Abstract
The expression of assembly-defective outer membrane proteins can confer lethality if they are not degraded by envelope proteases. We report here that the expression of a mutant OmpC protein, OmpC2Cys, which forms disulfide bonds in the periplasm due to the presence of two non-native cysteine residues, is lethal in cells lacking the major periplasmic protease, DegP. This lethality is not observed in dsbA strains that have diminished ability to form periplasmic disulfide bonds. Our data show that this OmpC2Cys-mediated lethality in a degP::Kmr dsbA+ background can be reversed by a DegP variant, DegPS210A, that is devoid of its proteolytic activity but retains its reported chaperone activity. However, DegPS210A does not reverse the lethal effect of OmpC2Cys by correcting its assembly but rather by capturing misfolded mutant OmpC polypeptides and thus removing them from the assembly pathway. Displacement of OmpC2Cys by DegPS210A also alleviates the negative effect that the mutant OmpC protein has on wild-type OmpF.
OmpC and OmpF represent a major class of Escherichia coli outer membrane proteins (OMPs). These proteins form water-filled channels that permit the diffusion of small solutes into the cell. Because of this property, OmpC and OmpF are called porins or sometimes “classical” porins, which refer to their stable β-barrel structure and nonspecific solute diffusion properties (17). Each porin monomer consists of a 16-stranded antiparallel β-barrel (22). These porins exist as homotrimers, in which three barrels assemble to produce three separate channels (22).
Besides their atomic structures, the assembly and targeting of the outer membrane β-barrel proteins have been an intense topic of study (for a recent review, please see reference 6). Based on studies conducted in several laboratories, it has been shown that the assembly of porin proteins begins with the proteolytic cleavage of an amino-terminal signal sequence that yields soluble nascent monomers. Rapid folding of these monomers is followed by their oligomerization into protease- and heat-sensitive trimers called metastable trimers, which slowly mature into protease- and heat-resistant trimers, the final product of assembly.
Although porins in vitro can be fully folded into their native trimeric structures from denatured monomers without the need of an accessory protein (7, 27), assembly in vivo is highly dependent on several cellular folding factors. Among them are specific periplasmic foldases such as SurA and FkpA, which catalyze cis-trans peptidyl-prolyl isomerization (16, 20), although it has never been demonstrated that this enzymatic activity is actually involved in OMP assembly. Consistent with this notion, Behrens et al. (3) have recently shown that SurA assists in OMP assembly independent of its isomerase activity. Another periplasmic foldase, DsbA, is involved in promoting correct disulfide bond formation of certain OMPs (2). The wild-type sequence of the classical porins, however, lacks cysteine residues and thus they assemble independently of DsbA's activity. In contrast to wild-type OmpC, the assembly of a mutant OmpC protein, OmpC2Cys, containing two non-native cysteine residues (C74 and C154) is drastically influenced by DsbA (11). Since OmpC2Cys is the subject of the present study, it is further discussed below.
Another periplasmic protein, Skp, is thought to assist in OMP assembly by acting as a general chaperone (5, 21). However, the skp-null mutant phenotype showing a dramatic reduction in OMP levels could not be repeated in other laboratories since it was first reported. Recent genetic studies have shown that the presence of skp- and surA-null alleles in a strain confers a synthetic lethal phenotype (19). Based on these studies, it has been proposed that Skp and SurA are components of separate folding pathways providing chaperone activities (19). Nonproteinaceous components, including phospholipids and lipopolysaccharide (which is exclusively localized on the outer leaflet of the outer membrane), are also important for OMP assembly (1, 9, 18).
Periplasmic and envelope proteases become particularly important when the machinery that ensures correct OMP assembly malfunctions or the primary sequence of an OMP endures an unacceptable alteration. One periplasmic protease, DegP, has received the most attention in connection with OMP assembly. degP-null mutants do not grow at temperatures above 39°C (10, 25, 26), presumably because of a greater need for the removal of misfolded proteins at elevated temperatures. However, expression of certain misfolded OMPs renders the DegP's presence essential even at lower growth temperatures (14). Elegant work carried out by Spiess et al. (25) showed that DegP, in addition to proteolysis, assists in the correct folding of a periplasmic protein. A recent genetic study also argued for DegP's involvement in providing periplasmic chaperone activities (19); however, this contention was not backed by direct biochemical evidence.
We have previously described the isolation of a novel class of OmpC mutants that affected either the channel or phage receptor property of the protein (13, 28). One such OmpC mutant carrying a R74C substitution functionally enlarged the channel to permit the entry of large sugars such as maltodextrins, which are normally excluded from the OmpC channel (13). This isolate was subsequently used for the isolation of phage-resistant mutants under conditions that demanded the retention of a functional OmpC channel but no phage receptor activity. A group of OmpC mutants from this selection contained a G154C substitution while retaining the original R74C alteration (11). This mutant OmpC protein, OmpC2Cys, which now had two non-native cysteine residues in its mature sequence, conferred phage resistance due to a biogenesis defect (11).
The OmpC2Cys biogenesis defect was shown to stem from the protein's ability to form disulfide bonds in DsbA+ cells (30); OmpC2Cys assembles normally in a genetic background lacking DsbA. OmpC2Cys produces a novel transdominant effect on OmpF (11). Since the effect on OmpF is specific, this suggests that either the inhibition was due to the mixing of mutant OmpC assembly intermediates with that of OmpF or that the two proteins share a common assembly factor.
Just as noted for several assembly-defective OmpF mutants (14), it was discovered that the expression of OmpC2Cys in a DsbA+ background obligated the presence of DegP at all growth temperatures. As anticipated, in a background lacking DsbA, and hence with a diminished disulfide forming ability in the periplasm, the absence of DegP produced no lethal effects at growth temperatures of ≤37°C.
We investigated here the effect of a protease-deficient DegP, DegPS210A, on OmpC2Cys-mediated lethality in a degP-null strain. Our findings provide direct biochemical evidence showing that DegPS210A alleviates OmpC2Cys's lethal effects not by improving its folding or assembly but rather by sequestering misfolded mutant OmpC polypeptides and thus removing them from the normal assembly pathway.
MATERIALS AND METHODS
Media, reagents, and bacterial strains.
Luria broth medium, Luria agar (1.5% [wt/vol]) medium, and M63 salt-based minimal medium were prepared according to the method of Silhavy et al. (23). When cultures were grown on minimal medium, 0.4% glycerol and 0.1 mM amino acid mixture lacking methionine were added. When required, ampicillin (50 μg/ml), kanamycin (25 μg/ml), chloramphenicol (25 μg/ml), tetracycline (25 μg/ml), and IPTG (isopropyl-β-d-thiogalactopyranoside; 0.4 mM) were added to the growth medium. Heat-killed Staphylococcus aureus (Pansorbin) was purchased from Calbiochem. [35S]methionine Expre protein labeling mixture was purchased from Du Pont-New England Nuclear. All other chemicals were of analytical grade. The bacterial strains used in the present study are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Relevant characteristicsa | Source or reference |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U139 rpsL150 flbB5301 ptsF25 deoC1 thi-1 rbsR relA | 4 |
| PLB3261 | MC4100 ΔlamB106 φompC′::lacZ+ | S. Benson |
| RAM412 | PLB3261 ompC234 (OmpC1Cys) zei::Tn10 (50% linkage to ompC by P1 transduction) | 12 |
| RAM415 | PLB3261 ompC402 (OmpC2Cys) zei::Tn10 | 12 |
| RAM1015 | MC4100 lamB677 (LamB amber) | Lab strain |
| RAM1025 | RAM1015 degP::Kmr | Lab strain |
| RAM1120 | RAM1025 pCS10 (DegPS210A) | This study |
| RAM1121 | RAM1025 pACYC184 | This study |
| RAM1122 | RAM1015 pCS10 | This study |
| RAM1123 | RAM1015 pACYC184 | This study |
| RAM1124 | PLB3261 degP::Kmr pCS10 | This study |
| RAM1125 | RAM412 degP::Kmr pCS10 | This study |
| RAM1126 | RAM415 degP::Kmr pCS10 | This study |
| RAM1127 | RAM415 recA::Kmr | This study |
| RAM1128 | RAM415 recA::Kmr pCS21 (DegPS210A-6His) | This study |
Kmr, kanamycin resistance.
Pulse-chase labeling.
Pulse-chase labeling was performed essentially as described previously (14). Cells were grown overnight at 37°C in glycerol minimal medium supplemented with 0.1 mM amino acid mixture. After a 1-in-30 dilution of overnight culture into fresh medium supplemented with 0.4 mM IPTG, cells were grown to an optical density at 600 nm of 0.3. Cells were pulse-labeled with [35S]methionine (20 μCi/ml) for 20 s and then chased with cold methionine (20 mM) in the presence of a protein synthesis inhibitor, spectinomycin (200 μg/ml). Samples (0.5 ml) were taken 0.5, 1, 2, 5, 10, and 20 min postchase and immediately mixed with 0.5 ml of ice-cold 15% trichloroacetic acid. Cells were centrifuged at 12,000 × g for 5 min at 4°C. Pellets were washed once with 0.5 ml of 5% trichloroacetic acid and once with 95% ethanol and then dried under a vacuum. Dried pellets were resuspended in 200 μl of sodium dodecyl sulfate (SDS) buffer (20 mM Tris-HCl, pH 7.5; 10 mM EDTA; 1% SDS) and boiled for 5 min. Cell lysates (100 μl) were transferred into tubes containing 1 ml of the immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5; 20 mM NaCl; 1% Triton X-100) and MalE and LamB antibodies. Tubes were incubated overnight at 4°C on a rocker. To bring down immunocomplexes, heat-killed S. aureus cells (50 μl) were added, and tubes were incubated at 4°C on a rocker for at least 4 h. Immunocomplexes were recovered by centrifugation and washed twice in immunoprecipitation buffer, twice in Tris buffer (10 mM Tris-HCl; pH 7.5), and once in water. Washed pellets were resuspended in sample buffer, boiled, and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Gels were fluorographed, dried, and exposed to X-ray films at −85°C. Radioactive protein bands were quantified by using a Bio-Rad gel analyzer.
Membrane analysis.
Whole-cell envelopes were extracted by the French cell lysis method as described previously (12). The detergent solubility of membrane proteins was tested by mixing an equal amount of membrane and 2% Sarkosyl in 20 mM Tris-HCl (pH 7.5). The mixture was incubated for 30 min at room temperature and then centrifuged in a Beckman benchtop ultracentrifuge at 105,000 × g for 30 min at 4°C with a TLA 120 rotor. Both the supernatant and the detergent-insoluble pellet were recovered for protein analysis.
Separation of inner and outer membranes from whole-cell envelopes was achieved by sucrose (30 to 55%) density gradients according to the method of Misra et al. (14). After centrifugation, fractions were collected and the refractive index for each fraction was determined at 25°C. Densities for each fraction were calculated by their respective refractive index. Samples from each fraction were analyzed by SDS-PAGE.
Interaction of OmpC2Cys with DegPS210A.
In vivo interactions between OmpC2Cys and DegPS210A were examined by purifying the soluble His tag variant of DegPS210A (DegPS210A-6His) through Ni2+ affinity chromatography. Subsequently, fractions containing DegPS210A-6His were examined for the presence of OmpC2Cys by Western blot analysis. Briefly, 5 ml of cells containing pCS21 (in which the expression of DegPS210A-6His is inducible by IPTG) were grown to an optical density at 600 nm of 0.2 in the presence of 0.4 mM IPTG. Cells were pelleted and resuspended in 2 ml of 10 mM imidazole containing 20 μl of freshly prepared lysozyme (100 mg/ml). Cells were lysed by a small French pressure cell and, after the removal of unlysed cells by low-speed centrifugation, lysates were centrifuged at 105,000 × g in a 50 Ti rotor for 1 h at 4°C to pellet the membranes. DegPS210A-6His was purified from the supernatant by affinity chromatography in a Ni2+-nitrilotriacetic acid column according to the Pharmacia Biotech HisTrap purification protocol, except that the elution of DegPS210A-6His was carried out in several steps involving a gradual increase in the imidazole concentration from 20 to 200 mM. The DegPS210A-6His protein eluted with 200 mM imidazole.
Proteins were analyzed by SDS-PAGE and transferred into Immuno Lite membranes by using a Mini Transblot electrophoretic transfer cell (Bio-Rad). Blotted proteins were incubated for 90 min with primary antibodies raised against DegP (1:10,000 dilution) and OmpC (1:10,000 dilution) and then with the secondary antibody (goat anti-rabbit immunoglobulin G) for 1 h. Detection was carried out according to the manufacturer's protocol (Bio-Rad). Anti-DegP antibodies were kindly provided by Jon Beckwith (Harvard).
Protein density levels from Coomassie blue-stained gels and Western blots were determined by using Scion Image Analysis software.
RESULTS AND DISCUSSION
Overexpression of a protease-deficient DegPS210A eliminates the toxic effect of OmpC2Cys.
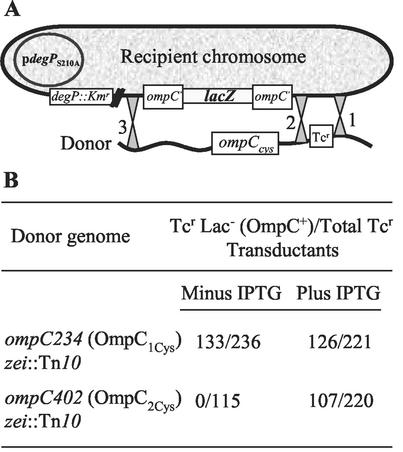
We have previously observed that the expression of assembly-defective OmpF mutants is lethal in a DegP− background (14). This is presumably because DegP degrades misfolded proteins, thus eliminating their toxic effects. As found for the assembly-defective OmpF mutants, the expression of OmpC2Cys also demanded the presence of DegP for cell viability. Initially, this conclusion was reached due to our inability to introduce, through P1 transduction, the ompC402 (OmpC2Cys) allele into a degP::Kmr strain. Subsequently, direct evidence for this synthetic lethality was obtained by demonstrating that the ompC402 allele could be transduced into a degP::Kmr background only when the expression of a protease-deficient DegPS210A protein was simultaneously induced from a plasmid replicon (Fig. 1). In contrast, the parental ompC234 (OmpC1Cys) allele was transducible into a degP::Kmr background at the expected frequency independent of DegPS210A's expression (Fig. 1B).
FIG. 1.
(A) Diagram showing two possible P1-mediated crossover events between the donor (ompC234 [OmpC1Cys] zei::Tn10 or (ompC402 [OmpC2Cys] zei::Tn10) and the recipient (ompC′::lacZ+ degP::Kmr/pdegPS210A) DNA. The cotransductional linkage between the zei::Tn10 (the selectable marker) and ompC is 50%. A crossover between 1 and 2 will yield tetracycline-resistant (Tcr) Lac+ (ompC′::lacZ+) transductants, whereas a crossover between 1 and 3 will yield Tcr Lac− (ompC) transductants. (B) P1 transduction results drawn from diagram presented in panel A.
DegPS210A does not stimulate OmpC2Cys degradation.
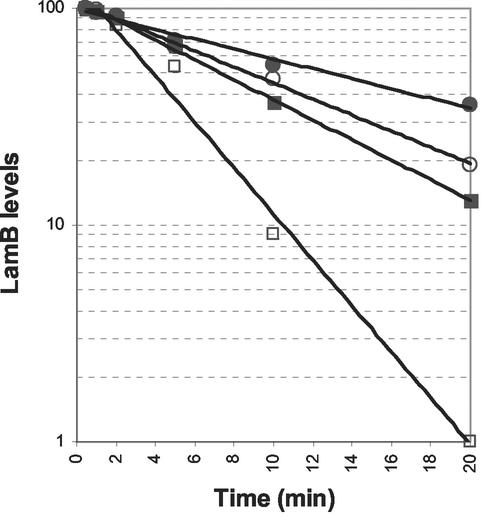
Since substitution of the only serine residue of DegP to alanine (S210A) is known to make it proteolytically inactive (29), it is unlikely that the rescue from OmpC2Cys-mediated lethality by DegPS210A involved degradation of the mutant protein. To substantiate this, we first confirmed that the overexpression of DegPS210A did not directly or indirectly induce proteolysis of mutant envelope proteins. For this we utilized a LamB amber fragment whose in vivo degradation is largely dependent on DegP (Fig. 2). Pulse-chase experiments showed that overexpression of DegPS210A did not increase the rate of degradation of the LamB amber fragment (Fig. 2). Instead, overexpression of DegPS210A in a DegP+ background increased the half-life of the LamB amber fragment (Fig. 2), thus suggesting a protective rather than a destructive role for the variant DegPS210A protein. This protective role of DegPS210A was most apparent in a background devoid of wild-type DegP. These results clearly demonstrated that the overexpression of DegPS210A did not involve increased proteolytic activity in vivo.
FIG. 2.
In vivo degradation of a LamB amber fragment in various genetic backgrounds. Freshly grown cells were subjected to pulse-chase analysis as described in Materials and Methods. LamB and MalE (as a gel loading control) were immunoprecipitated from [35S]methionine-labeled protein extracts obtained from DegP+ (□), DegP− (▪), DegP+/DegPS210A (○), and DegP−/DegPS210A (•). The highest LamB level was taken as 100, and the other LamB values were adjusted accordingly.
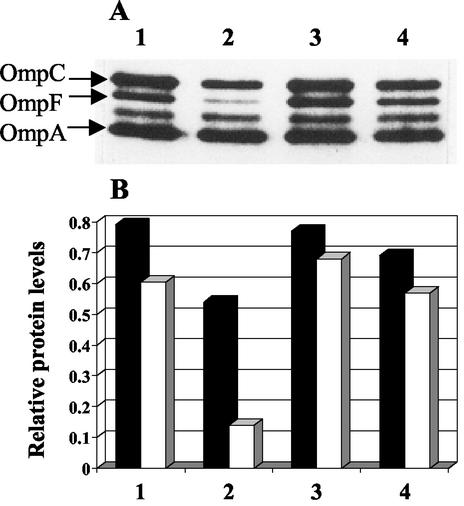
Next, we tested the effect of DegPS210A overexpression on mutant OmpC and wild-type OmpF. As found for the LamB amber fragment, the level of OmpC2Cys in the envelope increased when DegPS210A was overexpressed (Fig. 3). This result confirmed that the rescue from lethality did not involve degradation of the mutant protein. As mentioned earlier, the expression of OmpC2Cys imposes a negative effect on the assembly of the wild-type OmpF protein, presumably by interfering with the oligomerization of OmpF monomers or sequestering a common assembly factor (11). Under conditions in which DegPS210A overexpression elevated OmpC2Cys levels, it also elevated OmpF levels, thus abolishing the negative effect of OmpC2Cys on OmpF (Fig. 3, compare lanes 2 and 4).
FIG. 3.
Effect of DegPS210A on OmpC and OmpF. (A) Envelopes obtained from DegP+ OmpC1Cys (lane 1), DegP+ OmpC2Cys (lane 2), DegPS210A OmpC1Cys (lane 3), and DegPS210A OmpC2Cys (lane 4) were examined by SDS-PAGE. OmpC, OmpF, and OmpA bands were identified by Western blot analysis. (B) OmpC (solid bars) and OmpF (open bars) levels from panel A were quantified relative to OmpA.
DegPS210A increases the level of improperly assembled OmpC.
The elevation of OmpC2Cys levels and a concomitant increase in wild-type OmpF levels suggested that overexpression of DegPS210 improved the envelope's assembly or folding environment. Consistent with this notion, it was demonstrated by Spiess et al. (25) that the protease-deficient DegPS210A protein promoted proper folding of a periplasmic protein MalS. Also, based on genetic data, it has been recently argued that DegP may be involved as a general chaperone in the folding and assembly of OMPs (19). We therefore tested the possibility that the elevated OmpC2Cys levels caused by DegPS210A overexpression may be due to its improved assembly. For this we examined the solubility of membrane-bound OmpC in the anionic detergent Sarkosyl, which is known to solubilize inner membrane proteins and proteins that are loosely bound to the outer membrane. Since improperly assembled OMPs may not be correctly inserted in the outer membrane, they could be solubilized by Sarkosyl.
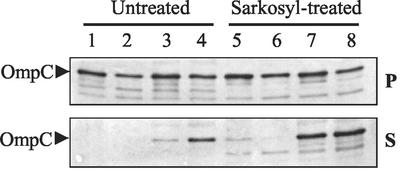
In the absence of DegPS210A overexpression, the parental and mutant OmpC proteins fractionated with the membrane pellet from untreated envelopes (Fig. 4, lanes 1 and 2). However, when DegPS210A was overexpressed, a significant portion of OmpC2Cys, and to some extent OmpC1Cys, became soluble even without detergent treatment (Fig. 4, lanes 3 and 4). When membranes obtained from cells not overexpressing DegPS210A were treated with Sarkosyl, OmpC2Cys, like the parental OmpC1Cys protein, was present primarily in the Sarkosyl-insoluble membrane pellet (Fig. 4, lanes 5 and 6 [top panel]), suggesting that at least as far as detergent solubility is concerned it was properly inserted in the outer membrane. Detergent treatment of membranes obtained from DegPS210A-overexpressing strains dislodged a large quantity of both OmpC species from the envelope (Fig. 4, lanes 7 and 8 [bottom panel]). These results showed that the DegPS210A-mediated increase in OmpC levels did not lead to the proper assembly or insertion of OmpC in the envelope. This finding was further supported by the observation that the increase in the OmpC2Cys level did not make cells sensitive to OmpC-specific bacteriophages, reflecting the continued presence of a conformationally defective OmpC2Cys. The observation that overexpression of DegPS210A causes the parental OmpC1Cys protein to become detergent soluble suggests that porin assembly intermediates in general can be targeted by DegPS210A. Consistent with this notion we found that a fraction of the assembly-competent wild-type OmpC protein was also subjected to DegPS210A-mediated detergent solubilization (data not shown). The lack of this effect on OmpA by DegPS210A overexpression may reflect distinct folding properties of its assembly intermediates compared to that of the trimeric porin proteins.
FIG. 4.
Sarkosyl solubility of membrane-associated OmpC. Purified whole-cell envelopes were treated either with just buffer (Untreated) or with Sarkosyl as indicated. After incubation, samples were centrifuged to separate Sarkosyl-soluble proteins (S, supernatant) from envelopes containing Sarkosyl-insoluble proteins (P, pellet). OmpC from each fraction was detected by Western blot analysis with polyclonal antibodies specific to OmpC. Lanes: 1 and 5, DegP+ OmpC1Cys; 2 and 6, DegP+ OmpC2Cys; 3 and 7, DegPS210A OmpC1Cys; 4 and 8, DegPS210A OmpC2Cys.
DegPS210A alters OmpC localization.
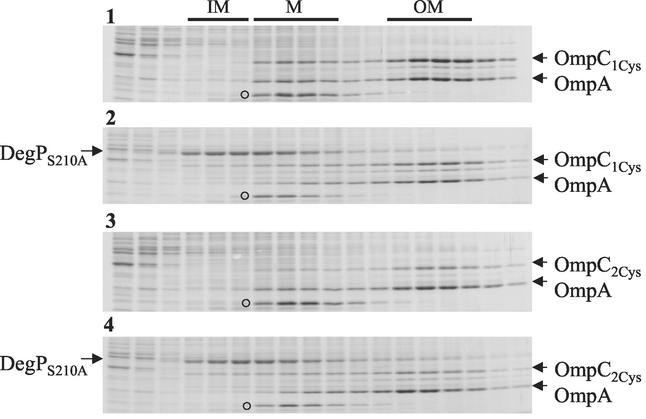
Since an increase in the mutant OmpC level by DegPS210A overexpression did not lead to its proper assembly or insertion, we were curious as to its localization in the cell envelope. This was examined by fractionating whole-cell envelopes into inner and outer membranes by using sucrose density gradients (Fig. 5). OmpC and OmpA (as a control protein) levels were quantified from the inner (buoyant densities of 1.15 to 1.16) and outer membrane (buoyant densities of 1.21 to 1.22) fractions (Table 2).
FIG. 5.
Sucrose density gradient analyses of whole-cell envelopes. Envelopes obtained from DegP+ OmpC1Cys, DegPS210A OmpC1Cys, DegP+ OmpC2Cys, and DegPS210A OmpC2Cys strains (panels 1 to 4, respectively) were fractionated into inner and outer membranes by sucrose density gradients. After centrifugation, fractions were collected from top and analyzed by SDS-PAGE. Protein bands were visualized by Coomassie blue staining. Fractions corresponding to inner membrane (IM), intermediate density material (M), and outer membrane (OM) are shown on top. Positions of OmpC, OmpA, DegPS210A, and a protein band (○) that primarily localizes with the intermediate-density material are indicated.
TABLE 2.
Distribution of OmpC and OmpA in outer and inner membrane fractions
| Relevant protein composition | Distribution of OmpC (%)
|
Distribution of OmpA (%)
|
||
|---|---|---|---|---|
| Outer membrane | Inner membrane | Outer membrane | Inner membrane | |
| OmpC1Cys DegP+ | 94 | 6 | 91 | 9 |
| OmpC1Cys DegPS210A | 82 | 18 | 94 | 6 |
| OmpC2Cys DegP+ | 92 | 8 | 90 | 10 |
| OmpC2Cys DegPS210A | 69 | 31 | 92 | 8 |
Remarkably, overexpression of DegPS210A caused the localization of some OmpC2Cys molecules to the inner membrane (Fig. 5) such that approximately four times more OmpC2Cys was present there compared to that in a DegP+ background (Table 2). This increase in the inner membrane was not due to an overall increase in mutant protein levels because, in contrast to the inner membrane, the level of OmpC2Cys decreased in the outer membrane (Table 2). Interestingly, the parental OmpC1Cys protein was also subjected to this effect of DegPS210A overexpression (Fig. 5), although a greater influence was observed for OmpC2Cys (Table 2). Unlike these variant OmpC proteins, overexpression of DegPS210A had no effect on OmpA's localization (Fig. 5 and Table 2); thus, this DegPS210A-mediated relocalization was specific to the mutant proteins.
Interestingly, fractionation studies revealed that a significant portion of DegPS210A was also localized in the inner membrane (Fig. 5). The inner membrane localization of DegPS210A is consistent with a detailed fractionation analysis carried out previously by Skorko-Glonek et al. (24). The colocalization of DegPS210A and OmpC in the inner membrane raises the possibility of direct interactions between these proteins. It is important to mention here that our fractionation studies showed that significant amounts of both DegPS210A and mutant OmpC proteins were also present in the soluble fraction; the data showing enhanced solubility of mutant OmpC due to increased DegPS210A expression are presented in Fig. 4. Experiments to examine a possible direct interaction between DegPS210A and OmpC2Cys from soluble fractions are described below.
Inner membrane-localized OmpC molecules have characteristics of unassembled polypeptides.
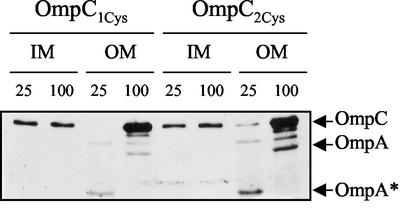
Properly assembled porin molecules are resistant to heat denaturation and detergent solubility. Heat sensitivity assays (Fig. 6) showed that the inner membrane-localized OmpC1Cys and OmpC2Cys denatured into monomers even when analyzed at room temperature (25°C). On the other hand, outer membrane-localized OmpC proteins displayed heat resistance that is characteristic of properly folded stable trimers (Fig. 6). Additionally, the inner membrane-localized OmpC molecules, in contrast to those localized in the outer membrane, could be completely solubilized by Sarkosyl (data not shown). Together, these results indicated that the inner membrane-localized OmpC molecules most likely represent unassembled polypeptides.
FIG. 6.
Heat sensitivity of OmpC1Cys and OmpC2Cys. Equal amounts of samples from the inner membrane (IM) and outer membrane (OM) were incubated at 25 or 100°C as indicated for 15 min prior to SDS-PAGE. OmpC was detected by Western blot analysis with a mixture of antibodies specific to OmpC and OmpA. OmpA* is a heat-modifiable form of OmpA.
DegPS210A directly interacts with OmpC2Cys.
As stated above, colocalization of subpopulations of OmpC2Cys and a fraction of DegPS210A in both the inner membrane and soluble fractions raised the possibility that these two proteins engage in direct interactions. To examine this, a variant DegPS210A containing a His6 tag was utilized. The His6-tagged DegPS210A protein (DegPS210A-6His) was expressed in a background also expressing OmpC2cys, and the soluble tagged DegP protein was purified through affinity chromatography with Ni2+ columns. Affinity-bound DegPS210A-6His was eluted with imidazole, and the eluted material was tested for the presence of mutant OmpC.
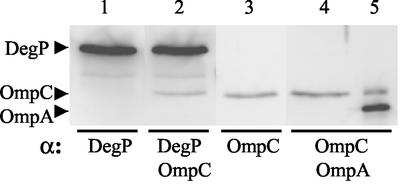
Four equal aliquots of the fraction containing eluted DegPS210A-6His were analyzed in separate lanes on a polyacrylamide gel and, after electroblotting, the filter was cut into strips corresponding to individual lanes and then hybridized with antibodies raised against DegP, OmpC, DegP plus OmpC, or OmpC plus OmpA (Fig. 7). The results showed unequivocally that the mutant OmpC2Cys polypeptides coeluted with the DegPS210A-6His protein (Fig. 7, lane 2). The absence of OmpA from the eluted DegPS210A signified the specificity of the DegPS210A-OmpC interaction (Fig. 7, lane 4). These results are consistent with the fractionation analysis (Table 2), which also showed a specific effect of DegPS210A on OmpC2Cys but not on OmpA.
FIG. 7.
Direct interaction between DegPS210A-6His and OmpC2Cys. Freshly grown cells expressing DegPS210A-6His and OmpC2Cys were lysed as described in Materials and Methods. Membrane-free cell lysates were subjected to Ni2+-nitrilotriacetic acid column chromatography to purify DegPS210A-6His. Four identical samples (lanes 1 to 4) of the eluted fraction containing DegPS210A-6His were subjected to SDS-PAGE and Western blot analyses with the indicated antibodies. An aliquot of outer membrane containing OmpC and OmpA proteins was analyzed in lane 5.
These results corroborated the notion that overexpression of DegPS210A relieved the toxicity of the mutant OmpC protein not by correcting assembly but rather by sequestering it. Capturing of the mutant OmpC2Cys protein by DegPS210A also alleviated the transdominant effect of mutant OmpC on OmpF's assembly (Fig. 3).
DegP and OMP assembly.
Both genetic (14, 15, 19) and biochemical (25) studies support the notion that DegP plays an important role in the assembly of envelope proteins. Initially, this was thought to be solely due to the degradation of mutant envelope proteins. However, the finding of Spiess et al. (25) suggested that DegP is also involved in the biogenesis of at least one periplasmic protein, MalS, as a general chaperone. Despite this exciting discovery, no other E. coli protein has been shown to be dependent on DegP's chaperone activity for its assembly; thus, the notion that DegP can act as a “general” chaperone still awaits further biochemical verification.
The data presented here and published earlier (14) showed that a protease-deficient DegPS210A, which retains its chaperone activity (25), stabilizes assembly-defective OMPs without actually improving their assembly. As demonstrated for OmpC2Cys in the present study, the stabilization of mutant proteins likely involves their sequestration by the polypeptide-binding cavity of DegPS210A (8). Such binding could improve the assembly of other OMPs indirectly by eliminating misfolded polypeptides that could directly interfere with the assembly of normal polypeptide chains. Also, misfolded polypeptides could potentially sequester folding factors and thus indirectly influence the assembly of normal proteins, so their removal by DegPS210A could prevent such a negative effect on overall assembly in the periplasm.
It can be argued that the overexpression of DegPS210A failed to improve OmpC2Cys assembly because the covalent disulfide bond formation would intrinsically disrupt normal assembly. Indeed, the absence of these bonds in a dsbA background allows normal OmpC2Cys assembly (11, 30). However, the assembly of OmpF mutants, in which the defect originates from noncovalent disruptions, was also not improved by DegPS210A overexpression (14). It thus remains to be seen whether DegPS210A assists OMP assembly by acting as a general chaperone. Elegant genetic studies carried out in Silhavy's lab showed that the synthetically lethal phenotype of degP::Tn10 surA::Kmr could be reversed by a plasmid expressing protease-deficient DegPS210A at a growth temperature of ≤37°C (19). Whether the observed reversibility by DegPS210A is through actually promoting assembly or sequestering misfolded polypeptides, as we have shown here, remained to be determined.
The recently solved crystal structure of protease-deficient DegP provides some exciting insights into the functioning of this protein (8). It suggests that partially folded polypeptides may be fed into DegP's central cavity which, in an open conformation, is accessible through the lateral pores. The presence of hydrophobic phenylalanine residues in the central cavity is likely to play an important role in substrate interaction. It is proposed that at lower temperatures, when the protease activity is dormant, unfolded substrates are released from DegP for refolding. In the case of substrates such as OmpC2Cys, where correct folding may be unattainable due to the covalent disulfide bond formation, the protein appears to be trapped eternally in this binding-release cycle.
Acknowledgments
We are grateful to Michael Ehrmann for providing plasmids expressing DegPS210A and its His tag derivative. We thank Leanne Misra for critically reading the manuscript.
This work was supported by a grant from the National Institutes of General Medical Sciences (GM48167). M.C. received funds from the Minority Graduate Education program.
REFERENCES
- 1.Ames, G. F.-L., E. N. Spudich, and H. Nikaido. 1974. Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. J. Bacteriol. 117:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardwell, J. C. A., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 3.Behrens, S., R. Maier, H. de Cock, H., F. X. Schmid, and C. A. Gross. 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 141:541-555. [DOI] [PubMed] [Google Scholar]
- 5.Chen, R., and U. Henning. 1996. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. 19:1287-1294. [DOI] [PubMed]
- 6.Danese, P. N., and T. Silhavy. 1999. Targeting and assembly of periplasmic and outer membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 7.Eisele, J.-L., and J. P. Rosenbusch. 1990. In vitro folding and oligomerization of a membrane protein. J. Biol. Chem. 265:10217-10220. [PubMed] [Google Scholar]
- 8.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 9.Laird, M. W., A. Kloser, and R. Misra. 1994. Assembly of LamB and OmpF in deep rough lipopolysaccharide mutants of Escherichia coli K-12. J. Bacteriol. 176:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misra, R. 1993. A novel ompC mutation of Escherichia coli K-12 that reduces OmpC and OmpF levels in the outer membrane. Mol. Microbiol. 10:1029-1035. [DOI] [PubMed] [Google Scholar]
- 12.Misra, R. 1993. OmpF assembly mutants of Escherichia coli K-12: isolation, characterization, and suppressor analysis. J. Bacteriol. 175:5049-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misra, R., and S. A. Benson. 1988. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J. Bacteriol. 170:3611-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misra, R., M. CastilloKeller, and M. Deng. 2000. Overexpression of protease-deficient DegPS210A rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP background. J. Bacteriol. 182:4882-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra, R., A. Peterson, T. Ferenci, and T. J. Silhavy. 1991. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J. Biol. Chem. 266:13592-13597. [PubMed] [Google Scholar]
- 16.Missiakas, D., J. M. Betton, and S. Raina. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA, and Skp/OmpH. Mol. Microbiol. 21:871-884. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido, H. 1994. Porins and specific diffusion channels in bacterial membranes. J. Biol. Chem. 269:3905-3908. [PubMed] [Google Scholar]
- 18.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizzitello, A. E., J. R. Harper, and T. J. Silhavy. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouvière, P. E., and C. A. Gross. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10:3170-3182. [DOI] [PubMed]
- 21.Schäfer, U., K. Beck, and M. Müller. 1999. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274:24567-24574. [DOI] [PubMed] [Google Scholar]
- 22.Schirmer, T. 1998. General and specific porins from bacterial outer membrane. J. Struct. Biol. 121:101-109. [DOI] [PubMed] [Google Scholar]
- 23.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Skorko-Glonek, J. B. Lipinska, K. Krzewski, G. Zolese, E. Bertoli, and F. Tanfani. 1997. HtrA heat shock protease interacts with phospholipid membranes and undergos coformational changes. J. Biol. Chem. 272:8974-8982. [DOI] [PubMed] [Google Scholar]
- 25.Spiess, C., A. Bell, and M. Ehrmann. 1999. A temperature-dependent switch from chaperones to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 26.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surrey, T., A. Schmid, and F. Jahnig. 1996. Folding and membrane insertion of the trimeric β-barrel protein OmpF. Biochmistry 35:2283-2288. [DOI] [PubMed] [Google Scholar]
- 28.Vakharia, H., and R. Misra. 1996. A genetic approach for analyzing surface-exposed regions of the OmpC protein of Escherichia coli K-12. Mol. Microbiol. 19:881-889. [DOI] [PubMed] [Google Scholar]
- 29.Waller, P. R. H., and R. T. Sauer. 1996. Characterization of degQ and degS. Escherichia coli genes encoding for homologs of the DegP protease. J. Bacteriol. 178:1146-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong, X., J. N. Deeter, and R. Misra. 1996. Assembly-defective OmpC mutants of Escherichia coli K-12. J. Bacteriol. 178:1213-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]