Abstract
The membrane topology of the ZntB Zn2+ transport protein of Salmonella enterica serovar Typhimurium was determined by constructing deletion derivatives of the protein and genetically fusing them to blaM or lacZ cassettes. The enzymatic activities of the hybrid proteins indicate that ZntB is a bitopic integral membrane protein consisting largely of two independent domains. The first 266 amino acids form a large, highly charged domain within the cytoplasm, while the remaining 61 residues form a small membrane domain containing two membrane-spanning segments. The overall orientation towards the cytoplasm is consistent with the ability of ZntB to facilitate zinc efflux.
Zinc is the second most abundant transition metal in biological systems (7, 11, 20). It is an essential element that is required to maintain the structural stability of macromolecules and to serve as a cofactor for more than 300 metabolic enzymes. Zinc also plays a prominent role in gene expression as a structural component in a large number of zinc-dependent transcription factors (3, 19). While the cellular requirement for zinc is absolute, excess concentrations of the cation are highly toxic. Consequently, the ability to maintain the intracellular concentrations of zinc within very narrow limits is a fundamental property of all living cells and must be achieved through the concerted actions of highly selective and highly regulated transport mechanisms. Our laboratory has recently identified ZntB as a novel zinc transport system in the enteric bacteria. Mutations in zntB render the cell hypersensitive to the cytotoxic effects of zinc and impair the cell's ability to extrude the cation (23). ZntB is homologous to the CorA family of transport proteins and, like CorA, appears to be widespread among the bacteria (13, 23). The protein is highly unusual for a transport protein in that it is relatively small and is predicted to possess a single membrane domain of minimal proportions. In this report, we use the well-developed gene fusion technique to determine the number and arrangement of membrane-spanning segments within this membrane domain and we determine the subcellular locations of the amino and carboxy termini.
Molecular characterization of zntB.
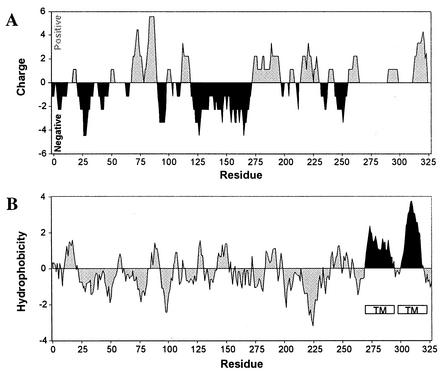
The physical structure of the zntB locus was determined by sequence analysis. The structural gene is situated between mcpA and dbpA at centisome 32. The mcpA gene encodes a homolog of the methyl-accepting chemotaxis receptor in Bacillus subtilis, and dbpA encodes a 23S rRNA helicase (8, 9). It is interesting that translation of both ZntB and DbpA initiates from an alternative GTG start codon. Fuller-Pace et al. have shown that changing the GTG start codon to ATG for translation of DbpA virtually abolishes expression from its native promoter (8). We have similarly mutated the ZntB start codon from GTG to ATG and found a 16-fold reduction in levels of ZntB expression (data not shown). The zntB structural gene encodes a protein consisting of 327 amino acids with a predicted molecular mass of 36 kDa. The amino acid sequence of ZntB is unusual for a membrane protein in that it is highly charged. Almost 27% of the amino acid residues carry a frank charge that is primarily distributed throughout the first 80% of the protein (Fig. 1A). Hydropathy profiles of the amino acid sequence were generated by the methods of Kyte and Doolittle, and of Rao and Argos, and by the use of other algorithms (1, 14). These plots predict a protein with a predominantly hydrophilic character except for two very hydrophobic regions at the C terminus with strong helix-forming potential sufficient to span the membrane bilayer (Fig. 1B). The preliminary model of topology that we have proposed describes ZntB as a bitopic protein that is integrated into the cell membrane by a single hydrophobic domain comprised of two membrane-spanning segments.
FIG. 1.
Charge distribution and hydropathy profile of the ZntB sequence. (A.) Regions of positive and negative charges were calculated by summing pK values of amino acids within a 15-residue sliding window. Regions with positive charge density are shaded grey, while regions with negative charge are shaded black. (B.) Hydropathy was determined by the algorithm of Kyte and Doolittle using a sliding window of 15 amino acids. Regions predicted to form transmembrane (TM) structures are denoted by open boxes.
Analysis of ZntB-BlaM and ZntB-LacZ chimeras.
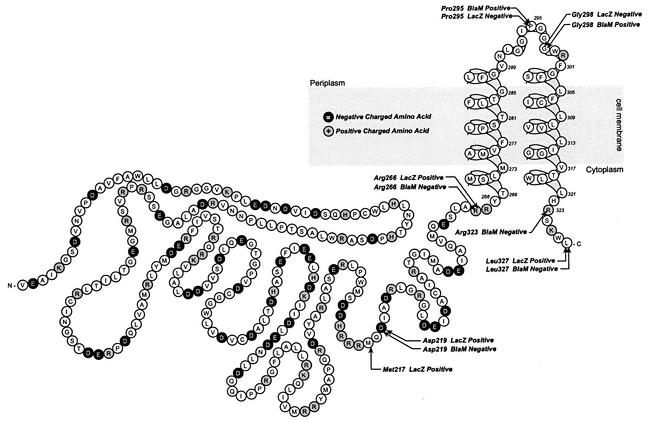
In order to validate the predictions of the Kyte-Doolittle hydropathy plots, a series of in-frame fusions of ZntB to BlaM and of ZntB to LacZ were constructed. The compartmental nature of BlaM and LacZ has been well established, and they are routinely used to investigate the topology of integral membrane proteins (4-6, 17). Deletion derivatives of ZntB were amplified from pAC5 by PCR using primers that generated unique restriction sites at the 5′ (HindIII) and 3′ (SalI) ends. The 3′ sites were situated to create an in-frame fusion to the reporter cassette upon ligation into either pAC23 (lacZ) or pAC31 (blaM). Twelve such fusions were constructed and sequenced to verify the fusion junctions. The MICs of ampicillin for strains expressing ZntB-BlaM fusions were determined by spotting 10 μl of a 10−6 dilution of an overnight culture onto Luria-Bertani agar plates containing antibiotic. The concentrations of ampicillin ranged from 0 to 450 μg/ml at 10-μg/ml intervals (24). Cells expressing the ZntB-BlaM fusion proteins were fractionated into periplasmic, cytosolic, and membrane components by the method of Harayama et al. (10). Western immunoblot analyses with anti-BlaM antibodies were performed on these fractions to determine the solubility and subcellular disposition of the chimeric proteins (2, 12, 15). β-Galactosidase specific activities of the ZntB-LacZ chimeras were determined as described by Miller (18). The locations of these fusions along with their enzymatic activities and subcellular locations are indicated in Table 1. LacZ fusions to residues Asp219, Met217, and Arg266 of ZntB conferred strong β-galactosidase activity, while BlaM fusions to the same residues failed to confer ampicillin resistance to the host strain. These combined activities are consistent with a cytoplasmic disposition of the N-terminal region of the protein. Moreover, the BlaM fusions to Asp219 and Arg266 failed to localize to the cell membrane when hybridized by Western blotting with the anti-BlaM antibody. Instead, these fusions remained soluble in the cytoplasmic fraction, indicating that no additional membrane-spanning segments exist within the first 266 amino acid residues of ZntB. BlaM fusions to Pro295 and Gly298 conferred resistance to ampicillin at a concentration of at least 450 μg/ml, while LacZ fusions to these residues failed to exhibit significant levels of β-galactosidase activity. The enzymatic activities of these fusion constructs are consistent with a periplasmic location for these residues, and they confirm the presence of a membrane-spanning segment situated between Arg266 and Pro295. The cellular location of the C terminus was determined by fusions to Arg323 and Leu327. BlaM fusions to these residues failed to confer ampicillin resistance, while a LacZ fusion at Leu327 displayed strong β-galactosidase activity. Moreover, BlaM fusions to Pro295, Gly298, and Leu327 produced hybrid proteins that localized to the membrane fraction. These data indicate that the C terminus of ZntB is located on the cytoplasmic surface of the cell membrane, and the data further confirm the presence of an additional transmembrane segment situated between Gly298 and Arg323.
TABLE 1.
Characteristics of ZntB chimeric proteins
| Fusion plasmid | Target residue | Reporter | Junction sequence
|
β-Lactamase activitya | β-Galactosidase activityb | Cellular dispositionc | ||
|---|---|---|---|---|---|---|---|---|
| ZntB | Spacer | Reporter | ||||||
| pJH122 | Met-217 | LacZ | HRRR | LEVP | SSNS | 324 | ||
| pAC27 | Asp-219 | LacZ | RMQD | VD | GPNS | 477 | ||
| pAC31 | Asp-219 | BlaM | RMQD | VD | HPET | 20 | Cyt | |
| pAC28 | Arg-266 | LacZ | LARR | VD | GPNS | 498 | ||
| pAC32 | Arg-266 | BlaM | LARR | VD | HPET | 20 | Cyt | |
| pAC29 | Pro-295 | LacZ | GGIP | VD | GPNS | 33 | ||
| pAC33 | Pro295 | BlaM | GGIP | VD | HPET | 450 | Mem | |
| pJH126 | Gly-298 | LacZ | PGGG | LEVP | SSNS | 00 | ||
| pJH127 | Gly-298 | BlaM | PGGG | LEVP | HPET | 450 | Mem | |
| pAC34 | Arg-323 | BlaM | WLHR | VD | HPET | 20 | Mem | |
| pAC39 | Leu327 | LacZ | SKWL | VD | GPNS | 230 | ||
| pAC45 | Leu327 | BlaM | SKWL | VD | HPET | 20 | Mem | |
β-Lactamase activity is expressed as the MIC of ampicillin (in micrograms per milliliter).
β-Galactosidase specific activity is expressed in Miller units.
Cellular disposition was determined by Western blot analysis of cellular fractions with anti-BlaM antibody. Mem, fusion proteins that localized to the membrane; Cyt, fusion proteins that remained soluble in the cytoplasmic fraction.
The combined enzymatic activities of the fusion constructs support a model which describes ZntB as a bitopic integral membrane protein consisting of two independent domains: a large, highly charged N-terminal domain and a small C-terminal membrane domain (Fig. 2). The membrane domain comprised of residues 267 to 327 contains two hydrophobic membrane-spanning segments that serve to orient both the C- and N-terminal regions of the protein in the cytoplasm. The precise positions of the helix boundaries cannot be resolved from the fusion techniques; however, their approximate positions can be estimated based on the distribution of charged residues in accordance with the observations of others (16, 21, 22). The two membrane-spanning segments are predicted by hydropathic profiling, and their boundaries seem fairly well defined by the positive charged residues on the cytoplasmic side. Other than an abundance of charge, the sequence and fusion data do not predict any prominent structural features of the N-terminal domain. It is likely that this domain serves to facilitate the acquisition and subsequent delivery of the cation to the transport channel. Given the minimal dimension of the ZntB membrane domain, it is not likely that a monomer of the protein would be sufficient to form a transport pore or channel. Therefore, the functional state of ZntB is likely to be that of an oligomer of unknown order and composition.
FIG. 2.
Model of the membrane topology of ZntB. Individual amino acids are depicted in single-letter code. Positively charged residues are enclosed in grey circles; negatively charged residues are enclosed in black circles. Locations of BlaM and LacZ fusions are labeled. The text following each fusion indicates the phenotype of the strain containing the plasmid encoding the chimera.
Acknowledgments
This work was supported by a grant from the Welch Foundation (Y1485) to R.L.S.
REFERENCES
- 1.Argos, P., and J. K. Rao. 1986. Prediction of protein structure. Methods Enzymol. 130:185-207. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 3.Bohm, S., D. Frishman, and H. W. Mewes. 1997. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 25:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., C. Manoil, and J. Beckwith. 1987. Determinants of membrane protein topology. Proc. Natl. Acad. Sci. USA 84:8525-8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broome-Smith, J. K., M. Tadayyon, and Y. Zhang. 1990. Beta-lactamase as a probe of membrane protein assembly and protein export. Mol. Microbiol. 4:1637-1644. [DOI] [PubMed] [Google Scholar]
- 6.Calamia, J., and C. Manoil. 1990. Lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc. Natl. Acad. Sci. USA 87:4937-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman, J. E. 1992. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu. Rev. Biochem. 61:897-946. [DOI] [PubMed] [Google Scholar]
- 8.Fuller-Pace, F. V., S. M. Nicol, A. D. Reid, and D. P. Lane. 1993. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 12:3619-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanlon, D. W., and G. W. Ordal. 1994. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J. Biol. Chem. 269:14038-14046. [PubMed] [Google Scholar]
- 10.Harayama, S., J. Bollinger, T. Iino, and G. L. Hazelbauer. 1983. Characterization of the mgl operon of Escherichia coli by transposon mutagenesis and molecular cloning. J. Bacteriol. 153:408-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, E. P. 1997. Metal ions and synaptic transmission: think zinc. Proc. Natl. Acad. Sci. USA 94:13386-13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, D. D., J. W. Gautsh, F. R. Sportsman, and J. H. Elder. 1984. Improved technique utilizing non-fat dry milk for analysis of proteins and nucleic acids. Gene Anal. Tech. 1:3-8. [Google Scholar]
- 13.Kehres, D. G., C. H. Lawyer, and M. E. Maguire. 1998. The CorA magnesium transporter gene family. Microb. Comp. Genomics 3:151-169. [DOI] [PubMed] [Google Scholar]
- 14.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J.-I., A. Kuhn, and R. E. Dalbey. 1992. Distinct domains of an oligotopic membrane protein are Sec-dependent and Sec-independent for membrane insertion. J. Biol. Chem. 267:938-943. [PubMed] [Google Scholar]
- 17.Manoil, C. 1990. Analysis of protein localization by use of gene fusions with complementary properties. J. Bacteriol. 172:1035-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24:4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suhy, D. A., K. D. Simon, D. I. H. Linzer, and T. V. O'Halloran. 1999. Metallothionein is part of a zinc-scavenging mechanism for cell survival under conditions of extreme zinc deprivation. J. Biol. Chem. 247:9183-9192. [DOI] [PubMed] [Google Scholar]
- 21.Von Heijne, G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 22.Von Heijne, G., and C. Manoil. 1990. Membrane proteins: from sequence to structure. Protein Eng. 4:109-112. [DOI] [PubMed] [Google Scholar]
- 23.Worlock, A. J., and R. L. Smith. 2002. ZntB is a novel Zn2+ transporter in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4369-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, J., L. S. Tisa, and B. P. Rosen. 1992. Membrane topology of the ArsB protein, the membrane subunit of an anion-translocating ATPase. J. Biol. Chem. 267:12570-12576. [PubMed] [Google Scholar]