Abstract
Communication based on autoinducer 2 (AI-2) is widespread among gram-negative and gram-positive bacteria, and the AI-2 pathway can control the expression of genes involved in a variety of metabolic pathways and pathogenic mechanisms. In the present study, we identified luxS, a gene responsible for the synthesis of AI-2, in Streptococcus gordonii, a major component of the dental plaque biofilm. S. gordonii conditioned medium induced bioluminescence in an AI-2 reporter strain of Vibrio harveyi. An isogenic mutant of S. gordonii, generated by insertional inactivation of the luxS gene, was unaffected in growth and in its ability to form biofilms on polystyrene surfaces. In contrast, the mutant strain failed to induce bioluminescence in V. harveyi and was unable to form a mixed species biofilm with a LuxS-null strain of the periodontal pathogen Porphyromonas gingivalis. Complementation of the luxS mutation in S. gordonii restored normal biofilm formation with the luxS-deficient P. gingivalis. Differential display PCR demonstrated that the inactivation of S. gordonii luxS downregulated the expression of a number of genes, including gtfG, encoding glucosyltransferase; fruA, encoding extracellular exo-β-d-fructosidase; and lacD encoding tagatose 1,6-diphosphate aldolase. However, S. gordonii cell surface expression of SspA and SspB proteins, previously implicated in mediating adhesion between S. gordonii and P. gingivalis, was unaffected by inactivation of luxS. The results suggest that S. gordonii produces an AI-2-like signaling molecule that regulates aspects of carbohydrate metabolism in the organism. Furthermore, LuxS-dependent intercellular communication is essential for biofilm formation between nongrowing cells of P. gingivalis and S. gordonii.
It has long been recognized that bacteria can regulate gene expression in response to cell density, and quorum-sensing systems based on acylhomoserine lactone or peptide autoinducer (AI) molecules are widespread among gram-negative and gram-positive bacterial species, respectively. Cellular functions regulated by quorum sensing include the expression of virulence factors, competence for genetic transformation, conjugal DNA transfer, the production of antibiotics and secondary metabolites, and biofilm formation (14, 16, 48, 69). More recently, a novel communication system was described in the marine bacterium Vibrio harveyi (61). Bioluminescence in V. harveyi is regulated by two distinct AI signaling molecules that are detected by independent signal transduction systems that subsequently converge in a common pathway to regulate gene expression (reviewed in reference 54). AI-1 is a well-characterized derivative of a homoserine lactone that is highly species specific for V. harveyi. In contrast, AI-2 is a furanosyl borate diester. AI-2 is formed chemically from 4,5-dihydroxy-2,3-pentanedione that is generated by the action of LuxS AI synthase on S-ribosylhomocysteine (8, 55). The luxS gene is highly conserved across a diverse range of gram-negative and gram-positive bacterial species, and AI-2 produced by many of these species, including Escherichia coli, Salmonella enterica serovar Typhimurium (61), Helicobacter pylori (19, 35), Mannheimia haemolytica, and Pasteurella spp. (45), can induce bioluminescence in V. harveyi. One interpretation of these data is that, whereas acylhomoserine lactone-based signaling by gram-negative bacteria and peptide pheromone-based signaling by gram-positive bacteria both exhibit, on the whole, a high degree of species specificity, AI-2-mediated signaling may resemble bacterial “esperanto,” allowing interspecies communication in natural environments (2). The extent to which AI-2-based signaling represents true quorum sensing or is dependent to some degree on the metabolic status of the bacterial cells remains to be determined (3).
Dental plaque is a complex biofilm community comprising more than 500 different bacterial species and normally exists in commensal harmony with the host. However, overrepresentation of a group of gram-negative, mostly anaerobic, organisms, including Porphyromonas gingivalis, is associated with the initiation and progression of periodontitis, one of the most prevalent of human diseases (57). Plaque formation and development on the tooth surface involves both physical and metabolic interactions between constituent species and follows a relatively well-defined and reproducible bacterial succession whereby colonization of commensals such as Streptococcus gordonii is antecedent to the arrival of pathogens such as P. gingivalis (reviewed in references 37 and 51). As a consequence, interspecies communication might be expected to play an important role in the formation of these ordered and relatively high cell density structures (37, 38).
Using the V. harveyi AI-2 reporter strains, Frias et al. (21) screened 33 oral bacterial species, derived from 12 different genera, and demonstrated bioluminescence-inducing activity in three different species, namely, Fusobacterium nucleatum, Prevotella intermedia, and P. gingivalis. Subsequently, functional luxS-based communication circuits were described in P. gingivalis (6, 9) and in Actinobacillus actinomycetemcomitans (18). Inactivation of the luxS gene resulted in alterations in the level of expression of genes encoding proteins involved in virulence and in hemin acquisition in P. gingivalis (9) and of LtxA leukotoxin in A. actinomycetemcomitans (18). An important observation made by Fong et al. (18) was that conditioned medium from E. coli DH5α expressing the A. actinomycetemcomitans luxS gene was able to complement, in trans, defects in gene expression in the P. gingivalis luxS mutant, indicating that LuxS-dependent signaling has the potential to mediate interspecies communication in mixed-species biofilms.
Members of the mitis group of streptococci (including S. gordonii, Streptococcus mitis, Streptococcus oralis, Streptococcus parasanguis and Streptococcus sanguis [36]) are prominent components of the human oral microbiota (20) and play a significant role as pioneer colonizers in the development of dental plaque (reviewed in references 51 and 70). It is well established that the development of competence for genetic transformation in oral streptococci is a density-dependent phenomenon requiring an extracellular factor (50). More recently, a quorum-sensing system regulating competence and utilizing peptide pheromones as the signaling molecule was described for members of the mitis group of streptococci (including S. gordonii and S. sanguis [23, 27]), anginosus group streptococci (including Streptococcus constellatus and Streptococcus milleri [28]), and Streptococcus mutans (39). Further, it was demonstrated that the transformation frequencies of biofilm-grown S. mutans were 10- to 600-fold higher than those of planktonic cells (40). Recent data suggest also that the development of acid tolerance by S. mutans (39), the formation of biofilms (42), and the regulation of expression of cell surface adhesins by S. gordonii (47) involve peptide-mediated density sensing.
The screen by Frias et al. (21) for AI-2 molecules produced by oral bacteria failed to demonstrate bioluminescence-inducing activity in the culture supernatants of the four oral streptococcal species tested: S. sanguis, S. oralis, S. mitis, and S. mutans. However, functional LuxS-based signaling does occur in Streptococcus pyogenes (43). S. pyogenes luxS mutants demonstrated aberrant expression of virulence factors (cysteine protease and streptolysin S hemolysin) and, additionally, were defective in growth in complex medium (43). In this work we report the identification of a LuxS protein in S. gordonii with 81% identity to the functional LuxS protein of S. pyogenes. Although conditioned medium from S. gordonii cultures only poorly induced bioluminescence in the AI-2 reporter strain of V. harveyi, the luxS mutant demonstrated altered expression of a number of genes, including a group involved in carbohydrate metabolism by S. gordonii. Moreover, the S. gordonii luxS mutant was unable to form normal biofilms with a LuxS-deficient strain of P. gingivalis. These data suggest that AI-2-mediated intercellular communication in S. gordonii may play a central role in the processing of carbohydrates and in mixed-species biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth media.
The bacterial strains and plasmids used are listed in Table 1. S. gordonii strains were grown at 37°C on Trypticase soy broth-yeast extract (TSBY) agar (34) in a candle jar. Liquid cultures were grown without shaking in screw-cap tubes or bottles at 37°C in TSBY or in brain heart infusion-yeast extract medium (34). A chemically defined medium, CDMT (5), was used to assess biofilm formation by S. gordonii on polystyrene surfaces. P. gingivalis strains were grown from frozen stocks at 37°C under anaerobic conditions (85% N2, 10% H2, 5% CO2) in Trypticase soy broth supplemented with 1 mg of yeast extract/ml, 5 μg of hemin/ml, and 1 μg of menadione/ml. V. harveyi reporter strain BB170 (sensor 1−, sensor 2+) was kindly provided by B. Bassler (Princeton University) and was grown aerobically at 30°C in AI bioassay (AB) medium (60). E. coli strains were grown aerobically in Luria-Bertani medium (53) or on Luria-Bertani medium supplemented with 15 g of agar. The concentrations of antibiotics used for selection were as follows: ampicillin, 100 μg/ml (E. coli); erythromycin, 200 μg/ml (E. coli) or 1 μg/ml (S. gordonii); and tetracycline, 5 μg/ml (E. coli or S. gordonii).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| S. gordonii | ||
| DL1 Challis | Wild type | 50 |
| 1.1L | luxS::ermAM (LuxS− Emr) | This study |
| 1.1LC | luxS::ermAM::pVA981luxS (LuxS+ Tcr Emr) | This study |
| P. gingivalis | ||
| ATCC 33277 | Wild type | ATCC type strain |
| PLM1 | luxS::pLR409 (LuxS− Emr) | 9 |
| E. coli | ||
| Top10 | F−mcrA (mrr-hsdRMS-mcrBC) φ80lacZΔM15 lac74 recA1 deoR araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| DH5α | F−deoR endA1 gyrA96 hsdR17 (rK− mK−) recA1 relA1 supE44 thi-1 Δ(lacZYA-argFV169) φ80lacZΔM15 | 26 |
| V. harveyi BB170 | luxN::Tn5lac (AI-2 reporter strain) | 1 |
| Plasmids | ||
| pVA736 | 7.6 kb; Emr, pVA380-Iori | 44 |
| pVA981 | 7.1 kb; Tcr, ColE1ori | 41 |
| pVALuxS | S. gordonii luxS in pVA981 | This study |
| pCRLux2 | S. gordonii luxS in pCR4-TOPO | This study |
| pCRLux2Em | S. gordonii luxS::ermAM in pCR4-TOPO | This study |
DNA techniques.
Routine molecular biology techniques were performed as described by Sambrook et al. (53). Plasmid DNA was prepared from E. coli by using QIAprep spin minipreps (Qiagen), and PCR products were purified by using QIAquick (Qiagen). Chromosomal DNA was isolated from S. gordonii as described previously (33), and S. gordonii cells were transformed with DNA according to the method of Haisman and Jenkinson (24). Restriction and modifying enzymes (from New England Biolabs) were used under the conditions recommended by the manufacturer.
Construction of the S. gordonii luxS mutant strain.
Degenerate primers used to PCR amplify the luxS gene from S. gordonii DL1 were derived from a CLUSTAL W alignment (http://clustalw.genome.ad.jp/) of luxS sequences from S. pyogenes, S. pneumoniae, and S. mutans genomic databases. Primers LuxF (5′-ATGWCAAAAGAAGTTA-3′) and LuxR (5′-ACRTCTGAAATSCCTTG-3′; where W = A or T, R = A or G, and S = G or C) were used at 0.25 μM concentrations in PCRs (50 μl) containing deoxynucleoside triphosphates (dNTPs; 250 μM), MgCl2 (2.5 mM), and 2 U of Taq polymerase (Bioline). The conditions for amplification were as follows: 94°C for 4 min (1 cycle); 94°C for 25 s, 42°C for 40 s, and 72°C for 1 min (34 cycles); and 72°C for 10 min (final cycle). The product (457 bp) was cloned into pCR4-TOPO (Invitrogen) to generate plasmid pCRLux, which was subsequently sequenced with T3 and T7 promoter primers to confirm that the insert was luxS. A unique NdeI site was located within the cloned DNA fragment. The ermAM gene (encoding erythromycin resistance [Emr]) was amplified from plasmid pVA736 (44) with the primers Erm5 (5′-GCACATATGCTTAGAAGCAAACTTAAGA-3′) and Erm3 (5′-GCCCATATGCTTGGAAGCTGTCAGTAGT-3′). A PCR product of the predicted size (1 kb) was purified and digested with NdeI (sites underlined in primer sequences) and ligated with NdeI-linearized pCRLux. The resultant plasmid (pCRLuxEm) was purified and transformed into S. gordonii with selection for Emr to generate mutant strain 1.1L. Insertion of ermAM within luxS on the S. gordonii chromosome was confirmed by PCR by using primers LuxF and LuxR.
During this work the full sequence of the S. gordonii luxS gene and flanking DNA became available and the entire luxS coding region, together with upstream and downstream sequences (764 bp), was PCR amplified (34 cycles, annealing temperature of 50°C) from S. gordonii genomic DNA with the primers DluxF2 (nucleotides 1 to 20; 5′-TAATTCGAAAATTCTTAATTA-3′) and DluxR2 (complementary to nucleotides 745 to 764; 5′-CGGATTCTCTAGATTATTAG-3′) and cloned into pCR4-TOPO to generate recombinant plasmid pCRLux2. The disrupted luxS locus (1.75 kb) was also amplified from S. gordonii mutant strain 1.1L genomic DNA with the above primers and cloned to generate plasmid pCRLux2Em. Recombinant plasmids were transformed into E. coli DH5α.
To complement the luxS mutation of S. gordonii 1.1L, the luxS gene was excised from pCRLux2 by EcoRI digestion and ligated with EcoRI-cut pVA981 (41). Recombinant plasmid pVALuxS was then used to transform S. gordonii 1.1L with selection for tetracycline resistance (Tcr) to generate strain 1.1LC. Correct integration of recombinant pVA981 at the disrupted luxS locus by a Campbell-like single-crossover was confirmed by PCR with primers DluxF2 and DluxR2.
AI assay.
Cell-free culture supernatants from S. gordonii wild-type or luxS mutant strains or from E. coli DH5α harboring pCRLux2 or pCRLux2Em were prepared by centrifugation (10,000 × g, 4°C, 10 min) and filtration (filter pore size of 0.22 μm) and then tested for the induction of signaling system 2 in V. harveyi BB170 by a previously described luminescence assay (60). Briefly, an overnight culture of V. harveyi BB170 was diluted 1:5,000 in AB medium, and 100 μl of cell-free S. gordonii or E. coli culture fluid was added to 900 μl of diluted V. harveyi cells. Cell-free culture fluid of V. harveyi BB170 was included as positive control, and sterile medium was included as a negative control. The reaction was carried out at 30°C, and light production was monitored with a Bio-Orbit 1251 luminometer.
Biofilm formation.
The formation of single species S. gordonii biofilms on polystyrene surfaces was measured according to the method of Loo et al. (42), as modified by Froeliger and Fives-Taylor (22). Briefly, cells from an exponential-phase culture of S. gordonii were harvested by centrifugation (6,000 × g, 4°C, 10 min), washed once in distilled H2O, and suspended at a density (A600) of 0.01 in fresh prewarmed medium. Aliquots (0.2 ml) were inoculated into wells on non-tissue-culture-treated polystyrene flat-bottom 96-well microtiter plates. Wells containing 0.2 ml of sterile growth medium were included as negative controls. Plates were incubated at 37°C for 16 h either aerobically in 5% CO2 or anaerobically. Biofilm formation was measured by crystal violet staining of adherent bacteria (22), except that dye released from cells by 30% (vol/vol) acetic acid was diluted 1:4 in distilled H2O prior to determining absorbance of the solution at A590 with a Dynex MRX TCII microplate reader.
Mixed-species biofilm formation by P. gingivalis and S. gordonii was determined essentially as previously described (10). P. gingivalis and S. gordonii were cultured as described above and washed twice in phosphate-buffered saline (PBS). Cells of S. gordonii wild-type (DL1), luxS mutant (1.1L), or luxS complemented (1.1LC) strains (107 cells/ml in PBS) were labeled with hexidium iodide and passed (x1) over a saliva-coated glass slide in a flow chamber for 4 h at a flow rate of 2 ml/h. After the deposition of streptococci, fluorescein-labeled P. gingivalis cells (107 cells/ml in PBS) were passed (x1) through the flow cell for 8 h at 2 ml/h. Both P. gingivalis and S. gordonii remain viable during this process and can be recovered by culture at the end of the assay. The resulting P. gingivalis-S. gordonii biofilm was visualized by using a Leica TCS-SP confocal scanning laser microscope with an Leica inverted DMRXE light microscope and a 40× water immersion objective lens. A total optical magnification of ×400 was then digitally zoomed ×4, resulting in a ×1,600 total digital magnification. The pixel resolution was 1024 × 1024. A representative area of the coverslip was then selected and observed under a reflected laser light of 488 nm. A series of fluorescent optical sections were collected, and the depth of the bacterial layers and/or microcolonies was determined by sagittal reconstruction of x-y plane images. The total z depth was acquired by imaging both the minimal and maximal x-y planes of focus. A three-dimensional view of the biofilms was assembled by using the Imaris Version 3.1.3 software imaging program.
RNA isolation and RT-PCR.
Total RNA was isolated from S. gordonii cells as previously described (47) by using TriPure reagent (Roche Applied Science), except that the incubation time with mutanolysin prior to cell disruption was reduced to 20 min to maximize the recovery of intact mRNA. For reverse transcription-PCR (RT-PCR), 1 μg of total RNA was first treated at 37°C for 30 min with 1 U of RQ1 DNase (Promega) in the presence of 12 U of RNasin RNase inhibitor (Promega) in a final volume of 20 μl. Stop solution (2 μl; 20 mM EGTA, pH 8.0) was added, and the samples were heated at 65°C for 10 min to inactivate the DNase. A portion of DNA-free RNA (11 μl, containing 0.5 μg of RNA) was annealed with random primers (0.5 μg; Promega) by heating at 70°C for 10 min and then cooled on ice. First-strand cDNA synthesis was performed by the addition of 200 U of Moloney murine leukemia virus reverse transcriptase (Promega) in the presence of 1× reaction buffer and 0.5 mM dNTPs (final volume of 20 μl), followed by incubation at 42°C for 60 min. Portions of cDNA (2 μl, containing 50 ng of RNA equivalent) were used in standard PCRs. Controls consisted of PCRs containing 1 μl of DNase-treated RNA (50 ng of RNA). To demonstrate the presence of luxS mRNA, the primers LuxF3 (nucleotides 228 to 247; 5′-TTTCGAGCTTGATCACACCA-3′) and LuxR3 (complementary to nucleotides 601 to 620; 5′-TCCTTGGCAGAAAAGAGGCT-3′) were used at an annealing temperature of 50°C.
DD-PCR.
For differential display RT-PCR (DD-PCR), parent and mutant strains were cultivated in TSBY medium to late exponential phase. Total RNA was isolated by using the RNA isolation Kit, Totally RNA (Ambion), and subjected to RT. The reaction mixture contained 2 μg of RNA, 1 μl of 10 mM dNTP, and 100 pmol of random hexamers and was incubated at 80°C for 10 min then cooled on ice. An enzyme mixture containing 40 U of Moloney murine leukemia virus reverse transcriptase (Ambion), 1× RT reaction buffer, and 1 μl of anti-RNase (Ambion) was added to a final volume of 20 μl, and the reaction was incubated at 42°C for 1 h, followed by inactivation of the enzyme at 92°C for 10 min. Controls without the reverse transcriptase enzyme were included to ensure that there was no chromosomal contamination. DD-PCR was performed with 5-μl portions of the synthesized cDNA (containing 0.5 μg of RNA equivalent) in a final volume of 100 μl with 1 U of Taq DNA polymerase (Promega), 1.5 mM MgCl2, 0.2 mM dNTP, and 100 pmol of arbitrary primers. The arbitrary primers used were as follows: 5′-GGCATGGGTCAGAAGGATT-3′, 5′-CTCAAGTTGGGGGACAAAAA-3′, 5′-CGGAACAGCTTCTTCCAATC-3′, 5′-AATCTTGCTCCGCCCTTATT-3′, 5′-CACCTGTGGTCCACCTGAC-3′, 5′-GCTACCCGTATTGCCAAGAA-3′, and 5′-TTCGGCAAGCGAATACTTT-3′. The thermal cycling parameters were 50 cycles of 94°C for 1 min, 34°C for 1 min, and 72°C for 2 min. Differentially expressed PCR products were excised from agarose gels and cloned into pCRII-TOPO, and the DNA sequence was obtained by the University of Washington DNA Sequencing Service. The DD-PCR results were further investigated by RT-PCR with RNA preparations identical to those described above and primers derived from the sequences of cloned products. Primer sequences and PCR conditions that were unique to each amplification reaction are provided in Table 2 and were designed to correspond to the linear range of amplification, i.e., before saturation had occurred. Amplification products were quantitated by using a Kodak DC290 digital camera and Kodak 1D image analysis software (v3.5). Expression of sca mRNA was used as a control for mRNA loading. Analysis of this gene product over the growth curve demonstrated no significant change in expression between the S. gordonii wild type and mutant. Similar results were obtained by using fbpA mRNA as a control (not shown).
TABLE 2.
Oligonucleotide primers and conditions for RT-PCR analysisa
| Gene | Primer | Sequence | Size of PCR product (bp) | RNA per reaction (ng) | No. of PCR cycles |
|---|---|---|---|---|---|
| sca | F | 5′-AACCTCTGCCCGAAGATGTC-3′ | 540 | 250 | 25 |
| R | 5′-GATGGGACTTTGGTCTTGCG-3′ | ||||
| fruA | F | 5′-ACTCTATCGCGGTCAGTATCATTATT-3′ | 1,010 | 250 | 25 |
| R | 5′-TCTTCTCACTAACTTCCACGTCTTTA-3′ | ||||
| gor | F | 5′-CTATCCCTTCTATTCCTGGCTCTG-3′ | 910 | 200 | 30 |
| R | 5′-CGTGAACGAACTCTTCTGATCC-3′ | ||||
| spo768 | F | 5′-ACATTTTCTACCAATGCTTTCTCG-3′ | 1,124 | 200 | 30 |
| R | 5′-AAAAGAAGCCAATCGTCACTTTGT-3′ | ||||
| gtfG | F | 5′-TACTAATAGTCTGAATGACCGCTCTG-3′ | 903 | 400 | 50 |
| R | 5′-TCAAGAGCAGGACTAGATTCATAAAC-3′ | ||||
| prmA | F | 5′-AAGCGACACTGGATGAGAAGATTAT-3′ | 667 | 200 | 25 |
| R | 5′-GTGACTTGATCGTCCTCTTTGAGTC-3′ | ||||
| abc-nbd | F | 5′-CCGGATTTGAGAAAATATCGAG-3′ | 2,112 | 250 | 25 |
| R | 5′-TCGCCTTATTTATGGGGAGGA-3′ | ||||
| accA | F | 5′-AAAAATCACTCGTTTGTTTGAGTATG-3′ | 1,296 | 300 | 50 |
| R | 5′-TTTTTGTCTATTTTATAGTCTATCTCC-3′ | ||||
| ylbN-like | F | 5′-CCGGAGGTATATTCAAAAGGATATT-3′ | 677 | 250 | 30 |
| R | 5′-ATGCCAGCTGCAACTGTAAATTC-3′ | ||||
| Rgg | F | 5′-CTTATCGTAAAGTCGTCGGGAAAA-3′ | 905 | 250 | 30 |
| R | 5′-TTTCAGAGGAGGTTCTATTCTATT-3′ | ||||
| lacD | F | 5′-AAACGCTTGCCAGACTGCTT-3′ | 572 | 175 | 30 |
| R | 5′-ACCATCTTTGATGTAGGCTTCAACT-3′ | ||||
| proWX | F | 5′-GCAGGATGGAGAGATTCGTC-3′ | 595 | 250 | 50 |
| R | 5′-AAGATAAAAGAACCCAGTCC-3′ | ||||
| tnpA | F | 5′-CTTGCGTAGCCAGATGTCTAAAGT-3′ | 490 | 200 | 50 |
| R | 5′-TTTATGCTTCCGGCTCGTATGT-3′ |
Forward (F) and reverse (R) primers were used to detect mRNA expression of the indicated genes by RT-PCR (results are shown in Fig. 5). The amount of total RNA added to each RT-PCR reaction mixture and the number of cycles were optimized for each gene to ensure amplification within the liner range. The annealing temperature was 50°C in all cases.
Antibodies and ELISA.
CshA-specific antibodies were raised to the N-terminal nonrepetitive amino acid sequence region of CshA (N-CshA [46]). Antiserum raised to purified P1 protein (SpaP) of S. mutans serotype c was provided by K. Knox (Institute of Dental Research, Sydney, Australia). These antibodies were used to determine the immunoreactivity of CshA or SspA and SspB, respectively, present on intact streptococcal cells by enzyme-linked immunosorbent assay (ELISA) (29) utilizing a Dynex microplate reader.
Determination of GTF activity.
GTF activity was measured by production of glucan in polyacrylamide gels as described by Tardif et al. (64). S. gordonii strains were cultured to late exponential phase, extracted with 1% sodium dodecyl sulfate, and equal volumes of cell-free culture supernatants or cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 8.5% gels. Gels were incubated overnight at 37°C in 10 mM sodium phosphate (pH 6.8) containing 3% sucrose and 0.5% Triton X-100, and the resulting glucan bands were stained with pararosaniline (64).
RESULTS
S. gordonii possesses a LuxS homologue.
Degenerate primers derived from a CLUSTAL W alignment of other streptococcal luxS gene sequences successfully amplified a portion of the luxS gene of S. gordonii DL1, and this was used to generate a luxS mutant strain (see below). However, while the present studies were in progress, the sequence of the complete S. gordonii luxS gene and flanking regions was provided by Paul Kolenbrander (National Institutes of Health) and is now available at www.tigr.org. LuxS polypeptide (160 amino acid [aa] residues) demonstrated 91 and 81% identities with the LuxS proteins of S. pneumoniae (GenBank accession no. AAK99112.1) and S. pyogenes (GenBank accession no. AAK34410.1), respectively. At the 5′ end of luxS there was a potential ribosome-binding site (GGAGA) and an extended −10 promoter sequence, comprising a −16 region (TTTG [66]) upstream of a canonical −10 region (TATAAT), positioned appropriately with respect to the luxS start codon. No recognizable −35 sequence was present at a suitable distance from the −10 promoter sequence. However, the extended −10 sequence can function naturally in the absence of a −35 site (52), and it was predicted that luxS was expressed from its own promoter. A potential stem-loop, followed by a run of 6 T residues, was identified 3′ to the luxS stop codon, indicating the presence of a ρ-independent transcriptional terminator and suggesting that luxS is expressed as a monocistronic message. RT-PCR on S. gordonii DL1 mRNA isolated from late-exponential-phase cells and utilizing the primer pairs LuxF3 and LuxR3 (both annealing within luxS coding region), DluxF2 (forward primer annealing 190 bp 5′ to luxS ATG start codon) and LuxR3, and LuxF3 and DluxR2 (reverse primer annealing 60 bp 3′ to luxS TGA stop codon) supported this hypothesis. Thus, an RT-PCR product was obtained only when the internal primer pair LuxF3 and LuxR3 was used (Fig. 1D and data not shown).
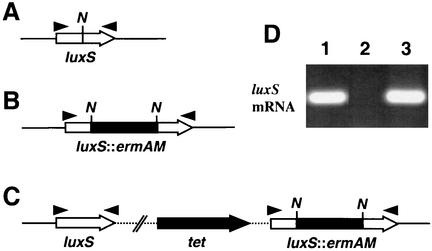
FIG. 1.
Schematic representation of the luxS locus of S. gordonii DL1 wild type (A), luxS mutant strain 1.1L (B), and complemented strain 1.1LC (C). The luxS gene was amplified from S. gordonii DL1 genomic DNA with the primers LuxF and LuxR; ermAM was ligated to a unique NdeI site (N) within the luxS gene, and the construct was transformed back onto the streptococcal chromosome (see Materials and Methods). Complemented strain 1.1LC was created by Campbell-like integration of plasmid pVALuxS carrying an intact copy of the luxS gene. The solid line indicates chromosomal DNA, and the broken line represents pVA981 DNA. (D) Expression of luxS by S. gordonii DL1 (lane 1) or mutant strains 1.1L (lane 2) or 1.1LC (lane 3). Total RNA was isolated from cells harvested at the late exponential phase of growth in TSBY medium and used in RT-PCR with luxS-specific primers LuxF3 and LuxR3.
Construction and characterization of luxS mutants.
The luxS gene on the S. gordonii DL1 chromosome was disrupted by insertion of the streptococcal Emr determinant ermAM (Fig. 1A and B). Correct integration of ermAM within luxS was confirmed by PCR with the primers DluxF2 and DluxR2. Thus, the luxS PCR product (764 bp) from S. gordonii DL1 genomic DNA was replaced by a product of ca. 1,750 bp (comprising luxS and the inserted ermAM gene) from the S. gordonii 1.1L mutant strain (data not shown). RT-PCR with luxS internal primers LuxF3 and LuxR3 confirmed that no luxS mRNA was produced by mutant strain 1.1L (Fig. 1D, lane 2).
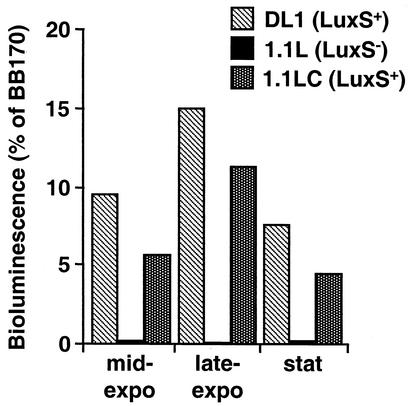
The mutant and wild type were unaffected in growth in a variety of media, including TSBY, TY-glucose, and the defined medium CDMT (not shown). To determine whether S. gordonii exhibited functional AI-2 signaling activity, cell-free culture supernatants from mid-exponential-, late-exponential-, or stationary-phase cultures of S. gordonii were assayed for induction of luminescence in the V. harveyi BB170 reporter strain (see Materials and Methods). The level of light induction exhibited by S. gordonii DL1 culture supernatants was weak compared to the V. harveyi BB170 positive control (Fig. 2) but exhibited growth-phase dependence with maximal light induction (15% of the BB170 positive control) being stimulated by late-exponential-phase culture supernatants. In contrast, light induction by luxS mutant strain was negligible at all phases of growth (Fig. 2). To eliminate the possibility that S. gordonii growth medium components or metabolic products were interfering with the luminescence assay, a plasmid carrying the luxS gene and flanking DNA (pCRLux2) was transformed into E. coli DH5α that contains a defective luxS gene (61). Light induction by conditioned medium from the resulting strain was relatively poor, despite the high copy number of the ColE1ori-based plasmid. Thus, conditioned medium from late-exponential-phase cultures of E. coli harboring pCRLux2 demonstrated light induction in the V. harveyi BB170 reporter strain of ca. 35% of the positive control (data not shown). Nevertheless, no light induction was observed for culture supernatants of recombinant E. coli harboring a disrupted copy of the S. gordonii luxS gene (on plasmid pCRLux2Em).
FIG. 2.
Induction of V. harveyi BB170 luminescence by cell-free supernatants of S. gordonii DL1 wild type, the luxS mutant strain 1.1L, and the complemented strain 1.1LC. Culture supernatants were prepared from the mid-exponential-, late-exponential-, or stationary-growth phase of growth in TY-glucose medium. Sterile medium and cell-free supernatants from V. harveyi BB170 served as negative and positive controls, respectively, and data are presented as the percent light induction relative to V. harveyi BB170 positive control. (The BB170 positive control gave a ca. 5,000-fold increase compared to the negative control.)
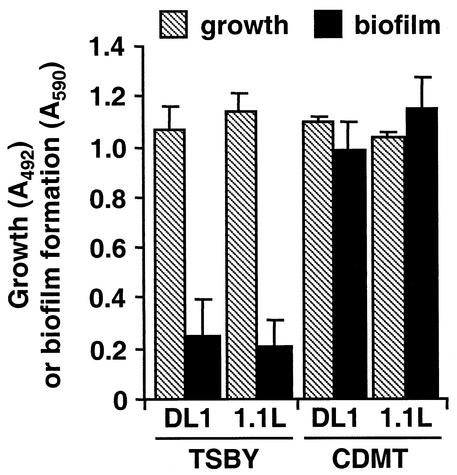
Wild-type and mutant S. gordonii cells expressed similar amounts of cell surface-anchored CshA, as well as SspA and SspB adhesins, as determined by whole-cell ELISA (Table 3). When grown in wells of polystyrene microtiter plates, the wild type and the 1.1L mutant strain reached the same optical density (OD) in TSBY or CDMT medium under anaerobic conditions (Fig. 3). Although biofilm formation was much lower for cells grown in TSBY medium compared to CDMT medium, there was no significant difference in biofilm formation between wild-type and mutant strains in either medium (Fig. 3). Similar data were obtained for microtiter plates incubated in an atmosphere of 5% CO2 in air (not shown).
TABLE 3.
luxS gene inactivation does not affect the expression of the coaggregation adhesins SspA, SspB, or CshA in S. gordonii
| Antiserum (dilution) | Mean ELISA reactivitya ± SDb with:
|
||
|---|---|---|---|
| DL1 (LuxS+) | 1.1L (LuxS−) | 1.1LC (LuxS+) | |
| P1c (1:500) | 0.165 ± 0.020 | 0.166 ± 0.018 | 0.182 ± 0.008 |
| CshA (1:1,000) | 0.278 ± 0.008 | 0.263 ± 0.007 | 0.276 ± 0.017 |
Input 2 × 107 cells per well. Results (A492) were corrected for background by subtracting values obtained with preimmune serum diluted appropriately.
Standard deviations of the mean (n = 4).
P1 antiserum reacts with both SspA and SspB.
FIG. 3.
Bacterial growth and biofilm formation of S. gordonii DL1 wild type or luxS mutant strain 1.1L. Cells from an exponential-phase culture of S. gordonii were washed once in sterile water and suspended at an OD of 600 nm (OD600) of 0.01 in fresh prewarmed medium. Cultures were then grown in the wells of polystyrene microtiter plates at 37°C for 16 h anaerobically. Growth and biofilm formation were quantified by determining the OD490 and OD562 values, respectively. The results are presented as the mean ± the standard deviation for quadruplicate determinations.
To confirm that the phenotypic characteristics of the 1.1L strain were a result of inactivation of the luxS gene, a complemented strain was constructed by integration of an intact copy of luxS (including 5′ and 3′ noncoding sequences) by a single-crossover homologous recombination event as described in Materials and Methods. Complemented strain 1.1LC carries both mutant and wild-type alleles of luxS (Fig. 1C), and complementation restored luxS mRNA expression (Fig. 1D, lane 3) and light induction in the bioluminescence assay (Fig. 2), although the complemented strain exhibited 25 to 40% lower AI-2 activity compared to S. gordonii wild-type strain.
LuxS is required for biofilm formation between S. gordonii and P. gingivalis.
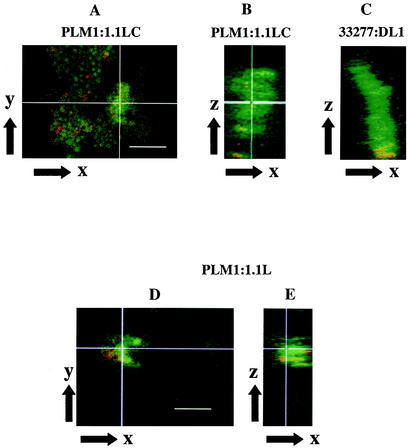
The role of LuxS in mixed-species biofilm formation between P. gingivalis and S. gordonii was investigated by using flow cells with substrata of streptococci attached to saliva-coated glass slides. A S. gordonii substratum supports adhesion of P. gingivalis, and P. gingivalis cells accrete progressively to form large, discrete, well-separated microcolonies (10, 38). The ability of luxS-null mutant strains to form a biofilm was first quantitated by microscopic counting of typical biofilm colonies. These microcolonies develop over a period of hours and can exceed 30 μm in depth. They are readily distinguishable from simple adhered microaggregates of P. gingivalis that do not exceed 10 μm in depth. Biofilm formation by either P. gingivalis PLM1 or S. gordonii 1.1L, the luxS-null mutants, was unimpaired when the partner species possessed a functional luxS gene (Table 4). This finding suggests that both P. gingivalis and S. gordonii can respond to AI-2 signal from the heterologous organism. In contrast, P. gingivalis biofilm microcolonies did not form between strains PLM1 and 1.1L in which LuxS expression was abolished in both partners (Table 4). Further support for the requirement of LuxS signal for biofilm formation was provided by the use of strain 1.1LC. P. gingivalis PLM1 formed biofilm microcolonies, to a degree similar to that of the parent strain 33277, on a substratum of the LuxS-producing S. gordonii 1.1LC (Table 4). Confocal microscopy was used to visualize the biofilm structures. A representative biofilm microcolony formed between P. gingivalis PLM1 and S. gordonii 1.1LC is shown in Fig. 4A. Rotation of the maximum projection of the x-y stacks to display the x-z perspective showed that the PLM1-1.1LC (Fig. 4B) and 33277-DL1 (Fig. 4C) biofilms developed to a similar depth. In contrast, there was little accumulation in the z dimension of PLM1 on 1.1L (Fig. 4E). Although significant intergeneric cell adhesion did occur between these luxS-null strains (Fig. 4D and E), the bound P. gingivalis cells did not exceed 10 μm and did not increase in depth over the assay period. Neither P. gingivalis strain PLM1 nor strain 33277 attached or accumulated to any significant degree to exposed areas of the saliva-coated glass slide (Fig. 4). Thus, the presence LuxS would appear to be required for the events subsequent to binding to S. gordonii cells that lead to accretion of P. gingivalis cells into microcolonies.
TABLE 4.
Biofilm formation with parent and engineered strains of P. gingivalis and S. gordonii
| Biofilm pair | Mean no. of biofilm coloniesa ± SD |
|---|---|
| P. gingivalis 33277, S. gordonii DL1 | 72 ± 12 |
| P. gingivalis 33277, S. gordonii 1.1L | 64 ± 9 |
| P. gingivalis PLM1, S. gordonii DL1 | 67 ± 11 |
| P. gingivalis PLM1, S. gordonii 1.1L | 0 |
| P. gingivalis PLM1, S. gordonii 1.1LC | 52 ± 4 |
| P. gingivalis 33277, S. gordonii 1.1LC | 47 ± 9 |
Number for P. gingivalis microcolonies greater than 10 μm in depth that formed on an area of 0.5 cm2 of the streptococcal substratum (n = 6).
FIG. 4.
Confocal microscopy of biofilm development on saliva-coated glass in flow cells with strains of P. gingivalis (green) and S. gordonii (red). P. gingivalis PLM1 adheres to S. gordonii 1.1LC (A) and accretes into biofilm microcolonies (B) similar to those formed by wild-type P. gingivalis 33277 and S. gordonii DL1 (C). P. gingivalis PLM1 and S. gordonii 1.1L coadhere (D); however accumulation of P. gingivalis PLM1 in the z dimension, to a depth greater than 10 μm, does not occur (E). The white lines in panels A and D represent the orientation in the x-y perspective, whereas B and E are the generated sagittal x-z perspectives. Bar, 10 μm.
S. gordonii genes are regulated by LuxS.
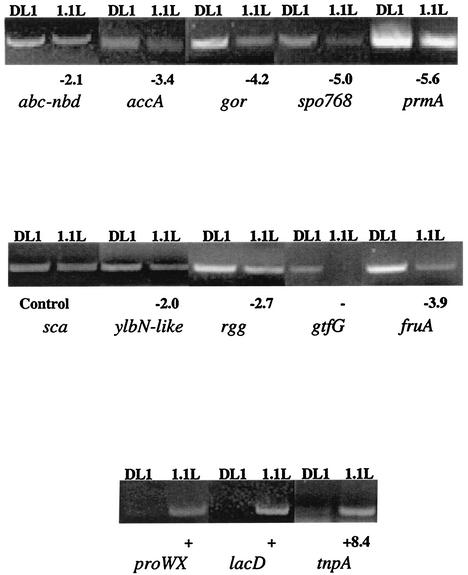
To investigate the role of LuxS in the regulation of S. gordonii gene expression, DD-PCR was performed. Amplification products that were differentially expressed between DL1 and 1.1L were sequenced and identified by a BLAST search of the GenBank database (http://www.ncbi.nlm.nih.gov). The S. gordonii database was incomplete and unannotated at the time of searching; thus, many sequences were identified on the basis of homology with annotated genes in the completed S. pneumoniae databases or the incomplete S. mutans database. The putative identities of proteins encoded by regulated genes are shown in Table 5. Although these proteins span a range of functional activities, four (glucosyltransferase [GTF], YlbN-like protein, fructanase, and tagatose 1,6-diphosphate aldolase) are involved in carbohydrate processing, indicating that LuxS regulates a variety of aspects of carbohydrate synthesis and metabolism in S. gordonii. Two regulated proteins (homologues of ABC-NBD and ProWX) may be involved in ABC-type transport. Similarly, Taga et al. (62) demonstrated that LuxS controls expression of an ABC-type transporter (Lsr) in Salmonella enterica serovar Typhimurium. The role of the Lsr apparatus was proposed to be the transport of AI-2 signaling molecules into the cells. The ability of either ABC-NBD or ProWX to function in a similar capacity in S. gordonii will require further analysis with gene knockout experiments. RT-PCR (Fig. 5) corroborated the differential expression of these genes, with the relative induction or repression in 1.1L ranging from 2.0 to absolute (gtfG, proWX, and lacD). Differences in the fold induction or repression as determined by RT-PCR compared to other global ex-pression procedures such as DD-PCR are not uncommon and are thought to result from differences in the dynamic ranges of the techniques (17). Since the S. gordonii gtfG gene is positively regulated by the upstream determinant rgg (59), steady-state levels of rgg mRNA were also investigated by RT-PCR. As shown in Fig. 5, expression of rgg was reduced in the 1.1L mutant strain. The downregulation of gtfG, may therefore result, at least in part, from the effects of LuxS on the activity of rgg.
TABLE 5.
Characterization of genes differentially regulated in S. gordonii 1.1L derived from DD-PCR
| Gene identification (species) | Homologous protein (GenBank accession no.) | e value | Expression in strain 1.1L |
|---|---|---|---|
| abc-nbd (S. pneumoniae)a | ABC transporter ATP-binding protein—unknown substrate (NP_357743) | 9e−89 | Absent |
| accA (S. pneumoniae)a | Acetyl coenzyme A carboxylase carboxyl transferase subunit alpha (NP_357981) | e−124 | Absent |
| gor (S. pneumoniae)a,b | Glutathione reductase (NP_345281) | e−117 | Absent |
| spo768 (S. pneumoniae) | Conserved hypothetical protein (NP_345266) | e−105 | Absent |
| prmA (S. pneumoniae)a | Ribosomal protein methyltransferase (NP_359200) | 2e−83 | Absent |
| ylbN-like (S. gordonii)b | YlbN-like hypothetical protein (AAG32547) | 6e−50 | Absent |
| gtfG (S. gordonii)b | Glucosyltransferase (AAC43483) | e−162 | Absent |
| fruA (S. mutans)a,b | Fructan β-fructosidase precursor (Q03174) | e−172 | Absent |
| proWX (S. pneumoniae)a | ABC transporter membrane-spanning permease—choline transporter (NP_359269) | 2e−60 | Present |
| lacD (S. pneumoniae)a | Tagatose 1,6-diphosphate aldolase (NP_358666) | 2e−97 | Present |
| tnpA (Lactococcus lactis)b | Transposase (CAA63529) | 5e−66 | Present |
Homologous nucleotide sequences were also present in the incomplete and unannotated S. gordonii genome database at www.tigr.org.
Multiple amplification products corresponding to these genes were identified.
FIG. 5.
RT-PCR analysis of mRNA expression of the indicated genes in S. gordonii DL1 and 1.1L. The results are representative of three separate preparations of total RNA. Fold induction or repression in 1.1L is indicated numerically or as “+” or “−” to show that mRNA expression could not be detected in either DL1 or 1.1L, respectively. The ratio of induction or repression in 1.1L was calculated from the relative intensity of linear range amplification products of RNA from DL1 and 1.1L normalized levels of sca mRNA (control) that was not regulated in 1.1L.
LuxS influences GTF activity.
The differential regulation of gtfG and rgg in the parent and luxS mutant indicated that GTF activity would differ between the two strains. GTF activity was measured by pararosaniline staining of in-gel sucrose-derived glucan bands. These GTF activity gels showed a significant reduction in extracellular GTF activity for strain 1.1L (Fig. 6). Complementation with luxS (strain 1.1LC) partially restored GTF activity (Fig. 6). Similar results were observed for cell-associated GTF activity (not shown). These data show that differences in expression levels of gtfG and rgg mRNA are reflected in GTF activity and support the concept that the levels of GTF are controlled by LuxS. The finding that 1.1LC is not completely restored in GTF activity is consistent with the bioluminescence induction data (Fig. 2) and may be due to the chromosomal configuration of luxS in this complemented strain. In particular, the presence of two functional luxS promoter regions, along with the presence of the tet element in the complemented strain, may modulate gene transcription.
FIG. 6.
In-gel GTF activities of culture supernatant of S. gordonii strains. The position of the 174-kDa native GTF protein band is indicated. The lower-molecular-mass forms of GTF are thought to result from proteolytic degradation of the native enzyme (65). The gel shown is representative of three independent experiments.
DISCUSSION
Numerous gram-positive and gram-negative bacteria produce and respond to AI-2 and, hence, LuxS-based signaling is thought to represent an important means of intergeneric communication. Furthermore, individual species regulate different aspects of metabolism and virulence factor expression in response to AI-2 (12, 15, 43, 58). In the present study, we show that S. gordonii possesses a functional luxS gene and that LuxS in this species controls aspects of carbohydrate metabolism and mixed-species biofilm formation with P. gingivalis. S. gordonii culture supernatant was capable of inducing bioluminescence in a V. harveyi AI-2 reporter strain; however, the level of induction was significantly lower than that obtained with homologous V. harveyi signal. This does not appear to be the result of medium-dependent interference but may instead be related to the levels of AI-2 production by S. gordonii. Alternatively, the structure of the LuxS-derived signal may differ between S. gordonii and V. harveyi. Similarly, the P. gingivalis LuxS-based signal was reported to induce bioluminescence at a level significantly lower than the control Vibrio signal (9). Organisms such as S. gordonii and P. gingivalis (which lack LuxI/LuxR-based signaling) may therefore produce modified AI-2 signals with distinct functional roles. This possibility can be addressed more fully once the structures of the signaling molecules from P. gingivalis and S. gordonii have been determined.
An analysis of the genes and pathways that may be controlled by LuxS in S. gordonii was undertaken by using differential display of the mRNA from parent and mutant strains. The absence of LuxS resulted in modulated expression of a variety of genes, several of which encode proteins involved in carbohydrate metabolism. The expression of GTF, YlbN-like protein, Rgg, and a homologue of exo-β-d-fructosidase (fructanase) was downregulated in the LuxS mutant, whereas expression of a homologue of tagatose 1,6-diphosphate aldolase was elevated. Since bacteria can control gene and protein expression on multiple levels, a reduction in the amount of mRNA cannot be assumed to produce a change in protein expression. However, in the case of GTF, an enzyme responsible for the production of glucan from sucrose, a correlation between protein activity and gene expression was observed. Furthermore, complementation with a single chromosomal copy of luxS partially restored GTF activity. It is reasonable to propose, therefore, that the downregulation of transcriptional activities observed by both DD-PCR and RT-PCR are likely to produce phenotypic effects. In S. gordonii, the transcription of gtfG is positively regulated by a trans-acting product of rgg (59). The gtfG and rgg genes are adjacent on the chromosome and ca. 2.5 kb upstream of the ylbN-like gene (65). A function for YlbN has not been determined; however, the gene is present in other streptococci, including S. mutans, S. pneumoniae, and S. pyogenes (65). Fructanse is an enzyme that degrades fructans [both β(2,6)-linked levan and β(2,1)-linked inulin] to fructose (7). GTF and fructanase can cleave sucrose into fructose or into glucose and fructose, the monosaccharide constituents of glucan and fructan. Downregulation of these proteins can thus be predicted to result in less-efficient utilization of sucrose and disruption of the ratios and amounts of glucan and fructan extracellular polymers. In contrast, tagatose 1,6-diphosphate aldolase is an enzymatic component of a pathway for galactose utilization (31). In S. mutans, this pathway is lactose inducible (25). Hence, the presence of AI-2 may favor the utilization of sucrose by S. gordonii, whereas in its absence lactose and other galactose-containing sugars are metabolized preferentially. In S. enterica serovar Typhimurium, AI-2 levels are maximal during exponential phase of growth, and this was shown to correlate not with transcriptional activity of luxS but with levels of expression of pfs encoding a second protein required for AI-2 synthesis (3). Expression of Pfs and levels of AI-2 are affected by carbohydrate source, leading to the hypothesis that AI-2-dependent signaling is a reflection of the metabolic state of the cell (3). The data presented here suggest that AI-2 signaling in S. gordonii may influence carbohydrate utilization, which will also reflect cellular metabolic activity. Although DD-PCR can theoretically provide total genome coverage, the degree of genome representation achieved in the present study is not known. Certainly, studies in E. coli indicate that expression of some 200 to 400 genes is affected by LuxS-mediated signaling (15, 58). It is possible, therefore, that there are additional carbohydrate-related genes that are also controlled by LuxS. A number of other LuxS-regulated protein homologues were identified, including ABC transporters, enzymes responsible for ATP-dependent carboxylation of acetyl coenzyme A (AccA) and the transfer of methyl groups from S-adenosyl-l-methionine to ribosomal protein L11 (PrmA), and glutathionine reductase (Gor). LuxS may therefore play a role in the regulation of a number of important metabolic properties of S. gordonii.
Although the importance of LuxI/LuxR-dependent signaling in biofilm formation by Pseudomonas aeruginosa has been established (11), the role of LuxS in biofilm formation has yet to be defined. Dental plaque is a complex mixed-species biofilm comprising commensals such as S. gordonii along with periodontal pathogens such as P. gingivalis. In vitro, accretion of P. gingivalis into biofilms on saliva-coated surfaces requires a conditioning layer of S. gordonii cells (10). The data from the present study demonstrate that mixed P. gingivalis-S. gordonii biofilms do not develop in the absence of LuxS signaling. The production of LuxS by either species is sufficient to allow biofilm formation, and complementation of luxS in a knockout of S. gordonii restored the biofilm phenotype. Collectively, these data suggest that both P. gingivalis and S. gordonii can sense and respond to heterologous LuxS signal and are consistent with the proposed role of LuxS in non-species-specific signaling (54). Nonetheless, the genes that are controlled through LuxS differ between P. gingivalis and S. gordonii. In P. gingivalis, genes relating to hemin acquisition are affected by LuxS (9), whereas differential regulation of carbohydrate metabolism and other non-iron-acquisition-related genes was observed in S. gordonii. Since it is possible that regulation of iron uptake genes was not detected by our conditions of DD-PCR, expression of the iron-regulated genes SGP50 (67) and comYA (iron regulated in the related species Streptococcus suis [56]) and of the iron-insensitive gene scaA (32) was investigated by RT-PCR. Steady-state mRNA levels of these genes were not altered in the S. gordonii LuxS-null mutant (laboratory observations). Thus, P. gingivalis and S. gordonii appear to generate unique responses to LuxS-based signaling, depending on the metabolic needs of the organism at different cell densities.
Biofilm formation between P. gingivalis and S. gordonii requires, as an initial event, adherence mediated through S. gordonii Ssp surface protein interactions with the P. gingivalis minor fimbriae (38). In the absence of either of these proteins biofilms do not accumulate, although the organisms can still bind to each other through redundant adhesion mechanisms (38). One possible function for LuxS in S. gordonii, therefore, would be regulation of the levels of the Ssp adhesins. However, expression of Ssp proteins was not affected in the LuxS-null mutant, and interbacterial binding between P. gingivalis and S. gordonii did not require LuxS. Hence, the influence of LuxS on biofilm formation occurs subsequent to initial adherence. The nature and amounts of extracellular polysaccharide produced by S. gordonii under LuxS control could affect the ability of P. gingivalis cells to accrete into microcolonies. Alternatively, or additionally, expression of other signaling molecules or adhesins responsible for autoaggregation in P. gingivalis could be controlled by LuxS-dependent pathways. It is interesting that P. gingivalis biofilm development in our model system occurs in the absence of significant growth and division. Biofilm-associated genes under the control of LuxS will, therefore, be involved in the early events of biofilm formation that result in the transition from a surface-adhered state to a nascent biofilm state. Adhesion of bacteria to surfaces and related surface-associated environmental signals are recognized to induce bacterial responses that facilitate longer-term, stable cell surface association. For example, Zhang and Normark (71) showed that after E. coli P-pili mediated binding to host cells there was transcriptional activation of a sensor-regulator protein (AirS) required for the iron starvation response. Also, in E. coli, initial adhesion to abiotic surfaces rapidly induces transcription of cpx-related genes and upregulation of the Cpx two-component signaling pathway, which leads to a greater stability of adhesion (49). In Neisseria meningitidis, there is upregulation of both the pilus-associated protein PilC1 and the transcriptional regulator CrgA after bacterial contact with host cells (13, 63). Moreover, Hudson and Curtis (30) demonstrated that transcriptional activity of the gtfB/C operon increases in S. mutans cells adsorbed to saliva-coated hydroxyapatite, a model of the tooth surface. The nature of the genes regulated by LuxS signaling in P. gingivalis during the process of interbacterial adhesion and biofilm accumulation remains to be determined.
A range of molecular techniques is being used successfully to investigate genes necessary for biofilm formation by many oral bacterial species, including the streptococci. For example, the inactivation of specific genes (4, 22, 68) or the use of random mutagenesis (42) has identified a number of loci required for single species biofilm formation on polystyrene surfaces, including genes encoding surface proteins (22), signal transduction system components (4, 42), or proteins involved in cell wall biosynthesis (42). However, the inactivation of luxS does not affect monospecies biofilm formation in S. mutans (68) or in S. gordonii (the present study). In this respect, it was interesting that none of the luxS-regulated S. gordonii genes identified here appeared on the list of genes required for monospecies biofilm formation. Taken together, these studies indicate that single- and mixed-species biofilm formation requires distinct sets of genes that are independently controlled. Investigation of the role of heterologous and homologous AI-2 signals, along with the interactions among LuxS-controlled and other regulatory pathways, is likely to provide significant insights into the development and pathogenic potential of the dental plaque biofilm.
Acknowledgments
We thank Don Demuth for much helpful advice, Paul Kolenbrander and David Blehert for providing S. gordonii luxS sequence information, Bonnie Bassler for the gift of V. harveyi strains, and Julia Ranford for critical evaluation of the manuscript.
This work was supported by DE12505 from the NIDCR.
REFERENCES
- 1.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:73-786. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. B. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 3.Beeston, A. L., and M. G. Surette. 2002. pfs-dependent regulation of autoinducer 2 production in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3450-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 5.Bouvet, A., I. van de Rijn, and M. McCarty. 1981. Nutritionally variant streptococci from patients with endocarditis: growth parameters in a semisynthetic medium and demonstration of a chromophore. J. Bacteriol. 146:1075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, et al. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 7.Burne, R. A., and J. E. C. Penders. 1992. Characterization of the Streptococcus mutans GS-5 fruA gene encoding exo-β-d-fructosidase. Infect. Immun. 60:4621-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X., S. Schauder, N. Potler, A. van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 9.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, G. S., J. W. Costerton, and R. J. Lamont. 1998. Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii. J. Periodontol. Res. 33:323-327. [DOI] [PubMed] [Google Scholar]
- 11.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 12.Day, W. A., and A. T. Maurelli. 2001. Shigella flexneri LuxS quorum-sensing system modulates virB expression but is not essential for virulence. Infect. Immun. 69:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deghmane, A. E., S. Petit, A. Topilko, Y. Pereira, D. Giorgini, M. Larribe, and M. K. Taha. 2000. Intimate adhesion of Neisseria meningitidis to human epithelial cells is under the control of the crgA gene, a novel LysR-type transcriptional regulator. EMBO J. 19:1068-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunny, G. M., and B. A. B. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 17.Eckmann, L., J. R. Smith, M. P. Housley, M. B. Dwinell, and M. F. Kagnoff. 2000. Analysis by high-density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella. J. Biol. Chem. 275:14084-14094. [DOI] [PubMed] [Google Scholar]
- 18.Fong, K. P., W. O. Chung, R. J. Lamont, and D. R. Demuth. 2001. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect. Immun. 69:7625-7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsyth, M. H., and T. L. Cover. 2000. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect. Immun. 68:3193-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frandsen, E. V., V. Pedrazzoli, and M. Kilian. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol. Immunol. 6:129-133. [DOI] [PubMed] [Google Scholar]
- 21.Frias, J., E. Olle, and M. Alsina. 2001. Periodontal pathogens produce quorum sensing molecules. Infect. Immun. 69:3431-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froeliger, E. H., and P. Fives-Taylor. 2001. Streptococcus parasanguis fimbria-associated adhesin Fap1 is required for biofilm formation. Infect. Immun. 69:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaustad, P., and L. S. Håvarstein. 1997. Competence-pheromone in Streptococcus sanguis: identification of the competence gene comC and the competence pheromone. Adv. Exp. Med. Biol. 418:1019-1021. [PubMed] [Google Scholar]
- 24.Haisman, R. J., and H. F. Jenkinson. 1991. Mutants of Streptococcus gordonii Challis overproducing glucosyltransferase. J. Gen. Microbiol. 137:483-489. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton, I. R., and H. Lebtag. 1979. Lactose metabolism by Streptococcus mutans: evidence for induction of the tagatose 6-phosphate pathway. J. Bacteriol. 140:1102-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning. IRL Press, Oxford, England.
- 27.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 28.Håvarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change phenotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes, A. R., P. K. Gopal, and H. F. Jenkinson. 1995. Adherence of Candida albicans to a cell surface polysaccharide receptor on Streptococcus gordonii. Infect. Immun. 63:1827-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson, M. C., and R. Curtiss. 1990. Regulation of expression of Streptococcus mutans genes important to virulence. Infect. Immun. 58:464-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagusztyn-Krynicka, E. K., J. B. Hansen, V. L. Crow, T. D. Thomas, A. L. Honeyman, and R. Curtiss. 1992. Streptococcus mutans serotype c tagatose 6-phosphate pathway gene cluster. J. Bacteriol. 174:6152-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 33.Jenkinson, H. F. 1987. Novobiocin-resistant mutants of Streptococcus sanguis with reduced cell hydrophobicity and defective in coaggregation. J. Gen. Microbiol. 133:1909-1918. [DOI] [PubMed] [Google Scholar]
- 34.Jenkinson, H. F., S. D. Terry, R. McNab, and G. W. Tannock. 1993. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect. Immun. 61:3199-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joyce, E. A., B. L. Bassler, and A. Wright. 2000. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J. Bacteriol. 182:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 37.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 38.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 39.Li, Y.-H., M. N. Hanna, G. Svensäter, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, Y.-H., P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindler, L. E., and F. L. Macrina. 1986. Characterization of genetic transformation in Streptococcus mutans by using a novel high-efficiency plasmid marker rescue system. J. Bacteriol. 166:658-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that encode for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lyon, W. R., J. C. Madden, J. C. Levin, J. L. Stein, and M. G. Caparon. 2001. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 42:145-157. [DOI] [PubMed] [Google Scholar]
- 44.Macrina, F. L., K. R. Jones, and P. H. Wood. 1980. Chimeric streptococcal plasmids and their use as molecular cloning vehicles in Streptococcus sanguis (Challis). J. Bacteriol. 143:1425-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malott, R. J., and R. Y. Lo. 2001. Studies on the production of quorum-sensing signal molecules in Mannheimia haemolytica and other Pasteurellaceae species. FEMS Microbiol. Lett. 206:25-30. [DOI] [PubMed] [Google Scholar]
- 46.McNab, R., A. R. Holmes, J. M. Clarke, G. W. Tannock, and H. F. Jenkinson. 1996. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect. Immun. 64:4204-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNab, R., and H. F. Jenkinson. 1998. Altered adherence properties of a Streptococcus gordonii hppA (oligopeptide permease) mutant result from transcriptional effects on cshA adhesion gene expression. Microbiology 144:127-136. [DOI] [PubMed] [Google Scholar]
- 48.Miller, M. B., and B. B. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 49.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pakula, R., and W. Walczak. 1963. On the nature of competence of transformable streptococci. J. Gen. Microbiol. 31:125-133. [DOI] [PubMed] [Google Scholar]
- 51.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 52.Sabelnikov, A. G., B. Greenberg, and S. A. Lacks. 1995. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J. Mol. Biol. 250:144-155. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Schauder, S., and B. B. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 55.Schauder, S., K. Shokat, M. G. Surette, and B. B. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 56.Smith, H. E., H. Buijs, R. de Vries, H. J. Wisselink, N. Stockhofe-Zurwieden, and M. A. Smits. 1999. Environmentally regulated genes of Streptococcus suis: identification by the use of iron-restricted conditions in vitro and by experimental infection of piglets. Microbiology 147:271-280. [DOI] [PubMed] [Google Scholar]
- 57.Socransky, S. S., and A. D. Haffajee. 1992. The bacterial etiology of destructive periodontal disease: current concepts. J. Periodontol. 63:322-331. [DOI] [PubMed] [Google Scholar]
- 58.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Surette, M. G., and B. B. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Surette, M. G., M. B. Miller, and B. B. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taga, M. E., J. L. Semmelhack, and B. B. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 63.Taha, M. K., P. C. Morand, Y. Pereira, E. Eugene, D. Giorgini, M. Larribe, and X. Nassif. 1998. Pilus-mediated adhesion of Neisseria meningitidis: the essential role of cell contact-dependent transcriptional upregulation of the PilC1 protein. Mol. Microbiol. 28:1153-1163. [DOI] [PubMed] [Google Scholar]
- 64.Tardif, G., M. Sulavik, G. W. Jones, and D. B. Clewell. 1989. Spontaneous switching of the sucrose-promoted colony phenotype in Streptococcus sanguis. Infect. Immun. 57:3945-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vickerman, M. M., P. E. Minick, and N. M. Mather. 2001. Characterization of the Streptococcus gordonii chromosomal region immediately downstream of the glucosyltransferase gene. Microbiology 147:3061-3070. [DOI] [PubMed] [Google Scholar]
- 66.Voskuil, M. I., and G. H. Chambliss. 1998. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 26:3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vriesema, A. J. M., J. Dankert, and S. A. J. Zaat. 1999. Isolation and characterization of promoter regions from Streptococcus gordonii CH1. Curr. Microbiol. 39:321-326. [DOI] [PubMed]
- 68.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 70.Whittaker, C. J., C. M. Klier, and P. E. Kolenbrander. 1996. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50:513-552. [DOI] [PubMed] [Google Scholar]
- 71.Zhang, J. P., and S. Normark. 1996. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science 273:1234-1236. [DOI] [PubMed] [Google Scholar]