Abstract
In Escherichia coli, the min system prevents division away from midcell through topological regulation of MinC, an inhibitor of Z-ring formation. The topological regulation involves oscillation of MinC between the poles of the cell under the direction of the MinDE oscillator. Since the mechanism of MinC involvement in the oscillation is unknown, we investigated the interaction of MinC with the other Min proteins. We observed that MinD dimerized in the presence of ATP and interacted with MinC. In the presence of a phospholipid bilayer, MinD bound to the bilayer and recruited MinC in an ATP-dependent manner. Addition of MinE to the MinCD-bilayer complex resulted in release of both MinC and MinD. The release of MinC did not require ATP hydrolysis, indicating that MinE could displace MinC from the MinD-bilayer complex. In contrast, MinC was unable to displace MinE bound to the MinD-bilayer complex. These results suggest that MinE induces a conformational change in MinD bound to the bilayer that results in the release of MinC. Also, it is argued that binding of MinD to the membrane activates MinC.
In Escherichia coli, division of a cell into two equal-sized progeny cells follows from assembly of the Z ring at midcell (2, 22, 23). Assembly of the Z ring at other locations within the cell is prevented by a combination of the nucleoid and the min system (6, 25, 34). An unknown mechanism prevents Z rings from forming on top of the nucleoids, and the min system prevents Z rings from assembling at the poles, thereby preventing minicell formation.
Initial characterization of the min system revealed that it consists of a bipartite inhibitor of division, encoded by minC and minD, which is topologically regulated by minE (6). Efficient division inhibition requires both minC and minD; however, overexpression of MinC but not MinD inhibits division, indicating that MinC is an inhibitor which is stimulated by MinD (7). Subsequent localization studies demonstrated that MinD recruits MinC to the membrane, raising the possibility that it activates MinC by recruiting it to the membrane (13, 28). More recent studies have shown that MinD can also target the C-terminal domain of MinC to the septum, indicating that the MinCD complex has a high affinity for some septal component (18). It is not clear if this targeting to the septum requires that the MinD-MinC complex first bind to the membrane.
MinC and MinD were initially reported to inhibit division by preventing Z-ring formation (1). A subsequent study suggested that MinC might act after Z-ring assembly by preventing FtsA from localizing to the Z ring (19). However, reexamination of this issue confirmed that MinC and MinD inhibit division by blocking Z-ring formation (25). Consistent with this, overexpression of a MalE-MinC fusion also inhibits division by preventing Z-ring formation (16). These results are also consistent with in vitro results which revealed that MinC is an antagonist of FtsZ assembly (16). Dissection of MinC revealed that it consists of two functional domains, an N-terminal domain that antagonizes FtsZ assembly and a C-terminal domain responsible for dimerization and interaction with MinD (14, 33). The crystal structure of MinC confirmed that the N- and C-terminal domains are structurally independent (3).
One of the most intriguing aspects of the min system is the topological regulation of the MinC inhibitor by MinD and MinE. This regulation allows Z ring formation at midcell but not at the poles and involves a remarkable oscillation of the Min proteins between the poles of the cell, with a period of about 50 s (13, 28, 29). The oscillation of MinC requires MinD and MinE, which also oscillate (8, 10). MinC is a passenger in the oscillation and is recruited to participate by MinD. During an oscillatory cycle, a GFP (green fluorescent protein)-MinD or GFP-MinC fusion protein appears as a horseshoe-shaped fluorescence (appearance in two dimensions) in one half of the cell. The arms of the horseshoe then recede towards the pole, a new horseshoe appears in the other half of the cell, and the cycle is repeated. MinE is mostly present in a ring at the receding edge of the horseshoe that appears to be coupled to the movement of MinD (8, 10, 27). Through this oscillation, the time-average concentration of the MinC inhibitor is lowest at midcell and highest at the poles (24, 29).
In vitro studies demonstrated that MinE stimulates the ATPase activity of MinD in the presence of phospholipid vesicles (15). This stimulatory activity of MinE correlates with the ability of MinE to induce oscillation of MinD and led to a model in which the MinE ring at the receding arms of the MinD horseshoe stimulates the MinD ATPase, releasing it from the membrane (15).
The requirement for phospholipid vesicles in the activation of MinD ATPase indicated that MinD interacts directly with phospholipids (15). Recent work confirmed that MinD bound to phospholipid vesicles in the presence of ATP and determined that it assembles into polymers that deform the vesicles into tubes (12). MinE addition induced ATP hydrolysis and the MinD tubes disassembled, releasing MinD from the vesicles. This reversible assembly of MinD on the surface of phospholipid vesicles, regulated by ATP and MinE, provides a biochemical mechanism for the reversible assembly of MinD on the membrane that is induced by MinE in vivo (29).
Although the MinDE oscillator positions MinC, it is not clear how MinC participation is regulated. Yeast two-hybrid studies have indicated that MinC and MinD interact and that MinE dampens this interaction (17). We therefore investigated interaction among the Min proteins to determine how MinC participation is regulated.
MATERIALS AND METHODS
Expression and purification of proteins.
The plasmids pJC90, pZH101, pZH111, pZH112, pZH115, pJPB216, and pZH216-4 were used for the expression of MalE-Lacα, MalE-MinC, MalE-MinC1-115, MalE-MinC116-231, MinD, MinE, and MinE4, respectively. These constructs have been described previously (14-16). Two buffers were used in this investigation. Pol buffer contained 50 mM MES (morpholineethanesulfonic acid, pH 6.5), 50 mM KCl, and 10 mM MgCl2. ATPase buffer consisted of 25 mM Tris-HCl (pH 7.5), 50 mM KCl, and 5 mM MgCl2 supplemented with 100 mM NaCl.
Phospholipid bicelles.
The effect of MinD on the recruitment of MinC and MinE to lipid bilayers was investigated by using bicelles. A bicelle solution (5 ml) was made by diluting a diheptanoylphosphatidylcholine solution (20 mg/ml) 1:10 in ATPase buffer at ≈40°C. Then 27 mg of dimyristoylphosphatidylcholine and 27 mg of dimyristoylphosphatidylglycerol were added, and the solution cycled several times with vortexing between 37°C and 4°C to accelerate hydration of the long-chain phospholipids. The bicelles were stored at 4°C and used at a concentration of 0.4% (4 mg/ml). All phospholipids were purchased from Avanti Polar Lipids (Alabaster, Ala.) and used according to instructions.
We found that bicelles with this composition supported the ability of MinE to stimulate MinD ATPase to the same extent as small unilamellar vesicles prepared from E. coli phospholipids that were used previously (12) (data not shown). Also, MinD appeared to bind to bicelles with the same affinity that it bound to small unilamellar vesicles. The bicelles were used in place of the small unilamellar vesicles because of their stability and ease of preparation.
Size-exclusion chromatography.
Proteins to be analyzed by size-exclusion chromatography on fast protein liquid chromatography were incubated in Pol buffer with or without 5 mM ADP or ATP. Samples incubated with nucleotide were chromatographed with Pol buffer containing 0.5 mM of the same nucleotide. The volume and amount of protein loaded onto a Superose 6 column (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) were as follows: MinD, 275 μl and 0.82 μg/μl; and MalE-MinC, 275 μl and 1.9 μg/μl. The same concentrations were used when the proteins were mixed prior to loading. Fractions (0.4 ml) were collected, and 10 μl was run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were stained with Coomassie brilliant blue and photographed.
Proteins binding to bicelles and protein quantitation.
The binding of proteins to bicelles was determined by sedimentation as described previously (12) except that bicelles were substituted for small unilamellar vesicles. In all experiments, MinD (4 μM) was incubated in ATPase buffer (150 μl final volume) at room temperature with ADP, ATP, or ATPγS at 1 mM. Bicelles were added at 4 mg/ml. MalE-MinC, MalE-MinC116-231, or other MalE fusions (MalE-MinC1-115 or MalE-Lacα) were added at 3 μM or as indicated in the figures. In some cases MinE or MinE4 was added (at 4 μM or as indicated in the figures). In control experiments, we determined that the time of incubation between additions of bicelles or various proteins did not affect the results. Therefore, additions were made in the order described, with about 1 min elapsing between additions.
Samples were centrifuged immediately at 80,000 rpm at 25°C in a Beckman TLA 100.2 rotor unless indicated otherwise. After centrifugation, the supernatants were carefully removed, and the pellets were resuspended in 150 μl of SDS sample buffer. A 10-μl aliquot of the sample was electrophoresed on SDS-12 or 14% PAGE and stained with Coomassie brilliant blue, and the bands were quantitated with digital imaging equipment from Alpha Innotech (San Leandro, Calif.). Standard curves with known amounts of the proteins were prepared to ensure that all determinations were within the linear range for densitometry.
The fraction of the total MinD, MinE, or MalE-MinC116-231 in the pellet after centrifugation was determined as follows. Two identical samples were prepared. One was centrifuged, and the pellet was resuspended to the original volume. Then 10-μl aliquots from both the centrifuged and noncentrifuged samples were mixed with 10 μl of SDS sample buffer and run on the same gel. Comparison of the amount of protein on the gel from these parallel samples allowed the fraction of the total protein in the pellet (bound to the bicelles) to be determined.
In these experiments, the ratio of the various Min proteins can be determined by comparing their spot densities. To determine the relative spot densities for the different proteins, a 10-μl aliquot of sample containing proteins at 4, 6, or 10 μM was analyzed by SDS-PAGE and stained with Coomassie. The average ratio of the spot densities from MalE-MinC116-231, MinD, and MinE was 1 to 0.57 to 0.15, respectively (the standard error for the different spot densities was less than 10% for each protein). This ratio corresponds closely to the ratio of their molecular weights (55,000 to 29,500 to 10,200, respectively, that is, 1 to 0.55 to 0.18), indicating that the binding of Coomassie by these proteins is proportional to their molecular size.
RESULTS
ATP-dependent interaction between MinC and MinD.
As an initial approach to examining the interaction between MinC and MinD, we took advantage of MalE-MinC binding to amylose resin. The MalE-MinC fusion, which can be readily purified, retains division-inhibitory activity and can be activated by MinD in vivo (16). MinD in the presence of ADP or ATP was incubated with amylose resin bound with MalE-MinC. We did not observe retention of MinD to the resin with either nucleotide, indicating that MinD did not bind to MalE-MinC or that the affinity was weak and MinD was removed during the wash steps (data not shown).
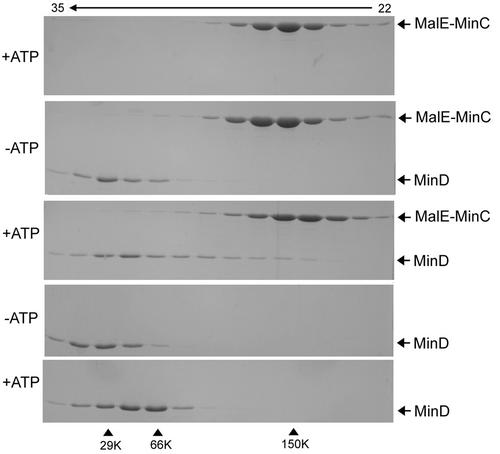
As an alternative approach to examining a possible interaction between MinC and MinD, we used size-exclusion chromatography. In the presence of ADP or in the absence of nucleotide, MinD eluted at the position expected for a monomer (Fig. 1, fourth panel [MinD is 29.6K], only the absence of nucleotide is shown; results with ADP were identical); however, in the presence of ATP, MinD was shifted to the dimer position (Fig. 1, bottom panel). Dimer formation by MinD has not been reported previously, although MinD has been shown to self-associate in the yeast two-hybrid system (32) and to assemble into polymers on the surface of vesicles (12). MalE-MinC eluted in the dimer to trimer range, as shown previously (14), and the elution profile was not affected by the addition of ATP (Fig. 1, top panel).
FIG. 1.
Size-exclusion chromatography of MalE-MinC and MinD. Proteins were incubated with or without nucleotide and analyzed by FPLC on a Superose 6 column. The elution buffer contained the same nucleotide as the preincubation mix at 0.5 mM. Aliquots of fractions were analyzed by SDS-PAGE. Top panel, MalE-MinC with ATP; second panel, MalE-MinC and MinD without nucleotide; third panel, MalE-MinC and MinD in the presence of ATP; fourth panel, MinD without nucleotide (the profile with ADP was the same); and last panel, MinD in the presence of ATP. The fraction numbers are indicated at the top. The size standards used were carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), and alcohol dehydrogenase (150 kDa).
Mixing MalE-MinC and MinD in the presence of ADP did not affect the positions at which they eluted (Fig. 1, second panel); each protein eluted at the same position as when chromatographed separately with ADP or without nucleotide. When MinD and MalE-MinC were cochromatographed in the presence of ATP, a portion of the MinD was shifted to an earlier elution than the dimer position (Fig. 1, third panel). Examination of the MalE-MinC elution profiles indicated that it was also shifted to a larger size in the presence of MinD and ATP. A likely explanation for the altered elution profiles is that MalE-MinC and MinD formed a complex in the presence of ATP, however, this complex is unstable and breaks down during the chromatography. A weak association between MalE-MinC and MinD is consistent with the failure of MinD to be retained on amylose resin containing MalE-MinC.
MinD recruits MinC to phospholipid bicelles.
Previously, we found that MinD bound to phospholipid vesicles in the presence of ATP (12). To determine if MinD could recruit MinC to vesicles, we used phospholipid bicelles (bilayered micelles), composed of a 50:50 mixture of dimyristoylphosphatidylcholine and dimyristoylphosphatidylglycerol for vesicles reconstituted from E. coli phospholipids that we used previously (15). We observed that bicelles with this composition were just as effective in supporting MinE's stimulation of MinD ATPase as phospholipid vesicles and were used because they are more stable and readily prepared (data not shown).
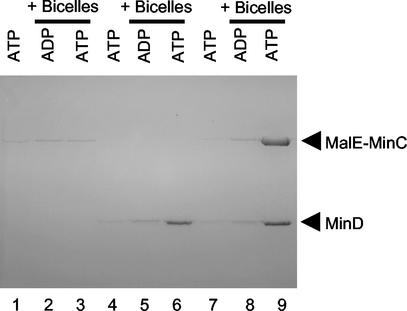
The control confirmed that under these buffer conditions (pH 7.5 and 150 mM salt [50 mM KCl plus 100 mM NaCl]), MinD pelleted with the bicelles if ATP was added but not if ADP was added (Fig. 2, lanes 5 and 6). MinD did not pellet with ATP in the absence of bicelles (Fig. 2, lane 4). MalE-MinC did not bind to bicelles in the presence of ADP or ATP (Fig. 2, lanes 2 and 3). However, addition of MalE-MinC and MinD to the bicelles led to the ATP-dependent appearance of MalE-MinC in the pellet along with MinD (Fig. 2, lanes 8 and 9). Quantitation revealed that 80% of the MalE-MinC and 88% of the MinD was associated with the bicelles (Table 1). These results demonstrate that MinC and MinD interact under these buffer conditions and that MinD recruits MinC to the bicelles in the presence of ATP. ATPγS also supported the appearance of these proteins in the pellet (although not to the same extent), whereas AMPPCP did not (Table 1). We previously argued that for MinD ATPγS is a better analogue of ATP than AMPPCP (12).
FIG. 2.
MinD recruits MalE-MinC to bicelles in the presence of ATP. MalE-MinC (3 μM) and MinD (4 μM) were incubated separately or together with or without bicelles in the presence of ATP or ADP. The reaction mixtures were immediately centrifuged, and the pellets were analyzed by SDS-PAGE. MalE-MinC was added to lanes 1 to 3, MinD was added to lanes 4 to 6, and both MalE-MinC and MinD were added to lanes 7 to 9. The samples with bicelles and nucleotide added are indicated at the top of the figure.
TABLE 1.
Effect of various nucleotides on binding of MinC and MinD to bicellesa
| Addition | % of protein in pelletb
|
|
|---|---|---|
| MalE-MinC | MinD | |
| ADP | 5 | 8 |
| ATP | 80 | 88 |
| ATP-γ-S | 53 | 63 |
| AMPPCP | 6 | 12 |
Nucleotides were present at 1 mM, and the protein concentrations were 3 μM MalE-MinC and 4 μM MinD. The proteins were mixed with bicelles (4 mg/ml) and 1 mM ATP. The samples were immediately centrifuged, and the amounts of MinD and MinC in the pellet were determined by SDS-PAGE and densitometry.
Calculated as the ratio of the amount of MinC and MinD in the pellet to total MinC and MinD in the sample. The ratio was determined by comparing the spot densities to that obtained from an aliquot of an uncentrifuged control.
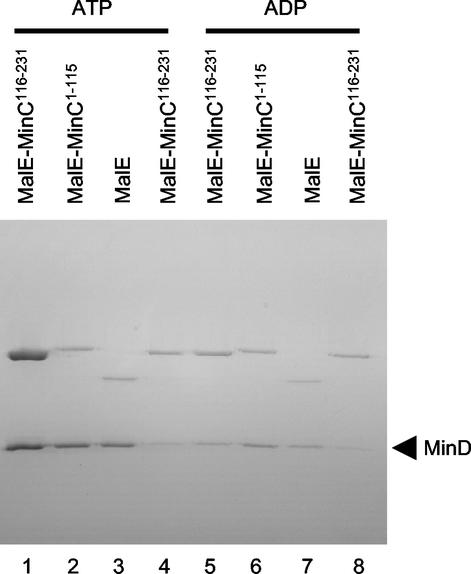
To further examine the specificity of the interaction between MinC and MinD in the presence of bicelles, we compared the ability of MinD to recruit the N-terminal and C-terminal domains of MinC to the bicelles. Yeast two-hybrid studies demonstrate that MinD interacts with the C-terminal domain of MinC (14, 18). As expected, only the C-terminal domain was recruited to the bicelles in an ATP-dependent manner (Fig. 3, compare lanes 1 and 5). The N-terminal domain of neither MinC nor MalE-Lacα, used as a control, was recruited to the bicelles by MinD (Fig. 3, lanes 2 and 3). As an additional control, we used MinD K16Q, which fails to bind vesicles in vitro and localize to the membrane in vivo (12). Neither MalE-MinC nor MinD K16Q was in the pellet, emphasizing the requirement for a functional MinD for the recruitment of MalE-MinC to the bicelles (Fig. 3, lanes 4 and 8).
FIG. 3.
MinD recruits MinC to bicelles through the C-terminal domain of MinC. MinD (4 μM) was incubated with bicelles and ATP. Various MalE fusions (4 μM) were added, the samples were immediately centrifuged, and the pellets were analyzed by SDS-PAGE. The specific MalE fusion is indicated at the top of the figure. The nucleotide used in the reaction is also indicated at the top of the figure. In lanes 4 and 8, MinD K16Q, which binds poorly to vesicles (12), was used in place of MinD.
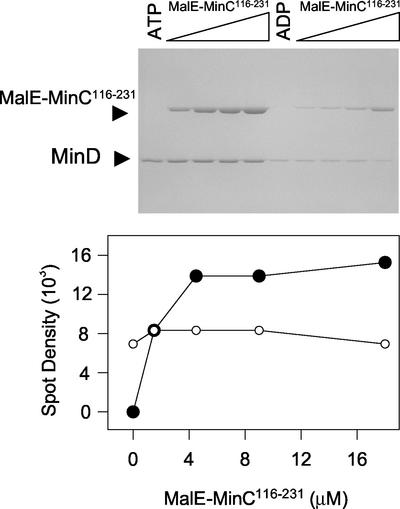
The stoichiometry of the binding of MinC to MinD in the presence of bicelles was determined. For this and subsequent experiments, we used MalE-MinC116-231 because it behaved similarly to the full-length MalE-MinC fusion. The concentration of MinD in the reaction was 4 μM, and MalE-MinC116-231 was varied from 0 to 18 μM (Fig. 4). The binding of MalE-MinC116-231 to the MinD-bicelle complex was saturable. The molar ratio of MalE-MinC116-231 to MinD in the pellet was determined to be 1 from the following data. The ratio of the spot densities of MalE-MinC116-231 to MinD at saturation in Fig. 4 is 1.9 ± 0.4, which is close to the value of 1.8 ± 0.3 obtained by determining the spot densities of equimolar amounts of these proteins run on a control gel, as described in Materials and Methods.
FIG. 4.
Saturable binding of MalE-MinC116-231 to the MinD-bicelle complex. MinD (4 μM) was incubated with bicelles in the presence of ATP or ADP. Increasing concentrations of MalE-MinC116-231 were added, the reactions were immediately centrifuged, and pellets were analyzed by SDS-PAGE (top panel). The amounts of MinD and MalE-MinC116-231 in the pellet were determined by densitometry. The amount of MalE-MinC116-231 and MinD in the pellet in the presence of ADP (lanes 6 to 10) was subtracted from the values obtained in the presence of ATP (lanes 1 to 5) and plotted (bottom panel). It was determined that the ratio of the spot densities for an equimolar mixture of MalE-MinC116-231 and MinD was 1.8 (described in Materials and Methods). This ratio is similar to that obtained from averaging the ratios of the spot densities of MalE-MinC to MinD in the last three lanes, 1.9, in which MalE-MinC116-231 is saturating.
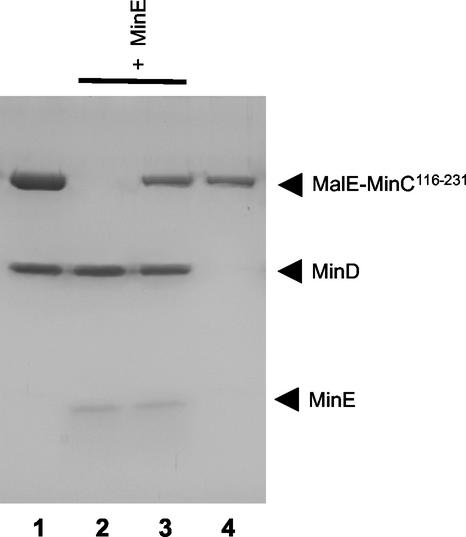
MinE removes MinC along with MinD from the bicelles.
Previously, we reported that MinE was able to remove MinD from phospholipid vesicles (12). This displacement required ATP hydrolysis, indicating that MinE's ability to stimulate MinD ATPase resulted in release of the ADP form of MinD. This result suggested that MinE should also cause release of MinC since, as we have shown, it is recruited to the membrane through MinD. As seen in Fig. 5 (lanes 1 and 2), addition of MinE to the bicelles incubated in the presence of MinD and ATP resulted in a decrease in the recovery of MinD in the pellet, as reported previously (12). Addition of MinE to the bicelles incubated with MinD, MalE-MinC116-231, and ATP resulted in decreased recovery of both MinD and MalE-MinC116-231 (Fig. 5, lanes 3 and 4).
FIG. 5.
MinE removes MinC along with MinD from bicelles. MinD (4 μM), bicelles, and ATP were incubated with (lanes 3 to 5) or without MalE-MinC116-231 (lanes 1 and 2) (3 μM). After 1 min, MinE (4 μM) was added to lanes 2 and 4, and MinE4 (4 μM) was added to lane 5. The reaction mixtures were immediately centrifuged, and the pellets were analyzed by SDS-PAGE.
As a control, we added MinE4, a mutant that is unable to stimulate MinD ATPase (15) (Fig. 5, lane 5). MinE4 did not remove MinD or MalE-MinC116-231 from the bicelles and was not recovered in the pellet. This is in contrast to MinE, which remained bound to the MinD-bicelle complex when ATP hydrolysis was prevented (by using ATPγS; also see Fig. 6, below) (12). This result demonstrates that MinE4 is deficient in binding to MinD, which would explain its inability to stimulate MinD ATPase. Thus, MinE is able to remove MinD and MalE-MinC116-231 from the bicelles in a reaction in which stimulation of MinD ATPase is occurring.
FIG. 6.
MinE displaces MinC from the MinCD-bicelle complex in the absence of ATP hydrolysis. MinD (4 μM) and bicelles were incubated in the presence of ATPγS. After 1 min, MalE-MinC116-231 (3 μM) was added to lanes 3 to 5. After an additional minute, MinE (lanes 2 and 4) or MinE4 (lane 5) was added (4 μM). The samples were immediately centrifuged and analyzed by SDS-PAGE.
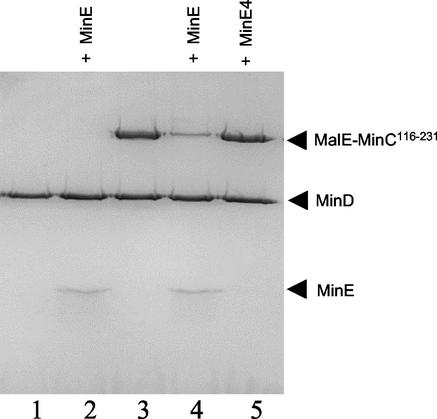
MinE displaces MinC from MinCD-bicelle complex in the absence of ATP hydrolysis.
The above experiments demonstrated that MinE caused the release of MinC from the bicelles along with the MinD. To test if ATP hydrolysis was required, the experiment in Fig. 5 was repeated with the nonhydrolyzable analog ATPγS. The addition of MinE to bicelles incubated with MinD or MinD and MalE-MinC116-231 did not reduce MinD binding to the bicelles (Fig. 6, lanes 1 and 2), confirming that ATP hydrolysis was necessary, as shown previously (12). Surprisingly, however, MalE-MinC116-231 was displaced by MinE (Fig. 6, lanes 3 and 4). In contrast, MinE4 was unable to bind to the complex or to displace MalE-MinC116-231 from the complex (Fig. 6, lane 5).
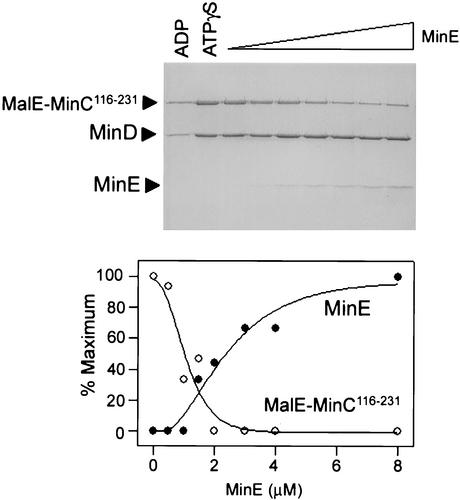
To further examine the displacement of MalE-MinC116-231 by MinE, a titration experiment was carried out (Fig. 7). MinD (4 μM) and MalE-MinC116-231 (3 μM) were mixed with bicelles in the presence of ATPγS. Increasing amounts of MinE were added, and the relative amounts of MalE-MinC116-231 and MinE bound to the bicelles were determined after centrifugation. The amount of MinD bound to bicelles was constant, varying less than 10% between the various lanes. Plotting the relative amounts of MinE and MalE-MinC116-231 bound to the MinD-bicelle complex demonstrated that MinE efficiently displaced MalE-MinC116-231 from the MinD-bicelle complex. At the highest concentration of MinE (the last lane in Fig. 7), the spot density ratio of MinD to MinE was 4.2. The spot density ratio of an equimolar mixture of MinD and MinE run on a gel and analyzed in the same way was 4. This result indicates that the ratio of MinE to MinD bound to the bicelles at saturating levels of MinE was 1, indicating that one molecule of MinD recruits one molecule of MinE to the bicelles.
FIG. 7.
Titration of the MinCD-bicelle complex with MinE. MinD (4 μM) and MinC116-231 (3 μM) were incubated with bicelles and ATPγS. After 1 min, MinE (0 to 12 μM) was added, the reaction mixtures were centrifuged immediately, and the pellets were analyzed by SDS-PAGE. The relative amounts of MinC, MinD, and MinE bound to bicelles were determined by densitometry and plotted after subtracting the amounts in the pellet observed with ADP (lane 1). The error for the amount of MinD in the pellet with ATPγS was 10%. For MinE, the amount in the last lane was set at 100%, whereas with MalE-MinC116-231, the amount in lane 2 (absence of MinE) was set at 100%. The ratio of the spot densities of MinD to MinE in the last lane, where MinE is in excess of MinD, is 4.2. This value is similar to that of a control, in which the ratio of spot densities for equimolar mixtures of MinD and MinE was found to be 4.0.
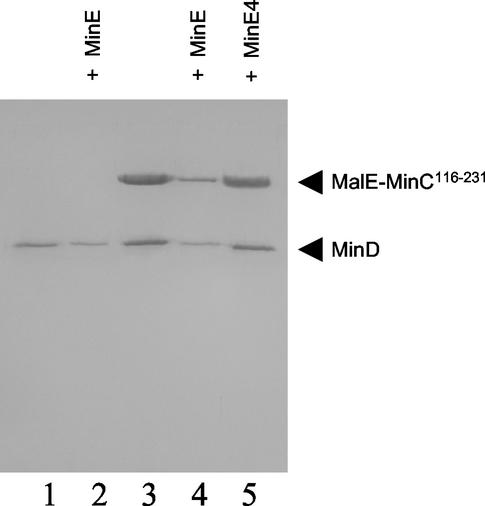
MinE could displace MinC from the MinD-bicelle complex by either of two mechanisms. MinE could compete with MinC for the same site on MinD, or the mechanism could be noncompetitive and MinE could bind to a distinct site on MinD, inducing a conformational change causing release of MinC. To try to distinguish between these two possibilities, the order of addition of MinC and MinE to the MinD-bicelle complex was reversed. MinD (4 μM) was mixed with bicelles, MinE (4 μM), and ATPγS. After a 5-min incubation, MalE-MinC116-231 was added at 8 μM and centrifuged. Analysis of the pellets (Fig. 8, lanes 2 and 3) revealed that the addition of MalE-MinC116-231 to a MinD-bicelle complex preloaded with MinE did not affect the level of bound MinE. In addition, the amount of MalE-MinC116-231 in the pellet was not increased over the background (Fig. 9, lane 4), which was higher than in earlier experiments due to the higher concentration used. This result indicates that MinE and MinC do not bind to the same site on MinD and that the binding of MinE to the MinD-bicelle complex induces a conformational change in MinD so that it no longer binds MinC.
FIG. 8.
MinC cannot displace MinE bound to MinD-bicelle complex. MinD (4 μM) was incubated with bicelles in the presence of ATPγS. After 1 min, MalE-MinC116-231 (8 μM) (lane 1) or MinE (4 μM) (lane 2) was added. In another reaction, MinD and MinE were incubated with ATPγS for 1 min, and then MalE-MinC was added (lane 3). A control (lane 4) contained only MalE-MinC116-231 (8 μM) and bicelles. The reaction mixtures were centrifuged, and the pellets were analyzed by SDS-PAGE.
FIG. 9.
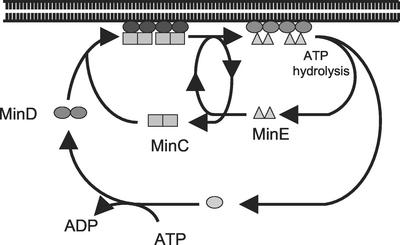
Model for regulation of the reversible interaction of Min proteins with the membrane. In the presence of ATP, MinD dimerizes and binds to MinC and the membrane. Upon binding to the membrane, MinD undergoes conformational changes leading to its assembly into filaments (12) and increased affinity for MinE. Binding of MinE to MinD results in displacement of MinC in a step that does not require ATP hydrolysis. MinE does not compete with MinC for binding to MinD but must alter MinD so that it has reduced affinity for MinC, since MinC cannot displace MinE. MinE stimulates MinD ATPase, causing release of MinE and the ADP form of MinD. MinC, MinD, and MinE are now dissociated in the cytoplasm. MinD undergoes nucleotide exchange, and the process is repeated.
DISCUSSION
The Min proteins oscillate rapidly between the poles of the cell (8, 10, 13, 28, 29). An entree into the biochemical basis for this oscillation was the observation that MinE stimulated MinD ATPase in the presence of phospholipid vesicles (15). This led to the discovery that MinD bound to the vesicles in the presence of ATP and recruited MinE (12). Subsequent MinE stimulation of the MinD ATPase resulted in release of MinD and MinE from the membrane.
In this report, we investigated the biochemical basis for the participation of MinC in the oscillation. We found that MinD dimerizes and interacts with MinC in an ATP-dependent manner. If a lipid bilayer is present, MinD recruits MinC to the bilayer. Subsequent addition of MinE displaces MinC from the MinCD-bilayer complex in a step that does not require ATP hydrolysis, although ATP hydrolysis is required for removal of MinD and MinE. These findings about the behavior of the Min proteins in vitro provide the underlying mechanism for their reversible membrane association in vivo and are summarized in Fig. 9.
MinD undergoes ATP-dependent dimerization.
One of the approaches we used to look for interaction between MinD and MinC was size-exclusion chromatography. This technique revealed that MinD is a monomer that undergoes ATP-dependent dimerization. Several MinDs (from several Archaea species) have been crystallized and occur as monomers, but the structures were obtained with ADP, AMPPCP, or no nucleotide bound (4, 11, 11). No structures of MinD with ATP, which supports dimerization, have been reported.
Interestingly, MinD is structurally similar to NifH (iron protein), which occurs as a dimer (9, 30). The monomers in the NifH dimer face each other, with the bound nucleotides (two per dimer) at the interface. Two monomers of MinD can be superimposed on the NifH dimer from Azotobacter vinelandii with an r.m.s. of 2.6 Å over 224 Cαs (per monomer). It is likely that ATP hydrolysis by NifH requires the participation of residues from each subunit within the dimer (30). Both lysine residues within the deviant Walker A motif (21), XKGGX2K[TS][UTS]X4[UTS] (where X is any residue and U is a bulky hydrophobic residue) make contact with the phosphates of the bound nucleotide. The second lysine within the motif contacts ATP bound to the same subunit, whereas the first lysine in the motif contacts the ATP bound to the other subunit. Modeling the structure of MinD on the structure of NifH from A. vinelandii locked in the transition state (PDB-1N2C) reveals that the bound nucleotides are superimposable with the lysine residues of the deviant Walker A motif in overlapping positions (K11 and K16 of MinD and K9 and K14 of NifH). Studies with MinD mutants indicate that both lysines are critical for MinD function (11). Thus, it is possible that MinD dimerizes like NifH.
Interaction between MinD and MinC requires ATP.
In the presence of MinE, GFP fusions to MinC and MinD oscillate with the same pattern and frequency, with the oscillation of MinC dependent upon MinD (13, 28). In the absence of MinE, MinC is located along the cell periphery, and this localization depends entirely upon MinD. These results, along with the yeast two-hybrid studies indicating interaction between these two proteins (17), led to the suggestion that MinD and MinC oscillate together (13, 28). The results of this study offer strong support for this suggestion, since MinC is recruited to lipid bicelles by MinD and stimulation of the MinD ATPase by MinE causes the release of both MinD and MinC.
Interaction between MinD and MinC was first suggested from genetic studies which demonstrated that both were required for efficient inhibition of division (6). Subsequent studies with the yeast two-hybrid system indicated a direct interaction and indicated that the interaction was between MinD and the C-terminal domain of MinC (14). Furthermore, the yeast two-hybrid studies indicated that ATP was probably important for the interaction, as a number of mutations that are likely to affect the interaction of MinD with ATP abolished the interaction (11). In this report, we found that ATP promoted the interaction of MinD and MinC in the presence and absence of lipid bicelles.
The association between MinC and MinD is relatively weak, as we were unable to observe MinD binding to a matrix containing MalE-MinC; the MinD was removed during the wash steps. During cochromatography, we observed a shift in the elution profiles of both MalE-MinC and MinD, indicating complex formation; however, we did not observe a new peak containing both proteins, indicating that the complex was breaking down during the chromatography.
MinD recruits MinC to bicelles.
The second assay that we employed for assessing interaction between MinC and MinD involved binding to phospholipid bicelles. We demonstrated previously that MinD bound to phospholipid vesicles in the presence of ATP or ATPγS (12). Here we showed that MinD recruited MinC to bicelles. This result is consistent with in vivo results which showed that GFP-MinC was recruited to the membrane by MinD (13, 28). Our results show clearly that ATP is required at two steps. It is required for MinD to bind to the vesicle surface. Interestingly, however, as shown by the chromatography results, MinD and MinC undergo an ATP-dependent interaction in the absence of the bicelles, suggesting that MinD and MinC can form a complex before binding to the membrane. This result indicates that ATP is required for MinD-MinC interaction as well as for MinD binding to the membrane. The binding of MinC to the MinD-bicelle complex was saturable, with a 1:1 stoichiometry. Since MinC is a dimer (14), it is likely that a dimer of MinD interacts with a dimer of MinC.
Two separable activities of MinE: displacement of MinC from the MinCD-membrane complex and stimulation of MinD ATPase.
We previously demonstrated that MinE could remove MinD from vesicles in a step requiring ATP hydrolysis (12) and therefore expected that MinE would also remove MinC. However, we found that MinE could displace MinC from the MinCD-bicelle complex even in the absence of ATP hydrolysis. One possibility is that MinE competes with MinC for binding to MinD. However, we found that MinC could not displace MinE from the MinE-MinD-bicelle complex. This argues that MinE and MinC have distinct binding sites on MinD. Furthermore, it argues that the binding of MinE to the MinCD-bicelle complex results in a conformational change in MinD that releases MinC. Interestingly, we observed that at saturating levels of MinE, the ratio of MinE to MinD was 1:1 in the MinDE-bicelle complex. Since MinE exists as a dimer (20), this raises the possibility that a dimer of MinE interacts with a dimer of MinD.
Why should MinE displace MinC from the MinCD-bicelle complex when MinC would eventually be released upon ATP hydrolysis? MinCD is a potent inhibitor of Z-ring formation, and its activity probably needs to be exquisitely controlled. We suggest that MinE displacing MinC from the MinCD-vesicle complex allows the MinCD inhibitor to be more sensitive to regulation by MinE.
Our studies also reveal why MinE4 is nonfunctional. MinE4 contains amino acid substitutions at positions 17 and 18, which lie within the N-terminal region of MinE required for suppression of the division-inhibitory activity of MinD and MinC (15, 26, 35). MinE4 is unable to support MinD oscillation or to stimulate MinD ATPase. In this study, we found that MinE4 was unable to bind to the MinD-bicelle complex.
Role of membrane in oscillation of Min proteins.
The main role of the MinDE oscillator is to direct the inhibitory activity of MinC to the membrane in polar regions of the cell. In this way, only the midcell is available for Z-ring assembly (23). How is this accomplished? We suggest that the membrane plays two roles in regulating the activity of MinC. First, it is likely that the membrane is involved in activating MinC. Although the MinCD complex can be targeted to the septum, as recently shown by Johnson et al. (18), such a mechanism would block cell division nonspecifically unless it is somehow regulated. We suggest that this regulation occurs through the activation of MinC upon MinCD binding to the membrane. The conformational changes associated with MinD binding to the membrane may be transmitted to MinC so that it has a high affinity for a septal component. In support of this, we have recently found that deletion of the C-terminal 10 amino acids of MinD has no effect on MinC binding but dramatically decreases membrane binding (15a). This MinD mutant is a poor activator of MinC, as it is unable to target it to the septum.
Second, the membrane plays a role in spatially restricting active MinC. Although our chromatography results showed that MinC and MinD can interact in the absence of the membrane, it is likely that the complex associates rapidly with the membrane, which in vivo is observed to start at the pole and extend towards midcell. Why it initiates at the pole is unknown. However, we have observed that MinD assembles into polymers on a vesicle surface and suggested that MinD in the polar zone was present in polymers (12). Assembly into polymers would help to restrict the diffusion of MinD, and therefore MinC, to prevent it from freely diffusing on the membrane and reaching the midcell Z ring.
Although MinC and MinD oscillate with the same pattern, that of MinE is somewhat different. MinE is mostly present as a ring at the edge of the MinD polar zone; however, some MinE appears to be present throughout the MinD polar zone (8, 10). Since there is less MinC than MinD in the cell (5), the MinD polar zone is unlikely to be saturated with MinC and so could be available to bind MinE not in the ring (33). This MinE appears to be insufficient to cause breakdown of the MinD polar zone, which instead appears to require the MinE ring located at the edge of the zone. Whether such a ring is essential is not clear, nor is the mechanism by which it is formed. Some MinE mutants that do not assemble into rings appear to support MinD oscillation (31).
The results presented here along with previous results (12, 15) provide the biochemical basis for the reversible membrane association of the Min proteins (Fig. 9). To extend these results to a full explanation of the oscillation of these proteins requires an understanding of how MinD assembles at the pole of the cell and how MinE primarily assembles at the edge of the MinD polar zone.
Acknowledgments
We thank M. Sundarsmoorthy for modeling the MinD structure on NifH.
This work was supported by grant GM 29764 from the National Institutes of Health.
REFERENCES
- 1.Bi, E., and J. Lutkenhaus. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, E. F., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 3.Cordell, S. C., R. E. Anderson, and J. Lowe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordell, S. C., and J. Lowe. 2001. Crystal structure of the bacterial cell division regulator MinD. FEBS Lett. 492:160-165. [DOI] [PubMed] [Google Scholar]
- 5.de Boer, P. A., R. E. Crossley, A. R. Hand, and L. I. Rothfield. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10:4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer, P. A., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641.err-649. [DOI] [PubMed]
- 7.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, X., Y. L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgiadis, M. M., H. Komiya, P. Chakrabarti, D. Woo, J. J. Kornuc, and D. C. Rees. 1992. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257:1653-1659. [DOI] [PubMed] [Google Scholar]
- 10.Hale, C. A., H. Meinhardt, and P. A. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi, I., T. Oyama, and K. Morikawa. 2001. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, Z., E. P. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinD. Proc. Natl. Acad. Sci. USA 99:6671-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, Z., and J. Lutkenhaus. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82-90. [DOI] [PubMed] [Google Scholar]
- 14.Hu, Z., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337-1343. [DOI] [PubMed] [Google Scholar]
- 15a.Hu, Z., and J. Lutkenhaus. A conserved sequence at the C-terminus of MinO is required for binding to the membrane and targeting MinC to the septum. Mol. Microbiol., in press. [DOI] [PubMed]
- 16.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, J., C. Cao, and J. Lutkenhaus. 1996. Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 178:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. E., L. L. Lackner, and P. A. de Boer. 2002. Targeting of ΔMinC/MinD and ΔMinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed]
- 19.Justice, S. S., J. Garcia-Lara, and L. I. Rothfield. 2000. Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol. Microbiol. 37:410-423. [DOI] [PubMed] [Google Scholar]
- 20.King, G. F., S. L. Rowland, B. Pan, J. P. Mackay, G. P. Mullen, and L. I. Rothfield. 1999. The dimerization and topological specificity functions of MinE reside in a structurally autonomous C-terminal domain. Mol. Microbiol. 31:1161-1169. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 229:1165-1174. [DOI] [PubMed] [Google Scholar]
- 22.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 23.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 24.Meinhardt, H., and P. A. de Boer. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA 98:14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pichoff, S., B. Vollrath, C. Touriol, and J. P. Bouche. 1995. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol. Microbiol. 18:321-329. [DOI] [PubMed] [Google Scholar]
- 27.Raskin, D. M., and P. A. de Boer. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E.coli. Cell 91:685-694. [DOI] [PubMed] [Google Scholar]
- 28.Raskin, D. M., and P. A. de Boer. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raskin, D. M., and P. A. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schindelin, H., C. Kisker, J. L. Schlessman, J. B. Howard, and D. C. Rees. 1997. Structure of ADP × AIF4(−)-stabilized nitrogenase complex and its implications for signal transduction. Nature 387:370-376. [DOI] [PubMed] [Google Scholar]
- 31.Shih, Y. L., X. Fu, G. F. King, T. Le, and L. Rothfield. 2002. Division site placement in E. coli: mutations that prevent formation of the MinE ring lead to loss of the normal midcell arrest of growth of polar MinD membrane domains. EMBO J. 21:3347-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szeto, J., S. Ramirez-Arcos, C. Raymond, L. D. Hicks, C. M. Kay, and J. A. Dillon. 2001. Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self-interaction. J. Bacteriol. 183:6253-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szeto, T. H., S. L. Rowland, and G. F. King. 2001. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J. Bacteriol. 183:6684-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, X. C., and W. Margolin. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32:315-326. [DOI] [PubMed] [Google Scholar]
- 35.Zhao, C. R., P. A. de Boer, and L. I. Rothfield. 1995. Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl. Acad. Sci. USA 92:4313-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]