Abstract
In the heat shock response of bacillary cells, HrcA repressor proteins negatively control the expression of the major heat shock genes, the groE and dnaK operons, by binding the CIRCE (controlling inverted repeat of chaperone expression) element. Studies on two critical but yet unresolved issues related to the structure and function of HrcA were performed using mainly the HrcA from the obligate thermophile Bacillus thermoglucosidasius KP1006. These two critical issues are (i) identifying the region at which HrcA binds to the CIRCE element and (ii) determining whether HrcA can play the role of a thermosensor. We identified the position of a helix-turn-helix (HTH) motif in B. thermoglucosidasius HrcA, which is typical of DNA-binding proteins, and indicated that two residues in the HTH motif are crucial for the binding of HrcA to the CIRCE element. Furthermore, we compared the thermostabilities of the HrcA-CIRCE complexes derived from Bacillus subtilis and B. thermoglucosidasius, which grow at vastly different ranges of temperature. The thermostability profiles of their HrcA-CIRCE complexes were quite consistent with the difference in the growth temperatures of B. thermoglucosidasius and B. subtilis and, thus, suggested that HrcA can function as a thermosensor to detect temperature changes in cells.
In the heat shock response of Bacillus subtilis, HrcA repressor proteins negatively control the expression of the major heat shock genes, the groE and dnaK operons, by binding the CIRCE (controlling inverted repeat of chaperone expression) element (9, 17). Studies of HrcA and hrcA, its gene in various microorganisms, have clarified that HrcA molecules have similar features, e.g., the primary sequences of HrcA are strikingly conserved among organisms; based on phylogenetic analysis, this characteristic has evolutionary implications (1, 22, 33). On the other hand, common and unfavorable characteristics are that HrcA is hardly soluble and easily forms aggregates. This disadvantage has prevented the clarification of the real nature of HrcA. To circumvent this disadvantage, it was demonstrated in a previous report that HrcA from the obligate thermophile Bacillus thermoglucosidasius KP1006 was most efficiently renatured by the addition of DNA, including the CIRCE element, and that the HrcA-CIRCE complex was stable in the soluble form in the absence of GroEL (29).
Two critical issues related to the structure and function of HrcA are outstanding. One is the identification of the region at which HrcA binds to the CIRCE element. Although three conserved regions have been previously pointed out (22, 33), no extended analysis of the DNA-binding region of HrcA has been conducted. The other issue is whether HrcA can play the role of a thermosensor. When bacillary cells are exposed to heat stress, the expression of genes controlled by the HrcA-CIRCE complex is turned on. This implies that the HrcA-CIRCE complex must be decomposed by heat stress to release the regulation system. However, no direct evidence that the HrcA-CIRCE complex is decomposed by heat stress, thus behaving as a thermosensor, has been presented. Because of the interest in characterizing the behavior of the HrcA-CIRCE complex under heat stress, we decided to address the thermostability of the HrcA-CIRCE complex.
To examine the first issue stated above, we searched for the position of a helix-turn-helix (HTH) motif in B. thermoglucosidasius HrcA, which is typical of DNA-binding proteins (30). In addition, to confirm the fact that the putative motif functions, we investigated the effect of mutations that altered the conserved basic amino acids in the helices of the motif on the DNA-binding activity of B. thermoglucosidasius HrcA to the CIRCE element. To examine the second issue, we compared the thermostabilities of the HrcA-CIRCE complexes derived from B. subtilis and B. thermoglucosidasius, which grow at vastly different ranges of temperature. We then investigated how the decomposition of the HrcA-CIRCE complex depends on temperature.
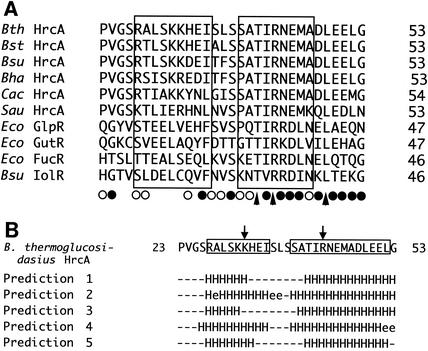
The primary sequence of B. thermoglucosidasius HrcA was compared with those of other microbial DNA-binding proteins by using BLAST software and taking into account the results of five secondary-structure predictions for B. thermoglucosidasius HrcA. As a result, the HTH DNA-binding region of a repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12 (GlpR) was found to have a distinctive homology, particularly in the second helix (the so-called recognition helix), to the amino-terminal region of HrcA (Fig. 1A) (34). GlpR, belonging to the repressor family of carbohydrate catabolic systems, possesses the DNA-binding motif near the amino terminus (residues 22 to 41) (34). In addition, other members of the family, the repressors GutR (glucitol operon, E. coli) (31), FucR (fucose operon, E. coli) (13), and IolR (inositol utilization operon, B. subtilis) (32), showed similar homologies in the recognition helix (Fig. 1A). The second helix of the HTH DNA-binding motif is highly conserved among the repressor family of carbohydrate catabolic systems and the HrcA family (Fig. 1A). The third to fifth positions (Thr-Ile-Arg) in the tentative recognition helix are all conserved among the repressors listed in Fig. 1A, except for the fifth position in the helix of B. subtilis IolR. Furthermore, Gln, Arg, or Lys, containing the amide or amino group in the side chain, occurs at the sixth position, and acidic amino acid (Glu or Asp) residues occur at the seventh position. There is also a significant homology in the residues following the seventh position. By contrast, there is very little conservation within the first helix of the HTH DNA-binding motif, not only among the repressor family of carbohydrate catabolic systems but also among the HrcA family.
FIG. 1.
(A) Alignment of amino acid sequences of various HrcAs with the repressors of carbohydrate catabolic systems. The symbols indicate identical or conserved residues (▴), conserved substitutions (•), and semiconserved substitutions (○). HrcAs are from B. thermoglucosidasius (Bth) (29), Bacillus stearothermophilus (Bst) (16), B. subtilis (Bsu) (22), Bacillus halodurans (Bha) (26), Clostridium acetobutylicum (Cac) (18), and Staphylococcus aureus (Sau) (2). Eco GlpR, Eco GutR, Eco FucR, and Bsu IolR are the repressors of the E. coli glp (34), E. coli gut (31), E. coli fuc (13), and B. subtilis iol (32) genes, respectively. Boxes indicate the tentative helices assigned based on the HTH of E. coli GlpR. Numbers represent amino acid residue positions. (B) Different secondary-structure predictions for B. thermoglucosidasius HrcA. The software used for secondary predictions was DSC (12) for prediction 1, GORIII (8) for prediction 2, PHD (20) for prediction 3, Predator (7) for prediction 4, and Jnet alignment prediction (6) for prediction 5. The symbols for the results from these predictions are as follows: H, α helix; e, strand; and −, others. The putative helices presumed in this study are indicated with boxes. The two mutated residues (Lys31 and Arg43) are shown with arrows.
However, three residues of the loop between two helices, which were identified as a turn in E. coli GlpR, are well conserved among the repressors listed in Fig. 1A. Serine or threonine occurs primarily at the first and third positions of the turn, and hydrophobic residues appear at the second position. This homology provides strong evidence for the idea that HrcA molecules have a turn structure at the position equivalent to that of the turn in the repressor family of carbohydrate catabolic systems. For additional evidence, the results from five types of secondary-structure predictions for B. thermoglucosidasius HrcA between residues 23 and 53 are shown in Fig. 1B. There are significantly common predictions for helices between residues 27 and 33 and residues 40 and 52. Both regions are located in the positions equivalent to two helices of the HTH DNA-binding motif of E. coli GlpR (Fig. 1A). The predictions for the boundaries between helices and loops are generally ambiguous due to the low accuracy of prediction (20). Therefore, there is a strong possibility that two helices correspond to the HTH motif between residues 27 and 35 and between residues 39 and 52. Furthermore, in many of the HTH-containing protein structures, a third helix flanking the HTH motif presumably exists to stabilize the motif as a compact globular domain (10). Such a stabilizing helix may occur between residues 67 and 77 of B. thermoglucosidasius HrcA, with a high probability of secondary-structure predictions (data not shown). Conclusively, the data in Fig. 1 suggest that the HrcA family should have the HTH motif between residues 27 and 52 of the N-terminal region, which can function in DNA binding.
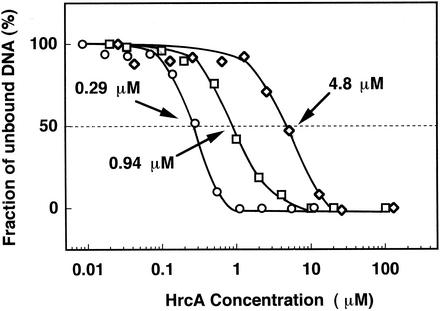
Based on the identification of the HTH motif, we constructed two mutant B. thermoglucosidasius HrcA proteins having single site-directed mutations on conserved or basic amino acid residues (Lys31 or Arg43) in two helices of the putative HTH motif and investigated the band shift mobility of the mutant HrcAs with the CIRCE element. Lys31 in the first helix and Arg43 in the second helix, which were presumed to interact with DNA, were replaced with Pro and Asp, respectively. The mutation of Lys31 to Pro was designed to break the helix in the middle by utilizing the structurally specific nature of proline (28), and that of Arg43 to Asp was designed to change the basic side chain to the acidic one, which failed to bind the DNA. Site-directed mutagenesis was carried out by using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) and the plasmid pETTY as a template (29). The pairs of primers used for each mutagenesis were GCGTTGTCACCAAAACATGAAATTTC (the degenerated bases are underlined) and GAAATTTCATGTTTTGGTGACAACGC for the mutant HrcA containing the Lys31→Pro mutation (designated K31P) and GGCGACGATCGACAATGAATG and CATTTCATTGTCGATCGTCGCC for the mutant HrcA containing the Arg43→Asp mutation (designated R43D). The mutagenesis protocol, including PCR by Pfu Turbo DNA polymerase, was performed according to the manufacturer's specifications. The resulting mutations were verified by sequencing. K31P and R43D mutant B. thermoglucosidasius HrcA proteins were overexpressed in the host E. coli BL21(DE3) [F− ompT hsdSB (rB− mB−) gal dem (DE3)] and purified to homogeneity by metal affinity chromatography (Clontech, Palo Alto, Calif.) under denaturing conditions with 6 M urea. The method was essentially the same as that for the wild-type B. thermoglucosidasius HrcA (29). The behaviors of mutant HrcA proteins in the purification processes were indistinguishable from that of the wild-type HrcA protein. However, the renaturation efficiency of mutant HrcAs by nonspecific DNA was rather low. Therefore, we compared the band shift abilities of wild-type and mutant B. thermoglucosidasius HrcA repressors for the DNA fragment containing the CIRCE element. Protein samples for band shift assays with the CIRCE element were prepared in the presence of 6 M urea. A 330-bp DNA fragment carrying the region (containing the CIRCE element) upstream of the start codon of B. thermoglucosidasius hrcA was prepared by PCR as described previously (29). The mixture for the binding-assay reactions contained a binding buffer (20 mM Tris-HCl, 5 mM EDTA, 2 mM MgCl2 [pH 7.5]), 0.75 μg of poly(dI-dC) · poly(dI-dC) (Amersham Pharmacia Biotech, Inc.), and 0.17 ng of the labeled probe in a volume of 15 μl. The HrcA concentration was varied from 0.01 to 100 nM. Binding was performed for 10 min at room temperature. Polyacrylamide gel electrophoresis and quantification of free and bound DNA were carried out by the same methods described previously (29). Kds were estimated to be the protein concentration at which 50% of the target DNA remained free. Figure 2 shows the difference in affinities of the wild-type and mutant HrcAs to the DNA fragment containing the CIRCE element. The data from these experiments were used to construct binding curves and the apparent equilibrium dissociation constants (Kds) of the HrcA-CIRCE complex. The results indicated that the Kds for the wild-type and two mutant HrcAs (mutants K31P and R43D) were 0.29, 0.94, and 4.8 μM, respectively. The apparent increase in Kds indicated that the in vitro affinity of HrcA to the CIRCE element was reduced 3.2-fold by the Lys31→Pro mutation in the first helix of the HTH motif but was reduced 16.6-fold by the Arg43→Asp mutation in the second helix. This suggests that two residues, Lys31 and Arg43, in two helices of the HTH motif play significant roles in the binding of the CIRCE element to the HTH motif. In addition, the difference in the changes in Kds reflected that the recognition helix is conserved more highly than the preceding helix; therefore, the recognition helix is more critical than the preceding helix in the binding of the HrcA molecule to the CIRCE element. In a previous study, Arg38 of E. coli GlpR, which is equivalent to Asn44 of B. thermoglucosidasius HrcA, was determined to be significant in recognizing the DNA (34).
FIG. 2.
Binding curve of wild-type and mutant (K31P and R43D) B. thermoglucosidasius HrcA proteins to the CIRCE element-containing probe. The curves indicating the results for wild-type HrcA (○), K31P mutant HrcA (□), and R43D mutant HrcA (⋄) are shown. The protein concentration at which 50% of the target DNA remained free for the apparent equilibrium dissociation constants of the HrcA-CIRCE complex is also shown (dotted line).
To compare the thermostability of the B. subtilis HrcA-CIRCE complex with that of the B. thermoglucosidasius HrcA-CIRCE complex, the B. subtilis hrcA gene was cloned from strain IFO13719 (ATCC 6051) by PCR. PCR was performed with LA Taq DNA polymerase (Takara Shuzo, Kyoto, Japan) for 30 cycles by using a program of denaturation (94°C, 1 min), annealing (55°C, 1 min), and extension (72°C, 2 min). Two primers, BSF (CCTGGATCCGATGTTAACAAATCGTCAG) and BSR (CCTGGATCCAAAGCGTAAGGAGAAGG), were used to amplify the DNA fragment containing the hrcA gene with the B. subtilis genomic DNA as the template. The primers corresponded to the nucleotide sequence of the B. subtilis hrcA gene reported previously (22, 33) and included the BamHI sites at the 5′ termini. The amplified DNA fragment was digested with BamHI and cloned into a BamHI site of the His-tagging vector pET15-b (Novagen, Madison, Wis.). With the use of the recombinant plasmid containing the cloned gene, His-tagged B. subtilis HrcA was expressed in E. coli BL21(DE3) and purified to homogeneity by the method described previously for B. thermoglucosidasius HrcA (29). The protein concentration was assessed by the Bradford method (3). The addition of nonspecific DNA, such as plasmid pUC119, to B. subtilis HrcA was scarcely effective in renaturing the HrcA molecule in the absence of urea, whereas plasmid pTY-1 containing the CIRCE element renatured HrcA satisfactorily. In the renaturation experiment, a quick method was used as follows. His-tagged B. subtilis HrcA (100 μg) dissolved in 20 mM Tris-HCl (pH 7.5)-6 M urea was added dropwise with vigorous stirring to 25 μg of pTY-1 in 1 ml of 20 mM Tris-HCl (pH 7.5). The obtained mixture was ultracentrifuged at 100,000 × g for 30 min, and the supernatant was then dialyzed extensively against 20 mM Tris-HCl (pH 7.5). B. subtilis HrcA solubilized with pTY-1 was as stable as B. thermoglucosidasius HrcA. Since B. thermoglucosidasius HrcA was renatured by the addition of specific and nonspecific DNA, this was determined to be the definitive difference between B. thermoglucosidasius and B. subtilis HrcAs. The difference proved that B. subtilis HrcA depends more on the CIRCE element for the soluble form. This characteristic is due to the fact that mesophilic B. subtilis HrcA is less stable than thermophilic B. thermoglucosidasius HrcA.
Light-scattering analysis for the renaturation of HrcAs with DNA was carried out with a Shimadzu RF5300PC fluorescence spectrophotometer equipped with a temperature-controlled cuvette holder and a magnetic stirrer. The setting conditions for the analysis were the same as those described in a previous report (29). Plasmid pTY-1 was prepared for this analysis by ultracentrifugation to equilibrium in cesium chloride-ethidium bromide density gradients (21).
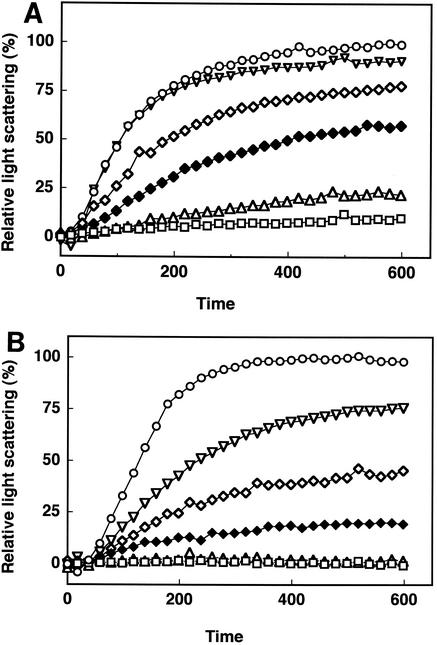
B. subtilis HrcA renatured with the plasmid was incubated in a cuvette maintained at various temperatures between 20 and 70°C (Fig. 3A). The complex of B. subtilis HrcA and pTY-1 was found to be significantly more denatured at temperatures above 40°C and far more thermolabile than B. thermoglucosidasius HrcA. At temperatures above 60°C, the complex was denatured rapidly, as in the absence of plasmid pTY-1 (data not shown). This temperature-dependent feature of B. subtilis HrcA in denaturation coincided with the observation that the heat induction of the HrcA-regulated genes took place at 48°C in B. subtilis (22). By contrast, the complex of B. thermoglucosidasius HrcA and pTY-1 was stable at temperatures up to 50°C but denatured at temperatures above 60°C, as shown in Fig. 3B. It was obvious that the complex of B. thermoglucosidasius HrcA and pTY-1 was much more thermostable than that of B. subtilis HrcA and pTY-1. However, this denaturation pattern shows that the complex of B. thermoglucosidasius HrcA and pTY-1 is rather thermally sensitive, compared with that of an enzyme protein, oligo-1,6-glucosidase, of the same cells (27). This thermal susceptibility of the HrcA-DNA complex seems to imply some physiological purpose. B. thermoglucosidasius grows at a higher range of temperatures (42 to 69°C) than B. subtilis (10 to 50°C), and the difference in growth temperatures satisfactorily reflects the results of light-scattering analysis at various temperatures. Interestingly, there is no difference in the nucleotide sequences of the stem regions of the CIRCE element in B. subtilis and B. thermoglucosidasius. Therefore, the difference in the degrees of temperature that are perceived as heat shock probably arises from the difference in thermal sensitivities between B. subtilis and B. thermoglucosidasius HrcA molecules. B. thermoglucosidasius HrcA consists of 344 amino acids and shares 63.1% identity with B. subtilis HrcA. The true mechanism for heat denaturation is probably hidden in HrcA molecules complexed with the CIRCE element. Studies of other repressors from thermophilic microorganisms have been reported (4, 11, 24). The thermostabilities of the DNA-binding protein HU from mesophilic, thermophilic, and extremely thermophilic bacteria were compared, and Christodoulou and Vorgias concluded that local interactions at the HTH region are responsible for the thermostabilities of HU proteins (4). However, in the case of HrcA, the primary sequence of its HTH region was highly conserved; therefore, it is difficult to attribute the difference in thermostabilities solely to the difference in the HTH structures. We have presumed that the difference in the thermostabilities of HrcAs is derived from other factors accumulated from the overall structures. Although the increase of proline residues can often contribute to protein thermostabilization (28), there was no increase in the proline content in B. thermoglucosidasius HrcA as there was in that of B. subtilis HrcA.
FIG. 3.
Thermal denaturation profiles of B. subtilis (A) and B. thermoglucosidasius (B) HrcA by light-scattering assays in the presence of the plasmid pTY-1 containing the CIRCE element. Twenty-five micrograms of B. thermoglucosidasius HrcA or B. subtilis HrcA in 50 μl of 20 mM Tris-HCl-5 mM EDTA-6 M urea (pH 7.5) was added to the solution (2.95 ml of 20 mM Tris-HCl-5 mM EDTA [pH 7.5]) at temperatures of 20 (□), 30 (▵), 40 (♦), 50 (⋄), 60 (▿), and 70°C (○).
GroEL had no effect on the denaturation of the HrcA-CIRCE complex in the light-scattering assay described in a previous report (29). Although the participation of the GroE chaperone machine as a major modulator in the regulation system with HrcA has been well documented (15), the involvement of GroEL in the release of HrcA from the CIRCE element as a result of heat stress has not been demonstrated. In light of the GroE function, the cooperation of GroE is reasonable due to the common characteristics of HrcA, i.e., HrcA is hardly soluble and easily forms aggregates in the absence of DNA (15, 17). Moreover, it has been reported that nonnative proteins expressed in cells lead to the induction of CIRCE-regulated genes due to the essential decrease of the GroEL molecules interacting with HrcA (14, 17). Therefore, there is no reason to exclude the possibility of a titration model in which the amount of active HrcA that is able to bind to the CIRCE element is directly correlated to the amount of available GroE chaperone molecules (17, 19). However, once HrcA molecules tightly bind to the CIRCE element with the aid of the GroE machinery in unstressed cells, the HrcA-CIRCE complex will not necessarily require the assistance of GroE chaperones to maintain itself in a cell. This is because the complex was shown to be stably intact in the absence of GroE molecules in vitro and the thermostability of the complex was indifferent whether in the presence or absence of GroEL (29). Therefore, it is more likely that the HrcA-CIRCE complex stays on chromosomal DNA to negatively regulate the heat shock genes, preparing to sense heat shock. On the other hand, the regeneration of the regulation system after the release of the HrcA from the CIRCE element by heat shock probably depends on the GroE concentration. Nevertheless, the thermal denaturation profiles of the HrcA-CIRCE complexes were very consistent with the growth temperatures of B. thermoglucosidasius and B. subtilis (5, 25) and, thus, suggested that HrcA can function as a thermosensor to detect temperature changes in cells. A similar heat shock-sensing function has been recently reported in the RheA repressor of the Hsp18 heat shock response in Streptomyces albus (23). RheA is rather small compared with HrcA, and therefore the mechanisms for thermosensing are probably different from each other.
REFERENCES
- 1.Ahmad, S., A. Selvapandiyan, and R. K. Bhantnagar. 1999. A protein-based phylogenetic tree for gram-positive bacteria derived from hrcA, a unique heat-shock regulatory gene. Int. J. Syst. Bacteriol. 49:1387-1394. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Christodoulou, E., and C. E. Vorgias. 2002. The thermostability of DNA-binding protein HU from mesophilic, thermophilic, and extreme thermophilic bacteria. Extremophiles 6:21-31. [DOI] [PubMed] [Google Scholar]
- 5.Claus, D., and R. C. W. Berkeley. 1986. Genus Bacillus Cohn 1872, p. 1105-1139. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md.
- 6.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 7.Frishman, D., and P. Argos. 1996. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 9:133-142. [DOI] [PubMed] [Google Scholar]
- 8.Gibrat, J. F., J. Garnier, and B. Robson. 1987. Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J. Mol. Biol. 198:425-443. [DOI] [PubMed] [Google Scholar]
- 9.Hecker, M., W. Schumann, and U. Völker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 10.Huffman, J. L., and R. G. Brennan. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 12:98-106. [DOI] [PubMed] [Google Scholar]
- 11.Karaivanova, I. M., P. Wiegel, M. Takahashi, C. Fort, A. Versavaud, G. V. Duyne, D. Charlier, J.-N. Hallet, N. Glansdorff, and V. Sakanyan. 1999. Mutation analysis of the thermostable arginine repressor from Bacillus stearothermophilus: dissecting residues involved in DNA binding properties. J. Mol. Biol. 291:843-855. [DOI] [PubMed] [Google Scholar]
- 12.King, R. D., and M. J. Sternberg. 1996. Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci. 5:2298-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu, Z., and E. C. Lin. 1989. The nucleotide sequence of Escherichia coli genes for l-fucose dissimilation. Nucleic Acids Res. 17:4883-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogk, A., A. V“lker, S. Engelmann, M. Hecker, W. Schumann, and U. Völker. 1998. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J. Bacteriol. 180:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mogk, A., and W. Schumann. 1997. Cloning and sequencing of the hrcA gene of Bacillus stearothermophilus. Gene 194:133-136. [DOI] [PubMed] [Google Scholar]
- 17.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, GTC Sequencing Center Production, Finishing, and Bioinformatics Teams, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reischl, S., T. Wiegert, and W. Schumann. 2002. Isolation and analysis of mutant alleles of the Bacillus subtilis HrcA repressor with reduced dependency on GroE function. J. Biol. Chem. 277:32659-32667. (First published 24 June 2002; 10.1074/jbc.M201372200.) [DOI] [PubMed] [Google Scholar]
- 20.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Servant, P., C. Grandvalet, and P. Mazodier. 2000. The RheA repressor is the thermosensor of the HSP18 heat shock response in Streptomyces albus. Proc. Natl. Acad. Sci. USA 97:3538-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song, H., H. Wang, D. Gigot, D. Dimova, V. Sakanyan, N. Glansdorff, and D. Charlier. 2002. Transcription regulation in thermophilic bacteria: high resolution contact probing of Bacillus stearothermophilus and Thermotoga neapolitana arginine repressor-operator interactions. J. Mol. Biol. 315:255-274. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, Y., T. Kishigami, K. Inoue, Y. Mizouchi, N. Eto, M. Takagi, and S. Abe. 1983. Bacillus thermoglucosidasius sp. nov., a new species of obligately thermophilic bacilli. Syst. Appl. Microbiol. 4:487-495. [DOI] [PubMed] [Google Scholar]
- 26.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe, K., K. Chishiro, K. Kitamura, and Y. Suzuki. 1991. Proline residues responsible for thermostability occur with high frequency in the loop regions of an extremely thermostable oligo-1,6-glucosidase from Bacillus thermoglucosidasius KP1006. J. Biol. Chem. 266:24287-24294. [PubMed] [Google Scholar]
- 28.Watanabe, K., and Y. Suzuki. 1998. Protein thermostabilization by proline substitutions. J. Mol. Catal. B 4:167-180. [Google Scholar]
- 29.Watanabe, K., T. Yamamoto, and Y. Suzuki. 2001. Renaturation of Bacillus thermoglucosidasius HrcA repressor by DNA and thermostability of HrcA-DNA complex in vitro. J. Bacteriol. 183:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wintjens, R., and M. Rooman. 1996. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262:294-313. [DOI] [PubMed] [Google Scholar]
- 31.Yamada, M., and M. H. J. Saier. 1988. Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J. Mol. Biol. 203:569-583. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida, K., S. Saeki, M. Fujimura, Y. Miwa, and Y. Fujita. 1995. Cloning and sequencing of a 36-kb region of the Bacillus subtilis genome between the gnt and iol operons. DNA Res. 2:61-69. [DOI] [PubMed] [Google Scholar]
- 33.Yuan, G., and S.-L. Wong. 1995. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 177:6462-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng, G., S. Ye, and T. J. Larson. 1996. Repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J. Bacteriol. 178:7080-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]