Abstract
AtCyp59 and its orthologs from different organisms belong to a family of modular proteins consisting of a peptidyl-prolyl cis–trans isomerase (PPIase) domain, followed by an RNA recognition motif (RRM), and a C-terminal domain enriched in charged amino acids. AtCyp59 was identified in a yeast two-hybrid screen as an interacting partner of the Arabidopsis SR protein SCL33/SR33. The interaction with SCL33/SR33 and with a majority of Arabidopsis SR proteins was confirmed by in vitro pull-down assays. Consistent with these interactions, AtCyp59 localizes to the cell nucleus, but it does not significantly colocalize with SR proteins in nuclear speckles. Rather, it shows a punctuate localization pattern resembling transcription sites. Indeed, by using yeast two-hybrid, in vitro pull-down, and immunoprecipitation assays, we found that AtCyp59 interacts with the C-terminal domain (CTD) of the largest subunit of RNA polymerase II. Ectopic expression of the tagged protein in Arabidopsis cell suspension resulted in highly reduced growth that is most probably due to reduced phosphorylation of the CTD. Together our data suggest a possible function of AtCyp59 in activities connecting transcription and pre-mRNA processing. We discuss our data in the context of a dynamic interplay between transcription and pre-mRNA processing.
Keywords: cyclophilin, SR proteins, CTD, RNA polymerase II, transcription, cellular localization
INTRODUCTION
Cyclophilins are ubiquitous proteins found in archea, bacteria, and eukarya, with the largest family described so far in the plant Arabidopsis thaliana (Hunter 1998; Schiene and Fischer 2000; Romano et al. 2004). They belong to a family of immunosuppressant receptor proteins called immunophilins, which in addition to cyclophilins includes the FK506 binding proteins and the parvulins (Schiene and Fischer 2000). The first described cyclophilin was cyclophilin A because of its high affinity for the immunosuppressive drug cyclosporine A (Handschumacher et al. 1984). Cyclophilins possess peptidyl–prolyl cis–trans isomerase (PPIase) activity; e.g., they catalyze cis–trans isomerization of peptide bonds preceding proline. Cis to trans isomerization of prolyl imide bonds takes place spontaneously, but is very slow (Schiene and Fischer 2000; Romano et al. 2004). As this is a rate limiting step in protein folding, the importance of these enzymes is best highlighted by the fact that over 90% of prolyl imide bonds are in trans conformation (Stewart et al. 1990).
In eukaryotic cells, cyclophilins have been found in all cellular compartments, with a variety of functions being ascribed to them. These include protein trafficking and maturation, receptor signaling, receptor complex stabilization, apoptosis, RNA processing, and spliceosome assembly (Hunter 1998; Schiene and Fischer 2000; Harrar et al. 2001; Lu et al. 2002 and references therein). However, the mechanisms of how cyclophilins contribute to these cellular events have been difficult to establish and are still largely unknown. Yet, several studies provided evidence for the mechanism of cyclophilin action. For example, the cyclo-philin A–cyclosporine A complex inhibits the phosphatase activity of calcineurin, which in turn results in inhibition of T-cell activation by blocking the expression of several immunosuppressive genes (Liu et al.1991). Another example is the parvulin Ess1/Pin1, which has been shown to interact with a number of phosphoproteins through recognition of a pSer/Thr–Pro motif by its N-terminal WW domain. By promoting the cis–trans isomerization of the prolyl peptide bond through its C-terminal PPIase domain, it regulates the activities of p53, tau, RNA polymerase II, and some mitotic proteins (Lu et al. 2002; Shaw 2002; Lu 2004; Lim and Lu 2005 and references therein). The best characterized plant cyclophilin is TLP40. This protein is located in the thylakoid lumen and is implicated in the turnover of the photosystem II protein D1 by regulating its dephosphorylation (Fulgosi et al. 1998; Vener et al. 1999).
The majority of cyclophilins are small proteins containing only a PPIase domain of about 120 amino acids. However, several multidomain cyclophilins from different organisms have been described as well. These include Ess1/Pin1 (Hanes et al. 1989; Lu et al. 1996), TLP40 (Fulgosi et al. 1998), hCyP33 (Mi et al. 1996), CyP-13 (Zorio and Blumenthal 1999), SRcyp (Bourquin et al. 1997), mocaCYP (Cavarec et al. 2002), NK-TR1 (Anderson et al. 1993; Rinfret et al. 1994), CypRS64 and CypRS92 (Lorković et al. 2004b), and Kin241p (Krzywicka et al. 2001), the last eight having domains characteristic for nuclear proteins being involved in pre-mRNA maturation. SRcyp, mocaCYP, NK-TR1, CypRS64, and CypRS92 are highly similar nuclear proteins consisting of an N-terminal PPIase domain followed by a charged domain and a C-terminal domain rich in arginine/serine (RS) dipeptides that are characteristic for splicing factors called SR proteins (Graveley 2000; Sanford et al. 2003). Human SRcyp has been identified as an interacting partner of the RNA polymerase II C-terminal domain (CTD) and the SR protein-specific kinase Clk/Sty (Nestel et al. 1996; Bourquin et al. 1997). Its rat and Drosophila homologs have been identified as components of the nuclear matrix (Mortillaro and Berezney 1998) and an interacting partner of the transcriptional regulator p300/CBP (Cavarec et al. 2002), respectively. Arabidopsis CypRS64 and CypRS92 are likewise nuclear proteins interacting with SR and snRNP-specific proteins. Interaction of the Arabidopsis SF2/ASF homolog, SRp34, with CypRS64 has been shown to be phosphorylation dependent, implicating this protein in spliceosomal dynamics (Lorković et al. 2004b). Alternatively, due to their interaction with the transcriptional machinery, members of the RS domain-containing cyclophilins could have a role in connecting transcription and pre-mRNA processing (Bourquin et al. 1997; Cavarec et al. 2002). An additional nuclear cyclophilin implicated in pre-mRNA splicing is the small 20-kDa USA-Cyp, a component of the U4/U6 di-snRNP complex (Horowitz et al. 1997, 2002; Teigelkamp et al. 1998).
The most complex multidomain cyclophilin is Kin241p, initially identified in Paramecium tetraurelia as a protein involved in cell morphogenesis. This protein consists of a PPIase domain at the N terminus, followed by RNA recognition motif (RRM) and a C-terminal domain enriched in charged amino acids and serines. Highly similar proteins have been found in the majority of eukaryotes (Krzywicka et al. 2001). Here, we show that the Arabidopsis homolog of Kin241p, AtCyp59, localizes to the nucleus, where it interacts with SR proteins and the CTD of the RNA pol II, which implicates this protein in the regulation of activities connecting transcription and pre-mRNA processing.
RESULTS
AtCyp59 cDNA cloning and characterization of its protein product
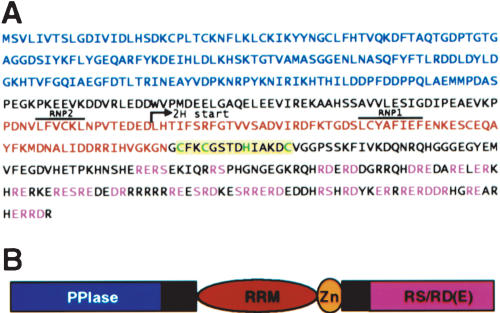
A partial cDNA clone encoding the C-terminal half of the Arabidopsis AtCyp59 (Fig. 1A ▶) was isolated in a two-hybrid screen with the SR protein SCL33/SR33 as the bait. The full-length cDNA was cloned by RT-PCR performed with total RNA isolated from mixed Arabidopsis tissues and oligonucleotides derived from the partial cDNA (3′ oligo) and genomic sequence (5′ oligo). Sequence analysis revealed an open reading frame encoding a protein of 506 amino acids. The N terminus of the encoded protein is occupied by the PPIase domain, followed by an RRM. The C-terminal domain is enriched in basic and acidic amino acids containing several RS, arginine/aspartate (RD), or arginine/glutamate (RE) dipeptides (Fig. 1A,B ▶). In addition to these three domains, AtCyp59 contains a CX2CX4HX4C zinc knuckle located immediately after the RRM (Fig. 1A,B ▶). As revealed by RT-PCR analysis, the gene encoding AtCyp59 is expressed in all major Arabidopsis organs (Romano et al. 2004).
FIGURE 1.
Sequence analysis of AtCyp59. (A) Protein sequence of Arabidopsis AtCyp59. The peptidyl-prolyl cis–trans isomerase (PPIase) domain is printed in blue letters, and the RNA recognition motif, (RRM) in red letters with RNP1 and RNP2 overlined. The CCHC zinc knuckle is printed on yellow background with CCHC in green letters. The RS, RD, and RE dipeptides are printed in pink letters. Position of the start of two-hybrid clone isolated with SCL33/SR33 is indicated by an arrow. (B) Schematic representation of AtCyp59 modular structure.
Database searches revealed that proteins with sequence and structure similarities to AtCyp59 are encoded in the majority of eukaryotic genomes. Proteins from different organisms show particularly high sequence conservation in the RRM domains and less so in the PPIase domain (Fig. 2 ▶; Table 1 ▶; data not shown), whereas the C-terminal domains are similar only in terms of being enriched in basic and acidic amino acids and in some cases in serines. The Arabidopsis protein and its plant orthologs contain a zinc knuckle between the RRM and the C-terminal domain. As this motif has not been found in any homologous protein from different organisms, this feature seems to be plant-specific. Although proteins similar to AtCyp59 are encoded in the majority of fungal genomes available at the NCBI BLAST server (Fig. 2 ▶; Table 1 ▶; data not shown) they are not found in the genome of Saccharomyces cerevisiae and other Saccharomyces species except S. kluyveri. Because AtCyp59 homologs are encoded in genomes of S. kluyveri, Eremothecium gossypii, Kluyveromyces waltii, Debaryomyces hansenii, and Yarrowia lipolytica (Table 1 ▶), this does not seem to be a characteristic of the order Saccharomycotina or genus Saccharomyces. However, the predicted proteins from genomic sequences of E. gossypii, K. waltii, S. kluyveri, and D. hansenii are shorter and do not contain the C-terminal charged domain (Table 1 ▶). Analyses of sequences downstream of the RRM from these organisms did not reveal a potential coding capacity for an RS/RD/RE-rich domain, indicating that the predicted proteins are correct. In contrast, the protein from Y. lipolytica, which also belongs to Saccharomycotina, does contain a C-terminal domain with many RS/RD/RE dipeptides (Table 1 ▶). Reasons for this divergence in fungi and consequences thereof are not clear. Phylogenetic analysis of all proteins listed in Table 1 ▶ revealed that the proteins from E. gossypii, K. waltii, S. kluyveri, D. hansenii, and Pichia angusta cluster together, whereas the Y. lipolytica protein appears as a separate branch but closest to the aforementioned group (data not shown). This is also obvious from alignments of the PPIase and RRM domains, which show that these two domains from E. gossypii, K. waltii, S. kluyveri, D. hansenii, P. angusta, and Y. lipolytica are the most divergent (Fig. 2 ▶; Table 1 ▶; data not shown). Similarly to Saccharomycotina, a mixed situation is found in proteins from Apicomplexa (Table 1 ▶). Proteins from Theileria annulata and Toxoplasma gondii do contain an RS/RD-rich domain, whereas proteins from Cryptosporidium hominis and Cryptosporidium parvum do not. In addition, proteins from Plasmodium falciparum and Plasmodium yoelii yoelii show very low sequence conservation in the PPIase domain compared to other proteins and they are much longer due to an extended C-terminal domain (Table 1 ▶).
FIGURE 2.
Sequence alignment of AtCyp59 RRM with corresponding domains from different organisms. Sequences were aligned by using ClustalW and shaded on BoxShade server. Amino acids identical or similar in 50% of sequences are shaded on black or on gray background, respectively. Order of the sequences is as appeared in ClustalW output. The RNP1 and RNP2 motifs of RRM are overlined. Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Ci, Cionia intestinalis; Dr, Danio rerio; Gg, Gallus gallus; Tn, Tetraodon nigrovirides; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Ag, Aenopheles gambiae; At, Arabidopsis thaliana; Os, Oryza sativa; Ch, Cryptosporidium hominis; Cp, Cryptosporidium parvum; Pf, Plasmodium falciparum; Pyy, Plasmodium yoelii yoelii; Dd, Dictyostelium discoideum; Eh, Entamoeba histolytica; Pt, Paramecium tetraurelia; Sp, SchizoSaccharomyces pombe; Nc, Neurospora crassa; Mg, Magnaportae grisea; Gz, Gibberella zeae; An, Aspergillus nidulans; Cpo, Coccidioides posadasii; Cim, Coccidioides immitis; Eg, Eremothecium gossypii; Dh, Debaryomyces hansenii; Kw, Kluyveromyces waltii; Sk, Saccharomyces kluyveri; Yl, Yarrowia lipolytica; Cc, Cryptococcus cinereea okayama; Cn, Cryptococcus neoformans; Pc, Phanerochaete chrysosporium; Um, Ustilago maydis; Pa, Pichia angusta.
TABLE 1.
Comparison of AtCyp59 with homologous proteins from different organisms
| Organism | Protein size (aa) | RS/RD or RE dipeptides | % S/Ia |
| Arabidopsis thaliana | 506 | 6/21 | |
| Oryza sativa | 564 | 7/18 | 86/94 |
| Homo sapiens | 492 | 6/7 | 80/89 |
| Mus musculus | 460 | 3/4 | 80/89 |
| Rattus norvegicus | 510 | 7/7 | 80/89 |
| Gallus gallus | 452 | 3/7 | 80/88 |
| Danio rerio | 454 | 9/15 | 79/88 |
| Tetraodon nigrovirides | 409 | 9/6 | 80/91 |
| Cionia intestinalis | 522 | 5/7 | 77/87 |
| Drosophila melanogaster | 653 | 17/16 | 75/85 |
| Aenopheles gambiae | 652 | 38/17 | 79/86 |
| Caenorhabditis elegans | 427 | 6/12 | 74/87 |
| Cryptosporidium hominis | 342 | 3/0 | 79/83 |
| Cryptosporidium parvum | 342 | 3/0 | 79/83 |
| Plasmodium falciparum | 958 | 70/80 | |
| Plasmodiem yoelii yoelii | 899 | 67/83 | |
| Dictiostelium discoideum | 489 | 2/29 | 64/81 |
| Paramecium tetraurelia | 695 | 1/25 | 64/83 |
| Entamoeba histolytica | 330 | 0/0 | 62/79 |
| Aspergilus nidulans | 461 | 8/12 | 76/87 |
| Coccidoides immitis | 443 | 10/2 | 74/82 |
| Coccidoides posadasii | 443 | 9/3 | 74/82 |
| Giberella zeae | 605 | 4/15 | 77/85 |
| Magnaporthe grisea | 505 | 6/19 | 77/86 |
| Neurospora crassa | 803 | 9/25 | 80/89 |
| Yarrowia lipolytica | 469 | 24/34 | 63/74 |
| Debaryomyces hansenii | 331 | 0/0 | 53/71 |
| Eremothecium gosypii | 310 | 0/0 | 62/76 |
| Kluyveromyces waltii | 330 | 0/0 | 63/75 |
| Pichia angusta | 301 | 0/0 | 60/77 |
| SchizoSaccharomyces pombe | 432 | 6/9 | 77/86 |
| Coprinopsis cinerea okayama | 438 | 10/11 | 74/82 |
| Cryptococcus neoformans | 501 | 7/33 | 71/87 |
| Phanerochaete chrysosporium | 446 | 5/17 | 74/83 |
| Ustilago maydis | 551 | 10/22 | 69/83 |
aPercentage identity/similarity of AtCyp59 RRM with corresponding sequences from other organisms as depicted in Figure 2 ▶.
Interactions of AtCyp59 with SR proteins in vitro
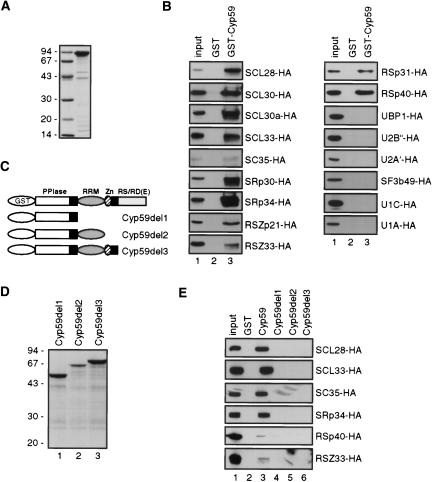
AtCyp59 was identified in a yeast two-hybrid screen as an interacting partner of the Arabidopsis SR protein SCL33/SR33 (data not shown). To examine whether it binds SCL33/SR33 in vitro and whether other Arabidopsis SR proteins are also able to interact with AtCyp59, pull-down experiments were performed. AtCyp59 was overexpressed in Escherichia coli as GST fusion and purified (Fig. 3A ▶). Protein extracts from Arabidopsis protoplasts expressing Arabidopsis HA-tagged SR proteins were incubated with glutathione Sepharose beads coated with GST-AtCyp59. After washing, bound proteins were analyzed by SDS-PAGE and Western blotting with anti-HA antibody. As AtCyp59 and SR proteins both contain RRMs, protein extracts were treated with RNase A in order to avoid indirect RNA-mediated interactions. Figure 3B ▶ (lanes 3) demonstrates that all tested Arabidopsis SR proteins, including SCL33/SR33, were able to interact with AtCyp59 in vitro. This, together with the fact that none of the SR proteins analyzed interacted with the beads coated with GST (Fig. 3B ▶, lanes 2) or with the beads only (data not shown), suggests that AtCyp59 could potentially interact with many SR proteins in vivo. Finally, to exclude the possibility that AtCyp59 is precipitating large aggregates of splicing factors, we performed pull-down experiments with several HA-tagged splicing factors (U1 and U2 snRNP-specific proteins) as well as with UBP1, an hnRNP-like protein from tobacco (Lambermon et al. 2000). As shown in Figure 3B ▶, none of these proteins was precipitated with GST-AtCyp59, further indicating that AtCyp59 interacts specifically with SR proteins.
FIGURE 3.
Interaction of AtCyp59 with SR proteins. (A) Coomassie blue–stained gel of purified recombinant GST-AtCyp59. Molecular mass standards in kilodaltons are indicated on the left. (B) AtCyp59 interacts with SR proteins in vitro. Whole cell extracts from protoplasts transiently expressing HA-tagged SR proteins were incubated with glutathione Sepharose beads coated with GST-AtCyp59. After washing, proteins retained on the beads were analyzed by SDS-PAGE and Western blotting with rat anti-HA monoclonal antibody. (Lanes 1) 1/10 of the input extract used for pull-downs in lanes 2 and 3; (lanes 2) pull-downs with beads coated with GST alone; (lanes 3) pull-downs with beads coated GST-AtCyp59. (C) Schematic representation of AtCyp59 deletion mutants fused to GST. (D) Coomassie blue–stained gel of purified recombinant GST-AtCyp59 deletion mutants. Molecular mass standards in kilodaltons are indicated on the left. (E) Interaction of AtCyp59 deletion mutants with SR proteins. Pull-down experiment performed with GST-AtCyp59 deletion mutants and one member of each Arabidopsis SR protein subfamilies. (Lane 1) 1/10 of the input extract used for pull-downs in lanes 2–6; (lane 2) pull-downs with beads alone; (lane 3) pull-downs with beads coated GST-AtCyp59; (lane 4) pull-downs with beads coated GST-AtCyp59D1; (lane 5) pull-downs with beads coated GST-AtCyp59D2; (lane 6) pull-downs with beads coated GST-AtCyp59D3.
As the major function of RS domains of SR proteins is in protein–protein interactions, and the length of RS domain and the number of RS repeats seems to be proportional to their splicing activation ability (Graveley 2000; Sanford et al. 2003), we asked which domain of AtCyp59 is necessary for the interactions with SR proteins. Three deletion mutants of AtCyp59 fused to GST (Fig. 3C ▶) were overexpressed in E. coli, purified (Fig. 3D ▶), and used for pull-down experiments with selected members of each Arabidopsis SR protein subfamily (Lorković et al. 2004a) expressed in Arabidopsis protoplasts. The PPIase domain alone (del1) (Fig. 3E ▶, lane 4) and in combination with either RRM (del2) (Fig. 3E ▶, lane 5) or RRM and zinc knuckle (del3) (Fig. 3E ▶, lane 6) were not able to interact with SR proteins in vitro (Fig. 3E ▶). This demonstrates that the C-terminal RS/RD(E) domain of AtCyp59 is necessary for efficient interaction with all tested SR proteins. Also, these results further indicate that interactions between SR proteins and AtCyp59 are not RNA-mediated.
AtCyp59 binds RNA in vitro
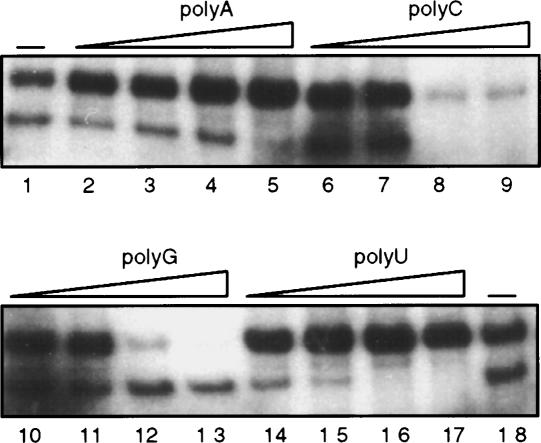
As AtCyp59 contains an RRM and a Zn knuckle, we asked whether it binds RNA in vitro. RNA binding properties of AtCyp59 were determined by UV cross-linking of a GST fusion protein (Fig. 3A ▶) to a synthetic RNA Syn7 (Goodall and Filipowicz 1989). Figure 4 ▶ (lane 1) demonstrates that AtCyp59 could be efficiently cross-linked to RNA in vitro. To establish the nucleotide specificity of binding, competition of RNA binding with four ribohomopolymers was performed. Poly(G) and poly(C) were able to compete with the RNA, whereas poly(A) and poly(U) did not show any effect on binding of AtCyp59 to RNA even at a 400-fold molar excess over radiolabeled RNA. From these studies we conclude that AtCyp59 is an RNA binding cyclophilin with preferences for GC-rich sequences. RNA binding is most probably mediated by RRM, although it is not excluded that Zn knuckle and RS/RD(E) domains might also contribute to RNA binding.
FIGURE 4.
Determination of RNA binding specificity of AtCyp59. Nucleotide binding specificity was measured by the UV cross-linking/homoribopolymer competition assay using 32P-labeled Syn7 RNA and recombinant GST-AtCyp59 protein. Different indicated polymers were added at 10-fold (lanes 2,6,10,14), 50-fold (lanes 3,7,11,15), 200-fold (lanes 4,8,12,16), and 400-fold (lanes 5,9,13,17) excess over Syn7 RNA (calculated in moles of nucleotides). Minus sign (–) (lanes 1,18), cross-linking without any competitor added.
Cellular localization of AtCyp59
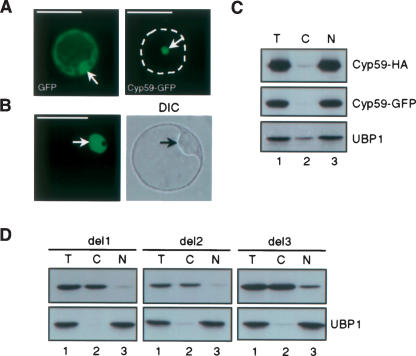
The interaction of AtCyp59 with SR proteins prompted us to investigate whether AtCyp59 is also a nuclear protein. Transient expression of the GFP- and HA-tagged protein in tobacco and Arabidopsis protoplasts was used to address this question. As shown in Figure 5A,B ▶, GFP-tagged AtCyp59 was found in nuclei of both tobacco leaf mesophyll- and Arabidopsis cell suspension-derived protoplasts with no GFP signal detected in the cytoplasm. In contrast, GFP alone was found in the cytoplasm and in the nucleus (Fig. 5A ▶, left panel). The intracellular distribution of AtCyp59 was also studied by cellular fractionation (Lambermon et al. 2000). Lysates prepared from Arabidopsis cell suspension protoplasts transiently expressing GFP- or HA-tagged AtCyp 59 were separated into nuclear and cytoplasmic fractions, and the distribution of AtCyp59-GFP and AtCyp59-HA was determined by Western blotting. As shown in Figure 5C ▶, both GFP- and HA-tagged AtCyp59 were predominantly found in the nuclear fraction. Analysis of the distribution of the nuclear RNA binding protein UBP1 (Lambermon et al. 2000) indicated that the fractionation procedure was quantitative, as UBP1 was found mainly in the nuclear fraction as reported previously (Lambermon et al. 2000; Fig. 5C ▶, bottom panel).
FIGURE 5.
Cellular localization of AtCyp59. (A) Localization of AtCyp59-GFP fusion protein and GFP alone in transiently transformed tobacco protoplasts. Dashed line delineates the shape of protoplasts. Arrows point to nuclei. (B) Localization of AtCyp59-GFP fusion protein in transiently transformed Arabidopsis protoplasts. Single confocal image with corresponding differential interference contrast (DIC) image of whole cell is shown. Arrows point to nuclei. Protoplasts were analyzed 24 h after transformation by using Zeiss Axiovert epifluorescence microscope (A) or Leica TCS confocal microscope (B). Bars, 50 μm (A,B). (C) Cellular localization of AtCyp59 studied by cellular fractionation of Arabidopsis protoplasts transiently expressing AtCyp59-GFP or AtCyp59-HA. Western blots were analyzed with monoclonal antibodies against GFP or HA tags. The distribution of endogenous nuclear UBP1 protein was used to control quality of the fractionation procedure. (Lane 1) Total protein extracts (T), (lane 2) cytoplasmic fraction (C), (lane 3) nuclear fraction (N). (D) Localization of AtCyp59-HA deletions in transiently transformed Arabidopsis protoplasts as determined by cellular fractionation. Details are as in C.
We also determined the sequence requirements for nuclear localization of AtCyp59 by constructing three deletion mutants (see Materials and Methods; Fig. 3C ▶) fused to GFP and HA tags. Cellular fractionation of protoplasts transiently expressing HA-tagged deletion mutants showed that the deletion of the RS/RD(E) domain results in a predominantly cytoplasmic localization of the protein, although some nuclear localization has also been observed (Fig. 5D ▶, del3 panel, lane 3). Further deletions from the C terminus (del2 and del1) resulted in cytoplasmic localization of tagged proteins (Fig. 5D ▶, cf. lanes 2 and 3 in del1 and del2 panels). Again, analysis of the distribution of the nuclear RNA binding protein UBP1 indicated that the fractionation procedure was quantitative as UBP1 was found in the nuclear fraction as reported previously (Fig. 5D ▶, lower panels). Confocal microscopy analyses of transiently transformed Arabidopsis protoplasts expressing GFP-tagged deletions revealed the same results (data not shown). Thus, we conclude that AtCyp59 is a nuclear RNA binding cyclophilin, and that for efficient nuclear localization the C-terminal RS/RD(E) domain is required.
AtCyp59 localizes in the nucleus in a pattern distinct from SR proteins
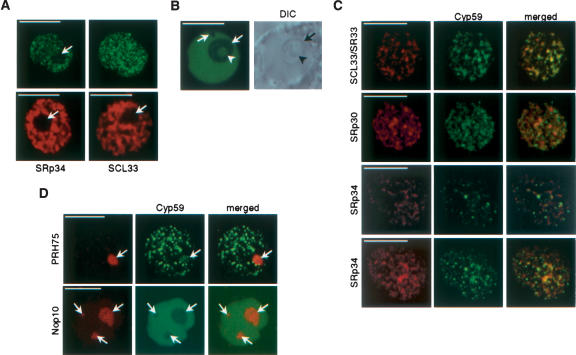
As revealed by laser scanning confocal microscopy, plant SR proteins, like other splicing factors, localize in the nucleus in a specific pattern called speckles (Ali et al. 2003; Docquier et al. 2004; Fang et al. 2004; Lorković and Barta 2004; Lorković et al. 2004a). AtCyp59 was found to interact with SR proteins; therefore we asked whether it is also localized in a speckled pattern. Confocal microscopy analysis of transiently expressed AtCyp59-GFP fusion protein in tobacco protoplasts revealed a punctuated nucleoplasmic pattern (Fig. 6A ▶). Several thousand dots have been observed per nucleus with an average size of 0.2–0.4 μm. In contrast to tobacco nuclei, AtCyp59 showed a diffuse localization in the nucleoplasm of Arabidopsis protoplasts that has been also observed for SR proteins in this cell type (Fig. 6B ▶). In addition, in both cell types we regularly observed low levels of nucleolar localization of AtCyp59 (Fig. 6A,B,D ▶). As these patterns clearly differed from that observed with SR proteins (Ali et al. 2003; Docquier et al. 2004; Fang et al. 2004; Lorković and Barta 2004; Lorković et al. 2004a) (Fig. 6A ▶, cf. upper and lower panels), we performed cotransformation of tobacco protoplasts with plasmids expressing AtCyp59-GFP and SR proteins fused to RFP. Confocal microscopy analysis of cotransformed protoplasts showed that AtCyp59 does not significantly colocalize with SR proteins in nuclear speckles. Partial colocalization has been observed with SCL33, however, not to the extent observed in cells coexpressing two different SR proteins (Z.J. Lorković , J. Hilscher, and A. Barta, unpubl.). In cells coexpressing SRp30-RFP or SRp34-RFP and AtCyp59-GFP only low levels of colocalization were observed. Rather, AtCyp59 dots have been found to localize on the periphery of speckles in which SR proteins are found (Fig. 6C ▶). Additional colocalization studies with fluorescent protein fusion markers for nucleoli or Cajal bodies (Lorković et al. 2004a) revealed that AtCyp59 does not localize into Cajal bodies, but it does, at least in some cells, localize to nucleoli (Fig. 6D ▶; data not shown). This is consistent with the observation that in some cells expressing AtCyp59-GFP alone we regularly observed nucleolar localization.
FIGURE 6.
Subnuclear localization of AtCyp59. (A) Cofocal images of nuclei from tobacco cells expressing AtCyp59-GFP (single confocal section, left; maximum intensity projection of all sections of the same nucleus, right) and two SR proteins, SRp34 and SCL33 (two lower panels; shown are only single confocal sections). Note that AtCyp59-GFP localizes into punctuate pattern, which is different from speckled pattern of SR proteins. Arrows point to nucleoli. Bars, 8 μm. (B) Single confocal section of Arabidopsis nucleus expressing AtCyp59-GFP (left) with corresponding DIC image (right). Arrows point to nucleoli and arrowhead to nucleolar cavity. Bar, 8 μm. (C) Colocalization studies of AtCyp59 with Arabidopsis SR proteins. Tobacco protoplasts were transiently cotransformed with plasmids expressing AtCyp59-GFP and indicated SR proteins fused to RFP. Maximum intensity projections of cotransformed nuclei are shown. For SRp34–AtCyp59 combination also a single confocal section is shown (second row from bottom). Merged images show superimposition of GFP and RFP signals. Bars, 7 μm. (D) Colocalization studies of AtCyp59 with markers for nucleoli [PRH75-RFP (tobacco protoplasts) and Nop10-RFP (Arabidopsis protoplasts)]. Only single confocal sections are shown. Arrows point to nucleoli. Bars, 5 μm.
AtCyp59 interacts with the CTD of RNA polymerase II
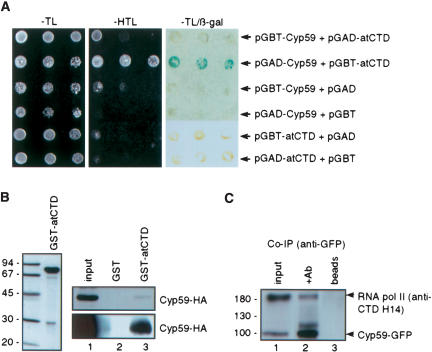
The punctuated nuclear localization pattern, where AtCyp59 localizes next to splicing speckles, resembles transcription sites. This observation raised the interesting possibility that AtCyp59 might interact with the transcriptional machinery as well. It is known that the CTD domain of the largest subunit of RNA polymerase II serves as an interacting platform for many proteins involved in RNA processing, thereby providing a link between transcription and RNA processing (Hirose and Manley 2000; Bentley 2002; Neugebauer 2002; Proudfoot et al. 2002; Kornblihtt et al. 2004; Zorio and Bentley 2004). This, together with our finding that AtCyp59 interacts with SR proteins in vitro, prompted the question of whether AtCyp59 interacts with the CTD of RNA pol II as well. The Arabidopsis CTD and AtCyp59 were cloned into yeast two-hybrid vectors and cotransformed into a yeast reporter strain. Possible interaction of the two proteins was monitored by the activation of His and β-galactosidase reporter genes. As shown in Figure 7A ▶, only cells coexpressing AtCyp59 fused to the activation domain, and CTD fused to the DNA binding domain were able to grow on medium lacking histidine and also showed activation of the second reporter gene, β-gal, indicative for a direct interaction between these two proteins. Control experiments were performed to exclude the possibility that CTD fused to the DNA binding domain activates reporter genes by itself. However, neither CTD fused to the activation domain nor CTD fused to the DNA binding domain alone was able to activate reporter genes, indicating that the interaction of AtCyp59 and CTD is specific.
FIGURE 7.
AtCyp59 interacts with the RNA polymerase II CTD. (A) Interaction between AtCyp59 and CTD in a yeast two-hybrid assay. Indicated combinations of plasmids were cotrans-formed into yeast reporter strain, and the interaction of AtCyp59 with the CTD was assessed by growth on plates lacking histidine (–HTL) or by analysis of the activation of the second reporter gene, β-galactosidase (–HTL/β-gal). (B) In vitro interaction between AtCyp59 and CTD studied by pull-down assay. Arabidopsis CTD was overexpressed as a GST fusion in E. coli and purified (left panel). Pull-downs with AtCyp59-HA expressed in Arabidopsis protoplasts (upper right panel) or in E. coli (lower right panel). (Lanes 1) 1/10 of the input protein extracts used in pull-downs in lanes 2 and 3. (Lanes 2) pull-down with GST alone. (Lanes 3) pull-down with GST-AtCyp59. (C) In vivo interaction of AtCyp59 and CTD determined by coimmunoprecipitation. Protein extract from protoplasts transiently expressing AtCyp59-GFP were immunoprecipitated with anti-GFP antibody. Blot was probed with anti-GFP and anti-CTD (H14) antibodies simultaneously. (Lane 1) 1/ 10 of the protein extract used in IPs in lanes 2 and 3; (lane 2) immunoprecipitation with anti-GFP antibody; (lane 3) protein extract incubated with Protein A beads only.
Interaction between the two proteins was also studied by in vitro pull-down assays. To do so, the Arabidopsis CTD was overexpressed in E. coli as GST fusion and purified on glutathione Sepharose beads (Fig. 7B ▶, left panel). Beads bound with GST-AtCTD were incubated with protein extracts prepared from either tobacco protoplasts transiently expressing AtCyp59-HA (Fig. 7B ▶, upper right panel, lane 3) or E. coli cells expressing AtCyp59-HA (Fig. 7B ▶, lower right panel, lane 3). As evident from Figure 7B ▶ (two right panels), AtCyp59-HA was able to bind recombinant CTD in vitro (lanes 3), thereby confirming interaction between these two proteins observed in the yeast two-hybrid assay.
We also investigated whether AtCyp59 and RNA pol II interact in vivo in plant cells. Arabidopsis protoplasts were transiently transformed with the plasmid expressing GFP-tagged AtCyp59, and the protein extracts were subjected to immunoprecipitation with the anti-GFP antibody. In order to avoid indirect RNA-mediated interaction, protein extracts were treated with RNase A (10 μg/100 μL). Proteins retained on the beads were simultaneously analyzed by Western blotting with antibodies against GFP and CTD. Clearly, AtCyp59-GFP coprecipitated significant amounts of RNA pol II (Fig. 7C ▶, lane 2). As neither AtCyp59-GFP nor CTD were found to bind to protein A Sepharose beads alone (Fig. 7C ▶, lane 3), we conclude that these interactions reflect the situation in vivo. Taken together with the yeast two-hybrid assay and the pull-down interactions described above, these results provide evidence for an interaction between AtCyp59 and RNA pol II (CTD).
Ectopic expression of AtCyp59 in Arabidopsis cell suspension results in decreased phosphorylation of the CTD
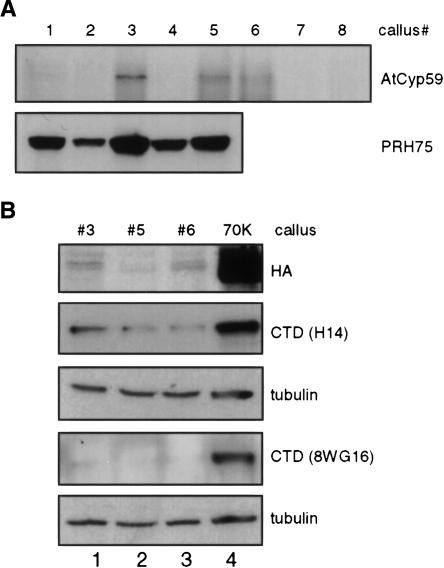
As we had shown that AtCyp59 interacts with SR proteins and the CTD of the RNA pol II we wanted to know whether it is a component of a larger protein complex. Therefore, we established transgenic Arabidopsis cell lines expressing TAP-tagged (Puig et al. 2001) AtCyp59 from the CaMV 35S promoter. This system should produce high levels of the protein of interest, which should allow purification of the putative protein complex and analysis of proteins by mass spectrometry (Z.J. Lorković and A. Barta, unpubl.). About 20 independent, kanamycin-resistant, transgenic cell lines were obtained, but only three of them expressed low levels of tagged AtCyp59 (Fig. 8A ▶; data not shown). This is in contrast to two other plant proteins, like PRH75 (Lorković et al. 2004a) and U1-snRNP-specific protein U1–70K, for which all tested cell lines expressed high amounts of the tagged protein (Fig. 8A ▶, lower panel; 8B, top panel, lane 4). In addition, we observed that all transgenic cell lines expressing AtCyp59 were growing very slowly.
FIGURE 8.
Stable expression of AtCyp59 in Arabidopsis cells results in cell growth arrest and decrease in CTD phosphorylation. (A) Western blot analysis of transgenic, kanamycin resistant Arabidopsis calli expressing TAP tagged AtCyp59 (upper panel). For comparison transgenic calli expressing TAP-tagged PRH75 (lower panel) were analyzed in parallel. Both proteins were detected with anti-HA antibody. (B) Analysis of the phosphorylation status of the RNA pol II CTD in Arabidopsis calli expressing TAP-tagged AtCyp59. (Top panel) Analysis of the expression of AtCyp59 (lanes 1–3). For comparison, protein extract from callus expressing Arabidopsis U1 snRNP-specific protein 70K was loaded as a control (lane 4). The same protein extracts were analyzed with antibodies against the CTD (H14, second panel, and 8WG16, fourth panel). These two membranes were subsequently analyzed with antibodies against α-tubulin (panels below H14 and 8WG16 panels).
The CTD of RNA pol II undergoes a complex pattern of phosphorylation/dephosphorylation steps during the transcription cycle (see Discussion). As cell lines expressing TAP-tagged AtCyp59 showed a growth defect and AtCyp59 interacts with the CTD, it was of interest to analyze the CTD phosphorylation status in these cell lines. Tagged proteins were detected with anti-HA antibody (Fig. 8B ▶, top panel), and RNA pol II was detected using antibodies specific to CTD phosphorylated at serine 5 (H14) ( Fig. 8B ▶, second panel) or CTD phsophorylated at serines 2 and 5 (8WG16) (Fig. 8B ▶, fourth panel). Both 8WG16 and H14 antibodies stained the RNA pol II much more weakly in cell lines expressing AtCyp59 (Fig. 8B ▶, lanes 1–3) compared to control cells expressing TAP-tagged U1–70K (Fig. 8B ▶, top panel, lane 4), indicating a reduced phosphorylation of the CTD in case of even a little overexpression of AtCyp59. A similar result was obtained with antibodies against phosphorylated serine 2 (data not shown). Analysis of the same membranes with the antibodies against α-tubulin revealed that equal amounts of proteins were loaded in each lane. These results indicate that AtCyp59 level is tightly regulated, as already small additional amounts of AtCyp59 are detrimental for cell growth, and that AtCyp59 somehow is influencing regulation of CTD phosphorylation.
DISCUSSION
AtCyp59 belongs to a family of proteins having combined a peptidyl-prolyl cis–trans isomerase domain and an RRM domain in one protein. This class of proteins was initially identified in Paramecium tetraurelia as a factor required for cell morphogenesis (Krzywicka et al. 2001). Both Arabidopsis AtCyp59 and Paramecium Kin241p are nucleus-localized proteins implicated in nuclear RNA metabolism. Consistent with this is our observation that AtCyp59 binds RNA in vitro and interacts with SR proteins, an important family of splicing regulators (Graveley 2000; Sanford et al. 2003). However, AtCyp59 does not colocalize significantly with SR proteins that localize in nuclear speckles in both plant and animal cells (Lamond and Spector 2003; Lorković and Barta 2004). Rather, in tobacco protoplasts AtCyp59 localizes into many, more regular dots throughout the nucleoplasm that are located at the periphery of speckles. We do not know why in Arabidopsis protoplasts AtCyp59 showed diffuse localization; however, it is possible that this is an intrinsic property of this cell type. Similarly, we also observed that SR proteins in these cells do not show a strong speckled pattern, and many more proteins were found in a diffused nucleoplasmic pool compared to tobacco protoplasts, possibly due to a higher transcriptional activity in Arabidopsis protoplasts (Lorković et al. 2004a). As the localization pattern observed in tobacco protoplasts resembled that of transcription sites we asked whether AtCyp59 interacts with the transcriptional machinery, in particular with the CTD of the RNA pol II largest subunit. It has been well documented that the CTD interacts with many proteins involved in pre-mRNA modification and processing, among them also SR proteins (Hirose and Manley 2000; Bentley 2002; Neugebauer 2002; Proudfoot et al. 2002; Kornblihtt et al. 2004; Zorio and Bentley 2004). Indeed, by using yeast two-hybrid, in vitro pull-down, and immunoprecipitation assays we demonstrate here a physical interaction between AtCyp59 and CTD. This, together with our observation that AtCyp59 interacts with SR proteins, allows us to hypothesize a role for AtCyp59 in connecting transcription and RNA processing. It is now well established that pre-mRNA processing takes place cotranscriptionally. By this model pre-mRNA modification and processing factors are recruited to nascent transcripts from speckles, which are considered a reservoir of pre-mRNA processing factors (Lamond and Spector 2003). The major player in this recruitment model is the CTD of RNA pol II, which functions as a loading pad for many processing proteins (Hirose and Manley 2000; Bentley 2002; Neugebauer 2002; Proudfoot et al. 2002; Kornblihtt et al. 2004; Zorio and Bentley 2004). Despite the cotranscriptionality of pre-mRNA processing, splicing factors do not accumulate at transcription sites (Lamond and Spector 2003), which might explain our observation that AtCyp59 and SR proteins do not colocalize significantly.
In addition to the PPIase domain, AtCyp59 contains an RRM and, consistent with this, binds RNA in vitro. Interestingly, alignment of Cyp59 RRMs from highly diverged organisms showed significantly higher sequence conservation when compared to the PPIase domain. This indicates an important role for the RRM in the overall function of AtCyp59. As AtCyp59 interacts with the CTD, it is possible that the CTD delivers it on nascent transcripts. Once bound to pre-mRNA AtCyp59 may prevent formation of unfavorable secondary structures that could interfere with splice site recognition and consequently block cotranscriptional assembly of the spliceosome. In this respect it is worth noting that AtCyp59 possesses an RNA chaperoning activity (as determined by an in vivo assay) and RNA annealing activity in vitro (data not shown). This activity of AtCyp59 may keep pre-mRNA in a more accessible conformation for binding of SR proteins and other factors involved in splice site recognition. Although hypothetical, this would be in agreement with the fact that no AtCyp59 homolog exists in budding yeast, which also does not encode SR proteins, but it does exist in fission yeast, which encodes at least two distinct SR proteins. In general, the splicing machinery of fission yeast seems to be more similar to that of mammals and plants than to budding yeast (Käufer and Potashkin 2000). Another possibility is that Cyp59 binds a noncoding regulatory RNA, like, for example, RNA pol II binding B2 RNA in humans, which is involved in regulation of transcription (Espinoza et al. 2004). The fact that AtCyp59 interacts with the CTD makes this possibility equally attractive. These two models may not be mutually exclusive, and both could be incorporated into a cycle of RNA polymerase II activity on intron-containing genes. Strong conservation of Cyp59 RRMs among different organisms would argue that the RNA bound by AtCyp59 is also conserved. However, additional experimentation is required to determine the exact RNA binding specificity of AtCyp59 and the nature of bound RNA(s).
An additional important question concerning Cyp59 is the substrate specificity of the PPIase domain, which catalyzes cis–trans isomerization of peptide bonds proceeding prolines. As AtCyp59 was shown to interact with SR proteins and with the CTD, both of them could be substrates for its PPIase activity. SR proteins have a C-terminal RS dipeptide-rich domain consisting of many RS repeats, with a major role in protein–protein interaction. An important feature of RS domains is their phosphorylation on serine residues, whereby the phosphorylation status of the RS domain influences protein–protein interactions and possibly the recruitment of SR proteins from speckles to actively transcribing genes (Misteli 1999; Graveley 2000; Lamond and Spector 2003; Sanford et al. 2003). In addition, RS domains often have prolines interrupting the RS repeats, and this is particularly widespread in plant SR proteins. Due to their repetitive sequence, RS domains are not structured and the PPIase activity of AtCyp59 may be necessary for putting them into a correct conformation for protein–protein interactions or, alternatively, for phosphorylation/dephosphorylation. This has already been proposed for two other RS domain-containing cyclophilins, CypRS64 and CypRS92 (Lorković et al. 2004b).
In all organisms the CTD consists of variable numbers of YSPTSPS heptapeptide repeats (Palancade and Bensaude 2003). Due to its high proline content (two prolines per heptapeptide repeat) it is, like an RS domain, largely unstructured, although upon binding of a particular protein it may adopt a specific secondary structure (Meinhart et al. 2005). Thus, the CTD might be a very good candidate substrate for AtCyp59. In addition, during ongoing transcription CTD undergoes a series of phosphorylation/dephosphorylation cycles on serines 2 and 5. In order to find out whether AtCyp59 is a component of a larger protein complex, we attempted to establish an Arabidopsis cell suspension line expressing TAP-tagged AtCyp59. However, despite the fact that we obtained about 20 transgenic lines that were kanamycin-resistant, in only three of them very low amounts of the protein could be detected. This is in contrast to the other two proteins for which TAP-tagging cell lines were obtained, as all of them expressed tagged protein in high amounts (Fig. 8 ▶). Analysis of the CTD phosphorylation status revealed that cells expressing very low amounts of AtCyp59 have a considerably reduced phosphorylation of the CTD. It seems that the level of AtCyp59 in the cell is under tight dosage control. This is further supported by our observation that the SchizoSaccharomyces pombe homolog of AtCyp59 is an essential gene and that even heterozygous cells show slow growth and cell size defects, presumably due to deregulation of CTD phosphorylation (M. Gullerova, A. Barta, and Z.J. Lorković , unpubl.). Another nuclear PPIase that modulates structure and function of the CTD is the multifunctional parvuline Pin1 (Xu et al. 2003). This, together with our data presented here, pinpoints the general importance of PPIases in the transcriptional cycle of RNA polymerase II. Several CTD kinases and two distinct CTD phosphatases have been shown to be involved in this process (Palancade and Bensaude 2003). While nothing is known about cis–trans specificities of CTD kinases and phosphatases for prolines adjacent to the phosphorylation site, one can speculate that some of them are specific for either cis or trans conformation. Alternatively, CTD kinases and phosphatatses could also be targets for AtCyp59, whereby AtCyp59 would introduce conformational changes that in turn could be a switch between their active and inactive forms. Several studies have indicated that regulation of phosphorylation/dephosphorylation of target proteins could be a key regulatory mechanism of PPIases (Vener et al. 1999; Xu et al. 2003; Calvo and Manley 2005; for reviews, see Lu et al. 2002; Shaw 2002; Lu 2004; Lim and Lu 2005; see also references therein). Whether this is the case for AtCyp59 remains to be determined.
Interaction of AtCyp59 with the CTD and SR proteins also suggests a direct or an indirect involvement in pre-mRNA splicing. Intron excision is mediated by a large ribonucleoprotein particle called spliceosome, which assembles stepwise anew at each intron. Spliceosome assembly requires many structural rearrangements that include changes in RNA–RNA base-pairings between snRNAs, snRNAs and pre-mRNA, as well as many changes in protein–protein interactions (Burge et al. 1999). Proteins responsible for disruption of RNA–RNA helices have been identified and they belong to a large family of RNA helicases or DEAD/DEAH-box proteins (Hamm and Lamond 1998; Staley and Guthrie 1998). However, it is not clear how specific protein–protein interactions are regulated. One possibility is that phosphorylation/dephosphorylation of splicing factors mediated by several kinases and phosphatases that specifically associate with the spliceosome regulates protein–protein interactions (Misteli 1999; Lorković et al. 2004b). Another possibility is that proteins with chaperone-like activity, for example, PPIases, are involved in the regulation of protein–protein interactions in assembling spliceosome. Interestingly, several PPIases, including the previously characterized USA-Cyp, have been found in proteomic characterization of human spliceosome (Rappsilber et al. 2002; Zhou et al. 2002). Although the properties of PPIases are highly suggestive of an active role in spliceosomal dynamics, functional support for this idea is still missing.
MATERIALS AND METHODS
Cloning of cDNAs encoding Arabidopsis cyclophilin AtCyp59
The cDNA clone encoding Arabidopsis AtCyp59 protein was amplified by RT-PCR from total RNA isolated from mixed flower and leaf tissue. Oligonucleotides ATGTCAGTTCTTATTGTGACG AGC and TATCTCTAGACTCTTCACGGCGT were derived from the genomic sequence (At1g53720) and from the partial cDNA sequence obtained in a two-hybrid screen with Arabidopsis SR protein SCL33/SR33. The PCR product was cloned into pGEM-T easy (Promega) and sequenced (see Results).
Plasmid constructions
Plasmid for overexpression of the AtCyp59 protein in bacteria was obtained by cloning the corresponding PCR product into pGEX-4T-1 (Amersham Pharmacia Biotech). cDNA was amplified by using the oligonucleotides TATTGCGGATCCATGTCAGTTCTTATTGTGAC G and GTCAGCGGATCCTCATCTATCCCTTCTCTCATG, which introduce a BamHI (in bold) site in front of the ATG codon and after the stop codon. The PCR product was cut with BamHI and ligated into the corresponding site of pGEX-4T-1, resulting in pGST-AtCyp59. Overexpression plasmids encoding GST-AtCyp59D1, GST-AtCyp59D2, and GST-AtCyp59D3 were created by cloning corresponding PCR EcoRI/Sal fragments into pGEX-4T-1. The 5′ oligonucleotide used for all three deletion mutants was GACTAGGAATTCAT GTCAGTTCTTATTGTGACG, and the 3′ oligonucleotides used were GACTAGGTCGACTCACAACAGCACTTGAATGGGC (del1), GAC TAGGTCGACTCAGGACACACTCTGACTGAAATC (del2), and GA CTAGGTCGACTCAATGACTATTATGCTTGGGGGT (del3).
To generate plant expression plasmids encoding hemagglutinine (HA), green fluorescent protein (GFP), and red fluorescent protein (RFP) tagged AtCyp59 protein, the coding region of AtCyp59 was amplified by using oligonucleotides: The 5′ oligo used was TGCGGGTCGACAATAAACCATGTCAGTTCTTATTGTGACGAG CCT, and the 3′ oligo used was TATTGCGGATCCTCTATCCCTTC TCTCATGTC, which introduces a SalI (bold) site and a plant translation consensus sequence (italics) in front of the ATG codon and a BamHI site in place of the stop codon, respectively. The PCR product was cut with SalI and BamHI and ligated into corresponding sites of pDEDH-HA, pDEDH-GFP, and pDEDH-RFP (Lorković et al. 2004a), resulting in pAtCyp59-HA, pAtCyp59-GFP, and pAt-Cyp59-RFP. Plant expression plasmids encoding AtCyp59 deletion mutants fused to HA and GFP were created as described above by using the above 5′ oligo for all three deletion mutants. Reverse oligos used were GACTAGGGATCCTACAACAGCACTTGAATG (Del1; PPIase domain only), GACTAGGGATCCGGACACACTCTGACT GAA (Del2; PPIase plus RRM), and GACTAGGGATCCATGAC TATTATGCTTGGG (Del3; PPIase, RRM, and zinc knuckle). PCR products were cloned into pDEDH-HA and pDEDH-GFP, resulting in pAtCyp59D1-HA, pAtCyp59D2-HA, pAtCyp59D3-HA, pAt-Cyp59D1-GFP, pAtCyp59D2-GFP, and pAtCyp59D3-GFP.
Plasmids pAD-AtCyp59 and pBD-AtCyp59 used in the yeast two-hybrid assay were created by cloning the PCR product, amplified by using oligos TATTGCGGATCCTGTCAGTTCTTATTGTGACG and GTCACGCGGATCCTCATCATCCCTTCTCTCAT, into the BamHI site of pGAD242 and pGBT9 (Clontech), respectively.
The yeast two-hybrid plasmids encoding Arabidopsis CTD were created by cloning the CTD fragment amplified from the cDNA library (Clontech) into EcoRI opened pGBT9 and pGAD242 resulting in pBD-AtCTD and pAD-AtCTD, respectively. Oligonucleotides used were 5′–TGCAGAATTCCCAATGTCAGATGCACAG and 3′–ACGTGAATTCAAGGGTTGCCTTTATC. For overexpression of GST-CTD fusion in E. coli, the CTD was amplified by using oligonucleotides 5′–TGCAGAATTCCAATGTCAGATGCACAG and the same 3′ oligo used for cloning into two-hybrid vectors, and ligated into the EcoRI site of pGEX-4T-1, resulting in pGST-AtCTD.
Plant expression plasmids pSRp34-GFP, pSRp34-RFP, pSRp34-HA, pU2B″-mRFP, U2A′-HA, U1A-HA, pNop10-mRFP, and pPRH-75-RFP encoding GFP-, RFP-, and HA-tagged Arabidopsis SRp34, U2B″, U2A′, U1A, and PRH75 proteins have been described (Lorković et al. 2004a). Plant expression plasmids pSCL28-HA, pSCL30-HA, pSCL30a-HA, pSCL33-HA, pSC35-HA, pSRp30-HA, pRSp31-HA, and pRSp40-HA for the expression of HA-tagged SR proteins have been described (Lorković et al. 2004a). Yeast expression plasmids pRSZp21-HA and pRSZ33-HA have been described earlier (Lopato et al. 2002).
Yeast two-hybrid assay
Yeast two-hybrid screening and direct assays have been performed in HF7c reporter strain according to the manufacturer’s instructions (Clontech) and Lopato et al. (2002). To avoid nonspecific activation of HIS3 reporter gene, 3-aminotriazole was used throughout at the 5 mM concentration.
Overexpression and purification of GST fusion proteins
The plasmids pGST-AtCyp59, pGST-AtCTD, GST-AtCyp59D1, GST-AtCyp59D2, and GST-AtCyp59D3 were transformed into the E. coli strain BL21 (DE3) CodonPlusRIL (Stratagene). Cell growth and protein purification was as described in Lorković et al. (2005). For each pull-down 50 μL of beads were used (~1 μg of bound protein). For in vitro RNA binding studies (UV cross-linking) GST-AtCyp59 was eluted with 5 mM glutathione in 100 mM HEPES-KOH (pH 7.9) and concentrated by using Amicon Ultra-4 centrifugal filter devices (Millipore). During concentration of the protein, the buffer was exchanged into 20 mM HEPES-KOH (pH 7.9) containing 100 mM KCl, 0.5 mM dithiothreitol, and 0.2 mM EDTA.
Preparation and transient transformation of tobacco and Arabidopsis protoplasts
Tobacco leaf mesophyll protoplasts were isolated and transformed with 20 μg of plasmid DNA per 106 protoplasts by the polyethylene glycol method and Arabidopsis cell suspension protoplasts were isolated and transformed as described (Lorković et al. 2004a). Transformed protoplasts were collected 24 h after transformation and stored at −80°C or were analyzed by laser scanning confocal microscope (Leica).
Preparation of whole-cell extracts from protoplasts and from yeast cells, immunoprecipitation, and pull-down assays
Protoplasts were collected by centrifugation 24 h after transformation (15 min, 70g), frozen in liquid nitrogen, and resuspended in protoplast extraction buffer (PEB400; 50 mM HEPES-KOH at pH 7.9, 400 mM KCl, 2.5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.1% Triton X-100), supplemented with EDTA-free protease inhibitor cocktail (Roche Applied Science), and further processed as described in Lorković et al. (2004a). After 15 min centrifugation in an Eppendorf centrifuge at maximum speed at 4°C, the supernatant was mixed with PEB without KCl to adjust KCl concentration to 150 mM (PEB150; for GST pull-down assay) or to 250 mM (PEB250; for immunoprecipitation). Immunoprecipitations were performed as described in Lorković et al. (2004a). Preparation of protein extracts from yeast cells expressing RSZp21-HA, RSZ33-HA, and SRp30-HA were as described in Lopato et al. (2002). For pull-down assays, protein extracts from transformed protoplasts or from yeast cells were mixed with glutathione Sepharose beads (Amersham Pharmacia Biotech) coated with recombinant GST-AtCyp59, GST-AtCTD, GST-AtCyp59D1, GST-AtCyp59D2, and GST-AtCyp59D3 proteins or with beads alone and incubated on a rotary shaker for 2 h at 4°C. After four washings with PEB150, the beads were resuspended in 50 μL of loading buffer, and 25 μL were analyzed by SDS-PAGE.
UV cross-linking
In vitro transcription, UV cross-linking, and ribohomopolymer competition assays with Syn7 RNA (Goodall and Filipowicz 1989) and GST-AtCyp59 were performed as described by Domon et al. (1998).
Generation and analysis of Arabidopsis cell suspension line expressing TAP-tagged AtCyp59
In short, expression cassette consisting of CaMV promoter followed by Protein A and HA tags was constructed in pBluescript II KS–(New England Biolabs). AtCyp59 or PRH75 and U1–70K were cloned in frame with the tags and the whole cassette was cut out and cloned into pBI121 (Clontech). Plasmids were transformed into Agrobacterium tumefaciens GV3101 and Arabidopsis cell suspension was transformed by cocultivation. Details about construction of TAP-tagging vectors for expression in plant cells, Arabidopsis cell suspension transformation, and protocol for protein extraction will be published elsewhere.
SDS-PAGE and Western blotting
SDS-PAGE (8% or 12% gels) was done according to standard procedure. Proteins were transferred onto PVDF membrane (Millipore) and Western blotting was performed according to standard procedure. Rat anti-HA (3F10; Roche Applied Science) and mouse anti-GFP (Roche Applied Science) monoclonal antibodies were used at 1:1000 dilution. Mouse anti-CTD (H14, H5, and 8WG16; Covance) monoclonal antibodies were used at 1:500 dilution. Mouse monoclonal antibody against α-tubulin (Sigma) was used at 1:1000 dilution. Secondary antibodies, goat anti-rat (Sigma), goat anti-mouse (Bio-Rad), and goat anti-rabbit (Bio-Rad) IgGs and goat anti-mouse IgM (Sigma), all conjugated with horseradish peroxidase were used at 1:10,000 dilutions. The blots were developed using an enhanced chemiluminescence kit (Amersham Pharmacia Biotech).
Acknowledgments
This work was supported by grants from the Austrian Science Foundation (FWF: SFB-F017C11 and SFB-F017C12) to A.B.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2226106.
REFERENCES
- Ali, G.S., Golovkin, M., and Reddy, A.S. 2003. Nuclear localization and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J. 36: 883–893. [DOI] [PubMed] [Google Scholar]
- Anderson, S.K., Gallinger, S., Roder, J., Frey, J., Young, H.A., and Ortaldo, J.R. 1993. A cyclophilin-related protein involved in the function of natural killer cells. Proc. Natl. Acad. Sci. 90: 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, D. 2002. The mRNA assembly line: Transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14: 336–342. [DOI] [PubMed] [Google Scholar]
- Bourquin, J.P., Stagljar, I., Meier, P., Moosmann, P., Silke, J., Baechi, T., Georgiev, O., and Schaffner, W. 1997. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 25: 2055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosome. In The RNA world (eds. R.F. Gesteland et al.), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Calvo, O. and Manley, J.L. 2005. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 24: 1009–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarec, L., Kamphausen, T., Dubourg, B., Callebaut, I., Lemeunier, F., Metivier, D., Feunteun, J., Fischer, G., and Modjtahedi, N. 2002. Identification and characterization of Mocacyp. A Drosophila melanogaster nuclear cyclophilin. J. Biol. Chem. 277: 41171–41182. [DOI] [PubMed] [Google Scholar]
- Docquier, S., Tillemans, V., Deltour, R., and Motte, P. 2004. Nuclear bodies and compartmentalization of pre-mRNA splicing factors in higher plants. Chromosoma 112: 255–266. [DOI] [PubMed] [Google Scholar]
- Domon, C., Lorković , Z.J., Valcarcel, J., and Filipowicz, W. 1998. Multiple forms of the U2 small nuclear ribonucleoprotein auxiliary factor U2AF subunits expressed in higher plants. J. Biol. Chem. 273: 34603–34610. [DOI] [PubMed] [Google Scholar]
- Espinoza, C.A., Allen, T.A., Hieb, A.R., Kugel, J.F., and Goodrich, J.A. 2004. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 11: 822–829. [DOI] [PubMed] [Google Scholar]
- Fang, Y., Hearn, S., and Spector, D.L. 2004. Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol. Biol. Cell 15: 2664–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulgosi, H., Vener, A.V., Altschmied, L., Herrmann, R.G., and Andersson, B. 1998. A novel multi-functional chloroplast protein: Identification of a 40 kDa immunophilin-like protein located in the thylakoid lumen. EMBO J. 17: 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall, G.J. and Filipowicz, W. 1989. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58: 473–483. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm, J. and Lamond, A.I. 1998. Spliceosome assembly: The unwinding role of DEAD-box proteins. Curr. Biol. 8: R532–R534. [DOI] [PubMed] [Google Scholar]
- Handschumacher, R.E., Harding, M.W., Rice, J., Drugge, R.J., and Speicher, D.W. 1984. Cyclophilin: A specific cytosolic binding protein for cyclosporin A. Science 226: 544–547. [DOI] [PubMed] [Google Scholar]
- Hanes, S.D., Shank, P.R., and Bostian, K.A. 1989. Sequence and mutational analysis of ESS1, a gene essential for growth in Saccharomyces cerevisiae. Yeast 5: 55–72. [DOI] [PubMed] [Google Scholar]
- Harrar, Y., Bellini, C., and Faure, J.-D. 2001. FKBPs: At the crossroads of folding and transduction. Trends Plant Sci. 6: 426–431. [DOI] [PubMed] [Google Scholar]
- Hirose, Y. and Manley, J.L. 2000. RNA polymerase II and the integration of nuclear events. Genes & Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Horowitz, D.S., Kobayashi, R., and Krainer, A.R. 1997. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA 3: 1374–1387. [PMC free article] [PubMed] [Google Scholar]
- Horowitz, D.S., Lee, E.J., Mabon, S.A., and Misteli, T. 2002. A cyclophilin functions in pre-mRNA splicing. EMBO J. 21: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, T. 1998. Prolyl isomerases and nuclear function. Cell 92: 141–143. [DOI] [PubMed] [Google Scholar]
- Käufer, N.F. and Potashkin, J. 2000. Analysis of the splicing machinery in fission yeast: A comparison with budding yeast and mammals. Nucleic Acids Res. 28: 3003–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt, A.R., de la Mata, M., Fededa, J.P., Munoz, M.J., and Nogues, G. 2004. Multiple links between transcription and splicing. RNA 10: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywicka, A., Beisson, J., Keller, A.M., Cohen, J., Jerka-Dziadosz, M., and Klotz, C. 2001. KIN241: A gene involved in cell morphogenesis in Paramecium tetraurelia reveals a novel protein family of cyclophilin-RNA interacting proteins (CRIPs) conserved from fission yeast to man. Mol. Microbiol. 42: 257–267. [DOI] [PubMed] [Google Scholar]
- Lambermon, M.H., Simpson, G.G., Wieczorek Kirk, D.A., Hemmings-Mieszczak, M., Klahre, U., and Filipowicz, W. 2000. UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19: 1638–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I. and Spector, D.L. 2003. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4: 605–612. [DOI] [PubMed] [Google Scholar]
- Lim, J. and Lu, K.P. 2005. Pinning down phosphorylated tau and tauopathies. Biochim. Biophys. Acta 1739: 311–322. [DOI] [PubMed] [Google Scholar]
- Liu, J., Farmer Jr., J.D., Lane, W.S., Friedman, J., Weissman, I., and Schreiber, S.L. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66: 807–815. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Forstner, C., Kalyna, M., Hilscher, J., Langhammer, U., Indrapichate, K., Lorković , Z.J., and Barta, A. 2002. Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J. Biol. Chem. 277: 39989–39998. [DOI] [PubMed] [Google Scholar]
- Lorković , Z.J. and Barta, A. 2004. Compartmentalization of the splicing machinery in plant cell nuclei. Trends Plant Sci. 9: 565–568. [DOI] [PubMed] [Google Scholar]
- Lorković , Z.J., Hilscher, J., and Barta, A. 2004a. Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol. Biol. Cell 15: 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković , Z.J., Lopato, S., Pexa, M., Lehner, R., and Barta, A. 2004b. Interactions of Arabidopsis RS domain containing cyclophilins with SR proteins and U1 and U11 small nuclear ribonucleo-protein- specific proteins suggest their involvement in pre-mRNA splicing. J. Biol. Chem. 279: 33890–33898. [DOI] [PubMed] [Google Scholar]
- Lorković , Z.J., Lehner, R., Forstner, C., and Barta, A. 2005. Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA 11: 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, K.P. 2004. Pinning down cell signaling, cancer and Alzheimer’s disease. Trends Biochem. Sci. 29: 200–209. [DOI] [PubMed] [Google Scholar]
- Lu, K.P., Hanes, S.D., and Hunter, T. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380: 544–547. [DOI] [PubMed] [Google Scholar]
- Lu, K.P., Liou, Y.C., and Zhou, X.Z. 2002. Pinning down prolinedirected phosphorylation signaling. Trends Cell Biol. 12: 164–172. [DOI] [PubMed] [Google Scholar]
- Meinhart, A., Kamenski, T., Hoeppner, S., Baumli, S., and Cramer,P.2005. A structural perspective of CTD function. Genes&Dev. 19: 1401–1415. [DOI] [PubMed] [Google Scholar]
- Mi, H., Kops, O., Zimmermann, E., Jaschke, A., and Tropschug, M. 1996. A nuclear RNA-binding cyclophilin in human T cells. FEBS Lett. 398: 201–205. [DOI] [PubMed] [Google Scholar]
- Misteli, T. 1999. RNA splicing: What has phosphorylation got to do with it? Curr. Biol. 9: R198–R200. [DOI] [PubMed] [Google Scholar]
- Mortillaro, M.J. and Berezney, R. 1998. Matrin CYP, an SR-rich cyclophilin that associates with the nuclear matrix and splicing factors. J. Biol. Chem. 273: 8183–8192. [DOI] [PubMed] [Google Scholar]
- Nestel, F.P., Colwill, K., Harper, S., Pawson, T., and Anderson, S.K. 1996. RS cyclophilins: Identification of an NK-TR1-related cyclophilin. Gene 180: 151–155. [DOI] [PubMed] [Google Scholar]
- Neugebauer, K.M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 115: 3865–3871. [DOI] [PubMed] [Google Scholar]
- Palancade, B. and Bensaude, O. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 270: 3859–3870. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501–512. [DOI] [PubMed] [Google Scholar]
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinfret, A., Collins, C., Menard, R., and Anderson, S.K. 1994. The N-terminal cyclophilin-homologous domain of a 150-kilodalton tumor recognition molecule exhibits both peptidyl-prolyl cis-trans-isomerase and chaperone activities. Biochemistry 33: 1668–1673. [DOI] [PubMed] [Google Scholar]
- Romano, P.G., Horton, P., and Gray, J.E. 2004. The Arabidopsis cyclophilin gene family. Plant Physiol. 134: 1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, J.R., Longman, D., and Caceres, J.F. 2003. Multiple roles of the SR protein family in splicing regulation. Prog. Mol. Subcell. Biol. 31: 33–58. [DOI] [PubMed] [Google Scholar]
- Schiene, C. and Fischer, G. 2000. Enzymes that catalyse the restructuring of proteins. Curr. Opin. Struct. Biol. 10: 40–45. [DOI] [PubMed] [Google Scholar]
- Shaw, P.E. 2002. Peptidyl-prolyl isomerases: A new twist to transcription. EMBO Rep. 3: 521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley, J.P. and Guthrie, C. 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Stewart, D.E., Sarkar, A., and Wampler, J.E. 1990. Occurrence and role of cis peptide bonds in protein structures. J. Mol. Biol. 214: 253–260. [DOI] [PubMed] [Google Scholar]
- Teigelkamp, S., Achsel, T., Mundt, C., Gothel, S.F., Cronshagen, U., Lane, W.S., Marahiel, M., and Luhrmann, R. 1998. The 20kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins. RNA 4: 127–141. [PMC free article] [PubMed] [Google Scholar]
- Vener, A.V., Rokka, A., Fulgosi, H., Andersson, B., and Herrmann, R.G. 1999. A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry 38: 14955–14965. [DOI] [PubMed] [Google Scholar]
- Xu, Y.X., Hirose, Y., Zhou, X.Z., Lu, K.P., and Manley, J.L. 2003. Pin1 modulates the structure and function of human RNA polymerase II. Genes & Dev. 17: 2765–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. [DOI] [PubMed] [Google Scholar]
- Zorio, D.A. and Bentley, D.L. 2004. The link between mRNA processing and transcription: Communication works both ways. Exp. Cell Res. 296: 91–97. [DOI] [PubMed] [Google Scholar]
- Zorio, D.A. and Blumenthal, T. 1999. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5: 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]