Abstract
Objective
To compare the frequency and spectrum of p53 gene mutations in adenocarcinomas of the esophagus and cardia and to compare clinical and pathologic features in patients with p53 mutant and nonmutant cancers.
Summary Background Data
The p53 gene is commonly mutated in human cancers, and a p53 mutation is reported to be present in more than 50% of esophageal adenocarcinomas. Although many studies have investigated the frequency of p53 protein overexpression in adenocarcinomas of the esophagus or esophagogastric junction, few studies have assessed the frequency and clinical significance of p53 mutations in these tumors. In particular, the prognostic importance of p53 mutation is uncertain. Adenocarcinomas of the esophagus and cardia share many epidemiologic and pathologic features, but it is controversial whether they represent the same tumor. A comparison of the frequency and spectrum of mutations in adenocarcinomas of the esophagus and cardia would test whether these tumors are also similar at the molecular level.
Methods
DNA was isolated from microdissected paraffin-embedded tumor tissues of patients who underwent esophagogastrectomy for adenocarcinoma of the esophagus (n = 19), cardia (esophagogastric junction, n = 12), or subcardia (n = 6). Exons 5 to 8 of the p53 gene were analyzed for the presence of mutations using the polymerase chain reaction with single-strand conformation polymorphism and DNA sequencing of bands showing abnormal mobility. The presence of mutation was confirmed by selective hybridization of a mutant-specific oligonucleotide to DNA isolated from the tumor.
Results
p53 mutations were identified in 18 of 37 (48.6%) tumors. Patients with p53 mutant tumors were significantly younger and had a significantly poorer prognosis. There was a similar prevalence of p53 mutations in adenocarcinomas of the esophagus (53%) and cardia (58%). In contrast, mutations were relatively uncommon in subcardia adenocarcinomas (one mutant tumor [17%]). The types of mutations found in the esophageal and the cardia cancers were also similar.
Conclusions
Adenocarcinomas of the esophagus and cardia have a similar frequency and spectrum of p53 gene mutations, suggesting that these tumors have a common pathogenesis. Patients with mutations are younger, have signs of more advanced disease, and have a poorer prognosis than patients without mutations.
p53 gene abnormalities are common in human tumors, 1 and involvement of p53, by mutation, 2–10 altered mRNA expression, 11,12 allelic loss, 8,13–18 or protein overexpression, is found in a majority of esophageal adenocarcinomas. Although numerous studies have investigated the role of p53 alterations in the development of esophageal adenocarcinoma and its precursor lesion Barrett esophagus, many of these studies measured immunohistochemical p53 protein overexpression as a surrogate for p53 gene mutation. The reported accuracy of p53 protein expression measurement for the detection of mutation has varied widely, 3 but significant false-positive and false-negative rates have been found for immunohistochemistry compared with genomic mutation analysis in studies of many cancers, 1 including studies of esophageal adenocarcinoma. 9,19
Despite the frequency and importance of p53 alterations in the development of Barrett cancers, the clinical importance of these findings remains uncertain. In particular, whether there is an association between the p53 mutation status of these cancers and the prognosis is uncertain. Studies that used immunohistochemical methods have reported varying results concerning p53 protein expression and clinical outcomes, 20–27 and few studies 9,28,29 have examined the prognostic value of p53 mutation analysis in patients with esophageal adenocarcinoma. As a result, assessment of p53 mutation, allelic loss, or p53 protein expression has not become part of the routine assessment of patients with Barrett esophagus or Barrett cancer. 30
Molecular analysis of cancer tissues can be used to investigate the etiologic or other similarities of different tumor types. Adenocarcinomas of the esophagus and cardia (with the term cardia here indicating the esophagogastric junction and not the gastric cardia region in the proximal stomach) have some significant pathologic and epidemiologic similarities, but whether cardia cancers should be grouped with esophageal adenocarcinomas is disputed. This study was undertaken to investigate the genetic similarity of adenocarcinomas arising in the esophagus, cardia, and subcardia by determining the frequency and type of mutations in the p53 gene in tumors from each site. A second aim of this study was to determine whether the presence of p53 mutation in adenocarcinomas of the esophagus or cardia has any prognostic or other clinical importance for patients with these cancers.
PATIENTS AND METHODS
DNA was extracted from microdissected paraffin-embedded tumor tissue sections from 37 patients with adenocarcinoma of the esophagus, cardia, or subcardia. The location of the tumor was judged by examination of the resected esophagogastrectomy specimen. Tumors with an epicenter above the anatomical esophagogastric junction were classified as esophageal (n = 19), those with an epicenter at or within 1 cm of the esophagogastric junction were classified as cardia tumors (n = 12), and those with an epicenter 1 cm or more distal to the junction were classified as subcardia tumors (n = 6). Matching histologically normal squamous esophagus tissues were studied for each patient.
Exons 5 to 8 of the p53 gene were amplified using the polymerase chain reaction (PCR). Amplified DNA from each tumor was compared with the other tumors and normal control DNA using single-strand conformation polymorphism (SSCP). The method of DNA isolation, primer sequences, PCR conditions, and method of autoradiography have been described previously. 31,32 PCR reactions were repeated on at least two occasions and SSCP gels were run under at least two different conditions: 6% acrylamide with and without 20% glycerol or Hydrolink (J.T. Baker, Phillipsberg, NJ) solution with or without 10% glycerol. Bands showing abnormal mobility on SSCP were excised and sequenced as previously described. 31,32 The sequence of the abnormal band was compared with the published wild-type p53 sequence to identify mutations. To confirm the presence of mutation and to check nontumor tissue for the mutant sequence, 17 nucleotide long wild-type and mutant allele-specific oligonucleotides (ASO) were designed so that the mutant ASO matched the mutant sequence and the wild-type ASO matched the wild-type sequence over the same coding region. Oligonucleotides were end-labeled with 32PγATP (0.1 mCi per microliter, NEN DuPont, Boston, MA), using T4 polynucleotide kinase (Boehringer Mannheim), and purified by separation on a 25G Sephadex column (Boehringer Mannheim), according to the manufacturer’s instructions. Tumor and control DNA were amplified using the same conditions used for the SSCP that showed the abnormal band. The resulting amplified DNA was dot-blotted onto a nylon membrane using an automated laboratory workstation (Beckman Instruments 1000, Fullerton, CA). Forty microliters of each DNA sample was denatured with an equivalent volume of a solution containing 5 mol/L NaOH and 5 mmol/L EDTA. Half of each sample was then dripped onto separate halves of the membrane to produce two identical dots from each sample. Each dot was washed four times with 100 μL 20× SSPE, and the DNA was fixed to the membrane by ultraviolet irradiation (UV Stratalinker, Stratagene, La Jolla, CA). The membrane was divided and each half hybridized with either the wild-type or the mutant probe. Hybridization was carried out in 8 mL of a salt solution consisting of formamide 100 mL, Denhart’s solution 50 mL, 20× SSC (Boehringer Mannheim) 100 mL, and dH2O 225 mL, to which 5 to 10 μL of the 32P-labeled probe was added. After hybridization, the membranes were washed in a salt solution consisting of 20× SSC 100 mL, 20% SDA (National Diagnostics) 5 mL, and dH2O 900 mL. Hybridization and washing temperatures were tailored empirically to the probe being used. After washing, the membranes were placed in a cassette for autoradiography at −70°C overnight. Mutations were confirmed by selective hybridization of the mutant ASO with the original tumor sample, without hybridization to normal DNA. The presence of amplified normal DNA was confirmed by hybridization of the wild-type ASO to the normal control sample.
The patients’ notes were reviewed for clinical and pathologic details. An advanced tumor was defined as a tumor with transmural invasion, five or more lymph node metastases, or distant metastasis. The surgical procedure was classified as en bloc if the patient underwent resection of the tumor with extensive lymph node dissection with the intent of cure. If the patient did not undergo an en bloc resection, the surgical procedure was classified depending on the surgical approach as either transthoracic esophagectomy or transhiatal esophagectomy.
Statistical Analysis
The chi-square test was used to compare proportions between more than two groups, Fisher’s exact test was used to compare proportions between individual groups, the Kruskal-Wallis test was used to compare continuous data between more than two groups, and the Mann-Whitney test was used to compare continuous data between individual groups. Survival curves were calculated using the Kaplan-Meier estimation method, and comparison between survival curves was with the log-rank test. P < .05 was accepted to denote statistical significance. Data are expressed as median with interquartile range (25th–75th percentiles) in parenthesis, unless indicated otherwise.
RESULTS
Comparison of Tumors by Location
The clinical characteristics of the patients, divided by location of tumor, are summarized in Table 1. Patients with tumors in the esophagus had a significantly longer history of heartburn than patients with tumors in the subcardia. Table 1 also shows the pathologic features of the tumors by location of tumor. Esophageal tumors were more likely to be associated with Barrett intestinal metaplasia than subcardia tumors (P = .01). Dysplastic Barrett epithelium was more frequently found adjacent to tumors located in the esophagus than tumors located in the cardia (P = .003). More distally located tumors tended to be more invasive, to have greater numbers of lymph node metastases, and to be less differentiated, but the small study population did not permit statistical analysis of these factors separately. However, if tumors with transmural invasion, five or more lymph node metastases, or distant metastasis were classified as advanced tumors (n = 27), then statistical analysis showed that advanced tumors were significantly less common in the esophagus than in the cardia or subcardia region.
Table 1. COMPARISON OF CLINICAL AND PATHOLOGIC FEATURES BY TUMOR LOCATION
NP, statistical analysis was not performed because of inadequate numbers.
p53 Mutations
p53 gene mutations were detected in 18 of the 37 tumors. Ten of the mutant tumors were esophageal cancers, seven were cardia cancers, and only one of the tumors in the subcardia region had a mutation. The epicenter of the mutant subcardia tumor was only 1 cm distal to the esophagogastric junction. The details of the mutations are summarized in Table 2. An example of an autoradiogram showing an abnormal SSCP is shown in Figure 1, examples of mutant sequences are shown in Figure 2, and a representative dot-blot hybridization confirming the presence of mutation is shown in Figure 3.
Table 2. LOCATION OF MUTATIONS IN p53 GENE, WITH NUCLEOTIDE AND AMINO ACID CHANGES
* Exon 5.5 refers to the first half of exon 5; 5.51 refers to the second half of exon 5.
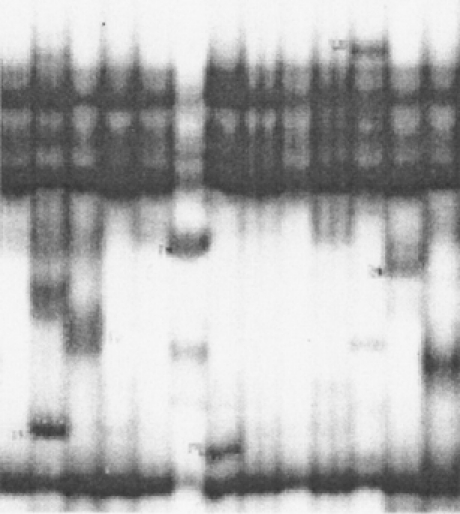
Figure 1. Autoradiogram from an single-strand conformation polymorphism gel of polymerase chain reaction products from exon 5 of the p53 gene.
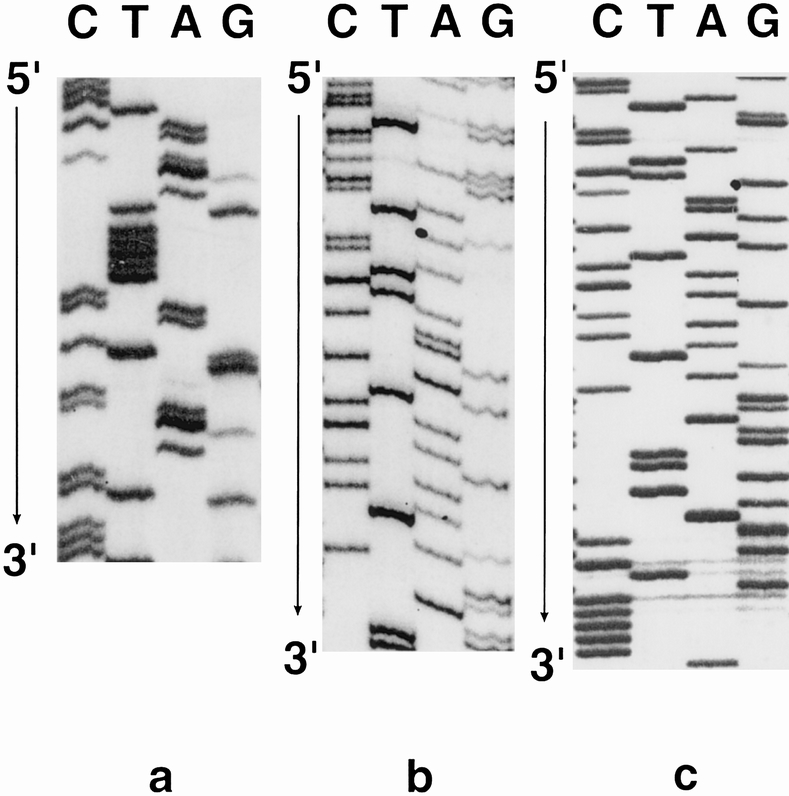
Figure 2. Sample autoradiograms of p53 mutant nucleotide sequences. (A) Patient F1, codon 135, T(G)C→T(T)C. (B) Patient F15, codon 161, (G)CC→(A)CC. (C) Patient F29, codon 163, T(A)C→T(G)C.
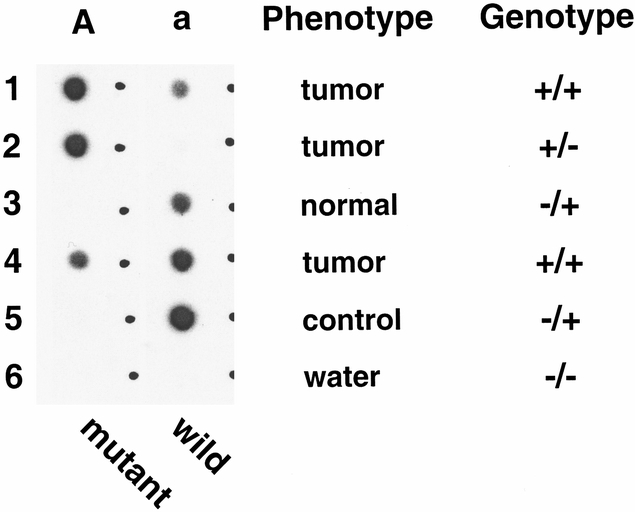
Figure 3. Autoradiogram of a dot-blot hybridization (patient F15) to confirm the presence of the mutation sequenced from an abnormal band seen on single-strand conformation polymorphism. DNA was isolated from tumor and normal tissues. Rows 1, 2, and 3 were obtained from microdissected tissue samples in an attempt to get as pure a tissue sample as possible. Row 4 was obtained from a larger dissection of the tumor and would contain more contamination of the tumor DNA with normal DNA from stromal elements. Row 4 acted as a positive control for the mutant because it was the source sample for the DNA used for the single-strand conformation polymorphism analysis. The DNA samples were amplified along with control normal DNA (positive control for wild-type DNA); then the polymerase chain reaction product was split and dripped onto two nylon membranes so that similar amplified DNA was present on each membrane. Membrane A was hybridized with the 32P-labeled mutant allele-specific oligonucleotide (ASO) and membrane a with the labeled wild-type ASO. Specific hybridization of the mutant ASO to tumor sample (row 4) without hybridization of the mutant probe to the control DNA (row 5) shows that the mutant DNA sequence is present in the tumor sample but not in the control. Absence of the mutant DNA sequence in the patients’ normal tissue (row 3) shows that the mutant DNA sequence is not present in the germ line. Row 2 shows that there is practically no wild-type DNA in the microdissected tumor sample, whereas strong hybridization of the mutant ASO confirms the presence of amplified DNA, in effect showing the “two hits” mutant DNA with loss of the wild-type allele.
No patient had more than one p53 mutation detected. The mutant sequence was not detectable in normal nontumor tissue from any of the patients, indicating that the mutation was a somatic event. Most of the mutations were missense point mutations. There was one deletion of a single base pair that resulted in a frame shift. There were two insertions, one of three base pairs and the other of nine base pairs. In both cases the inserted sequence duplicated an immediately adjacent nucleotide sequence. Mutations were most commonly found in exons 5 and 7. Two patients had a mutation in codon 154, two in codon 175, and three codon 248 (see Table 2).
The most common mutations identified were transitions. G→A transitions were found in six tumors (three esophageal and three cardia); five of these were at CpG dinucleotides. A→G transitions were found in four patients (two esophageal and two cardia tumors). G→T transversions were less common, being found in four patients (two esophageal and two cardia). There was a marked similarity between the prevalence and mutation spectrum of tumors located in the esophagus and tumors located in the cardia.
Comparison of Clinicopathologic Factors in Patients With Mutant and Nonmutant Tumors
Patients with tumors in the esophagus and cardia were combined to permit examination of the relation between p53 mutation and clinicopathologic features. The clinical features of patients with and without p53 mutation are summarized in Table 3. Patients with p53 gene mutations were 14.5 years younger on average than patients without p53 mutations (Fig. 4, P = .01).
Table 3. COMPARISON OF CLINICAL AND PATHOLOGIC FEATURES BETWEEN PATIENTS WITH AND WITHOUT p53 GENE MUTATIONS FOR PATIENTS WITH TUMORS IN ESOPHAGUS OR CARDIA
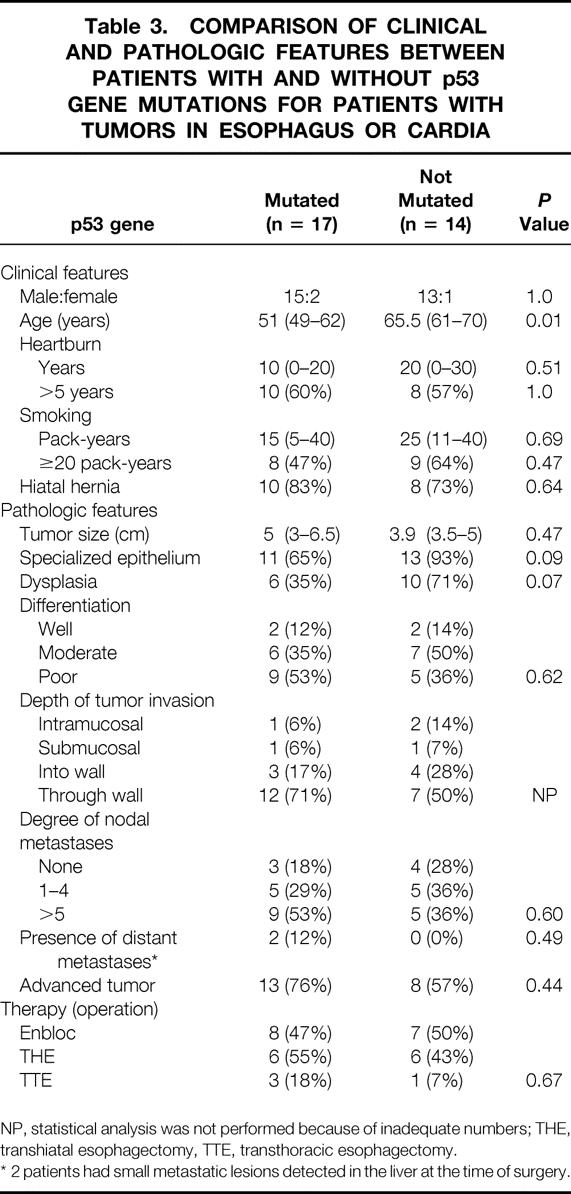
NP, statistical analysis was not performed because of inadequate numbers; THE, transhiatal esophagectomy, TTE, transthoracic esophagectomy.
* 2 patients had small metastatic lesions detected in the liver at the time of surgery.
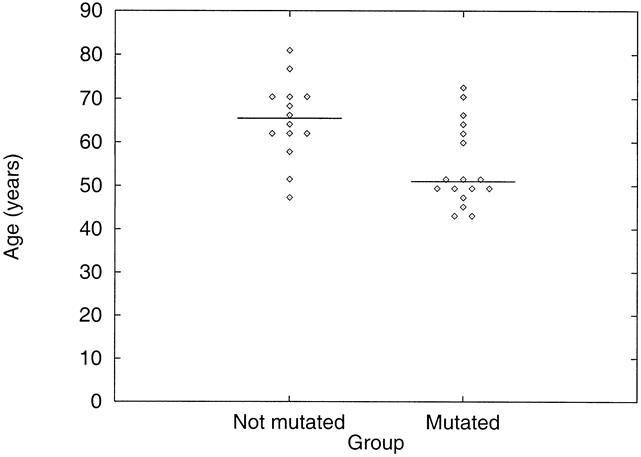
Figure 4. Relation between p53 mutation status and age. Age of patients with p53 mutant tumors is compared with that of patients with p53 wild-type tumors. The horizontal bar represents the median value and each point represents the age of one patient. Patients with p53 mutant tumors were significantly younger than patients with wild-type tumors.
Patients with p53 mutations tended to have more advanced tumors with respect to tumor size, differentiation, depth of invasion, degree of nodal metastasis, and presence of distant metastasis. However, there were no statistically significant differences in these factors. Curative resections with en bloc excision of the tumor and lymph nodes were performed in eight (47%) of the patients with mutation and in seven (50%) of the patients without mutation. Despite these nonsignificant differences in tumor stage and surgical treatment, patients with p53 mutations had a significantly poorer prognosis than patients without p53 mutations (Fig. 5). Two of the surviving four patients with p53 mutations are disease-free, even though all were initially free of nodal disease. In contrast, five of the seven surviving patients without p53 mutations are disease-free, even though three of the five initially had node metastasis. As expected from the data, patients with recurrent disease who had nonmutant tumors had a better survival prognosis than patients with recurrent disease who had mutant tumors.
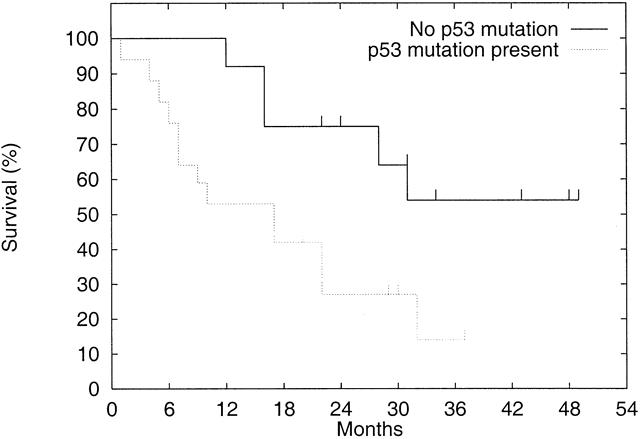
Figure 5. Effect of p53 mutation status on survival. Actuarial survival of patients with p53 mutations is compared with that of patients without p53 mutations. Patients with subcardia tumors and patients who died of postoperative complications are excluded. Patients with mutant tumors had significantly worse survival than patients with wild-type tumors. Censored values are denoted by a tick.
DISCUSSION
This study demonstrates that, in common with other tumors such as lung and colon cancers, approximately 50% of patients with adenocarcinoma of the esophagus or cardia (esophagogastric junction) have a p53 mutation. p53 mutations were detected in patients with all stages of tumor, including one patient who had a tumor confined to the lamina propria. This suggests that p53 mutations occur early in the tumorigenic process, probably before the development of tumor. If this is so, then the presence of p53 mutation may be a useful marker for identifying patients with Barrett esophagus who are at a greater risk of progression to adenocarcinoma. The results of several studies that showed an increased likelihood of disease progression in patients with Barrett esophagus with p53 abnormalities support this possibility. 14,16,33,34 However, as shown in this study, not all patients with adenocarcinoma have a p53 mutation, and the absence of p53 mutation in patients with Barrett esophagus does not indicate that there is no risk of cancer development. Some authors have described p53 mutations in nondysplastic Barrett metaplasia, 5,26,35,36 but the fate of patients with mutations in nondysplastic Barrett is unknown and will need to be assessed in prospective longitudinal studies. At present, the diagnosis of high-grade dysplasia on biopsy remains the best marker for the prediction of patients with occult cancer or those at highest risk of developing cancer.
The prevalence of p53 mutations in adenocarcinomas of the esophagus and cardia in this study was higher than the mutation prevalence in patients with tumors of the subcardia region. These results are in keeping with those of Strickler et al, 31 who found p53 mutations in 6 of 14 (43%) patients with tumors at the gastroesophageal junction compared with 2 of 26 (8%) patients with adenocarcinoma of the distal stomach (P = .01). The similar spectrum and prevalence of mutation in patients with tumors of the esophagus and cardia provide strong genetic support for the concept that these tumors have a common pathophysiology. Other similarities between esophagus and cardia cancers are a similar age at diagnosis, 37–40 a similar frequency of presentation at late stages, and a similarly poor prognosis. Pathologic features of both tumors are similar, with similar gross morphologic features, and an association with the specialized intestinal metaplasia that characterizes Barrett esophagus has been shown in morphologic acid mucin staining studies. 37,38,40–42 Because of this, both esophageal and cardia cancers can be considered Barrett cancers.
Our group has also used the tissue and tumor specificity of retinoic acid receptor expression 43,44 to test the genetic similarity of esophagus and cardia adenocarcinomas. The retinoic acid receptor mRNA expression profile of these two tumors was similar, 45 and it was distinctly different from that reported for both noncardia gastric cancers 46–48 and esophageal squamous cell cancers. 47,49
Further supporting evidence for a common pathophysiology comes from epidemiologic studies that report a rapid increase in the incidence of both esophageal and cardia adenocarcinomas during the same period that the incidence of noncardia gastric cancer was decreasing. 50–52 Epidemiologic surveys also show that in contrast to noncardia gastric cancers, both esophageal and cardia cancers are more common in white men. 37,41,50,51 An analysis of four studies 37,38,40,53 shows that the number of men for each woman with cardia adenocarcinoma varies from 2.3 39 to 11, 40 with an average male:female ratio of 5.5:1. The number of men for each woman with esophageal adenocarcinoma varies from 3.6 38 to 15, 37 with an average male:female ratio of 9.2:1. In tumors of the distal stomach, in contrast, there is an almost equal sex distribution (male:female ratio 1.1–1.3). 39,53
Epidemiologic studies have also shown that risk factors for adenocarcinomas of the esophagus and cardia are similar, and that they are often significantly different from the known risk factors for noncardia cancers. 54–57 Chow et al 56 found that colonization with the cag+ genetic variant of Helicobacter pylori was significantly inversely associated with development of both esophageal and cardia adenocarcinomas. That study found no significant association between infection with this H. pylori subtype and development of noncardia gastric cancer, but other studies have found that cag+H. pylori infection is significantly positively associated with development of gastric adenocarcinoma, including distal gastric cancers. 58–60 An inverse association between H. pylori infection and the presence of Barrett esophagus has also been found. 61–64 It is thus possible that certain types of H. pylori infection may promote noncardia gastric adenocarcinoma development but may protect against the development of Barrett esophagus and the Barrett-associated adenocarcinomas of the esophagus and cardia.
The mutation spectrum for adenocarcinoma of the esophagus or cardia can be examined further by combining the 17 mutations in the present study with the 36 reported in the literature. 2,3,5,11,36 There is a high prevalence of transition mutations—that is, replacement of the purine adenine (A) with the purine guanidine (G) or vice versa, or of the pyrimidine thymidine (T) with the pyrimidine cytosine (C) or vice versa. Overall, 38 (73%) of the 53 reported mutations are transitions either from G→A or from A→G. This type of mutation is commonly believed to be the effect of an external carcinogen. 1 In particular, G→A transitions may be due to the formation of O6 guanidine DNA adducts by methylating nitrosamines. 65–68 Indeed, 30/38 (79%) of these transitions are G→A, which indicates that a G:C base pair has changed to an A:T base pairing. The change could be due to either G→A on one DNA strand or from C→T on the opposite strand. The main cause of C→T transitions is thought to be “spontaneous” deamination of 5-methyl cytosine to thymine. 69–72 In a recent review, 451 of 960 (47%) p53 mutations in colon cancer were C→T transitions at CpG dinucleotides. 1 This prevalence is similar to the 49% prevalence of this type of mutation in esophageal adenocarcinoma, indicating that the spectrum of mutations in esophageal adenocarcinoma is similar to that in adenocarcinoma of the colon.
A significant finding in our study was that patients with p53 mutations were younger than those without a p53 mutation (see Fig. 4). This age difference may indicate that the p53 route to tumorigenesis represents a shortcut, in that patients with a p53 mutation progressed to invasive tumors 15 years on average before patients without a p53 mutation. This clinical association has not been previously described for adenocarcinoma of the esophagus or cardia, but Zheng et al 73 reported that patients with squamous cancer of the lung in whom a p53 mutation was detected were 11 years younger on average than patients in whom mutation was not detected (54 vs. 65 years, P < .001).
In the present study, in addition to being younger, patients with p53 mutations tended to have more advanced tumors (see Table 3), and the prognosis was significantly worse in patients with p53 mutant tumors than in those with wild-type tumors (see Fig. 5). Reduced survival in patients with esophageal adenocarcinoma with p53 alterations has also been reported in two other recent studies that included DNA sequencing analysis. 28,29 If the results of these studies and the present study are consistently observed in other large studies, these findings will indicate that “molecular staging” of these tumors by analysis of the p53 gene (and other genes that significantly distinguish between different Barrett stages 45,74–78) may be worthwhile. However, at present the best prognostic indicator for patients with these cancers remains tumor stage.
Footnotes
Correspondence: Tom R. DeMeester, MD, Dept. of Surgery, University of Southern California, 1510 San Pablo St., Suite 514, Los Angeles, CA 90033-4612.
Supported by grant #CA 58704 from the National Cancer Institute (to DKS).
Accepted for publication March 12, 1999.
References
- 1.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res 1994; 54:4855–4878. [PubMed] [Google Scholar]
- 2.Neshat K, Sanchez CA, Galipeau PC, et al. p53 mutations in Barrett’s adenocarcinoma and high-grade dysplasia. Gastroenterology 1994; 106:1589–1595. [DOI] [PubMed] [Google Scholar]
- 3.Hamelin R, Flejou JF, Muzeau F, et al. TP53 gene mutations and p53 protein immunoreactivity in malignant and premalignant Barrett’s esophagus. Gastroenterology 1994; 107:1012–1018. [DOI] [PubMed] [Google Scholar]
- 4.Gleeson CM, Sloan JM, McGuigan JA, Ritchie AJ, Russell SE. Base transitions at CpG dinucleotides in the p53 gene are common in esophageal adenocarcinoma. Cancer Res 1995; 55:3406–3411. [PubMed] [Google Scholar]
- 5.Schneider PM, Casson AG, Levin B, et al. Mutations of p53 in Barrett’s esophagus and Barrett’s cancer: a prospective study of ninety-eight cases. J Thorac Cardiovasc Surg 1996; 111:323–331. [DOI] [PubMed] [Google Scholar]
- 6.Ireland AP, Clark GW, DeMeester TR. Barrett’s esophagus. The significance of p53 in clinical practice. Ann Surg 1997; 225:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore JH, Lesser EJ, Erdody DH, Natale RB, Orringer MB, Beer DG. Intestinal differentiation and p53 gene alterations in Barrett’s esophagus and esophageal adenocarcinoma. Int J Cancer 1994; 56:487–493. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez MV, Artimez ML, Rodrigo L, et al. Mutation analysis of the p53, APC, and p16 genes in the Barrett’s oesophagus, dysplasia, and adenocarcinoma. J Clin Pathol 1997; 50:212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coggi G, Bosari S, Roncalli M, et al. p53 protein accumulation and p53 gene mutation in esophageal carcinoma. A molecular and immunohistochemical study with clinicopathologic correlations. Cancer 1997; 79:425–432. [DOI] [PubMed] [Google Scholar]
- 10.Galiana C, Lozano JC, Bancel B, Nakazawa H, Yamasaki H. High frequency of Ki-ras amplification and p53 gene mutations in adenocarcinomas of the human esophagus. Mol Carcinogenesis 1995; 14:286–293. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Meltzer SJ, Yin J, et al. Altered messenger RNA and unique mutational profiles of p53 and Rb in human esophageal carcinomas. Cancer Res 1993; 53:1889–1894. [PubMed] [Google Scholar]
- 12.Sorsdahl K, Casson AG, Troster M, Van MD, Inculet R, Chambers AF. p53 and ras gene expression in human esophageal cancer and Barrett’s epithelium: a prospective study. Cancer Detect Prevent 1994; 18:179–185. [PubMed] [Google Scholar]
- 13.Wu TT, Watanabe T, Heitmiller R, Zahurak M, Forastiere AA, Hamilton SR. Genetic alterations in Barrett esophagus and adenocarcinomas of the esophagus and esophagogastric junction region. Am J Path 1998; 153:287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blount PL, Galipeau PC, Sanchez CA, et al. 17p allelic losses in diploid cells of patients with Barrett’s esophagus who develop aneuploidy. Cancer Res 1994; 54:2292–2295. [PubMed] [Google Scholar]
- 15.Blount PL, Meltzer SJ, Yin J, Huang Y, Krasna MJ, Reid BJ. Clonal ordering of 17p and 5q allelic losses in Barrett dysplasia and adenocarcinoma. Proc Natl Acad Sci USA 1993; 90:3221–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galipeau PC, Cowan DS, Sanchez CA, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci USA 1996; 93:7081–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett MT, Galipeau PC, Sanchez CA, Emond MJ, Reid BJ. Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene 1996; 12:1873–1878. [PubMed] [Google Scholar]
- 18.Barrett MT, Sanchez CA, Prevo LJ, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet 1999; 22:106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blount PL, Ramel S, Raskind WH, et al. 17p allelic deletions and p53 protein overexpression in Barrett’s adenocarcinoma. Cancer Res 1991; 51:5482–5486. [PubMed] [Google Scholar]
- 20.Polkowski W, van Lanschot JJ, Ten Kate FJ, et al. The value of p53 and Ki67 as markers for tumour progression in the Barrett’s dysplasia-carcinoma sequence. Surg Oncol 1995; 4:163–171. [DOI] [PubMed] [Google Scholar]
- 21.Yang B, Rice TW, Adelstein DJ, Rybicki LA, Goldblum JR. Overexpression of p53 protein associates decreased response to chemoradiotherapy in patients with esophageal carcinoma. Mod Pathol 1999; 12:251–256. [PubMed] [Google Scholar]
- 22.Sauter ER, Keller SM, Erner SM. p53 correlates with improved survival in patients with esophageal adenocarcinoma. J Surg Oncol 1995; 58:269–273. [DOI] [PubMed] [Google Scholar]
- 23.Flejou JF, Paraf F, Potet F, Muzeau F, Fekete F, Henin D. p53 protein expression in Barrett’s adenocarcinoma: a frequent event with no prognostic significance. Histopathology 1994; 24:487–489. [DOI] [PubMed] [Google Scholar]
- 24.Duhaylongsod FG, Gottfried MR, Iglehart JD, Vaughn AL, Wolfe WG. The significance of c-erb B-2 and p53 immunoreactivity in patients with adenocarcinoma of the esophagus. Ann Surg 1995; 221:677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vijeyasingam R, Darnton SJ, Jenner K, Allen CA, Billingham C, Matthews HR. Expression of p53 protein in oesophageal carcinoma: clinicopathological correlation and prognostic significance. Br J Surg 1994; 81:1623–1626. [DOI] [PubMed] [Google Scholar]
- 26.Casson AG, Kerkvliet N, O’Malley F. Prognostic value of p53 protein in esophageal adenocarcinoma. J Surg Oncol 1995; 60:5–11. [DOI] [PubMed] [Google Scholar]
- 27.Moskaluk CA, Heitmiller R, Zahurak M, Schwab D, Sidransky D, Hamilton SR. p53 and p21(WAF1/CIP1/SDI1) gene products in Barrett esophagus and adenocarcinoma of the esophagus and esophagogastric junction. Hum Pathol 1996; 27:1211–1220. [DOI] [PubMed] [Google Scholar]
- 28.Casson AG, Tammemagi M, Eskandarian S, Redston M, McLaughlin J, Ozcelik H. p53 alterations in oesophageal cancer: association with clinicopathological features, risk factors, and survival. Mol Pathol 1998; 51:71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribeiro UJ, Finkelstein SD, Safatle-Ribeiro AV, et al. p53 sequence analysis predicts treatment response and outcome of patients with esophageal carcinoma. Cancer 1998; 83:7–18. [PubMed] [Google Scholar]
- 30.Kubba AK, Poole NA, Watson A. Role of p53 assessment in management of Barrett’s esophagus. Dig Dis Sci 1999; 44:659–667. [DOI] [PubMed] [Google Scholar]
- 31.Strickler JG, Zheng J, Shu Q, Burgart LJ, Alberts SR, Shibata D. p53 mutations and microsatellite instability in sporadic gastric cancer: when guardians fail. Cancer Res 1994; 54:4750–4755. [PubMed] [Google Scholar]
- 32.Li ZH, Zheng J, Weiss LM, Shibata D. c-k-ras and p53 mutations occur very early in adenocarcinoma of the lung. Am J Path 1994; 144:303–309. [PMC free article] [PubMed] [Google Scholar]
- 33.Younes M, Ertan A, Lechago LV, Somoano JR, Lechago J. p53 protein accumulation is a specific marker of malignant potential in Barrett’s metaplasia. Dig Dis Sci 1997; 42:697–701. [DOI] [PubMed] [Google Scholar]
- 34.Younes M, Lebovitz RM, Lechago LV, Lechago J. p53 protein accumulation in Barrett’s metaplasia, dysplasia, and carcinoma: a follow-up study. Gastroenterology 1993; 105:1637–1642. [DOI] [PubMed] [Google Scholar]
- 35.Campomenosi P, Conio M, Bogliolo M, et al. p53 is frequently mutated in Barrett’s metaplasia of the intestinal type. Cancer Epidemiol Biomarkers Prev 1996; 5:559–565. [PubMed] [Google Scholar]
- 36.Casson AG, Mukhopadhyay T, Cleary KR, Ro JY, Levin B, Roth JA. p53 gene mutations in Barrett’s epithelium and esophageal cancer. Cancer Res 1991; 51:4495–4499. [PubMed] [Google Scholar]
- 37.Clark GW, Smyrk TC, Burdiles P, et al. Is Barrett’s metaplasia the source of adenocarcinomas of the cardia? Arch Surg 1994; 129:609–614. [DOI] [PubMed] [Google Scholar]
- 38.Kalish RJ, Clancy PE, Orringer MB, Appelman HD. Clinical, epidemiologic, and morphologic comparison between adenocarcinomas arising in Barrett’s esophageal mucosa and in the gastric cardia. Gastroenterology 1984; 86:461–467. [PubMed] [Google Scholar]
- 39.Wang HH, Antonioli DA, Goldman H. Comparative features of esophageal and gastric adenocarcinomas: recent changes in type and frequency. Hum Pathol 1986; 17:482–487. [DOI] [PubMed] [Google Scholar]
- 40.Cameron AJ, Lomboy CT, Pera M, Carpenter HA. Adenocarcinoma of the esophagogastric junction and Barrett’s esophagus. Gastroenterology 1995; 109:1541–1546. [DOI] [PubMed] [Google Scholar]
- 41.Hamilton SR, Smith RR, Cameron JL. Prevalence and characteristics of Barrett esophagus in patients with adenocarcinoma of the esophagus or esophagogastric junction. Hum Pathol 1988; 19:942–948. [DOI] [PubMed] [Google Scholar]
- 42.Schnell TG, Sontag SJ, Chejfec G. Adenocarcinomas arising in tongues or short segments of Barrett’s esophagus. Dig Dis Sci 1992; 37:137–143. [DOI] [PubMed] [Google Scholar]
- 43.Hong WK, Itri LM. Retinoids and human cancer. In: Sporn MB, Roberts AB, Goodman DS, eds. The Retinoids. New York: Raven; 1994: 597–630.
- 44.Lippman SM, Davies PJ. Retinoids, neoplasia and differentiation therapy. In: Pinedo HM, Longo DL, Chabner BA, eds. Elsevier Science B.V.; 1997: 349–622. [PubMed]
- 45.Lord RV, Tsai P, Danenberg KD, Peters JH, DeMeester TR, Danenberg PV. Increase in retinoic acid receptor-a expression and decrease in retinoic acid receptor-g expression in the Barrett’s metaplasia, dysplasia, adenocarcinoma sequence. Cancer Res (submitted).
- 46.Chao TY, Jiang SY, Shyu RY, Yeh MY, Chu TM. All-trans retinoic acid decreases susceptibility of a gastric cancer cell line to lymphokine-activated killer cytotoxicity. Br J Cancer 1997; 75:1284–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu G, Wu M, Levi G, Ferrari N. Inhibition of cancer cell growth by all-trans retinoic acid and its analog N-(4-hydroxyphenyl) retinamide: a possible mechanism of action via regulation of retinoid receptors expression. Int J Cancer 1998; 78:248–254. [DOI] [PubMed] [Google Scholar]
- 48.Naka K, Yokozaki H, Domen T, et al. Growth inhibition of cultured human gastric cancer cells by 9-cis-retinoic acid with induction of cdk inhibitor Waf1/Cip1/Sdi1/p21 protein. Differentiation 1997; 61:313–320. [DOI] [PubMed] [Google Scholar]
- 49.Xu X-C, Liu X, Tahara E, Lippman SM, Lotan R. Expression and up-regulation of retinoic acid receptor-b is associated with retinoid sensitivity and colony formation in esophageal cancer cell lines. Cancer Res 1999; 59:2477–2483. [PubMed] [Google Scholar]
- 50.Devesa SS, Blot WJ, Fraumeni JF Jr. Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer 1998; 83:2049–2053. [PubMed] [Google Scholar]
- 51.Lord RV, Law MG, Ward RL, Giles GG, Thomas RJ, Thursfield V. Rising incidence of oesophageal adenocarcinoma in men in Australia. J Gastroenterol Hepatol 1998; 13:356–362. [DOI] [PubMed] [Google Scholar]
- 52.Pera M, Cameron AJ, Trastek VF, Carpenter HA, Zinsmeister AR. Increasing incidence of adenocarcinoma of the esophagus and esophagogastric junction. Gastroenterology 1993; 104:510–513. [DOI] [PubMed] [Google Scholar]
- 53.MacDonald WC. Clinical and pathologic features of adenocarcinoma of the gastric cardia. Cancer 1972; 29:724–732. [DOI] [PubMed] [Google Scholar]
- 54.Brown LM, Silverman DT, Pottern LM, et al. Adenocarcinoma of the esophagus and esophagogastric junction in white men in the United States: alcohol, tobacco, and socioeconomic factors. Cancer Causes & Control 1994; 5:333–340. [DOI] [PubMed] [Google Scholar]
- 55.Chow WH, Blot WJ, Vaughan TL, et al. Body mass index and risk of adenocarcinomas of the esophagus and gastric cardia. J Natl Cancer Inst 1998; 90:150–155. [DOI] [PubMed] [Google Scholar]
- 56.Chow WH, Blaser MJ, Blot WJ, et al. An inverse relation between cagA+ strains of Helicobacter pylori infection and risk of esophageal and gastric cardia adenocarcinoma. Cancer Res 1998; 58:588–590. [PubMed] [Google Scholar]
- 57.Gammon MD, Schoenberg JB, Ahsan H, et al. Tobacco, alcohol, and socioeconomic status and adenocarcinomas of the esophagus and gastric cardia. J Natl Cancer Inst 1997; 89:1277–1284. [DOI] [PubMed] [Google Scholar]
- 58.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA-positive or CagA-negative Helicobacter pylori infection. Gut 1997; 40:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blaser MJ, Perez-Perez GI, Kleanthous H, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res 1995; 55:2111–2115. [PubMed] [Google Scholar]
- 60.Queiroz DM, Mendes EN, Rocha GA, et al. cagA-positive Helicobacter pylori and risk for developing gastric carcinoma in Brazil. Int J Cancer 1998; 78:135–139. [DOI] [PubMed] [Google Scholar]
- 61.Werdmuller BF, Loffeld RJ. Helicobacter pylori infection has no role in the pathogenesis of reflux esophagitis. Dig Dis Sci 1997; 42:103–105. [DOI] [PubMed] [Google Scholar]
- 62.Quddus MR, Henley JD, Sulaiman RA, Palumbo TC, Gnepp DR. Helicobacter pylori infection and adenocarcinoma arising in Barrett’s esophagus. Hum Pathol 1997; 28:1007–1009. [DOI] [PubMed] [Google Scholar]
- 63.Vicari JJ, Peek RM, Falk GW, et al. The seroprevalence of cagA-positive Helicobacter pylori strains in the spectrum of gastroesophageal reflux disease. Gastroenterology 1998; 115:50–57. [DOI] [PubMed] [Google Scholar]
- 64.Lord RV, Frommer DJ, Inder S, Tran D, Ward RL. Helicobacter pylori infection in patients with Barrett’s oesophagus or Barrett’s adenocarcinoma. Aust NZ J Surg (in press). [DOI] [PubMed]
- 65.Wang Y, You M, Reynolds SH, Stoner GD, Anderson MW. Mutational activation of the cellular Harvey ras oncogene in rat esophageal papillomas induced by methylbenzylnitrosamine. Cancer Res 1990; 50:1591–1595. [PubMed] [Google Scholar]
- 66.Ohgaki H, Hard GC, Hirota N, Maekawa A, Takahashi M, Kleihues P. Selective mutation of codons 204 and 213 of the p53 gene in rat tumors induced by alkylating N-nitroso compounds. Cancer Res 1992; 52:2995–2998. [PubMed] [Google Scholar]
- 67.Lozano JC, Nakazawa H, Cros MP, Cabral R, Yamasaki H. G→A mutations in p53 and Ha-ras genes in esophageal papillomas induced by N-nitrosomethylbenzylamine in two strains of rats. Mol Carcinogenesis 1994; 9:33–39. [DOI] [PubMed] [Google Scholar]
- 68.Mirvish SS. Role of N-nitroso compounds (NOC) and N-nitrosation in etiology of gastric, esophageal, nasopharyngeal and bladder cancer and contribution to cancer of known exposures to NOC. Cancer Lett 1995; 93:17–48. [DOI] [PubMed] [Google Scholar]
- 69.Schmutte C, Yang AS, Beart RW, Jones PA. Base excision repair of U: G mismatches at a mutational hotspot in the p53 gene is more efficient than base excision repair of T:G mismatches in extracts of human colon tumors. Cancer Res 1995; 55:3742–3746. [PubMed] [Google Scholar]
- 70.Schmutte C, Rideout WM3, Shen JC, Jones PA. Mutagenicity of nitric oxide is not caused by deamination of cytosine or 5-methylcytosine in double-stranded DNA. Carcinogenesis 1994; 15:2899–2903. [DOI] [PubMed] [Google Scholar]
- 71.Magewu AN, Jones PA. Ubiquitous and tenacious methylation of the CpG site in codon 248 of the p53 gene may explain its frequent appearance as a mutational hot spot in human cancer. Mol Cell Biol 1994; 14:4225–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen JC, Rideout WM 3rd, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res 1994; 22:972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng J, Shu Q, Li ZH, Tsao JI, Weiss LM, Shibata D. Patterns of p53 mutations in squamous cell carcinoma of the lung. Acquisition at a relatively early age. Am J Path 1994; 145:1444–1449. [PMC free article] [PubMed] [Google Scholar]
- 74.Lord RV, Danenberg K, Peters JH, et al. Increased COX-2 and iNOS expression and decreased COX-1 expression in Barrett’s esophagus and Barrett’s associated adenocarcinomas. Proceedings of the Annual Meeting of the American Association for Cancer Research, 1999; 40:A2109.
- 75.Theisen J, Danenberg KD, DeMeester TR, et al. Effect of acid and bile salts on COX-2 gene expression in an esophageal adenocarcinoma cell line. Proceedings of the Annual Meeting of the American Association for Cancer Research, 1999; 40:A3333.
- 76.Lord RV, O’Grady R, Sheehan C, Field AF, Ward RL. K-ras codon 12 mutations in Barrett’s oesophagus and Barrett’s associated adenocarcinomas. J Gastroenterol Hepatol (in press). [DOI] [PubMed]
- 77.Lord RV, Salonga D, Danenberg KD, DeMeester TR, Peters JH, Danenberg PV. Telomerase reverse transcriptase expression is increased early in the Barrett’s metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg (submitted). [DOI] [PubMed]
- 78.Park JM, Danenberg KD, Lord RV, Peters JH, DeMeester TR, Danenberg PV. Induction of vascular endothelial growth factor and basic fibroblast growth factor in Barrett’s esophagus and Barrett’s associated adenocarcinomas. Proceedings of the Annual Meeting of the American Association for Cancer Research. 1999; 40:A1519.