Abstract
The aim of the RNA Ontology Consortium (ROC) is to create an integrated conceptual framework—an RNA Ontology (RO)—with a common, dynamic, controlled, and structured vocabulary to describe and characterize RNA sequences, secondary structures, three-dimensional structures, and dynamics pertaining to RNA function. The RO should produce tools for clear communication about RNA structure and function for multiple uses, including the integration of RNA electronic resources into the Semantic Web. These tools should allow the accurate description in computer-interpretable form of the coupling between RNA architecture, function, and evolution. The purposes for creating the RO are, therefore, (1) to integrate sequence and structural databases; (2) to allow different computational tools to interoperate; (3) to create powerful software tools that bring advanced computational methods to the bench scientist; and (4) to facilitate precise searches for all relevant information pertaining to RNA. For example, one initial objective of the ROC is to define, identify, and classify RNA structural motifs described in the literature or appearing in databases and to agree on a computer-interpretable definition for each of these motifs. To achieve these aims, the ROC will foster communication and promote collaboration among RNA scientists by coordinating frequent face-to-face workshops to discuss, debate, and resolve difficult conceptual issues. These meeting opportunities will create new directions at various levels of RNA research. The ROC will work closely with the PDB/NDB structural databases and the Gene, Sequence, and Open Biomedical Ontology Consortia to integrate the RO with existing biological ontologies to extend existing content while maintaining interoperability.
Keywords: biological ontology, RNA, sequence alignments, 3D structure, RNA motifs
INTRODUCTION
Biological sciences are knowledge-intensive disciplines: Prior knowledge is applied to newly discovered or unknown entities to extend understanding. The current challenge is therefore to organize and integrate the deluge of disparate data to gain access to new biological knowledge. Until recently, bioinformatics efforts have focused on organizing fragments of biological information into large electronic databases accessible through the World Wide Web. Large amounts of data are available to the end user, who is understood to be another scientific specialist. In such endeavors, one analyzes macromolecules and their complexes, identifies and classifies their components (base pairs, elements of secondary structure, motifs), dissects how component parts interact with each other, and identifies their interactions with ions, solvent, small molecules, or other macromolecules. Web servers are used to mediate between databases or to access information contained in databases. For example, the Protein Data Bank (Berman et al. 2000) and the Nucleic Acid Database (Berman et al. 1992) provide 3D macromolecular structure data, sequences, and functional data. Structural entities are also classified in terms of their components (base pairs, base-pair stacking, secondary structure, and motifs). In parallel, large sequence databases provide access to individual genes, gene clusters, or complete genomes (NCBI, TIGR, EMBL) or collections of aligned homologous sequences such as PFAM (Bateman et al. 2004) and RFAM (Griffiths-Jones et al. 2005).
Nowadays, biological investigations span the huge distance between two extremes: from understanding biological catalysts and molecular machines with atomic precision to complete genomes and the study of complex networks, culminating in the study of whole organisms, in health and disease, in the context of the ecosystems they form. Clearly, the integration of data produced by diverse approaches requires precise and coherent descriptions at the appropriate granularity (Kumar et al. 2005). In recent years, ontologies have emerged as the key mechanism for encoding structured knowledge (Stevens et al. 2000). Applied in the context of bioinformatics and structural databases, they open the possibility of more automated and integrated use of biological data.
WHAT IS AN ONTOLOGY?
Ontology is defined as the science of what exists, of the kinds and structures of objects, events, and processes, their properties and relations in all domains of reality (Smith 2004b). For information scientists, ontologies comprise shared, common taxonomies of relevant entities and the relationships between them, within an application domain. Ontologies provide a “representation of a shared conceptualization of a particular domain” by defining a common, controlled, and structured vocabulary to enable people and computer applications to share information (Gruber 1993). Thus, ontologies include human-understandable and machine-interpretable definitions of entities in the knowledge domain, their properties, and the relations that exist among them, as well as constraints on those relations, formalized axiomatically (Smith et al. 2005). Biological ontologies are intended therefore to serve as computable representations of the underlying biological reality that will enable computers to reason over data in some of the ways humans do, but potentially more systematically and drawing on much larger amounts of data than individual humans can possibly grasp (Stevens et al. 2000). In order to achieve these ambitious goals, ontologies should ideally represent, as accurately as possible, the underlying reality that is being modeled and should be constructed with logical rigor (Smith 2004a).
The evolution of data representation in structural biology provides a useful example of the collaborative development of ontologies. The macromolecular Crystallographic Information Framework (Fitzgerald et al. 2005), originally developed to describe experimental crystallography, has subsequently been extended to represent structures and methodology obtained by NMR, cryo-electron microscopy, and computational modeling. The ontology has been further extended to provide a description of the experimental protein production steps preceding the structure determination. Collectively these terms and relationships are used by the Protein Data Bank to manage and disseminate data (Westbrook et al. 2005). RNAML (Waugh et al. 2002) is derived from this ontology and provides detailed descriptions of RNA structural features that can be used as input in computer programs that search sequences (Gautheret et al. 1990) or that model 3D structures (Major et al. 1991). Similar collaborative activities have developed in other areas of biology such as the Gene Ontology (Lewis 2005) and BioPAX (Luciano 2005).
These ontologies, produced by different groups and pertaining to different aspects of biological reality and knowledge, need to work together in a coherent manner—they need to “interoperate” to define the semantic links among the available bioinformatics data that will make it possible to create the new generation of network called the “Semantic Web” (Hendler 2003; Neumann 2005). The Semantic Web Initiative (http://www.w3.org/2001/sw) defines technologies and methodologies that map directly onto many of the challenges in the life sciences, including collecting and representing complex forms of information in an intelligent, flexible form that is interpretable by software as well as viewable by humans.
Another challenge is the ability to make critical decisions based on an aggregation of information that may share common entities, such as molecules, diseases, and intellectual property. Ontologies should provide the “lingua franca” necessary to allow scientists to attach meaning to their data (“semantic markup”) and should aid in the integration of experimental results and the attached conceptual interpretations within a coherent semantic framework. More than simple data integration, such ontologies produce knowledge aggregation and diffusion and ultimately lead to further discovery (Neumann 2005). Quite simply, these developments will make it possible for everyone to search by meaning (semantics) rather than simply matching strings of characters. A final point is that ontologies are built to be shared: Ontology development is inherently a collaborative activity. Therefore, this article aims to describe the challenges of representing, integrating, and merging RNA structural and biological data at various levels of complexity and to attract fellow RNA scientists, bioinformaticians, and ontologists to join us in this endeavor.
WHAT ARE ONTOLOGIES GOOD FOR?
Knowledge engineers and information scientists have emphasized the following major purposes for which ontologies are intended:
express and share community knowledge,
express the meaning of information in databases,
support intelligent querying over multiple databases,
enable reuse of domain knowledge, and
support automated reasoning and inference over domain knowledge.
The first and perhaps most important purpose for developing ontologies is to express and share common understanding of the structure of information among people and software agents (Musen 1992; Gruber 1993). For example, suppose several different Web sites contain information about an RNA molecule or family of RNA molecules, including the structure of a part of the molecule as determined in solution by NMR; a crystal structure showing the interaction of the molecule with other molecules; microarray data regarding its expression at different time points of the cell cycle or in different tissues; and an alignment of related sequences or 2D structure in graphical form. If these Web sites share the same electronically available, underlying ontology of the terms they use to semantically annotate their data, then computer agents can automatically identify, extract, analyze, and aggregate information from these different sites. Moreover, the agents can use this aggregated information to answer high-level user queries or to provide input data to other applications to integrate heterogeneous data sources.
Enabling reuse of domain knowledge is another driving force behind the recent surge in ontology research. Ontologies model entities in a domain of reality, their properties, and the relations between them. Thus, a general ontology of biologically relevant relations (i.e., the OBO Relations Ontology), once formulated, can be reused when formulating more specific biological ontologies (Smith et al. 2005). To create an RNA Ontology that integrates 1D, 2D, and 3D structural information about RNA, the ROC will draw on the OBO Relations Ontology; the entities and properties defined in the Sequence Ontology (Eilbeck et al. 2005), which models the primary (1D) structure of DNA and RNA; and the structural ontology used by PDB/NDB (Westbrook et al. 2005).
PROPOSED PLAN FOR DEVELOPING THE RNA ONTOLOGY
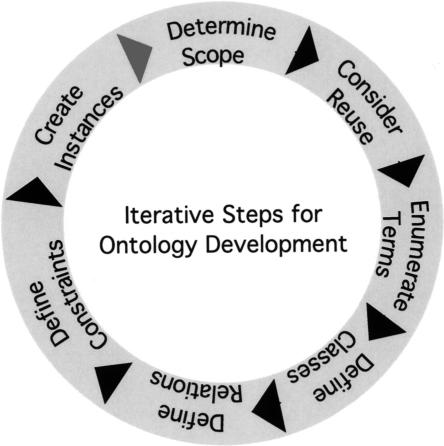
The steps for developing ontologies are shown in Figure 1 ▶ as a cycle, to emphasize that the process is an ongoing and iterative one. The first step is defining the scope of the ontology. As new ontologies should be orthogonal to existing ones, one begins by identifying related ontologies with partially overlapping knowledge domains. One strives to seamlessly integrate the new ontology with the existing related ones, so as to avoid duplication, conflict, and confusion.
FIGURE 1.
Ontology development (Noy and McGuiness 2001): The process of developing and deploying biological ontologies is a community effort that requires frequent iteration of the basic steps represented here as a cycle. The process begins by determining the scope of the ontology and proceeds next to considering how the ontology will be used. This is followed by enumeration of terms and definition of classes and relations, and the constraints that operate on these. Finally, the classes are populated with instances to create knowledge bases.
At the first meeting of the ROC in May 2005, three issues relating to defining the scope of the RNA Ontology were considered:
What are the domains that the RO should cover?
Who are the potential users of the RO?
What types of questions should the RO be intended to answer?
The following topics were therefore proposed for the domains of the RO:
RNA sequences (1D): coding and noncoding, and their identification in genomes (to be incorporated within the Sequence Ontology).
RNA secondary structures and Watson-Crick base-pairing.
RNA 3D structures and recurrent motifs: backbone conformations, base stacking, and tertiary interactions.
Alignments of homologous RNA sequences.
Relationships between alignments and 3D structures.
RNA–RNA, RNA–protein, and RNA–ligand (metabolite, drug, metal and other ion, and water) interactions.
RNA conformational changes and dynamics of functional significance.
Molecular biology of RNA (processing, maturation, splicing, etc.).
Biochemical and biophysical experimental data relating to RNA structure and structure–function relationships.
RNA as regulator of biological networks and pathways.
The potential users of the RO were defined functionally to include anyone who has a need to exchange or use RNA information. The following examples of queries that the RO should be able to support were identified:
Have we seen this particular RNA sequence, secondary structure, or 3D structural motif before?
If yes, in what structural and functional contexts?
Can we deduce a possible 3D structure for a particular RNA element?
What other RNA or protein elements does a given RNA element interact with?
How do we relate most efficiently the precise molecular details to RNA molecular biology, function, and evolution?
The ROC also identified the following research topics related to the issues of reuse of the RNA Ontology (Fig. 1 ▶, step 2):
How do we improve sequence alignments of homologous RNAs?
How do we identify and annotate ncRNA genes in genomes?
How do we improve prediction of RNA 3D structure and dynamics?
How are RNA 3D structure, function, and evolution coupled?
How is RNA evolution coupled to biological evolution?
How do we make new functionality (e.g., software) integrating RNA sequence, structure, and function maximally user-friendly for non-experts?
In discussing the scope of the RNA Ontology, the ROC members identified three immediate, concrete goals to achieve during the first year of the project:
To define the relationships of the RNA Ontology to other biological ontologies.
To create adequate and concise definitions of RNA structural motifs.
To specify the sequence signatures of RNA structural motifs.
We have entered into discussions with members of the National Center for Biomedical Ontology to achieve the first goal. We have also established working groups to address issues related to goals 2 and 3.
The first working group is addressing the classification of the most important backbone conformations observed in recurrent RNA structural motifs, extending and reconciling three extant systems for describing RNA backbone conformations (Gautheret et al. 1993; Hershkovitz et al. 2003; Murray et al. 2003; Schneider et al. 2004; Wadley and Pyle 2004). A second working group is dealing with base-stacking and base-pairing relations in RNA structural motifs. As there is general agreement to use the previously proposed geometric classification of base pairs to describe base-pairing interactions in RNA structural motifs (Leontis and Westhof 2001), the second group is focusing on establishing parallel conventions to describe base-stacking. A third working group is addressing how to reconceptualize RNA alignments to account for evolutionary changes at the level of secondary structure and 3D structural motifs. Attributes specific to the multiple alignment of RNA molecules are currently being defined in collaboration with the members of the Multiple Alignment Ontology (Thompson et al. 2005).
The third step in Figure 1 ▶ is to enumerate terms relevant to the domain of the RNA Ontology. This stage involves collecting and collating terms as they appear in the literature and the daily discourse of scientists working on RNA structure and function. Firstly, this includes terms that refer to physical entities, which range from individual atoms and ions, to bonded moieties (e.g., sugar, phosphate, and base), to whole molecules and finally complexes of molecules. Ontologies already exist to describe RNA and other biological molecules at this level and will be reused for the purposes of the RO, as discussed above (Berman and Westbrook 2003; Westbrook et al. 2005).
A second category of terms refers to qualities or properties of individual physical entities. Examples include acid or base dissociation constants of functional groups, polariz-abilities of individual bases, and thermodynamic parameters for helix formation. A third category of terms refers to relations between physical entities. Relations may be considered generalizations of properties, where properties inhere in one entity at a time, while relations inhere in two or more entities at a time. Examples of binary relations pertaining to RNA entities are base-pairing or base-stacking interactions, which involve two nucleotides at a time. For example, MC-Annotate is a computer program that infers binary relations between nucleotides in RNA 3D structures (Gendron et al. 2001; Lemieux and Major 2002). Larger collections of nucleotides are identified in the literature as various kinds of RNA structural motifs, and these may be considered n-ary relations, where n is the number of nucleotides (Major et al. 2005).
The next steps referred to in Figure 1 ▶ are to determine the classes into which to group the entities enumerated in the previous step and to specify the relations between them. Not only may physical entities be classified, but also properties and relations. Classes are ways of organizing recurrences found in the natural world, and are also referred to as “concepts” or “universals” in the ontological literature. The relation that holds between individual entities, properties, or relations and the classes to which they belong is called instantiation. Once the classes of relevant physical entities and their properties and relations have been identified and classified, the relevant constraints that apply can be worked out (Fig. 1 ▶). For example, nucleotides in RNA are generally constrained to A, C, G, and U, but modified nucleotides will also have to be admitted for describing certain molecules like tRNAs. The last step shown in Figure 1 ▶ is to create instances. Strictly speaking, this refers to the use of the ontology to build integrated databases; it is included in Figure 1 ▶ to indicate that ontology building is an iterative process; the experience one gains in using the ontology to instantiate real data (i.e., assign instances to classes) informs the next cycle of ontology development and refinement.
In addition to representing physical entities and their properties and relations, there is also an opportunity to capture the experimental structural data obtained, among others, from X-ray crystallography, NMR, cross-linking studies, footprinting studies, and fluorescence energy transfer (FRET) experiments. The “assay” upon which a structural annotation is based is useful to record—particularly when there is a chance that different assays may produce different results, and a computer system may need to present alternative hypotheses to help users decide what to believe. The RiboWEB system was an ontology-based system for describing the experimental data related to the structure of the ribosome (ontology available in Protégé format at https://simtk.org/home/ribosomalkb) (Altman et al. 1999). The RiboWEB Ontology contained a subontology for describing experiments that measure structural features. The experiments were organized in a hierarchy based on how they made measurements (local probes vs. global probes, distance measurements vs. surface/buried measurements) and were associated with key properties that needed to be specified in order to minimally describe the experiment. The resulting system allowed the creation of computer programs that supported the discovery of conflicting measurements and that allowed the information content of different experiment types to be assessed (Whirl-Carrillo et al. 2002).
DATABASE INTEGRATION
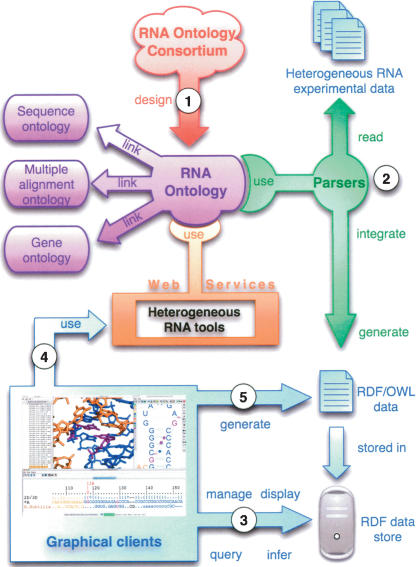
A central scientific aim for creating the RNA Ontology is to describe accurately the coupling between RNA structure and RNA evolution and the role of RNA evolution in biological evolution. The RO will make it possible to structure knowledge databases that integrate diverse primary data (for example, genomic sequences, 3D structures, and RNA secondary structures) and to generate new problem-solving methods and domain-independent applications and software agents. Once the RO is developed and implemented (Fig. 2 ▶, step 1), it will make possible the integration of heterogeneous data from diverse depositories to produce semantically marked-up RDF/OWL files that can be centrally stored (Fig. 2 ▶, step 2). Furthermore, the RO will make it possible to write new graphical clients that intelligently query the RDF/OWL data stores, manage and display the data in useful forms, and draw inferences from the data (Fig. 2 ▶, step 3). The RO will make it possible to transparently access online computational tools that transform primary data, for example, predicting secondary structures or 3D structures from sequences and other experimental data, or automatically generating alignments (Fig. 2 ▶, step 4). Finally, these new, derived data can also be incorporated into the data stores using the same RDF/OWL format (Fig. 2 ▶, step 5).
FIGURE 2.
Ontologies formalize the structure of data by providing domain conceptualization to allow databases and software tools to interoperate. (1) The RNA Ontology (RO) is designed, written, and linked with related ontologies (Gene, Sequence, and Multiple Alignment Ontology). (2) The RO makes it possible to write parsers that can read heterogeneous experimental data and integrate these data to generate RDF/OWL data and make it available through RDF data stores. (3) New graphical clients can be written that use the RO to query RDF data store, manage and display data in useful ways, and infer new knowledge. (4) Graphical clients can also use the RO to access heterogeneous RNA tools to calculate new data (e.g., predict secondary or 3D structures, align sequences, find new genes). (5) New knowledge is added to RDF/OWL data and stored.
RNA architecture is more conserved than secondary structure, and RNA secondary structure is more conserved than primary sequence. Therefore, for accurate sequence alignments to be carried out, homologous motifs and tertiary interactions must be identified and classified at each hierarchical level. The RO will need to precisely describe recurrent modular motifs, their roles and positions in the RNA structural hierarchy, and their relationships to each other. Taxonomies of relationships that exist between entities at different levels of the structural hierarchy are therefore key components of the RO.
First, taxonomies of nucleotide interactions that form and stabilize RNA 3D structure are needed. The geometric classification of RNA base pairs (Leontis and Westhof 2001; Lemieux and Major 2002; Leontis et al. 2002; Lee and Gutell 2004) appears to represent a well-formed taxonomy that can serve the RO to classify base pairs. Well-formed taxonomies comprise classes that are jointly exhaustive and pairwise disjoint (JEPD); they have desirable logical properties (Smith 2004b). At the level of individual interactions, taxonomies will be required that describe base-stacking interactions (Major and Thibault 2005), including the less frequent perpendicular base-to-base interactions, and base–backbone interactions. Separate ontologies of RNA–protein and RNA–ligand interactions will be developed by extension.
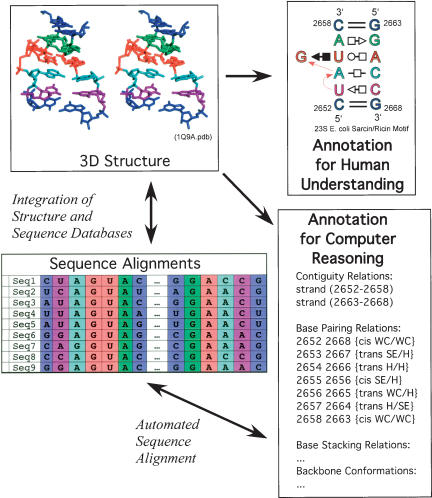
Second, taxonomies of RNA motifs are needed to efficiently define sequence signatures of motifs. These will be based on the taxonomies for base-pairing, base-stacking, backbone conformations, and base–backbone interactions, which combine to form more complex motifs. Such taxonomies of RNA motifs will allow one to quickly find all motifs in a 3D database that share common structural features of interest. Then one can identify homologous aligned sequences for each molecule having the motif, and highlight the relevant portions of the alignment and determine in which sequences the motif is conserved. As new sequences are catalogued, they too will become available for analysis. The sequences can then be analyzed to obtain (or update) substitution or transition probabilities for each nucleotide or base-pairing position in a motif. These data are crucial for improving RNA statistical models of motifs for sequence analysis and evolutionary modeling. Figure 3 ▶ summarizes the integration of structure and sequence data to achieve automated sequence alignment.
FIGURE 3.
The RNA Ontology will facilitate the integration of heterogeneous RNA data including experimental 3D structures (upper left) and nucleotide sequences (lower left). The RNA Ontology will represent information about RNA in ways that are comprehensible by humans (upper right) and machines (lower right). (Here the sarcin motif is annotated for strand continuity and base-pairing relations as for input scripts for the programs MC-Sym—for 3D modeling—and MC-Search—for 3D motif searching [Gautheret et al. 1993; Hoffmann et al. 2003].) Machine annotation will facilitate automation of processes such as sequence alignment, by exploiting all relevant 3D structure data.
As the main components of the RNA Ontology fall into place, we will attempt to formulate a functional ontology that will be used to describe the biological capacities of individual RNA motifs. First steps in this direction have been taken at the SCOR database (http://scor.lbl.gov/function.jsp?parse=new). We will coordinate this work with the Gene Ontology (GO) Consortium to support underlying cross-links between the RO and the GO so that—while externally they will look like independent ontologies—under the hood, they will share a subset of terms by way of unique identifiers. Recurrent motifs occur in different contexts in evolutionarily unrelated molecules and arise independently by virtue of their distinct, sometimes unique biophysical properties. For example, in proteins, the Rossmann fold is found in many different environments for binding nucleotides, and the TIM barrel is a recurrent fold observed in many enzymes with different catalytic activities (Branden and Tooze 1999). In RNA, certain recurrent hairpin loops are particularly stable and may have evolved to provide nucleation centers for RNA folding. Other hairpin loops are particularly well suited for interacting with RNA helical elements, and in fact recur in nonhomologous sites of many different structured RNA molecules playing exactly this same role. Certain internal loops can substitute for hairpin loops to mediate very similar loop–helix interactions. Other recurrent internal loops form platform motifs and exhibit increased affinity for particular hairpin loops. These “loop receptors,” for example, the 11-nt motif that binds the GAAA loop (Costa and Michel 1995), appear to have evolved independently within helical elements to further stabilize tertiary interactions. Still other hairpin loops such as T-loops or “kissing” hairpin loops interact specifically with other hairpin loops. It is not presently known how many distinct RNA motifs exist or are possible (Ferre-D’Amare and Doudna 1999; Moore 1999; Leontis and Westhof 2003; Holbrook 2005; Noller 2005). Therefore, every new motif observed in new RNA 3D structures provides potentially useful information for structurally aligning new RNA sequences and must be included in the motif taxonomy.
CONCLUSIONS
Ontology development is a collaborative and collective activity. The process of ontology development is therefore an iterative and ongoing process, since ontologies can only be improved as they are applied to actual instances of data and when these data are used to answer research questions. Thus, ontologies require mechanisms for change and mechanisms for broad community input. The RNA Ontology will be developed so as to be orthogonal to and inter-operable with related ontologies such as the Gene Ontology (GO), Sequence Ontology (SO), and the structural ontology used by the PDB. The RNA Ontology will be developed according to current principles and standards to facilitate automated reasoning and computability, as well as transparent integration with other biological and biomedical ontologies. Learning from previous efforts, we shall endeavor first to identify the doable aims (for example, shared descriptions and nomenclature of motifs), and second we shall attempt to find the right balance between ontological perfection and practical and useful tools for the community at large. The broader RNA community is invited to join the ROC and to provide comments, suggestions, and criticisms of this plan for the work of the ROC through the Community Discussion Board accessible on the ROC Web site (http://roc.bgsu.edu) and by participating in ROC meetings. A more detailed version of this paper is available on the ROC Web site.
TOOLS FOR ONTOLOGY CONSTRUCTION
The Open Biomedical Ontologies (OBO) consortium is an umbrella Web address for ontologies for shared use across different biological and medical domains (http://obo.sourceforge.net), managed by the National Center for Biomedical Ontology (http://www.bioontology.org). To be included in the OBO ontology library, ontologies must be open (i.e., accessible to all users) and must meet these additional criteria: First, they must include clear and precise textual definitions of terms used within the particular ontology, intelligible to human readers; second, they must use a standard syntax, interpretable by computers; third, they must be orthogonal to other ontologies already included within OBO (http://obo.sourceforge.net/crit.html). Orthogonality means that people working on neighboring ontologies should coordinate their efforts to cover the knowledge domains while minimizing overlap. Recently, an additional criterion was introduced as part of the current reform efforts of the OBO consortium: Relations that are used to connect terms in the ontology must be applied in ways consistent with definitions set forth in the recently published OBO Relations Ontology (Smith et al. 2005).
Specific computational tools exist for developing and editing ontologies, including, among others, DAG-EDIT, OILEd, Chimaera, COBra, and Protégé (Noy et al. 2003). The tool chosen for ontology development should support the Ontology Web Language (OWL; http://www.w3.org/TR/owl-features), a standard language defined for representing and exchanging ontological data. OWL is an ontology language developed by the World Wide Web Consortium (W3C) for the Semantic Web (http://www.w3.org/2004/OWL). Ontologies are expected to play an important role in realizing the Semantic Web by helping automated processes (“intelligent agents”) to access and process all kinds of information. They will make it possible to “semantically mark up” Web pages so that, for example, pages with related meaning (semantics) can be found even when they use different words or phrases (i.e., different syntax). Thus OWL was designed to represent information about categories of objects and how they are interrelated. It is constructed on the Resource Description Format (RDF), which can describe in a machine-readable format objects and relations between them (http://www.w3.org/RDF), and RDF Schema (RDFS), an extension of RDF that can declare classes and properties and structure them in a “subsumption” hierarchy. OWL extends the limited capabilities of RDFS: In OWL, classes can be specified as logical combinations of other classes (intersections, unions, or complements) or as enumerations of specified objects; properties can be declared and organized into subsumption hierarchies (subproperties); domains and ranges for properties can be declared as classes; properties can be defined as transitive, symmetric, functional, or as the inverse of other properties; classes can be defined such that particular properties of their instances are restricted in a variety of ways (Horrocks et al. 2003). The BioPAX, a collaborative effort to create a data exchange format for biological pathway data (http://www.biopax.org), and uniprot-RDF (http://www.isb-sib.ch/~ejain/rdf) are examples of ontologies based on OWL.
In addition to adopting a standard ontology representation language and using state-of-the-art ontology development tools, appropriate tools for automatic reasoning must be selected. FaCT (Fast Classification of Terminologies) is a Description Logic (DL) classifier that can also be used for testing modal logic satisfiability (Horrocks 1998). RACER (Renamed ABox and Concept Expression Reasoner) is a core reasoning agent for the Semantic Web that currently supports a wide range of inference services about ontologies specified in OWL (Haarslev and Möller 2003). Description Logics (DL) are a family of class-based knowledge representation formalisms that include logic-based semantics that use first-order predicate logic (Baader et al. 2003). Using DL makes it possible to deploy sound, complete, and tractable reasoning services. For example, with DL, one can check and reason over the classes, properties, and instances of an ontology. By using appropriate tools from the very start, we hope to increase the likelihood that shared development of an RNA Ontology proceeds with minimal inconsistencies and maximal portability.
Acknowledgments
The ROC is supported by a Research Coordination Network (RCN) grant from the National Science Foundation (grant no. 0443508) and its annual general workshop takes place as part of the RNA Society meeting, where it was initiated in 2004.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2343206.
REFERENCES
- Altman, R.B., Bada, M., Chai, X.J., Carillon, M.W., Chen, R.O., and Abernethy, N.F. 1999. RiboWEB: An ontology-based system for collaborative molecular biology. IEEE Intelligent Systems 14: 68–76. [Google Scholar]
- Baader, F., Calvanese, D., McGuinness, D., Nardi, D., and Patel-Schneider, P. 2003. The description logic handbook: Theory, implementation and applications. Cambridge University Press, New York.
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M. and Westbrook, J. 2003. The need for dictionaries, ontologies, and controlled vocabularies. OMICS 7: 9–10. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Olson, W.K., Beveridge, D.L., Westbrook, J., Gelbin, A., Demeny, T., Hsieh, S.H., Srinivasan, A.R., and Schneider, B. 1992. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys. J. 63: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branden, C. and Tooze, J. 1999. Introduction to protein structure. Garland, New York.
- Costa, M. and Michel, F. 1995. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 14: 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck, K., Lewis, S.E., Mungall, C.J., Yandell, M., Stein, L., Durbin, R., and Ashburner, M. 2005. The Sequence Ontology: A tool for the unification of genome annotations. Genome Biol. 6: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D’Amare, A.R. and Doudna, J.A. 1999. RNA folds: Insights from recent crystal structures. Annu. Rev. Biophys. Biomol. Struct. 28: 57–73. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, P.M.D., Westbrook, J.D., Bourne, P.E., McMahon, B., Watenpaugh, K., and Berman, H.M. 2005. Macromolecular Dictionary (mmCIF). In International tables for crystallography (eds. S. Hall and B. McMahon), pp. 295–443. Springer, Dordrecht, The Netherlands.
- Gautheret, D., Major, F., and Cedergren, R. 1990. Pattern searching/alignment with RNA primary and secondary structures: An effective descriptor for tRNA. Comput. Appl. Biosci. 6: 325–331. [DOI] [PubMed] [Google Scholar]
- ———. 1993. Modeling the three-dimensional structure of RNA using discrete nucleotide conformational sets. J. Mol. Biol. 229: 1049–1064. [DOI] [PubMed] [Google Scholar]
- Gendron, P., Lemieux, S., and Major, F. 2001. Quantitative analysis of nucleic acid three-dimensional structures. J. Mol. Biol. 308: 919–936. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Moxon, S., Marshall, M., Khanna, A., Eddy, S.R., and Bateman, A. 2005. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33: D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, T.R. 1993. Ontolingua: A translation approach to portable ontologies. Knowledge Acquisition 5: 199–220. [Google Scholar]
- Haarslev, V. and Möller, R. 2003. Racer: An OWL reasoning agent for the semantic web. In Proceedings of the International Workshop on Applications, Products and Services of Web-Based Support Systems, in conjunction with the 2003 IEEE/WIC International Conference on Web Intelligence, pp. 91–95. Halifax, Canada.
- Hendler, J. 2003. Communication. Science and the semantic web. Science 299: 520–521. [DOI] [PubMed] [Google Scholar]
- Hershkovitz, E., Tannenbaum, E., Howerton, S.B., Sheth, A., Tannenbaum, A., and Williams, L.D. 2003. Automated identification of RNA conformational motifs: Theory and application to the HM LSU 23S rRNA. Nucleic Acids Res. 31: 6249–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, B., Mitchell, G.T., Gendron, P., Major, F., Andersen, A.A., Collins, R.A., and Legault, P. 2003. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc. Natl. Acad. Sci. 100: 7003–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook, S.R. 2005. RNA structure: The long and the short of it. Curr. Opin. Struct. Biol. 15: 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks, I. 1998. Using an expressive description logic: FaCT or fiction? In Proceedings of the Sixth International Conference on the Principles of Knowledge Representation and Reasoning. Trento, Italy.
- Horrocks, I., Patel-Schneider, P.F., and van Harmelen, F. 2003. From SHIQ and RDF to OWL: The making of a web ontology language. J. Web Semantics 1: 7–26. [Google Scholar]
- Kumar, A., Smith, B., and Novotny, D. 2005. Biomedical informatics and granularity. Comp. Funct. Genomics 5: 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.C. and Gutell, R.R. 2004. Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J. Mol. Biol. 344: 1225–1249. [DOI] [PubMed] [Google Scholar]
- Lemieux, S. and Major, F. 2002. RNA canonical and non-canonical base pairing types: A recognition method and complete repertoire. Nucleic Acids Res. 30: 4250–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis, N.B. and Westhof, E. 2001. Geometric nomenclature and classification of RNA base pairs. RNA 7: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13: 300–308. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B., Stombaugh, J., and Westhof, E. 2002. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 30: 3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S.E. 2005. Gene Ontology: Looking backwards and forwards. Genome Biol. 6: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano, J.S. 2005. PAX of mind for pathway researchers. Drug Discov. Today 10: 937–942. [DOI] [PubMed] [Google Scholar]
- Major, F. and Thibault, P. 2005. Computer modeling of RNA 3D structure. In Encyclopedia of molecular biology and molecular medicine (ed. R.A. Meyers), pp. 605–636. VCH Publishers, Inc., New York.
- Major, F., Turcotte, M., Gautheret, D., Lapalme, G., Fillion, E., and Cedergren, R. 1991. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science 253: 1255–1260. [DOI] [PubMed] [Google Scholar]
- Major, F., Lemieux, S., Larose, M., and Thibault, P. 2005. Modelling RNA three-dimensional structure by combining short nucleotide interaction cycles. Eur. Biophys. J. 34: 560. [Google Scholar]
- Moore, P.B. 1999. Structural motifs in RNA. Annu. Rev. Biochem. 68: 287–300. [DOI] [PubMed] [Google Scholar]
- Murray, L.J., Arendall III, W.B., Richardson, D.C., and Richardson, J.S. 2003. RNA backbone is rotameric. Proc. Natl. Acad. Sci. 100: 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musen, M.A. 1992. Dimensions of knowledge sharing and reuse. Comput. Biomed. Res. 25: 435–467. [DOI] [PubMed] [Google Scholar]
- Neumann, E. 2005. A life science Semantic Web: Are we there yet? Sci. STKE http://stke.sciencemag.stke.com. [DOI] [PubMed]
- Noller, H.F. 2005. RNA structure: Reading the ribosome. Science 309: 1508–1514. [DOI] [PubMed] [Google Scholar]
- Noy, N.F. and McGuinness, D.L. 2001. “Ontology development 101: A guide to creating your first ontology.” Stanford Knowledge Systems Laboratory Technical Report KSL-01-05/Stanford Medical Informatics Technical Report SMI-2001-0880, Stanford University, Stanford, CA.
- Noy, N.F., Crubezy, M., Fergerson, R.W., Knublauch, H., Tu, S.W., Vendetti, J., and Musen, M.A. 2003. Protege-2000: An open-source ontology-development and knowledge-acquisition environment. AMIA Annu. Symp. Proc. 2003: 953. [PMC free article] [PubMed] [Google Scholar]
- Schneider, B., Zdenek, M., and Berman, H.M. 2004. RNA conformational classes. Nucleic Acids Res. 32: 1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, B. 2004a. Beyond concepts: Ontology as reality representation. In Proceedings of FOIS 2004 International Conference on Formal Ontology and Information Systems (eds. A. Varzi and L. Vieu). IOS Press, Amsterdam.
- ———. 2004b. The logic of biological classification and the foundations of biomedical ontology. In Invited papers from the 10th International Conference in Logic Methodology and Philosophy of Science (ed. D. Westerstahl). Elsevier, North-Holland.
- Smith, B., Ceusters, W., Klagges, B., Kohler, J., Kumar, A., Lomax, J., Mungall, C., Neuhaus, F., Rector, A.L., and Rosse, C. 2005. Relations in biomedical ontologies. Genome Biol. 6: R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, R., Goble, C.A., and Bechhofer, S. 2000. Ontology-based knowledge representation for bioinformatics. Brief. Bioinform. 1: 398–414. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Holbrook, S.R., Katoh, K., Koehl, P., Moras, D., Westhof, E., and Poch, O. 2005. MAO: A Multiple Alignment Ontology for nucleic acid and protein sequences. Nucleic Acids Res. 33: 4164–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadley, L.M. and Pyle, A.M. 2004. The identification of novel RNA structural motifs using COMPADRES: An automated approach to structural discovery. Nucleic Acids Res. 32: 6650–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh, A., Gendron, P., Altman, R., Brown, J.W., Case, D., Gautheret, D., Harvey, S.C., Leontis, N., Westbrook, J., Westhof, E., et al. 2002. RNAML: A standard syntax for exchanging RNA information. RNA 8: 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook, J., Ito, N., Nakamura, H., Henrick, K., and Berman, H.M. 2005. PDBML: The representation of archival macromolecular structure data in XML. Bioinformatics 21: 988–992. [DOI] [PubMed] [Google Scholar]
- Whirl-Carrillo, M., Gabashvili, I.S., Bada, M., Banatao, D.R., and Altman, R.B. 2002. Mining biochemical information: Lessons taught by the ribosome. RNA 8: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]