Abstract
Poly(A) signals are required for efficient 3′ end formation and transcriptional termination of most protein-encoding genes transcribed by RNA polymerase II. However, transcription can extend far beyond the poly(A) site before termination occurs. This implies the existence of further downstream termination signals. In mammals, a variety of sequence elements, in addition to the poly(A) site, have been implicated in the termination process. For example, termination of the human β- and ɛ-globin genes is mediated by a sequence downstream of the poly(A) site that promotes an RNA cotranscriptional cleavage (CoTC). Here we report the identification of multiple termination sequences in the mouse serum albumin (MSA) 3′ flanking region. Many transcripts from this region are cleaved cotranscriptionally, implying that such cleavage of pre-mRNA may be a more general feature of transcriptional termination.
Keywords: albumin, transcriptional termination, poly(A) signal, cotranscriptional cleavage
INTRODUCTION
Transcriptional termination by RNA polymerase II (Pol II) is an important step in gene expression. It serves to maintain an active pool of Pol II and to insulate downstream promoters from nonterminated polymerases originating from upstream genes (Greger and Proudfoot 1998). Transcriptional termination proceeds via cessation of RNA synthesis followed by Pol II-DNA dissociation. The 3′ end of protein encoding genes, with the exception of replication-dependent histone genes, is defined by a poly(A) signal, which is required for efficient 3′ end formation. It consists of an upstream, largely invariant, hexanucleotide sequence (AATAAA) followed by a more variable GU-rich tract (Proudfoot and Brownlee 1976; Gil and Proudfoot 1984; McLauchlan et al. 1988; Zhao et al. 1999). The poly(A) signal provides a binding platform for various trans-acting proteins, which participate in cleavage of the primary transcript. The actual site of transcript cleavage lies between the AAUAAA and GU-rich elements, commonly after a CA di-nucleotide (Sheets et al. 1990). The upstream product of cleavage is subject to a polyadenylation reaction, which acts to protect the transcript from exonucleases, promote its export to the cytoplasm (Huang and Carmichael 1996), and enhance its translation (Kahvejian et al. 2005). The poly(A) signal was shown to be vital for transcriptional termination (Whitelaw and Proudfoot 1986; Connelly and Manley 1988; Edwalds-Gilbert et al. 1993). Many of the proteins identified as having a role in cleavage and polyadenylation are also essential for transcriptional termination (Birse et al. 1998; Proudfoot 2004).
In yeast, Pol II transcriptional termination occurs shortly after the poly(A) signal, perhaps reflecting the close spacing of genes in this organism (Hyman and Moore 1993; Birse et al. 1998). However, in higher eukaryotes transcriptional termination occurs at varying distances beyond the poly(A) signal (Proudfoot 1989). For example, transcriptional termination of the liver-specific C2 complement gene occurs very close to the poly(A) site and involves Pol II pausing (Ashfield et al. 1991). Pause sequences have also been shown to aid in termination of both human γ-globin genes (Plant et al. 2005) and the human α-globin gene (Enriquez-Harris et al. 1991). In contrast, termination of the human β- and ɛ-globin genes occurs hundreds of base pairs downstream of the poly(A) site and requires cotranscriptional cleavage (CoTC) of 3′ flanking RNA sequences (Dye and Proudfoot 2001). In this case, a combination of a functional poly(A) site and the downstream CoTC sequences elicits termination.
Two prevailing models are used to explain transcriptional termination: the anti-termination and the torpedo hypotheses. The anti-termination model postulates the existence of proteins, associated with elongating Pol II, that normally function to prevent transcriptional termination. Dissociation of anti-termination proteins, following transcription of a functional poly(A) site, results in Pol II-DNA destabilization and subsequent termination (Logan et al. 1987). Alternatively, association of a termination factor at the poly(A) site could also bring about termination. The torpedo model proposes that the downstream product of poly(A) site cleavage acts as a substrate for 5′ → 3′ exonuclease-mediated degradation. Transcriptional termination then occurs, in part, through the degrading exonuclease catching elongating Pol II and causing it to release from the DNA template (Connelly and Manley 1988; Proudfoot 1989).
Accumulating experimental results show that the actual mechanism of termination is likely to be a synthesis of both models (Proudfoot 2004). For instance, the human protein PC4 has been shown to act as a suppressor of termination (Calvo and Manley 2001). Dissociation of PC4 upon transcription of the poly(A) site renders Pol II prone to transcriptional termination in a way analogous to the anti-termination model. Furthermore, multiple studies have revealed differences in the protein components of the elongating polymerase upstream and downstream of the poly(A) site (Ahn et al. 2004; Kim et al. 2004a). One such protein, Pcf11, was shown to promote release of stalled Pol II complexes in vitro, indicating that it may play a role in promoting transcriptional termination in vivo (Zhang et al. 2005). Two recent parallel studies have provided support for a torpedo component of the termination process in both yeast and man. We have shown that the 5′ → 3′ exonuclease, Xrn2, promotes transcriptional termination of the human β-globin gene by targeting sites of CoTC (West et al. 2004). Degradation of the nascent RNA by Xrn2 contributes to efficient termination. These observations differ from the original torpedo model in that the exonuclease torpedo is likely to degrade RNA 5′ ends generated by CoTC sites rather than the poly(A) signal. However, consistent with the original model, studies using a transcriptional pause site show that Xrn2 might also promote termination through degradation of the 3′ product of poly(A) site cleavage (N. Gromak, S. West, and N.J. Proudfoot, unpubl.). Similarly, in yeast it has been demonstrated that the 5′ → 3′ exonuclease, Rat1, degrades the downstream, Pol II-associated, product of poly(A) site cleavage leading to termination (Kim et al. 2004b). In summary, transcription of the poly(A) site renders Pol II termination-competent. Interplay between anti-termination factors acting on elongating Pol II and exonucleolytic degradation of RNA brings about termination. The existence of both CoTC and pause site termination signals as well as alternative exonuclease entry points in mammals points to a greater complexity in the termination mechanism of mammals as compared to yeast. Principally, there is clear necessity for auxiliary termination signals downstream of the poly(A) signal in mammals.
Very few downstream termination elements have been characterized in higher eukaryotes. Even so, they are expected to be widespread since most detailed studies on transcriptional termination of individual genes imply the presence of such termination sequences (Citron et al. 1984; Hagenbuchle et al. 1984; Ashfield et al. 1991; Enriquez-Harris et al. 1991; Dye and Proudfoot 2001; Peterson et al. 2002). However, there appears to be extensive heterogeneity among the defined termination elements and the potential ways that they exert their effect on Pol II. In order to begin to carry out genome-wide searches for terminator sequences, further examples must be characterized. Here we define termination sequences for the mouse serum albumin (MSA) gene. Analysis of endogenous MSA transcription, together with a plasmid-based transfection, reveals sequence elements that promote transcriptional termination. Moreover, we have crudely mapped the regions responsible for the termination activity and show that they function in the absence of surrounding sequences. We also demonstrate that many transcripts emanating from this region are cleaved cotranscriptionally. Our study supports a view that termination sequences are widespread in higher eukaryotes and that cleavage of 3′ flanking region RNAs may be a feature of many such elements.
RESULTS
RT-PCR analysis of MSA 3′ flank transcripts
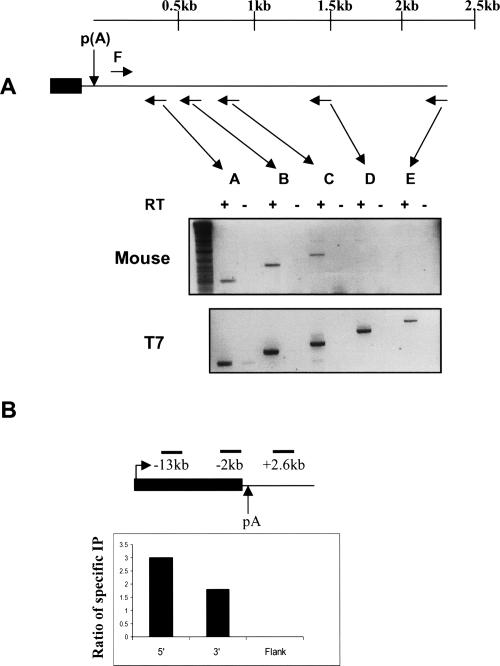
Albumin is highly expressed, mainly in the liver, and is an important component of serum. In view of its high expression levels, we reasoned that transcriptional termination may occur in a regulated fashion. Early data indicated that transcription of MSA terminated within a broad region, 1–2.5 kb downstream of the poly(A) site (Liu et al. 1988). We therefore investigated the possible existence of termination elements within the MSA 3′ flanking region. First, RT-PCR was used to map the site of transcriptional termination. RNA isolated from mouse liver was used as a template for the synthesis of five separate cDNAs (Fig. 1A ▶). Individual cDNAs were synthesized using primers positioned at increasing distances from the poly(A) site (Fig. 1A, A–E ▶). Subsequently, the cDNA primer and an upstream primer (F), positioned just downstream of the poly(A) site were used for PCR amplification. Products were obtained from cDNAs A–C but not D and E (Fig. 1A ▶, top panel). This indicates that transcriptional termination occurs between the positions of primers C and D (0.8–1.4 kp downstream of the poly(A) site). An alternative explanation (discussed later) could be that transcripts exist in the 3′ flank region that primer D binds to but are not continuous with the position of primer F. As a control, the equivalent 3′ flank region was transcribed in vitro and analyzed in an identical fashion. In this case PCR bands were observed from all cDNAs (Fig. 1A ▶, lower panel). This suggests that the lack of products D and E from the mouse liver RNA is not due to an inability of primers D and E to prime cDNA synthesis or subsequent PCR amplification. To check that Pol II was released within the MSA 3′ flanking region, its levels were examined by chromatin immunoprecipitation (ChIP) (Fig. 1B ▶). Real-time PCR analysis was employed to allow quantitative analysis of these data. The density of Pol II was analyzed over three regions of the MSA gene (see diagram in Fig. 1B ▶). Although Pol II pull-down efficiency was low, significant levels were detectable within the ORF but not within the selected portion of the 3′ flanking region (downstream of the termination site mapped by RT-PCR). Therefore, the absence of a signal from RT-PCR analysis correlates with Pol II release from the template.
FIGURE 1.
(A) RT-PCR analysis of endogenous MSA transcriptional termination: Diagram shows the final MSA exon (black box), poly(A) site and 3′ flank (line). Arrows are shown, corresponding to primers A–E, and forward primer used for subsequent PCR (F) is also labeled. A scale bar is provided indicating the distance from the poly(A) site. Exact primer coordinates are provided in the Materials and Methods section. (Top panel) Agarose gel electrophoretic analysis of PCR products made from cDNAs A–F using mouse liver RNA as a template. Presence or absence of reverse transcriptase for cDNA synthesis is indicated by + or −, respectively. (Lower panel) Identical RT-PCR analysis of an in vitro transcribed, full-length MSA 3′ flanking region RNA. (B) ChIP analysis of Pol II occupancy in 5′ and 3′ regions of the MSA ORF and 3′ flank. The accompanying diagram shows the MSA gene (black bar), poly(A) site and 3′ flank (black line). The positions of the 5′, 3′, and flank primer pairs are indicated above the diagram. Distances upstream (–) and downstream (+) of the poly(A) site are also shown. Exact coordinates are given in Materials and Methods. Quantitation is represented as the fold extra signal obtained by using a Pol II-specific antibody to pull down chromatin as opposed to no antibody.
Nuclear run on analysis of MSA gene termination
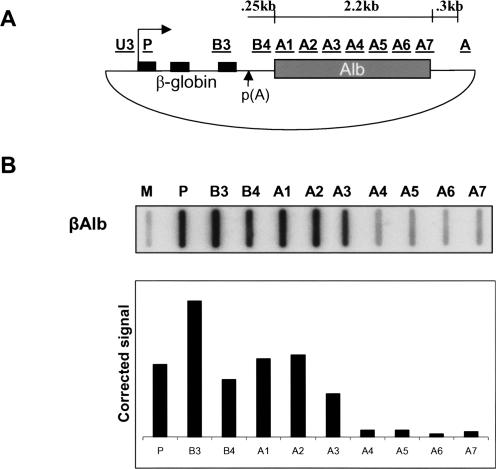
To examine MSA transcriptional termination more precisely, we performed nuclear run on (NRO) analysis using a plasmid transfection system incorporating the MSA 3′ flank. The high transcription levels achieved from such a plasmid transfection system is vital to obtaining adequate NRO signals. We utilized a human β-globin gene construct previously employed (Ashe et al. 1997; Dye and Proudfoot 1999 Dye and Proudfoot 2001), since the β-globin termination sequences are well defined. Thus, termination effects observed upon insertion of foreign DNA are easy to assess. The β-globin CoTC termination element was deleted and replaced with the first ~2.4 kb of MSA 3′ flank, containing the putative termination element (Fig. 2A ▶). HeLa cells were then transfected with the construct (βAlb) and Pol II transcription on the plasmid was examined by NRO (Fig. 2B ▶). To analyze transcription over the MSA 3′ flank insert more precisely, a panel of single-stranded M13 probes (A1–A7) were constructed (see diagram in Fig. 2A ▶). Further probes were used to analyze transcription over the HIV promoter (P), β-globin exon 3 (B3), and between the β-globin poly(A) site and MSA insert (B4). Also, M13 alone (M) was employed to give a measure of background hybridization of NRO transcripts to the vector backbone. Positive hybridization signal (above M background) is indicative of actively transcribing Pol II at that position on the template. Significant signals were seen over probes P–A3 but not over probes A4–A7, indicating that transcriptional termination occurs within probe A3 (see graph in Fig. 2B ▶). This is in agreement with the data obtained in Figure 1A ▶ since probe A3 spans a region between primers C and D. As a control for hybridization efficiency, a radiolabeled, in vitro transcribed, MSA 3′ flank-specific RNA was hybridized to probes A3–A7 (Fig. 2C ▶). In contrast to the in vivo result, high signals were observed over all probes. This clearly shows that an absence of signal over these probes, in vivo, was not due to inefficient hybridization of RNA to the M13 probes specific to regions A3–A7. Importantly, because equal amounts of RNA specific to each probe were used in this analysis (Fig. 2C ▶), the ratios of signals obtained are representative of the profile that would be observed in the absence of termination in vivo. As a final control, NRO analysis was carried out on untransfected HeLa cells (Fig. 2D ▶). In this case, no above background signal was seen over any of the MSA specific probes, showing that there is no cross-hybridization of cellular transcripts to these probes. In summary, the transient transfection system appeared to be an accurate representation of events at the endogenous locus. Termination occurred between 800 bp and 1.2 kb downstream of the poly(A) site as seen in the previous RT-PCR experiment.
FIGURE 2.
(A) Diagrammatic representation of the βAlb plasmid used for transient transfection-based analysis of transcriptional termination. β-Globin exons (black boxes) and intervening introns are followed by the poly(A) site and MSA 3′ flank (gray box). NRO probes (underlined) are above their positions on the plasmid. Construction of each probe is described in Materials and Methods. A scale bar is provided showing approximate distances from the β-globin poly(A) site. (B) NRO analysis of βAlb: The M slot represents background hybridization to single-stranded, empty M13 vector. Hybridization signals are shown below their respective probes. Graph shows above-background hybridization signals. (C) Hybridization of synthetic MSA 3′ flanking region transcript to probes A3–A7. (D) NRO analysis of mock transfected HeLa cells. All probe signals are close to background (M).
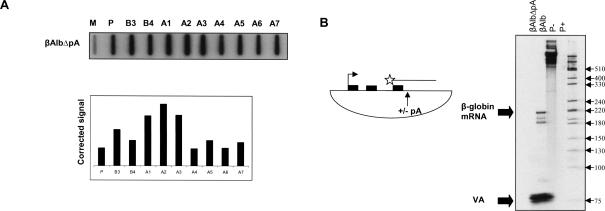
In all genes tested, a poly(A) site has been shown to be a critical determinant of Pol II transcriptional termination. We thus deleted the β-globin gene poly(A) site from βAlb, forming βAlbΔpA, and examined the effect of this deletion on termination by NRO analysis, using the same M13 probes employed in Figure 2B ▶ (Fig. 3A ▶). In line with previous data, significant hybridization signals were observed over all probes. This shows that the full activity of the MSA terminator is dependent on the presence of a poly(A) site. In order to prove that the poly(A) site deletion abolished all 3′ end processing, HeLa cells were transfected with βAlb or βAlbΔpA, and RNA was analyzed by S1 nuclease analysis. An end-labeled probe that detects cleaved and polyadenylated mRNA was employed (Fig. 3B ▶). The expected band pattern was detected from the βAlb transfection but not the βAlbΔpA transfection. As a control for transfection efficiency, a plasmid encoding the RNA polymerase III (Pol III) transcribed VA gene was cotransfected. In both cases the signal from the VA cotransfection control was the same. This shows that the poly(A) site deletion abolished all cleavage and polyadenylation, resulting in unstable (not detected) readthrough transcripts.
FIGURE 3.
(A) NRO analysis of βAlbΔpA: High signal is observed over all probes relative to background (again, M). Graph shows above-background hybridization signals. (B) S1 analysis of poly(A) site cleavage: Position of S1 probe (star and line) is shown in the diagram. It spans the final exon of β-globin and the poly(A) site. β-Globin and VA specific signals are indicated by arrows. The triplet band pattern obtained in βAlb for the β-globin poly(A) site reflects some over digestion by S1 nuclease at this AT-rich 3′–end sequence in addition to cleavage at an upstream, cryptic, poly(A) site. P– and P+ are control lanes showing probes either undigested or digested with S1 nuclease, respectively.
Dissection of MSA gene termination signals
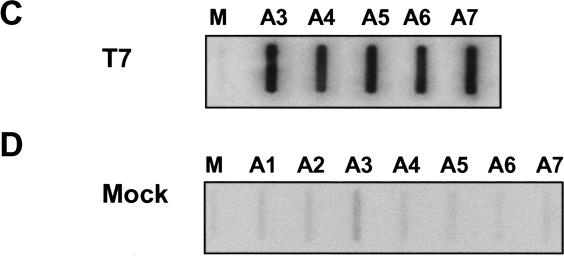
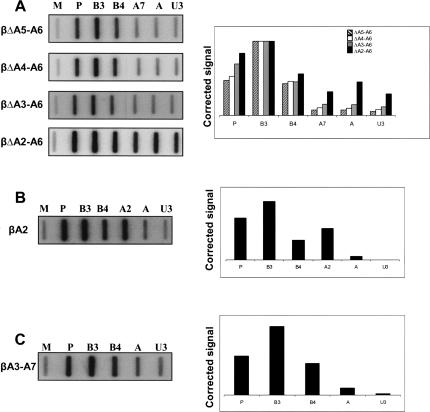
To identify the minimal sequence(s) forming the termination element, a series of deletions was made within the MSA 3′ flank insert. Four additional constructs were made, from which regions A5 and A6 (βΔA5–A6), A4–A6 (βΔA4–A6), A3–A6 (βΔA3–A6), or, finally, the 3′ part of A2–A6 (βΔA2–A6) were deleted. The effect of these deletions on termination efficiency was assessed by NRO analysis of HeLa cells transfected with the individual constructs (Fig. 4A ▶). In this case two further M13 probes were introduced to detect transcriptional readthrough: probe A, positioned ~300 bp downstream of the MSA 3′ flank insert, and probe U3, positioned upstream of the HIV promoter (~2.5 kb downstream of MSA 3′ flank insert) (see Fig. 2A ▶). Deletion of either regions A5–A6 or regions A4–A6 had no significant effect on termination because signals over probes A7, A, and U3 remained close to the background (Fig. 4A ▶, panels 1,2). This was expected, as termination was shown to occur upstream of these regions by NRO (Fig. 2B ▶). However, removal of A3 (in βΔA3–A6) did not abolish termination (Fig. 4A ▶, panel 3). This was somewhat surprising because the NRO in Figure 2B ▶ showed that this was the point at which active transcription ceased. Termination is, however, lost upon the additional deletion of ~200 nt (βΔA2–A6) (Fig. 4A ▶, panel 4). This suggests that the signal to terminate is conferred by transcription of region A2, but the effect on Pol II transcription is delayed for a few hundred base pairs. This conclusion is supported by the observation of slightly higher signal over probes A7, A, and U3 with the βΔA3–A6 construct when compared to the signals seen over these probes with both βΔA5–A6 and βΔA4–A6 (see graph in Fig. 4A ▶). The U richness of the A3 RNA sequence may also account for an exaggerated Pol II signal intensity over this region in Figure 2B ▶. To examine the effect of region A1 on termination, we deleted it from βΔA3–A6 to leave only A2 (forming βA2). NRO analysis on cells transfected with βA2 resulted in low signals over probes A and U3 (Fig. 4B ▶). This shows that the majority of the termination activity is encoded by A2 and that there is no significant contribution by A1. Importantly, this result shows that the A2 termination element functions at varying distances from the poly(A) site. This is analogous to the CoTC termination elements found in the human β-globin gene.
FIGURE 4.
(A) NRO analysis on constructs from which sections of MSA 3′ flank had been deleted. βAlb was used as a background for the deletion of various sequences (deleted sequence indicated to the left of each NRO blot). (B) NRO analysis of βA2. Probes are indicated above each slot. (C) NRO analysis of βA3–A7. Again, probes are indicated above their representative slots. Corrected hybridization signals for A–C are represented in the accompanying graphs.
In the human β-globin gene 3′ flank there are multiple termination elements that are able to terminate transcription independently. We wanted to look for similar sequences beyond the normal site of termination in the MSA 3′ flank. We therefore deleted regions A1–A2 from βAlb to form βA3–A7. NRO analysis was then carried out on HeLa cells transfected with this construct (Fig. 4C ▶), and very efficient transcriptional termination was observed, shown by low signals over probes A and U3. This shows that the MSA 3′ flank (like that of β-globin) contains multiple sequence elements that promote transcriptional termination. These additional elements may act as part of a “fail-safe” mechanism to guard against transcription beyond the major site of termination.
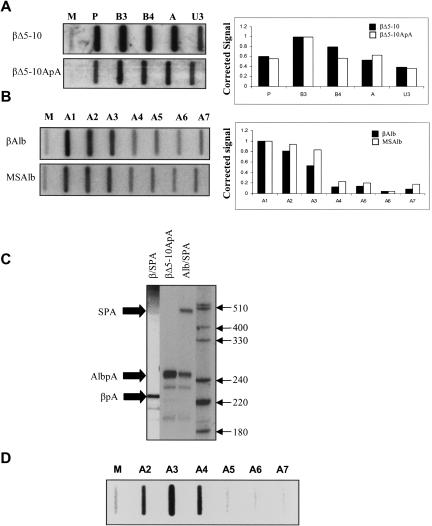
In Saccharomyces cerevisiae, the poly(A) signal is usually the only apparent cis-acting element that influences termination. In this light, it is possible that the MSA poly(A) site is sufficiently strong to terminate transcription of the endogenous MSA gene without the aid of downstream sequences. Therefore, although the sequences isolated in Figure 2 ▶ are necessary to terminate transcription in the presence of the β-globin poly(A) site, in the presence of the MSA poly(A) site they may be redundant. To test this possibility, an additional clone was made in which the β-globin poly(A) site was removed and replaced with the MSA poly(A) site in the absence of the MSA 3′ flank, forming βΔ5–10ApA (Fig. 5A ▶). NRO was performed on cells transfected with either βΔ5–10ApA or, as a control, the previously described βΔ5–10 construct (Dye and Proudfoot 2001). βΔ5–10 contains the β-globin gene and poly(A) site but no CoTC sequence and thus supports no significant transcriptional termination. Comparison of NRO results obtained from βΔ5–10 with those from βΔ5–10ApA revealed that the MSA poly(A) site had no effect on termination in the absence of a termination sequence, as observed by high signals over the readthrough probes A and U3 in NRO analysis (Fig. 5A ▶, lower panel). This result shows that additional sequences to the poly(A) site are required for termination of the endogenous MSA gene, a conclusion supported by data presented in Figures 1 ▶–4 ▶ ▶ ▶. To reaffirm this conclusion the MSA 3′ flanking region was inserted downstream of the MSA poly(A) site, within the βΔ5–10ApA clone, to form MSAlb. Pol II density over regions A1–A7 was then assayed by NRO (Fig. 5B ▶). The same experiment was also carried out on βAlb. By comparing βAlb to MSAlb, the relative influence of the β-globin and MSA poly(A) sites on termination could be assessed. Termination is relatively efficient in the MSAlb clone, as seen by the reduced signal over probes A4–A7. This illustrates the necessity for the A1–A3 termination region. Interestingly, Pol II density within the MSA 3′ flanking region is slightly higher in the MSAlb clone as compared to the βAlb clone. This may be due to the MSA poly(A) site being weaker than the β-globin poly(A) site (see below).
FIGURE 5.
(A) NRO analysis of βΔ5–10 and βΔ5–10ApA. Probes are indicated above their respective slots. Accompanying quantitation is also shown. (B) NRO analysis of βAlb and MSAlb. Probes are indicated above their respective slots. Accompanying quantitation is also shown. (C) Poly(A) site competition assay: Constructs used are indicated above their respective lanes, and various products are indicated by black arrows on the left-hand side. Size markers are also shown. (D) NRO analysis of transcription over the endogenous MSA 3′ flank using the HEPA1–6 cell line. Probes are indicated above their respective slots.
To examine the relative strengths of the MSA and β-globin poly(A) sites we performed a poly(A) site competition assay (Fig. 5C ▶). Two additional clones were made whereby a synthetic poly(A) site (SPA) was inserted ~200 bp downstream of either the β-globin poly(A) site in the βΔ5–10 construct (forming β/SPA) or the MSA poly(A) site in the βΔ5–10ApA construct (forming Alb/SPA). Cytoplasmic RNA, isolated from cells transfected with these constructs, was probed with homologous, end-labeled, DNA probes and subjected to S1 nuclease analysis to detect cleavage at the various poly(A) sites. In the Alb/SPA transfection, cleavage at the MSA poly(A) site and the SPA was observed, showing that a significant proportion of the MSA poly(A) site was out-competed by the downstream SPA. In contrast, RNA from the β/SPA transfection yielded products corresponding exclusively to cleavage at the β-globin poly(A) site since there was no apparent competition from the downstream SPA. This experiment shows that the β-globin poly(A) site is significantly stronger than the MSA poly(A) site. An extra lane is present, showing the result of probing RNA from a βΔ5–10ApA transfection with a probe specific to the MSA poly(A) site. In this case, a strong MSA poly(A) site-specific band is seen, further confirming that the MSA poly(A) site is functional. We predict that, because the MSA poly(A) site is relatively weak, the additional termination elements within regions A3–A7 are normally required to fully terminate transcription of the MSA gene. The RT-PCR of endogenous MSA expression and ChIP analysis (Fig. 1 ▶) did not provide the necessary resolution to test this possibility. We therefore employed a mouse liver cell line (HEPA1–6) and analyzed active Pol II transcription over the endogenous MSA 3′ flank using NRO (Fig. 5D ▶). Although signals were low, the profile observed was slightly different from that obtained from the transfected βAlb plasmid in Figure 2B ▶, with transcription continuing into A4 before termination takes place. We predict that this extended position of Pol II termination reflects the weaker activity of the MSA poly(A) signal. The signal observed over A4 is greater than that seen in the MSAlb NRO in Figure 5B ▶. This may be due to differences in transcription levels between the endogenous MSA gene and the transfected β-globin construct. Alternatively, other signals required for termination, such as the 3′ splice site in the final intron (which, in MSAlb, derives from the β-globin gene), might be stronger in the β-globin gene than in the MSA gene. These data also suggest that in the endogenous situation the downstream transcriptional terminators described above (A3–A7) are required to elicit efficient termination.
Some MSA termination region transcripts are cleaved cotranscriptionally
Previous RT-PCR analysis of β-globin 3′ flanking region transcripts failed to detect products from positions more than ~900 nt from the poly(A) site despite positive NRO signals over these regions (Dye and Proudfoot 2001). This was because transcripts were cotranscriptionally cleaved within these regions prior to downstream Pol II termination. Interestingly, region A4, which gives a positive NRO signal in liver cells, is beyond the annealing site of primer D in Figure 1A ▶. Even so, a D-specific product from that position in the RT-PCR analysis was not observed, suggesting the possible presence of CoTC activity. We therefore examined the MSA 3′ flank for potential CoTC activity. βAlb was transfected into HeLa cells and hybrid selection NRO analysis was performed. This technique allows analysis of transcript continuity between the transcribing Pol II and a chosen upstream point. Nascent nuclear transcripts are hybridized to a biotinylated anti-sense RNA probe specific to this chosen point. RNA hybrids are magnetically selected using streptavidin-coated magnetic beads. Selected transcripts are then released from the beads by hydrolysis and hybridized to a nitrocellulose filter containing gene-specific M13 probes. This profile of selected transcripts is then compared to that of unselected transcripts that do not bind the biotinylated anti-sense RNA probe.
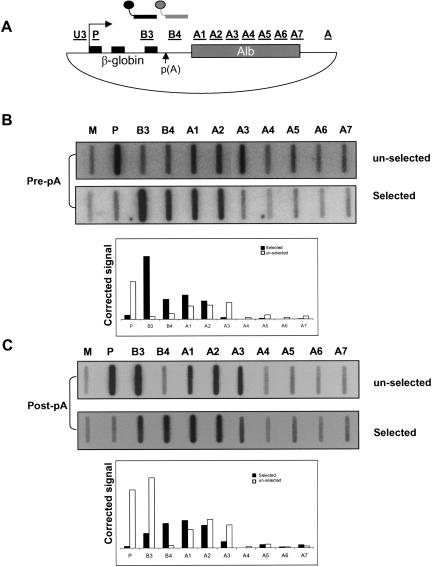
Radiolabeled nascent transcripts from the βAlb transfection were hybridized to a biotinylated anti-sense probe (pre-pA) specific to a region immediately upstream of the β-globin poly(A) site corresponding to probe B3 (Fig. 6A ▶). Transcripts, not cleaved at the poly(A) site, were found to be intact up to region A2, but no signal above background was observed over probe A3 or beyond (Fig. 6B ▶, lower panel). This is in contrast to the unselected Pol II density profile, which detects actively transcribing Pol II up to and within region A3 (Fig. 2B ▶). Confirmation for the specificity of the anti-sense pre-pA site probe can be seen by the unselected NRO profile (i.e., the RNA that escaped selection with the pre-pA site probe), which shows a high P signal (5′ of the selection probe) but low signal over probe B3, corresponding to the selection probe. These data indicate that RNA transcribed from A2 is cleaved prior to transcription of region A3. However, the possibility remained that poly(A) site cleavage separated A3 transcripts from the pre-pA site selection probe. We therefore repeated the above experiment but this time used a biotinylated RNA probe specific to a region downstream of the poly(A) site (post-pA) (Fig. 6C ▶). In the selected RNA fraction we observed a reduced signal over A3 again recovered in the nonselected RNA (Fig. 6B ▶, lower and upper panels, respectively). The specificity of the anti-sense post-pA site probe is again evident as the unselected NRO profile gives high P and B3 signals (upstream of the selection probe) but low signals over B4, corresponding to the selection probe. We were able to select transcripts extending up to 2.5 kb from the selection probe upon hybrid selection NRO analysis of βΔ5–10 with the post-pA probe (data not shown), indicating that the drop in signal over the MSA 3′ flank observed here is not due to random RNA degradation. These selection experiments demonstrate that there is cleavage of some A2 transcripts. Consequently these results might explain the presence of Pol II on regions of the endogenous gene from which transcripts are undetectable by RT-PCR (cf. Figs. 5C ▶ and 1A ▶).
FIGURE 6.
Hybrid selection analysis of βAlb. (A) Diagram of βAlb showing the positions of the pre- and post-pA biotinylated selection probes (black line and connected circle, and gray line and connected circle, respectively). (B) Pre-pA site selection of βAlb transcripts showing selected and unselected fractions. Probes are indicated. Quantitation is shown in the graph. (C) Post-pA site selection of βAlb transcripts. Again selected and unselected fractions are shown. Probes are indicated and quantitation is shown in the graph.
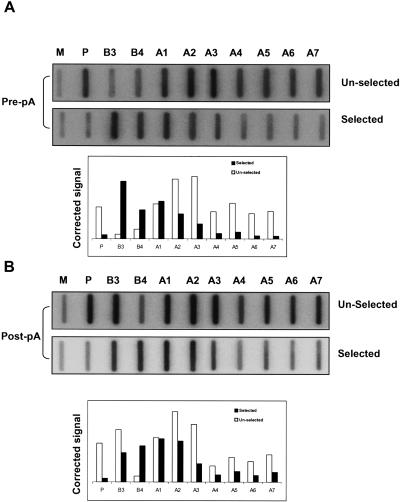
We finally examined the dependence of the MSA transcript cleavage on a poly(A) site, since β-globin CoTC was shown to occur in the absence of the poly(A) site. βAlbΔpA was transfected and hybrid selection NRO analysis was performed using the biotinylated pre- and post-pA selection probes (Fig. 7 ▶). Selection of transcripts with the pre-pA site probe reveals a decline in signal intensity over A2–A7 as compared to the unselected profile (Fig. 7A ▶). This indicates that some cleavage of MSA 3′ flanking region RNA occurs independently of the poly(A) site. Hybrid selection NRO analysis of βAlbΔpA with the post-pA probe again resulted in a decline in signals, which over probes A4–A7, (Fig. 7B ▶, lower panel). However, these signals were somewhat higher than observed with the pre-pA site probe. Nevertheless, a higher percentage of the selected signal for probes A4–A7 was present in the unselected fraction (Fig. 7B ▶, upper panel) as compared to the fraction. The finding that some A3–A7 transcripts are not cleaved upon mutation of the poly(A) site may reflect a less efficient mechanism as compared to human β-globin CoTC. Alternatively, some of the termination activity of region A2 might not depend on transcript cleavage. Overall, hybrid selection analysis, as shown in Figures 6 ▶ and 7 ▶, for βAlb and βAlbΔpA reveal that transcript cleavage is associated with regions A2–A4, and correlates with positions of Pol II termination on transfected gene constructs and on the endogenous MSA gene.
FIGURE 7.
Hybrid selection analysis of βAlbΔpA. (A) Pre-pA site selection of βAlbΔpA transcripts showing selected and unselected fractions. Probes are indicated. Quantitation is shown in the graph. (B) Post-pA site selection of βAlbΔpA transcripts. Again selected and unselected fractions are shown. Probes are indicated and quantitation is shown in the graph.
DISCUSSION
Transcriptional termination in mammals requires coordination of specific sequence elements and proteins. Here we have identified multiple sequence elements in the MSA 3′ flanking region that are necessary for transcriptional termination. Using hybrid selection NRO analysis, we have shown that many MSA 3′ flanking transcripts are cleaved cotranscriptionally. RT-PCR data in Figure 1A ▶ indicate this transcript cleavage activity is also a feature of the endogenous gene. However, the lower NRO signals achieved from the endogenous MSA gene preclude the use of hybrid selection NRO analysis. Overall our data show that some MSA transcripts are cleaved cotranscriptionally and that sequences associated with this activity also promote termination.
The human β-globin gene CoTC element functions to terminate transcription irrespective of its distance from the poly(A) site, as does the MSA termination element described here. The 5′ → 3′ exonuclease Xrn2 promotes termination from sites of β-globin CoTC (West et al. 2004). Given the observations described here, it will be interesting to determine any role(s) of cleavage in the MSA 3′ flank with regard to Xrn2-mediated termination.
The β-globin CoTC termination region is significantly AU-rich. However, the MSA 3′ termination sequence appears not to be significantly enriched in A and U. A previous study on the human β-globin CoTC element pointed toward the importance of secondary structure (Teixeira et al. 2004). Here it was suggested that autocatalytic cleavage by this structure brought about transcriptional termination. However, the low cleavage efficiency observed in vitro implies that, in vivo, further protein cofactors are required, either for the cleavage or for the maintenance of the secondary structure. Preliminary attempts to find autocatalytic cleavage sites in the MSA 3′ flanking region did not yield any activity, although we cannot exclude the possibility that residual activity might exist if the correct fragment were analyzed. In light of these observations, the transcript instability in both the MSA and perhaps β-globin 3′ flanking regions may be highly dependent on proteins.
Cotranscriptional packaging of RNA into RNP structures is essential to preserve genomic stability (Huertas and Aguilera 2003; Li and Manley 2005). In yeast, mRNP packaging proteins such as Tho/TREX were shown to cross-link throughout genes but not downstream of poly(A) sites (Kim et al. 2004a). We note that MSA 3′ flanking region transcript cleavage is less robust than β-globin CoTC since mutation of the poly(A) site reduced the cleavage efficiency, as seen in Figure 7B ▶. This may be due to perturbation of RNA structure and/or protein–RNA interactions if the ongoing mRNP packaging process is defective in the absence of a functional poly(A) signal. It is clear from analysis of both β-globin and MSA termination sequences that they comprise multiple redundant elements. Additional studies will be required to pinpoint the exact nature of both the RNA sequence involved and their potential protein partners.
MATERIALS AND METHODS
Constructs
βΔ5–10 and the Tat plasmid have been described previously (Adams et al. 1988; Dye and Proudfoot 2001). βAlb was made by insertion of an EcoR1/PvuII restriction fragment of MSA genomic DNA into a vector prepared by PCR amplification of βΔ5–10 with oligos VF (5′-CAGGAAACTATTACTCAAAGGGT A-3′) and VR (5′-CTTGAATCCTTTTCTGAGGGATG-3′). βAlbΔpA was made by ligation of a PCR amplification of βAlb using oligos ΔpA5′ (5′-CATTGCAATGATGTATTTAAA-3′) and ΔpA3′ (5′-A ATCCAGATGCTCAAGGCCC-3′). βΔA5–A6 was made by ligation of a PCR amplification of βAlb using oligos M13105′ and M1373′. βΔA4–A6 was made by ligation of a PCR product, generated by amplification of βAlb with oligos M13105′ and M1363′. βΔA3–A6 was made by ligation of a PCR product, generated by amplification of βAlb with oligos M13105′ and C2. βΔA2–A6 was made by ligation of a PCR product, generated by amplification of βAlb with oligos M13105′ and M1333′. βA2 was made by insertion of a PCR product generated by amplification of βAlb with oligos M1335′ and C2. βA3–A7 was made by inserting a Sac1/PvuII generated restriction fragment of MSA genomic DNA into a vector generated by PCR amplification of βΔ5–10 using oligos VF and VR. β5–10ApA was generated by insertion of a PCR product obtained by amplification of mouse genomic DNA with oligos ApA5′ and ApA3′ into a vector prepared by PCR amplification of βΔ5–10 using oligos ΔpA3′ and ΔbGU3′ (5′-CCTTGGGAAAATACACTATATC-3′). Alb/SPA was made by insertion of the annealed oligo pair SPAF (5′- GATCCAATAAAAGATCTTTATTTTCATTAGATCTGTGTGTTGGTTTTTTGTGTGGATC-3′) and SPAR (5′-GATCCACACAAAAAACCAACACACAGATCTAATGAAAATAAAGATCTTTTATTGGATC-3′) into a vector prepared by PCR amplification of βΔ5–10ApA using oligos VF and VR. β/SPA was made by insertion of the annealed SPAF/SPAR oligo pair into a vector prepared by PCR amplification of βΔ5–10 using oligos VF and VR. MSAlb was made by insertion of a product made by PCR amplification of βΔ5–10ApA using oligos ApA5′ and ApA3′ into a vector generated by PCR amplification of βAlb, using oligos ΔpA3′ and ΔbGU3′.
M13 probes
Probes P, B3, B4, and U3 have been described previously (Dye and Proudfoot 2001) as has the A probe (West et al. 2004). In generating probes A1–A7, all inserts were cloned into M13mp19 (New England Biolabs), digested with HinCII. Inserts were prepared by PCR amplification using βAlb as a template. A1 was made by PCR amplification using oligos M1315′ and M1313′. A2 was made by PCR amplification using oligos M1335′ and C2. A3 was made by PCR amplification using oligos C3 and M1363′. A4 was made by PCR amplification using oligos M1375′ and M1373′. A5 was made by PCR amplification with oligos M1385′ and M1383′. A6 was made by PCR amplification using oligos M1395′ and M1393′. A7 was made by PCR amplification using oligos M13105′ and C4.
Biotinylated RNA selection probes
Preparation of the pre- and post-pA site probes are described in Dye and Proudfoot (2001).
HeLa Cell transfections
Fifteen-centimeter-diameter plates of subconfluent HeLa cells were transfected with 15 μg of reporter plasmid and 1.5 μg of tat plasmid using Lipofectamine 2000 (Invitrogen) following the manufacturer’s guidelines. For nascent analysis, RNA was isolated 12–16 h post-transfection, and for steady-state analysis RNA was isolated 24 h post-transfection.
NRO analysis
NRO analysis of transiently transfected HeLa cells was carried out as previously described (Ashe et al. 1997). NRO analysis of endogenous MSA transcriptional termination was carried out using the same protocol. To quantitate the NRO signals the value obtained for M was taken away from the value of each of the probes on the same filter. These values (corrected signal) were then plotted graphically. Alternatively, (in Fig. 4 ▶) when comparing a number of filters, the B3 value on the filter was taken to be 1 and all other values were expressed as a percentage of B3 in order to equalize potentially differing signal intensities on different filters.
Hybrid selection NRO
Hybrid selection NRO analysis was carried out as described previously (Dye and Proudfoot 2001).
S1 nuclease analysis
For analysis of cleavage and polyadenylation in wild-type and mutant β-globin poly(A) site-containing constructs and βΔ5–10ApA, end labeled DNA probes were prepared from EcoR1 restriction fragments of βΔ5–7 and βAlbΔpA in the same manner described in Ashe et al. (1995). For poly(A) site competition analysis end-labeled DNA probes were made in the same way but using an EcoR1 digest of either Alb/SPA or β/SPA. The S1 nuclease analysis was performed as previously described (Ashe et al. 1995).
Chromatin immunoprecipitation (ChIP)
The ChIP protocol and Pol II-specific antibody have been described previously (West et al. 2004). Primer pairs used to detect Pol II over the 5′ region were alb55′ and alb53′. Primers for the 3′ region were alb35′ and alb33′. Primers for the 3′ flank were albflank5′ and albflank3′.
RNA isolation from mouse tissue
To isolate RNA from mouse liver, frozen liver tissue (in liquid nitrogen) was crushed with a mortar and pestle. RNA was isolated from 100 mg of tissue with 1ml of Trizol following the manufacturer’s guidelines.
In vitro transcription
The template from which the MSA 3′ flanking region (Fig. 2C ▶) was transcribed was made by PCR amplification of βAlb using primers AtcF (5′-TAATACGACTCACTATAGGGCCCTGATGCCTATGCCTTATT-3′) and C4.
RT-PCR
Two micrograms of mouse liver RNA were used as a template for reverse transcription. Oligos used were F (M1315′), A (M1313′), B (M1323′), C (M1333′), D (M1363′), and E (M13103′). SuperScrip-tIII reverse transcriptase (Invitrogen) was used to synthesize the cDNA following manufacturer’s guidelines. cDNA was amplified by Taq under standard conditions (25 cycles of PCR).
MSA oligonucleotides
Sequences and their coordinates (using GenBank no. AC135240):
Table t1.
| ApA5′ | (5′-CCCTAAGGAACACAAATTTCTTTA-3′) | 182701–182678 |
| ApA3′ | (5′-AAAGGCAGGGATTCCTCTGAGCC-3′) | 182545–182567 |
| M1315′ | (5′-TAAAAGCTTATGAACTGTGGC-3′) | 182409–182389 |
| M1313′ | (5′-TCAATCCAGCACCCCTCCCGA-3′) | 182199–182219 |
| M1335′ | (5′-GTGCACATTATTCTTCAACCA-3′) | 181989–181969 |
| M1333′ | (5′-ATCTCCATTCTACCCTCCCAG-3′) | 181788–181808 |
| C2 | (5′-CAGAGCTCACTGGGCTCTTTA-3) | 181540–181560 |
| C3 | (5′-TTTTTGAGAGGGCATCCATGG-3′) | 181539–181519 |
| M1363′ | (5′-CAATACCATTCTCTAATGAAA-3′) | 181115–181135 |
| M1375′ | (5′-CAAAGATCTGTTTACTGGCTC-3′) | 181114–181094 |
| M1373′ | (5′-AACTCTTATGTGCATAGATAG-3′) | 180902–180922 |
| M1385′ | (5′-TCAACTGATAATTCTGACTCT-3′) | 180901–180881 |
| M1383′ | (5′-GTTCTTATGGAAGTATGAGGT-3′) | 180700–180720 |
| M1395′ | (5′-CATCAACAACTAGGATATAGT-3′) | 180699–180679 |
| M1393′ | (5′-TGCAGTATAAACCAAAGAATC-3′) | 180481–180501 |
| M13105′ | (5′-CAGATCGGTCCATTGTGTCTT-3′) | 180480–180460 |
| M13103′ | (5′-TTCAAATACCTGGGATGAAGT-3′) | 180269–180289 |
| C4 | (5′-AAGAGTAGTTACACAGCTGAC-3′) | 180119–180139 |
| alb55′ | (5′-CTGCTCCTCAAATTCCCTTG-3′) | 195727–195708 |
| alb53′ | (5′-GCAAAGAGCTAATGGCCTTG-3′) | 195616–195635 |
| alb35′ | (5′-AAACTGTGGCCTCACATTCC-3′) | 184747–184728 |
| alb33′ | (5′-CTTGGCTTCACCGCTCAG-3′) | 184632–184651 |
| albflank5′ | (5′-TGGCAAGATGATTTGTTTGC-3′) | 179841–179822 |
| albflank3′ | (5′-TCTTCCGTGTCAAGTGCATC-3′) | 179710–179729 |
Acknowledgments
We thank all members of the NJP laboratory for helpful discussion. Special thanks go to Olivia Rissland for critical reading of the manuscript. These studies were supported by a Programme Grant from the Welcome Trust. S.W. is supported by an EPA research studentship.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2232406.
REFERENCES
- Adams, S.E., Johnson, I.D., Braddock, M., Kingsman, A.J., Kingsman, S.M., and Edwards, R.M. 1988. Synthesis of a gene for the HIV transactivator protein TAT by a novel single stranded approach involving in vivo gap repair. Nucleic Acids Res. 16: 4287–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S.H., Kim, M., and Buratowski, S. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13: 67– 76. [DOI] [PubMed] [Google Scholar]
- Ashe, M.P., Griffin, P., James, W., and Proudfoot, N.J. 1995. Poly(A) site selection in the HIV-1 provirus: Inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes & Dev. 9: 3008–3025. [DOI] [PubMed] [Google Scholar]
- Ashe, H.L., Monks, J., Wijgerde, M., Fraser, P., and Proudfoot, N.J. 1997. Intergenic transcription and transinduction of the human β-globin locus. Genes & Dev. 11: 2494–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield, R., Enriquez-Harris, P., and Proudfoot, N.J. 1991. Transcriptional termination between the closely linked human complement genes C2 and factor B: Common termination factor for C2 and c-myc? EMBO J. 10: 4197–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse, C.E., Minvielle-Sebastia, L., Lee, B.A., Keller, W., and Proudfoot, N.J. 1998. Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301. [DOI] [PubMed] [Google Scholar]
- Calvo, O. and Manley, J.L. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7: 1013–1023. [DOI] [PubMed] [Google Scholar]
- Citron, B., Falck-Pedersen, E., Salditt-Georgieff, M., and Darnell Jr., J.E. 1984. Transcription termination occurs within a 1000 base pair region downstream from the poly(A) site of the mouse β-globin (major) gene. Nucleic Acids Res. 12: 8723–8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, S. and Manley, J.L. 1988. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Dev. 2: 440–452. [DOI] [PubMed] [Google Scholar]
- Dye, M.J. and Proudfoot, N.J. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3: 371–378. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Multiple transcript cleavage precedes polymerase release in termination by RNA polymerase II. Cell 105: 669– 681. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert, G., Prescott, J., and Falck-Pedersen, E. 1993. 3′ RNA processing efficiency plays a primary role in generating termination- competent RNA polymerase II elongation complexes. Mol. Cell. Biol. 13: 3472–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez-Harris, P., Levitt, N., Briggs, D., and Proudfoot, N.J. 1991. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 10: 1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, A. and Proudfoot, N.J. 1984. A sequence downstream of AAUAAA is required for rabbit β-globin mRNA 3′-end formation. Nature 312: 473–474. [DOI] [PubMed] [Google Scholar]
- Greger, I.H. and Proudfoot, N.J. 1998. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17: 4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuchle, O., Wellauer, P.K., Cribbs, D.L., and Schibler, U. 1984. Termination of transcription in the mouse α-amylase gene Amy- 2a occurs at multiple sites downstream of the polyadenylation site. Cell 38: 737–744. [DOI] [PubMed] [Google Scholar]
- Huang, Y. and Carmichael, G.C. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas, P. and Aguilera, A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12: 711–721. [DOI] [PubMed] [Google Scholar]
- Hyman, L.E. and Moore, C.L. 1993. Termination and pausing of RNA polymerase II downstream of yeast polyadenylation sites. Mol. Cell. Biol. 13: 5159–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian, A., Svitkin, Y.V., Sukarieh, R., M’Boutchou, M.N., and Sonenberg, N. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & Dev. 19: 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Ahn, S.H., Krogan, N.J., Greenblatt, J.F., and Buratowski, S. 2004a. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23: 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M., Krogan, N.J., Vasiljeva, L., Rando, O.J., Nedea, E., Greenblatt, J.F., and Buratowski, S. 2004b. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522. [DOI] [PubMed] [Google Scholar]
- Li, X. and Manley, J.L. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378. [DOI] [PubMed] [Google Scholar]
- Liu, J.K., Bergman, Y., and Zaret, K.S. 1988. The mouse albumin promoter and a distal upstream site are simultaneously DNase I hypersensitive in liver chromatin and bind similar liver-abundant factors in vitro. Genes & Dev. 2: 528–541. [DOI] [PubMed] [Google Scholar]
- Logan, J., Falck-Pedersen, E., Darnell Jr., J.E., and Shenk, T. 1987. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc. Natl. Acad. Sci. 84: 8306–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan, J., Moore, C.L., Simpson, S., and Clements, J.B. 1988. Components required for in vitro cleavage and polyadenylation of eukaryotic mRNA. Nucleic Acids Res. 16: 5323–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, M.L., Bertolino, S., and Davis, F. 2002. An RNA polymerase pause site is associated with the immunoglobulin mus poly(A) site. Mol. Cell. Biol. 22: 5606–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant, K.E., Dye, M.J., Lafaille, C., and Proudfoot, N.J. 2005. Strong polyadenylation and weak pausing combine to cause efficient termination of transcription in the human Gγ-globin gene. Mol. Cell. Biol. 25: 3276–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J. 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 14: 105–110. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16: 272–278. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J. and Brownlee, G.G. 1976. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature 263: 211–214. [DOI] [PubMed] [Google Scholar]
- Sheets, M.D., Ogg, S.C., and Wickens, M.P. 1990. Point mutations in AAUAAA and the poly (A) addition site: Effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18: 5799–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, A., Tahiri-Alaoui, A., West, S., Thomas, B., Ramadass, A., Martianov, I., Dye, M., James, W., Proudfoot, N.J., and Akoulitchev, A. 2004. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature 432: 526–530. [DOI] [PubMed] [Google Scholar]
- West, S., Gromak, N., and Proudfoot, N.J. 2004. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature 432: 522–525. [DOI] [PubMed] [Google Scholar]
- Whitelaw, E. and Proudfoot, N. 1986. α-Thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α 2 globin gene. EMBO J. 5: 2915–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Fu, J., and Gilmour, D.S. 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes & Dev. 19: 1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405– 445. [DOI] [PMC free article] [PubMed] [Google Scholar]