Abstract
The mRNA processing body (P-body) is a cellular structure that has an important role in mRNA degradation. P-bodies have also been implicated in RNAi-mediated post-transcriptional gene silencing. The objective of this study was to identify and characterize novel components of the mammalian P-body. Approximately 5% of patients with the autoimmune disease primary biliary cirrhosis have antibodies directed against this structure. Serum from one of these patients was used to identify a cDNA encoding RAP55, a 463–amino acid protein. RAP55 colocalized with previously identified P-body components DCP1a and Ge-1. RAP55 contains an N-terminal Sm-like domain and two C-terminal RGG-rich domains separated by an FDF motif. The two RGG domains and the FDF domain were necessary and sufficient to target the protein to P-bodies. A fragment of RAP55 consisting of the FDF and the second RGG domains did not localize to P-bodies, but was able to displace other P-body components from this structure. After cells were subjected to arsenite-induced stress, RAP55 was detected in TIA-containing stress granules. The second RGG domain was necessary and sufficient for stress granule localization. siRNA-mediated knock-down of RAP55 resulted in loss of P-bodies, suggesting that RAP55 acts prior to the 5′-decapping step in mRNA degradation. The results of this study show that RAP55 is a component of P-bodies in cells at rest and localizes in stress granules in arsenite-treated cells. RAP55 may serve to shuttle mRNAs between P-bodies and stress granules.
Keywords: mRNA processing body, stress granule, mRNA decay, autoantigen
INTRODUCTION
Some of the enzymes involved in mRNA degradation are concentrated in discrete locations in the cell known as cytoplasmic mRNA processing bodies (P-bodies, also known as “cytoplasmic foci” and “GW182 bodies”) (Eystathioy et al. 2003; Sheth and Parker 2003; Cougot et al. 2004). These structures contain decapping enzymes (DCP1a and DCP2) and a 5′-to-3′ exoribonuclease (XRN1). P-bodies also contain seven “Sm-like” proteins (LSm1–7), which form a heptameric complex that binds mRNA, and the DEAD-box family helicase Rck/p54, which increases the efficiency of mRNA decapping (Bouveret et al. 2000; Coller et al. 2001; Eystathioy et al. 2003; Cougot et al. 2004). Two autoantigens, GW182 and Ge-1, also localize to P-bodies (Eystathioy et al. 2003; Yu et al. 2005).
The size and the number of P-bodies are related to the amount of mRNA that is available to undergo 5′-decapping (Teixeira et al. 2005). For example, treatment of eukaryotic cells with cyclohexamide, which inhibits translation elongation and traps mRNAs on polysomes, decreases the flow of mRNA to P-bodies and causes rapid loss of these structures. Inhibition of XRN1, which blocks 5′-to-3′ mRNA degradation, increases the amount of mRNA at the decapping step and increases the size and number of P-bodies (Sheth and Parker 2003; Cougot et al. 2004). The observation that P-bodies are modified by changes in mRNA metabolism suggests that these structures are actively involved in mRNA decay, rather than storage sites for components of the decapping and degradation complex.
Recent studies showed that components of the RNA interference machinery, including Argonaute proteins (Ago) 1 and 2, localize to P-bodies (Ding et al. 2005; Jakymiw et al. 2005; Liu et al. 2005; Rehwinkel et al. 2005; Sen and Blau 2005). The presence of Ago proteins in P-bodies suggests that these structures may have a role in RNAi-mediated, post-transcriptional gene silencing.
In mammalian cells, exposure to environmental stress results in the formation of cytoplasmic structures known as “stress granules,” which act as storage domains for “stalled” mRNAs (for review, see Kedersha and Anderson 2002). Stress granules contain mRNAs, translation initiation factors, the mRNA-binding proteins TIA and TIAR, and 40S ribosome subunits. There is a complex relationship between P-bodies and stress granules. During the course of arsenite-induced oxidative stress and during the immediate recovery period, stress granules form in the cytoplasm, and P-bodies are observed adjacent to these structures (Kedersha et al. 2005; Wilczynska et al. 2005; Yu et al. 2005). The physical relationship between stress granules and P-bodies suggests that these structures may function together. For example, a subset of mRNAs that is stored in stress granules during oxidative stress may subsequently be transferred to P-bodies for degradation. The mechanisms by which mRNA may be transferred between these two structures are unknown.
Primary biliary cirrhosis (PBC) is an autoimmune disease of unknown etiology characterized by the progressive destruction of intrahepatic biliary ductules, leading to hepatic fibrosis and liver failure (for review, see Kaplan 1996). Patients with PBC may develop autoantibodies directed against a spectrum of cellular structures including mitochondria, PML nuclear bodies, and nuclear pore complexes. Approximately 5% of PBC patients have antibodies directed against P-bodies (Bloch et al. 2005). In the present study, we used serum from a patient with PBC to show that RAP55 is an autoantigen that localizes to P-bodies. RAP55 also localizes in stress granules induced by arsenite. We delineated the portions of RAP55 that localize the protein to P-bodies and stress granules, and suggest that RAP55 may have a role in shuttling mRNAs between these two structures.
RESULTS AND DISCUSSION
PBC autoantibodies react with a component of P-bodies and stress granules
Using indirect immunofluorescence and the human Hep-2 cell line as substrate, we observed that antibodies in serum from PBC patient 0080 reacted with 5–20 cytoplasmic foci per cell (Fig. 1 ▶, panel I, A). To confirm that these foci were P-bodies, Hep-2 cells were treated with cyclohexamide and then stained with human serum. Treatment of cells with cyclohexamide, which disrupts P-bodies, resulted in loss of cytoplasmic dot staining by 0080 serum (Fig. 1 ▶, panel I, C). Direct confirmation of anti-P-body antibodies in 0080 serum was obtained using two-color indirect immunofluorescence. Rabbit anti-DCP1a antibodies and human antibodies colocalized in cytoplasmic foci (Fig. 1 ▶, panel I, E–H). These results suggest that at least one constituent of P-bodies is recognized by autoantibodies in 0080 serum.
FIGURE 1.
Antibodies in the serum of a primary biliary cirrhosis patient react with P-bodies and stress granules. (Panel I) (A,E) Patient 0080’s serum contained antibodies that reacted with 5–20 dots in the cytoplasm of Hep-2 cells as determined by indirect immunofluorescence. The patient’s serum also contained antibodies directed against E2-pyruvate dehydrogenase complex (E2-PDC), which produced granular, filamentous (mitochondrial) cytoplasmic staining. (C) Treatment of Hep-2 cells with cyclohexamide resulted in disappearance of cytoplasmic dots with no effect on the staining of mitochondria. (E) Patient antibodies colocalized with (F) anti-DCP1a antibodies. To examine the effect of stress on the cellular location of the P-body antigen, cells were treated with arsenite and stained with (I) human serum and (J) anti-TIA antiserum. Human serum 0080 reacted with one or more proteins that colocalized with TIA in stress granules. Overlap of red and green staining is shown in yellow in G and K. DAPI staining in B, D, H, and L indicates the location of cell nuclei. (Panel II) Immunoblotting was used to characterize the putative P-body/stress granule autoantigen. (A) Antibodies in patient 0080’s serum reacted with 70-kDa (E2-PDC) and 60-kDa proteins in an extract prepared from Hep-2 cells. In addition, the serum reacted weakly with a 50-kDa band, which may be a breakdown product of one of the larger proteins (lane 1). Serum from patient 0012 reacted with the 60-kDa protein, but not with E2 PDC. Serum from a third patient (Ge) reacted with a 160-kDa protein, previously shown to be Ge-1, as well as E2 PDC. In addition, Ge serum produced weak bands at 60 kDa and 50 kDa. Serum from patient 0050 reacted with Ge-1 and an unknown 110-kDa protein (a black arrow indicates the location of this protein). (B) To determine the cellular location of the 110-kDa protein, serum 0050 was used to probe an immunoblot prepared using extracts from cytoplasmic (Cyt) or nuclear (Nuc) fractions of Hep-2 cells. Ge-1 was detected in the cytoplasmic fraction, and the unidentified 110-kDa autoantigen was present in the nuclear fraction.
Because of the known relationship between P-bodies and stress granules, we determined the cellular location of the 0080 autoantigen(s) relative to TIA-containing stress granules. Hep-2 cells were exposed to arsenite for 1 h and subsequently fixed and stained with patient serum and anti-TIA antiserum. The human serum reacted with one or more antigens that localized to stress granules in arsen-itetreated cells (Fig. 1, I–L ▶).
To characterize the P-body/stress granule antigen(s) recognized by the patient’s serum, an immunoblot was prepared using Hep-2 cell lysates. Antibodies in 0080 serum reacted with the 70-kDa PBC autoantigen E2 pyruvate dehydrogenase complex (E2-PDC) and with a 60-kDa protein in Hep-2 cells (Fig. 1 ▶, panel II, A). Serum from a second patient with anti-P-body antibodies (0012) reacted only with the 60-kDa protein. Serum from a previously described patient (Ge) (Bloch et al. 1994; Yu et al. 2005) reacted with autoantigens Ge-1 and E2 PDC, as well as the 60-kDa protein. Note that although P-body autoantigens Ge-1 and GW182 have similar molecular weights, we previously showed that the Hep-2 cells used in this study do not contain detectable GW182 (Bloch et al. 2005; Yu et al. 2005). A fourth patient’s serum (0050) reacted with Ge-1 and an unidentified 110-kDa protein but did not react with the 60-kDa protein. Because antibodies from patient 0050 reacted with Ge-1 but not with the 60-kDa protein, serum from this patient was used in the studies described below to identify Ge-1 in P-bodies. To consider the possibility that the 110-kDa protein was an additional, as yet unidentified, P-body autoantigen, we used cell fractionation studies to determine the cellular location of this protein. Antibodies in 0050 serum reacted with a 110-kDa protein in Hep-2 cell extracts prepared from nuclear, but not cytoplasmic, proteins, suggesting that the 110-kDa protein is not a P-body component (Fig. 1 ▶, panel II, B). As expected, serum 0050 stained Hep-2 cells with cytoplasmic dot and diffuse nuclear staining patterns (see Fig. 2 ▶, panel II, F,H,J below). Taken together, these studies show that some patient sera contain antibodies directed against a 60-kDa P-body auto-antigen. Patient sera 0012 and 0080 reacted with the 60-kDa protein but not with Ge-1. Patient serum 0050 reacted with autoantigen Ge-1 but not with the 60-kDa protein.
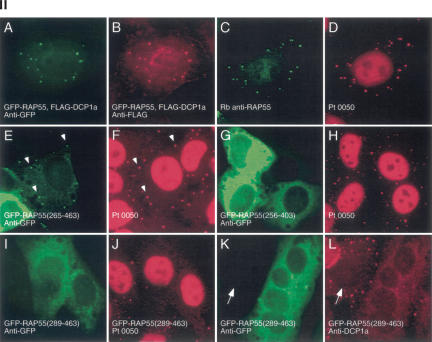
FIGURE 2.
Structure of RAP55 and identification of the portions of RAP55 that mediate localization to P-bodies and stress granules. (Panel I) The predicted amino acid sequence of RAP55 contains an N-terminal LSm domain and two C-terminal RGG-rich domains. Between the two RGG domains is an FDF motif. The portions of RAP55 that were capable of localizing green fluorescent protein (GFP) to P-bodies (PB) or stress granules (SG) are indicated by +. Representative indirect immunofluorescence results are shown in panel II. (Panel II) Two-color immunofluorescence was used to identify the portions of RAP55 that were necessary and sufficient for localization to P-bodies. (A) Full-length GFP-RAP55 localized to cytoplasmic dots and (B) colocalized with cotransfected DCP1a. (C) Rabbit anti-RAP55 antibodies colocalized with Ge-1-containing P-bodies as determined (D) using human serum 0050, which reacts with Ge-1, but not RAP55. A C-terminal portion of RAP55 (amino acids 265–463) (E) localized to P-bodies and (F) colocalized with Ge-1. White arrows indicate examples of overlapping cytoplasmic foci. (G) RAP55(256–403), which lacks the RGG2 domain, was present diffusely throughout the cytoplasm and did not localize to P-bodies. (H) RAP55(256–403) did not alter the P-body location of Ge-1. (I) RAP55(289–463) also did not localize to P-bodies, but this portion of RAP55 (J) displaced Ge-1 from P-bodies. Similarly, (K) RAP55(289–463) displaced DCP1a from (L) P-bodies in transfected cells. Note that DCP1a-containing P-bodies were detected in nontransfected cells (indicated by white arrows in K and L). Successful production of the GFP-RAP55 fragment fusion proteins used in these studies was confirmed by immunoblot using anti-GFP antiserum (data not shown).
To identify the 60-kDa P-body autoantigen, serum from patient 0012 was used to screen a λGT11 cDNA expression library. A single clone producing immunoreactive recombinant protein was identified and plaque-purified. This clone contained nucleotides 9–874 of a cDNA encoding the human homolog of Pleurodeles waltl RNA-associated protein, 55 kDa (RAP55; also known as LSm14) (Anantharaman and Aravind 2004; Khusial et al. 2005). RAP55 was previously identified as a cytoplasmic, oocyte-specific protein in Pleurodeles (Lieb et al. 1998). The cDNA encoding human RAP55 (NM_015578) predicts a 463–amino acid polypeptide with a molecular weight of 50.5kDa. The N-terminal portion of RAP55 (aminoacids1–98) contains an “Sm-like” (LSm) domain. The C terminus contains a short RGG-rich domain (RGG1, 268–280), followed by an FDF motif (290–400; see below) and a second, larger RGG domain (RGG2, 404–447). The structural features of RAP55 are shown schematically in Figure 2 ▶, panel I.
RAP55 is a member of the LSm family of proteins, which is characterized by the presence of an N-terminal α-helical domain followed by five anti-parallel β-pleated strands (for review, see Khusial et al. 2005). LSm proteins 1–7 form a heptameric, ring-shaped structure that binds RNA. The LSm1–7 complex localizes to P-bodies and enhances mRNA decapping. A second heptameric complex lacks LSm1 and includes LSm8. The LSm2–8 complex localizes to the nucleus and functions in pre-mRNA splicing. The presence of the LSm domain in RAP55 suggests that this protein has a role in binding mRNA. RAP55, like LSm1 and LSm8, may function as an alternative component of the LSm complex.
The RGG domain was described by Kiledjian and Dreyfus (1992) and is characterized by repeating, closely spaced arginine–lysine residues. This motif is found in RNA-binding proteins including hnRNP U, fibrillarin, nucleolin, and RNA helicase. The RGG domain may directly bind RNA or may mediate interaction with other RNA-binding proteins.
The FDF motif was named for three conserved amino acids in the N terminus of the domain (Anantharaman and Aravind 2004). The FDF domain in RAP55 contains many polar and charged residues that are predicted to form an α-helical structure with exposed hydrophilic loops. The large number of charged residues in this domain may explain the slow migration of RAP55 in SDS gels (apparent molecular weight of 60 kDa compared with the predicted 50 kDa). Anatharaman and Aravind (2004) anticipated that the FDF domain interacts with RNA or with highly charged peptides that are present in RNP complexes.
RAP55 localizes to P-bodies
To define the cellular location of RAP55, a fusion protein containing GFP attached to the N terminus of full-length RAP55 was coexpressed with Flag-DCP1a in Hep-2 cells. Antibodies directed against GFP localized to cytoplasmic dots and colocalized with anti-Flag antibodies, confirming that RAP55 is a component of P-bodies (Fig. 2 ▶, panel II, A,B). A polyclonal antiserum directed against RAP55 was prepared in rabbits and was found to react with cytoplasmic dots in Hep-2 cells. Rabbit anti-RAP55 antibodies colocalized with Ge-1, as determined using human serum 0050, further confirming that RAP55 is a component of P-bodies (Fig. 2 ▶, panel II, C,D).
To identify the portion of RAP55 that directs the protein to P-bodies, expression vectors encoding GFP fused to various segments of RAP55 were transfected into Hep-2 cells. The results are summarized in Figure 2 ▶, panel I, and representative photomicrographs are shown in Figure 2 ▶, panel II. A GFP fusion protein containing the RAP55 N terminus (1–266) did not localize to cytoplasmic dots but was distributed diffusely throughout the cytoplasm (data not shown), demonstrating that the LSm domain was not sufficient to direct RAP55 to P-bodies. In contrast, the RAP55 C terminus (265–463) localized to cytoplasmic foci and colocalized with Ge-1 (Fig. 2 ▶, panel II, E,F). This portion of RAP55 contains the two RGG-rich domains and the FDF motif. Segments of RAP55 that lacked either the RGG2 domain (Fig. 2 ▶, panel II, G) or the RGG1 domain (Fig. 2 ▶, panel II, letter I) failed to localize to P-bodies, suggesting that both RGG domains are required to target the protein to cytoplasmic foci. Interestingly, although the portion of RAP55 that contained the FDF motif and RGG2 (amino acids 289–463) was unable to localize to P-bodies, it displaced other P-body components, including Ge-1 and DCP1a, from these structures (Fig. 2 ▶, panel II, I–L). Ge-1 was displaced from P-bodies in 96% of Hep-2 cells expressing RAP55(289–463). Shorter segments of RAP55, which contained progressively smaller portions of the FDF domain, were less able to disrupt P-bodies. Ge-1 was displaced from P-bodies in 58% and 36% of cells expressing RAP55(332–463) and RAP55(355–463), respectively. A segment of RAP55 that lacked the FDF domain and contained only RGG2 (amino acids 396–463) displaced Ge-1 from P-bodies in <25% of transfected cells.
The results of these structure–function studies show that both RGG domains and the FDF motif are required for localization of RAP55 to P-bodies. In addition, segments of RAP55 that contain the FDF domain and RGG2, but not RGG2 alone, displaced P-body proteins from these structures. Possibly, the FDF domain interacts with, and displaces, one or more central component of P-bodies, thereby leading to dissolution of the structure.
RAP55 shuttles between P-bodies and stress granules
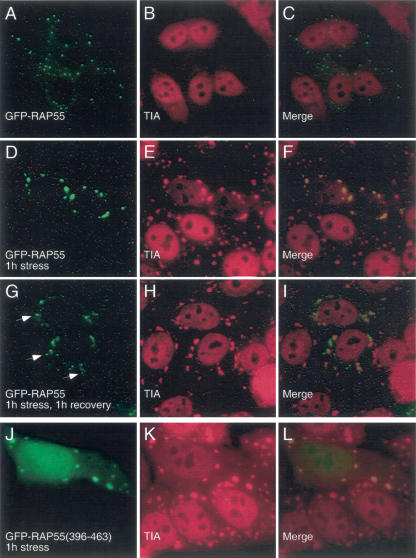
To further investigate the relationship between RAP55 and P-bodies and stress granules, we transfected GFP-RAP55 into Hep-2 cells and exposed the cells to arsenite. Cells were permitted to recover in culture medium for varying intervals and were subsequently fixed and stained with anti-GFP and anti-TIA antisera. Prior to treatment with arsenite, TIA localized to nuclei (but not nucleoli), and RAP55 was detected in P-bodies that were distributed diffusely throughout the cytoplasm (Fig. 3A–C ▶). After the cells were exposed to arsenite for 60 min, RAP55 and TIA were present together in stress granules (Fig. 3D–F ▶). When cells were exposed to arsenite for 1 h and then permitted to recover for 1 h, RAP55 was detected both in TIA-containing stress granules and in adjacent P-bodies (Fig. 3G–I ▶). Twelve hours after treatment with arsenite, stress granules were no longer present in the cytoplasm, and TIA was again detected in nuclei. RAP55-containing P-bodies were detected in the cytoplasm (data not shown).
FIGURE 3.
The effect of arsenite-induced oxidative stress on the cellular location of RAP55. (A) In resting Hep-2 cells, RAP55-containing P-bodies were distributed throughout the cytoplasm, and (B) TIA was detected diffusely throughout the nucleus, but excluded from nucleoli. After 60 min of exposure to sodium arsenite, both (D) RAP55 and (E) TIA were detected in cytoplasmic granules. (H) After 1 h exposure to arsenite and 1 h recovery in culture medium, TIA-containing stress granules persisted in the cell cytoplasm. RAP55 was detected both within stress granules and in adjacent P-bodies (indicated by white arrows in G). To identify the portion of RAP55 that mediated localization to stress granules, GFP-RAP55 fragment fusion proteins were expressed in Hep-2 cells, which were then subjected to arsenite-induced stress for 1 h. (J) RAP55 amino acids 396–463 were necessary and sufficient to direct GFP to (K) TIA-containing stress granules. Overlap of green and red staining appears yellow in C, F, I, and L.
To determine the portion of RAP55 that directs the protein to stress granules, Hep-2 cells were transfected with plasmids encoding GFP-RAP55-fragment fusion proteins, stressed for 1 h with arsenite, and then stained with anti-GFP and anti-TIA antisera. The results of these studies are summarized in Figure 2 ▶, panel I. In the setting of arsenite-induced oxidative stress, RGG2(396–463) was necessary and sufficient to target the protein to stress granules (Fig. 3J–L ▶).
Conservation of RAP55 during evolution
Proteins that are homologous to human RAP55 exist in Caenorhabditis elegans (CAR-1) and Drosophila (Tra1), as well as Pleurodeles and Xenopus (Lieb et al. 1998; Audhya et al. 2005; Boag et al. 2005). In these species, RAP55 expression is limited to germ cell development and early embryogenesis. In C. elegans and Drosophila, Car-1 and Tra1 localize to P-granules, structures that are similar to P-bodies and function as storage sites for maternal mRNA. CAR-1 and Tra1 interact with homologs of the human P-body component Rck (CGH-1 in C. elegans and Me31B in Drosophila), supporting the observation that RAP55 associates with components of P-body-like structures (Audhya et al. 2005; Boag et al. 2005; Wilhelm et al. 2005). However, in contrast to CAR-1, Tra1, and Xenopus RAP55, human RAP55 is expressed in nearly all tissues and during all developmental stages (http://www.ncbi.nlm.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.407368). We have detected immunoreactive RAP55 in several human cell lines, including lymphocytes (Hut78, BJAB), monocytes (HL60, U937), and others (Hep-2, A549, Be2C, and U2OS) (data not shown). The widespread expression of human RAP55 in tissues and cell lines and at all stages of development suggests that, during the course of evolution, human RAP55 acquired a more general role in the control of mRNA translation, storage, and/or degradation.
Decreasing the cellular level of RAP55 alters the structure of P-bodies
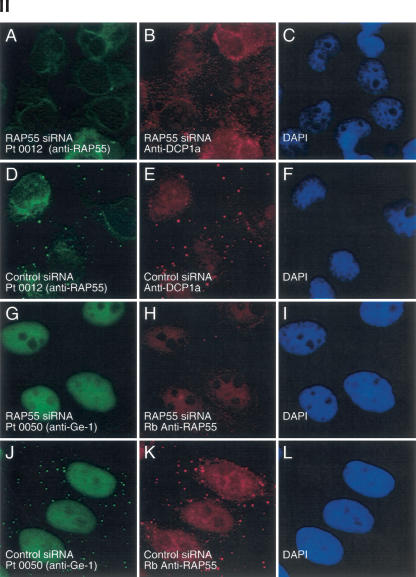
To investigate the effect of lowering the cellular level of RAP55 on the composition of P-bodies, siRNA specific for RAP55 was introduced into Hep-2 cells. RAP55 siRNA decreased the level of RAP55 protein in Hep-2 cells, as determined by immunoblot (Fig. 4 ▶, panel I, A). Note that the level of P-body component DCP1a was not altered by RAP55 siRNA (Fig. 4 ▶, panel I, B). On examination by indirect immunofluorescence, >80% of RAP55 siRNA-transfected cells lacked RAP55-containing P-bodies (Fig. 4 ▶, panel II, A,H). A second RAP55 siRNA produced similar results (data not shown). Control siRNA did not decrease the cellular level of RAP55 (Fig. 4 ▶, panel I) and did not alter RAP55-containing P-bodies (Fig. 4 ▶, panel II, D,K). To determine the effect of decreasing RAP55 on the cellular location of other P-body components, RAP55 siRNA-transfected cells were stained for DCP1a and Ge-1. In cells that lacked RAP55, DCP1a and Ge-1 were no longer detected in P-bodies (Fig. 4 ▶, panel II, B,G).
FIGURE 4.
The effect of RAP55 knock-down on P-bodies. (Panel I) Immunoblotting was used to determine the effect of siRNAs on the levels of RAP55 and DCP1a in Hep-2 cells. Compared with control siRNA, RAP55 siRNA decreased the level of (A) RAP55 but not (B) DCP1a in Hep-2 cells. As indicated, siRNA knock-down was performed in duplicate. RAP55 and DCP1a were detected using patient serum 0080 and rabbit anti-DCP1a antiserum, respectively. The human serum reacted weakly with E2 PDC (70 kDa). (Panel II) RAP55 siRNA decreased the level of RAP55, DCP1a, and Ge-1 in P-bodies. After transfection of RAP55 siRNA into Hep-2 cells, neither (A) RAP55 nor (B) DCP1a was detected in P-bodies. (D,E) Control siRNA did not alter the P-body location of either protein. (G,H) Transfection of RAP55 siRNA into Hep-2 cells also displaced Ge-1 from P-bodies. (J,K) Control siRNA had no effect on the P-body localization of Ge-1. Patient 0012 serum or rabbit anti-RAP55 antiserum was used to detect RAP55, as indicated. Patient 0050 serum was used to detect Ge-1. DAPI staining indicates the location of nuclei in C, F, I, and L.
Previous investigators showed that the loss of an early component of the 5′ → 3′ mRNA decay pathway disrupted the entire structure (Sheth and Parker 2003; Andrei et al. 2005). siRNA-mediated knock-down of components involved in the early steps of 5′ → 3′ mRNA degradation, including LSm1 and Rck/p54, prevented the accumulation of other degradation factors in P-bodies (Yang et al. 2004b; Andrei et al. 2005). Because RAP55 knock-down prevented DCP1a from accumulating in P-bodies, it is possible that RAP55 acts prior to decapping of mRNA and exonuclease activity.
The mechanism by which knock-down of RAP55 results in loss of P-bodies is uncertain. One possibility is that P-bodies are physically unable to assemble in the absence of RAP55. Alternatively, the lack of RAP55 may decrease the delivery of mRNA to P-bodies, potentially favoring disassembly of these structures.
Conclusions
In this study, we identified RAP55 as a component of mammalian P-bodies that localizes in stress granules in response to arsenite-induced oxidative stress. The RGG domains and FDF motif are necessary and sufficient to target the protein to P-bodies. The second RGG domain, by itself, is sufficient to target the protein to stress granules. The N-terminal Sm-like domain is not required to localize the protein to either P-bodies or stress granules. Through interaction with other LSm proteins, the N terminus of RAP55 may bind mRNA, while the C terminus of RAP55 may transport the mRNA between P-bodies and stress granules. We speculate that RAP55 acts as a shuttle, perhaps targeting irreversibly damaged mRNA present in stress granules, for transport to, and degradation in, P-bodies.
MATERIALS AND METHODS
Plasmids and antisera
A plasmid encoding RAP55 (IMAGE no. 3902704) was obtained from American Type Culture Collection (ATCC). A cDNA encoding full-length RAP55 was prepared by PCR and ligated in pEGFP (Clontech) to produce a green fluorescent protein (GFP) fusion protein (GFP-RAP55). DNA containing RAP55 nucleotides 9–874 was obtained by expression cloning (see below) and ligated into the EcoRI site of pEGFP to produce GFP-RAP55(1–266). The full-length RAP55 cDNA was treated with BglII and EcoRI or BstYI and EcoRI, to prepare pEGFP constructs expressing RAP55(172–463) and RAP55(355–463), respectively. PCR was used to prepare DNA encoding RAP55 amino acids 265–463, 289–463, 332–403, 332–463, and 396–463.
DNA encoding RAP55 amino acids 1–266 was ligated into the prokaryotic expression vector pGEX (Pharmacia) so as to encode the GST-RAP55(1–266) fusion protein. Recombinant GST-RAP55 was prepared in E. coli and purified as described (Smith and Johnson 1988). Rabbit polyclonal anti-RAP55 antiserum was prepared at Cocalico Biologics, Inc. following a standard protocol.
A plasmid encoding the Flag epitope fused to DCP1a and rabbit anti-DCP1a antiserum were provided by J. Lykke-Andersen (University of Colorado, Boulder). Mouse anti-Flag antibodies were purchased from Sigma-Aldrich. Goat anti-TIA antiserum was obtained from Santa Cruz Biotechnology. FITC- or rhodamine-conjugated, species-specific donkey anti-human, anti-rabbit, and anti-mouse IgG antisera were obtained from Jackson Immuno-Research Laboratories.
Serum from PBC patient Ge was previously described (Bloch et al. 1994; Yu et al. 2005). Sera from PBC patients 0012, 0080, and 0050 were identified in a study to determine the clinical significance of autoantibodies in PBC (Yang et al. 2004a).
Cell culture and immunohistochemistry
Hep-2 cells were obtained from ATCC and maintained in DMEM supplemented with 10% fetal calf serum, L-glutamine (2 mM), penicillin (200 U/mL), and streptomycin (200 μg/mL). To induce stress granule formation, Hep-2 cells were exposed to sodium arsenite (0.5 mM) for 1 h at 37°C. For immunofluorescent staining, Hep-2 cells were grown in tissue culture chambers (Nunc Inc.), fixed with 2% paraformaldehyde in PBS, and per-meabilized with 0.1% Triton X-100 in PBS. Cells were stained with primary and secondary antisera as previously described (Bloch et al. 2005).
Expression library screening
Serum from patient 0012 was diluted 1:1000 in blocking solution (PBS containing 5% nonfat dry milk) and used to screen a λGT11 cDNA expression library prepared from HepG2 (human hepatocellular carcinoma) cells (Clontech) according to the method of Young and Davis (1983). Bound antibodies were detected using chemiluminescence. A single clone encoding immunoreactive protein was identified from among 1 million bacteriophages and was isolated by the method of plaque purification.
SDS-PAGE and immunoblotting
Hep-2 cell extracts were fractionated in SDS–8% polyacrylamide gels and transferred to nitrocellulose membranes. In cell fractionation studies, nuclear and cytoplasmic fractions of Hep-2 cells were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce Biotechnology) as directed by the manufacturer. Nitrocellu-lose membranes were incubated in blocking solution and then with human serum or rabbit anti-DCP1a antiserum diluted in blocking solution. Bound human or rabbit antibodies were detected using HRP-conjugated goat anti-human IgG or anti-rabbit IgG antiserum (Jackson ImmunoResearch) and chemiluminescence.
Mammalian cell transfection
To identify the portion of RAP55 that mediates localization to P-bodies, plasmids encoding GFP fused to segments of RAP55 were transfected into Hep-2 cells using the Effectene transfection system (QIAGEN) as directed by the manufacturer. Cells were fixed and stained 24 h after transfection.
siRNAs were used to decrease the level of RAP55 in Hep-2 cells. Two siRNAs for RAP55 were obtained from Dharmacon Research Inc. (catalog nos. D-013920-01 and D-013920-03). Control siRNA was obtained from Ambion, Inc (catalog no. 4605). Double-stranded siRNA was transfected into Hep-2 cells using oligofectamine (Invitrogen) as directed by the manufacturer, and cells were fixed and stained 48 h after transfection.
Acknowledgments
We thank J. Lykke-Andersen for providing rabbit anti-DCP1a antiserum and a plasmid encoding Flag-tagged DCP1a. We also thank P. Anderson, N. Kadersha, and K.J. Bloch for advice. K.D.B. was supported by grants from the NIH (HL070896 and HL074352). D.B.B. was supported by grants from the Arthritis Foundation, the Milton Fund, and the NIH (DK051179). D.B.B. is a recipient of an Established Investigator Grant from the American Heart Association.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2302706.
REFERENCES
- Anantharaman, V. and Aravind, L. 2004. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics 5: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei, M.A., Ingelfinger, D., Heintzmann, R., Achsel, T., Rivera-Pomar, R., and Luhrmann, R. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya, A., Hyndman, F., McLeod, I.X., Maddox, A.S., Yates III, J.R., Desai, A., and Oegema, K. 2005. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 171: 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, D.B., Rabkina, D., Quertermous, T., and Bloch, K.D. 1994. The immunoreactive region in a novel autoantigen contains a nuclear localization sequence. Clin. Immunol. Immunopathol. 72: 380–389. [DOI] [PubMed] [Google Scholar]
- Bloch, D.B., Yu, J.H., Yang, W.H., Graeme-Cook, F., Lindor, K.D., Viswanathan, A., Bloch, K.D., and Nakajima, A. 2005. The cytoplasmic dot staining pattern is detected in a subgroup of patients with primary biliary cirrhosis. J. Rheumatol. 32: 477–483. [PubMed] [Google Scholar]
- Boag, P.R., Nakamura, A., and Blackwell, T.K. 2005. A conserved RNA–protein complex component involved in physiological germ-line apoptosis regulation in C. elegans. Development 132: 4975–4986. [DOI] [PubMed] [Google Scholar]
- Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M., and Seraphin, B. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19: 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A., and Parker, R. 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., and Seraphin, B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165: 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, L., Spencer, A., Morita, K., and Han, M. 2005. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 19: 437–447. [DOI] [PubMed] [Google Scholar]
- Eystathioy, T., Jakymiw, A., Chan, E.K., Seraphin, B., Cougot, N., and Fritzler, M.J. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakymiw, A., Lian, S., Eystathioy, T., Li, S., Satoh, M., Hamel, J.C., Fritzler, M.J., and Chan, E.K. 2005. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 7: 1167–1174. [DOI] [PubMed] [Google Scholar]
- Kaplan, M.M. 1996. Primary biliary cirrhosis. N. Engl. J. Med. 335: 1570–1580. [DOI] [PubMed] [Google Scholar]
- Kedersha, N. and Anderson, P. 2002. Stress granules: Sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30: 963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fitzler, M.J., Scheuner, D., Kaufman, R.J., Golan, D.E., and Anderson, P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khusial, P., Plaag, R., and Zieve, G.W. 2005. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci. 30: 522–528. [DOI] [PubMed] [Google Scholar]
- Kiledjian, M. and Dreyfuss, G. 1992. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 11: 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb, B., Carl, M., Hock, R., Gebauer, D., and Scheer, U. 1998. Identification of a novel mRNA-associated protein in oocytes of Pleurodeles waltl and Xenopus laevis. Exp. Cell Res. 245: 272–281. [DOI] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., and Parker, R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel, J., Behm-Ansmant, I., Gatfield, D., and Izaurralde, E. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11: 1640–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, G.L. and Blau, H.M. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7: 633–636. [DOI] [PubMed] [Google Scholar]
- Sheth, U. and Parker, R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.B. and Johnson, K.S. 1988. Single-step purification of poly-peptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40. [DOI] [PubMed] [Google Scholar]
- Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M., and Parker, R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska, A., Aigueperse, C., Kress, M., Dautry, F., and Weil, D. 2005. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 118: 981–992. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., Buszczak, M., and Sayles, S. 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell 9: 675–685. [DOI] [PubMed] [Google Scholar]
- Yang, W.H., Yu, J.H., Nakajima, A., Neuberg, D., Lindor, K., and Bloch, D.B. 2004a. Do antinuclear antibodies in primary biliary cirrhosis patients identify increased risk for liver failure? Clin. Gastroenterol. Hepatol. 2: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Jakymiw, A., Wood, M.R., Eystathioy, T., Rubin, R.L., Fritzler, M.J., and Chan, E.K. 2004b. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell Sci. 117: 5567–5578. [DOI] [PubMed] [Google Scholar]
- Young, R.A. and Davis, R.W. 1983. Efficient isolation of genes by using antibody probes. Proc. Natl. Acad. Sci. 80: 1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.H., Yang, W.H., Gulick, T., Bloch, K.D., and Bloch, D.B. 2005. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11: 1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]