host immune response. Infect. Immun. 71:2634-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y., M. R. Smith, K. Thirumalai, and A. Zychlinsky. 1996. A bacterial invasin induces macrophage apoptosis by directly binding ICE. EMBO J. 15:3853-3860. [PMC free article] [PubMed] [Google Scholar]
- 6.Connor, S. J., N. Paraskevopoulos, R. Newman, N. Cuan, T. Hampartzoumian, A. R. Lloyd, and M. C. Grimm. 2004. CCR2 expressing CD4+ T lymphocytes are preferentially recruited to the ileum in Crohn's disease. Gut 53:1287-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, A., S. Lopez-Briones, H. Ong, C. O'Neil-Marshall, F. A. Lemonnier, K. Nagaraju, E. S. Metcalf, and M. J. Soloski. 2004. Infection-induced expansion of a MHC class Ib-dependent intestinal intraepithelial gammadelta T cell subset. J. Immunol. 172:6828-6837. [DOI] [PubMed] [Google Scholar]
- 8.Falkow, S., H. Schneider, L. S. Baron, and S. B. Formal. 1963. Virulence of Escherichia-Shigella genetic hybrids for the guinea pig. J. Bacteriol. 86:1251-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay, B. B., and S. Falkow. 1988. Comparison of the invasion strategies used by Salmonella cholerae-suis, Shigella flexneri and Yersinia enterocolitica to enter cultured animal cells: endosome acidification is not required for bacterial invasion or intracellular replication. Biochimie 70:1089-1099. [DOI] [PubMed] [Google Scholar]
- 10.Hayday, A., E. Theodoridis, E. Ramsburg, and J. Shires. 2001. Intraepithelial lymphocytes: exploring the third way in immunology. Nat. Immunol. 2:997-1003. [DOI] [PubMed] [Google Scholar]
- 11.Hayday, A., and R. Tigelaar. 2003. Immunoregulation in the tissues by gammadelta T cells. Nat. Rev. Immunol. 3:233-242. [DOI] [PubMed] [Google Scholar]
- 12.Hilbi, H., Y. Chen, K. Thirumalai, and A. Zychlinsky. 1997. The interleukin 1β-converting enzyme, caspase 1, is activated during Shigella flexneri-induced apoptosis in human monocyte-derived macrophages. Infect. Immun. 65:5165-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilbi, H., J. E. Moss, D. Hersh, Y. Chen, J. Arondel, S. Banerjee, R. A. Flavell, J. Yuan, P. J. Sansonetti, and A. Zychlinsky. 1998. Shigella-induced apoptosis is dependent on caspase-1 which binds to IpaB. J. Biol. Chem. 273:32895-32900. [DOI] [PubMed] [Google Scholar]
- 14.Ingersoll, M. A., J. E. Moss, Y. Weinrauch, P. E. Fisher, E. A. Groisman, and A. Zychlinsky. 2003. The ShiA protein encoded by the Shigella flexneri SHI-2 pathogenicity island attenuates inflammation. Cell. Microbiol. 5:797-807. [DOI] [PubMed] [Google Scholar]
- 15.Islam, D., and B. Christensson. 2000. Disease-dependent changes in T-cell populations in patients with shigellosis. APMIS 108:251-260. [DOI] [PubMed] [Google Scholar]
- 16.Jarry, A., M. Robaszkiewicz, N. Brousse, and F. Potet. 1989. Immune cells associated with M cells in the follicle-associated epithelium of Peyer's patches in the rat. Cell Tissue Res. 255:293-298. [DOI] [PubMed] [Google Scholar]
- 17.Jovanovic, D. V., J. A. Di Battista, J. Martel-Pelletier, F. C. Jolicoeur, Y. He, M. Zhang, F. Mineau, and J. P. Pelletier. 1998. IL-17 stimulates the production and expression of proinflammatory cytokines, IL-beta and TNF-alpha, by human macrophages. J. Immunol. 160:3513-3521. [PubMed] [Google Scholar]
- 18.Kolls, J. K., and A. Linden. 2004. Interleukin-17 family members and inflammation. Immunity 21:467-476. [DOI] [PubMed] [Google Scholar]
- 19.Kotloff, K. L., J. P. Winickoff, B. Ivanoff, J. D. Clemens, D. L. Swerdlow, P. J. Sansonetti, G. K. Adak, and M. M. Levine. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. W. H. O. 77:651-666. [PMC free article] [PubMed] [Google Scholar]
- 20.Le-Barillec, K., J. G. Magalhaes, E. Corcuff, A. Thuizat, P. J. Sansonetti, A. Phalipon, and J. P. Di Santo. 2005. Roles for T and NK cells in the innate immune response to Shigella flexneri. J. Immunol. 175:1735-1740. [DOI] [PubMed] [Google Scholar]
- 21.Linden, A. 2001. Role of interleukin-17 and the neutrophil in asthma. Int. Arch. Allergy Immunol. 126:179-184. [DOI] [PubMed] [Google Scholar]
- 22.Mandic-Mulec, I., J. Weiss, and A. Zychlinsky. 1997. Shigella flexneri is trapped in polymorphonuclear leukocyte vacuoles and efficiently killed. Infect. Immun. 65:110-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurelli, A. T., and P. J. Sansonetti. 1988. Genetic determinants of Shigella pathogenicity. Annu. Rev. Microbiol. 42:127-150. [DOI] [PubMed] [Google Scholar]
- 24.McLoughlin, R. M., B. J. Jenkins, D. Grail, A. S. Williams, C. A. Fielding, C. R. Parker, M. Ernst, N. Topley, and S. A. Jones. 2005. IL-6 trans-signaling via STAT3 directs T cell infiltration in acute inflammation. Proc. Natl. Acad. Sci. USA 102:9589-9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto, M., O. Prause, M. Sjostrand, M. Laan, J. Lotvall, and A. Linden. 2003. Endogenous IL-17 as a mediator of neutrophil recruitment caused by endotoxin exposure in mouse airways. J. Immunol. 170:4665-4672. [DOI] [PubMed] [Google Scholar]
- 26.Moss, J. E., T. J. Cardozo, A. Zychlinsky, and E. A. Groisman. 1999. The selC-associated SHI-2 pathogenicity island of Shigella flexneri. Mol. Microbiol. 33:74-83. [DOI] [PubMed] [Google Scholar]
- 27.Mounier, J., T. Vasselon, R. Hellio, M. Lesourd, and P. J. Sansonetti. 1992. Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect. Immun. 60:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller, K., S. Bischof, F. Sommer, M. Lohoff, W. Solbach, and T. Laskay. 2003. Differential production of macrophage inflammatory protein 1γ (MIP-1γ), lymphotactin, and MIP-2 by CD4+ Th subsets polarized in vitro and in vivo. Infect. Immun. 71:6178-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.New, J. Y., B. Li, W. P. Koh, H. K. Ng, S. Y. Tan, E. H. Yap, S. H. Chan, and H. Z. Hu. 2002. T cell infiltration and chemokine expression: relevance to the disease localization in murine graft-versus-host disease. Bone Marrow Transplant. 29:979-986. [DOI] [PubMed] [Google Scholar]
- 30.Niess, J. H., S. Brand, X. Gu, L. Landsman, S. Jung, B. A. McCormick, J. M. Vyas, M. Boes, H. L. Ploegh, J. G. Fox, D. R. Littman, and H. C. Reinecker. 2005. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307:254-258. [DOI] [PubMed] [Google Scholar]
- 31.Numasaki, M., M. T. Lotze, and H. Sasaki. 2004. Interleukin-17 augments tumor necrosis factor-alpha-induced elaboration of proangiogenic factors from fibroblasts. Immunol. Lett. 93:39-43. [DOI] [PubMed] [Google Scholar]
- 32.Oppmann, B., R. Lesley, B. Blom, J. C. Timans, Y. Xu, B. Hunte, F. Vega, N. Yu, J. Wang, K. Singh, F. Zonin, E. Vaisberg, T. Churakova, M. Liu, D. Gorman, J. Wagner, S. Zurawski, Y. Liu, J. S. Abrams, K. W. Moore, D. Rennick, R. de Waal-Malefyt, C. Hannum, J. F. Bazan, and R. A. Kastelein. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 13:715-725. [DOI] [PubMed] [Google Scholar]
- 33.Perdomo, J. J., P. Gounon, and P. J. Sansonetti. 1994. Polymorphonuclear leukocyte transmigration promotes invasion of colonic epithelial monolayer by Shigella flexneri. J. Clin. Investig. 93:633-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perdomo, O. J., J. M. Cavaillon, M. Huerre, H. Ohayon, P. Gounon, and P. J. Sansonetti. 1994. Acute inflammation causes epithelial invasion and mucosal destruction in experimental shigellosis. J. Exp. Med. 180:1307-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sansonetti, P., A. Phalipon, J. Arondel, K. Thirumalai, S. Banerjee, S. Akira, K. Takeda, and A. Zychlinsky. 2000. Caspase-1 activation of IL-1β and IL-18 are essential for Shigella flexneri induced inflammation. Immunity 12:581-590. [DOI] [PubMed] [Google Scholar]
- 36.Sansonetti, P. J. 1992. Molecular and cellular biology of Shigella flexneri invasiveness: from cell assay systems to shigellosis. Curr. Top. Microbiol. Immunol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 37.Sansonetti, P. J., J. Arondel, J. M. Cavaillon, and M. Huerre. 1995. Role of interleukin-1 in the pathogenesis of experimental shigellosis. J. Clin. Investig. 96:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sansonetti, P. J., J. Arondel, M. Huerre, A. Harada, and K. Matsushima. 1999. Interleukin-8 controls bacterial transepithelial translocation at the cost of epithelial destruction in experimental shigellosis. Infect. Immun. 67:1471-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sansonetti, P. J., T. L. Hale, G. J. Dammin, C. Kapfer, H. H. Collins, and S. B. Formal. 1983. Alterations in the pathogenicity of Escherichia coli K-12 after transfer of plasmid and chromosomal genes from Shigella flexneri. Infect. Immun. 39:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sansonetti, P. J., and A. Phalipon. 1999. M cells as ports of entry for enteroinvasive pathogens: mechanisms of interaction, consequences for the disease process. Semin. Immunol. 11:193-203. [DOI] [PubMed] [Google Scholar]
- 42.Soesatyo, M., J. Biewenga, G. Kraal, and T. Sminia. 1990. The localization of macrophage subsets and dendritic cells in the gastrointestinal tract of the mouse with special reference to the presence of high endothelial venules. An immuno- and enzyme-histochemical study. Cell Tissue Res. 259:587-593. [DOI] [PubMed] [Google Scholar]
- 43.Thirumalai, K., K.-S. Kim, and A. Zychlinsky. 1997. IpaB, a Shigella flexneri invasin, colocalizes with interleukin-1β-converting enzyme in the cytoplasm of macrophages. Infect. Immun. 65:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Damme, J., A. Wuyts, G. Froyen, E. Van Coillie, S. Struyf, A. Billiau, P. Proost, J. M. Wang, and G. Opdenakker. 1997. Granulocyte chemotactic protein-2 and related CXC chemokines: from gene regulation to receptor usage. J. Leukoc. Biol. 62:563-569. [DOI] [PubMed] [Google Scholar]
- 45.van de Verg, L. L., C. P. Mallet, H. H. Collins, T. Larsen, C. Hammack, and T. L. Hale. 1995. Antibody and cytokine responses in a mouse pulmonary model of Shigella flexneri serotype 2a infection. Infect. Immun. 63:1947-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voino-Yasenetsky, M. V., and M. K. Voino-Yasenetska. 1961. Experimental pneumonia caused by bacteria of the Shigella group. Acta Morphol. X:440-454. [PubMed] [Google Scholar]
- 47.Vokes, S. A., S. A. Reeves, A. G. Torres, and S. M. Payne. 1999. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol. Microbiol. 33:63-73. [DOI] [PubMed] [Google Scholar]
- 48.Wassef, J. S., D. F. Keren, and J. L. Mailloux. 1989. Role of M cells in initial antigen uptake and in ulcer formation in rabbit intestinal loop model of shigellosis. Infect. Immun. 57:858-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Witowski, J., K. Pawlaczyk, A. Breborowicz, A. Scheuren, M. Kuzlan-Pawlaczyk, J. Wisniewska, A. Polubinska, H. Friess, G. M. Gahl, U. Frei, and A. Jorres. 2000. IL-17 stimulates intraperitoneal neutrophil infiltration through the release of GRO alpha chemokine from mesothelial cells. J. Immunol. 165:5814-5821. [DOI] [PubMed] [Google Scholar]
- 50.World Health Organization. 2003. State of the art of new vaccines: research and development. [Online.] http://www.who.int/vaccine_research/documents/stateoftheart/en/.
- 51.Yao, Z., M. K. Spriggs, J. M. Derry, L. Strockbine, L. S. Park, T. VandenBos, J. D. Zappone, S. L. Painter, and R. J. Armitage. 1997. Molecular characterization of the human interleukin (IL)-17 receptor. Cytokine 9:794-800. [DOI] [PubMed] [Google Scholar]
- 52.Ye, P., F. H. Rodriguez, S. Kanaly, K. L. Stocking, J. Schurr, P. Schwarzenberger, P. Oliver, W. Huang, P. Zhang, J. Zhang, J. E. Shellito, G. J. Bagby, S. Nelson, K. Charrier, J. J. Peschon, and J. K. Kolls. 2001. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J. Exp. Med. 194:519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.