Abstract
Here we describe a procedure for the rapid enrichment of RNA from cell extracts and the subsequent fractionation and analysis of the “small RNA” population by ion pair reverse phase chromatography. Solid phase extraction procedures have been developed utilizing nonporous alkylated poly(styrene-divinylbenzene) particles in conjunction with ion pair reagents to enrich total RNA. This approach facilitates the selective enrichment and separation of the relatively lower abundance small RNAs, from the more abundant higher molecular weight rRNA species. We also describe the application of monolithic capillaries in conjunction with ion pair reverse phase chromatography to bring increased sensitivity in the analysis of very low abundance RNAs. These approaches will simplify the biochemical analysis of this class of molecules, which are emerging as important regulators of global gene expression in higher organisms.
Keywords: ion pair reverse phase chromatography, solid phase extraction, total RNA, small RNAs and miRNA
INTRODUCTION
Our appreciation of the relative importance of RNA in numerous biological processes has increased substantially over recent years. The discoveries of catalytic RNA (Kruger et al. 1982; Guerrier-Takada et al. 1983), RNA interference and noncoding regulatory RNAs (for review, see Ambros 2004; Bartel 2004) have impacted a broad range of disciplines. The development of methodology for the isolation and characterization of biological RNA has had a somewhat checkered history alongside comparable methods for the analysis of its macromolecular counterparts, DNA and proteins. Moreover, the extraction, isolation, and analysis of RNA is routinely more difficult in comparison to that required for DNA. In approaching the problem of RNA isolation, the stability and molecular heterogeneity are of immediate concern. RNA is susceptible to endo- and exo-nuclease mediated degradation, rendering the initial stages of extraction and the downstream storage of the purified material more challenging than for DNA. Furthermore, downstream analysis of RNA requires separations to be typically performed under harsh denaturing conditions.
Current total RNA isolation procedures use a combination of denaturing agents, acid phenol chloroform extraction followed by precipitation of the nucleic acids (Chomczynski and Sacchi 1987). This procedure is time-consuming and is particularly inefficient with respect to the recovery of “small” RNAs. In addition, the use of glass filters that bind RNA in the presence of chaotropic salts have been used to isolate and purify total RNA from cell extracts and tissues (Boom et al. 1990). Proteins and DNA are removed by washing the filter, and RNA is subsequently eluted. Low RNA yields are often obtained due to overloading of the column, which can cause the column to clog or can prevent the RNA from binding efficiently. Moreover, the recovery of small RNAs using traditional approaches is inefficient and the fractionation of RNA species is not always possible.
Following enrichment of an RNA species, downstream analysis is necessary to determine such properties as molecular weight and primary, secondary, and tertiary structure of the RNA. A variety of approaches have been developed centered around gel electrophoresis under denaturing conditions, with the aim of disrupting RNA secondary structure to provide accurate molecular weight determinations (Sambrook et al. 1989). Many of the current methodologies utilizing denaturing conditions incorporate the use of toxic, hazardous chemicals such as glyoxal/dimethyl sulfoxide and methyl mercury hydroxide (Bailey and Davidson 1976; McMaster and Carmichael 1977; Sebo and Schmit 1982). Alternative denaturants include urea, formamide, and formaldehyde (Floyd et al. 1974; Pinder et al. 1974; Lehrach et al. 1977). There are additional problems associated with the use of such reagents, including difficulty in staining the gels and interference with the electrophoretic mobility of the RNA species (Tsang et al. 1993).
Ion pair reverse phase chromatography
Ion pair reverse phase high-performance liquid chromatography (IP RP HPLC) on nonporous alkylated poly(styrene-divinylbenzene) particles combines very high-resolution separation of nucleic acids with very short analysis times, therefore offering significant advantages and opportunities for the analysis of nucleic acids (Gjerde et al. 2002) over electrophoresis methods. The nonporous polymeric media, in conjunction with the highly monodisperse nature of the particles, results in the minimization of the diffusion paths (Huber et al. 1993; Huber 1998). Furthermore, the media is robust, resistant to a broad range of temperatures and pH. When a sample is injected, the anions from the nucleic acid combine with the oppositely charged ammonium cations of the ion pair reagent. The most common ion pair reagent is triethylammonium acetate (TEAA). The alkyl groups of such ion pair reagents enable the nucleic acids to become hydrophobic and the molecule adsorbs to the stationary phase. The longer the alkyl chain used, the more hydrophobic the nucleic acid and the stronger the interaction with the stationary phase. In addition, larger nucleic acids have a stronger interaction with the stationary phase as the increased length enables an increased number of ion pair molecules to be associated with the nucleic acid. During the HPLC a gradient of acetonitrile is started. As the acetonitrile concentration is increased this causes the smaller nucleic acids to desorb from the stationary phase first. Finally as the acetonitrile concentration is further increased the larger nucleic acids are desorbed and travel down the column to the detector.
Given the complexity of the RNA population from even the simplest of organisms, it is critical that measures be taken to preserve that population prior to separation (or indeed any form of analysis). Separation and analysis using ion pair reverse phase chromatography is performed using a combination of ion pairing reagent (TEAA at pH 7.4) and acetonitrile at elevated temperatures. Under such conditions RNA maintains its chemical integrity and biological activity (Azarani and Hecker 2001). Furthermore, when the effect of acetonitrile upon the activity of a typical ribonuclease enzyme was investigated at a range of solvent concentrations and temperatures, a marked effect upon the stability of the enzyme was observed. Thus at 60°C in 20% acetonitrile, Bovine RNase is reversibly unfolded, while at 60% acetonitrile at the same temperature the enzyme is unfolded but in a reversible manner (M.J. Conroy, D.P. Hornby, and M.J. Dickman, unpubl.). In addition, the mechanism of separation using IP RP HPLC also serves to eliminate contaminating RNases during the chromatography. Under the conditions used, the RNA is retained on the stationary phase allowing separation from the RNases. These observations suggest that the elimination of the deleterious effects of contaminating RNases enables the purification and analysis of high-quality RNA using IP RP methodology.
Here we describe the enrichment of RNA, including the subsequent fractionation of small RNAs in conjunction with the rapid analysis of the isolated purified RNA. The methodology and analysis is centered on ion pair reverse phase chromatography. Solid phase extraction procedures have been developed utilizing nonporous alkylated poly-(styrene-divinylbenzene) particles in conjunction with ion pair reagents to enrich total RNA. This methodology enables the selective enrichment of the lower abundance small RNAs from the larger, more abundant rRNA species. In addition, rapid analysis of the RNA fractions isolated using the solid phase extraction procedures using ion pair reverse phase chromatography has been performed. These results demonstrate the successful enrichment of small RNAs and in addition facilitate the analysis of small RNAs and larger rRNA species during a single chromatographic analysis. RNA analysis has also been extended using monolithic capillaries in conjunction with ion pair reverse phase chromatography, demonstrating higher sensitivity, and is ideal for the analysis of low concentrations of RNA samples and the subsequent downstream fractionation of the RNA species. Using such approaches allows the analysis of low nanogram quantities of RNA and furthermore enables the subsequent fraction of RNA in small volumes.
RESULTS AND DISCUSSION
Ion pair reverse phase chromatography of RNA under denaturing conditions
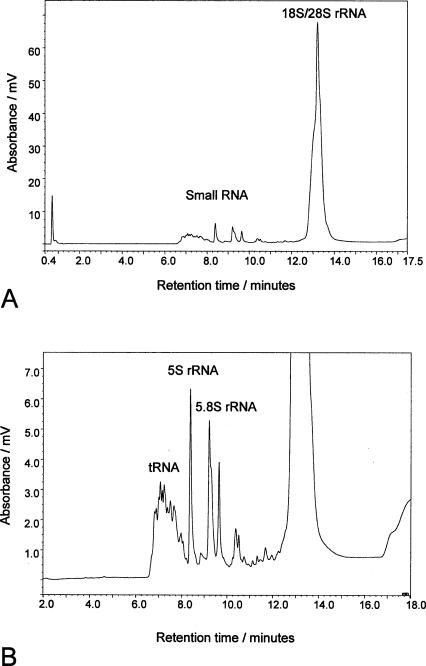
When a total RNA preparation extracted from both prokaryotic and eukaryotic cells is subjected to IP RP HPLC the resulting chromatogram is typified by that shown in Figure 1 ▶. There are several distinct features within the profile; the earliest eluting species shown in the chromatogram are the tRNA (including possible mature miRNA), followed by the elution of the rRNA. Verification of the RNA species was performed by comparison with molecular weight standards, purified tRNA and rRNA standards and using specific primers for RT PCR (data not shown). Standard gel electrophoretic analysis of total RNA samples often requires the combined use of denaturing agarose and denaturing polyacrylamide gels. The separation of the larger rRNA species is performed using denaturing agarose gel electrophoresis, while the separation and analysis of the small RNAs requires denaturing polyacrylamide gels. These results demonstrate that ion pair reverse phase chromatography enables the rapid analysis of total RNA, including the rapid separation and fractionation of tRNA, small rRNA, and large rRNA species during a single chromatographic analysis.
FIGURE 1.
IP RP HPLC chromatogram of eukaryotic total RNA. The chromatogram shows the analysis of (3 μg) total RNA extracted from Hela cells using gradient 2 (see Materials and Methods). A shows the typical profile of the early eluting small RNA species and the later coeluting 18S and 23S rRNA. B shows an enhanced view of the smaller RNA species. The tRNA, 5.8S, and 5S rRNA are highlighted.
Solid phase extraction and enrichment of small RNAs
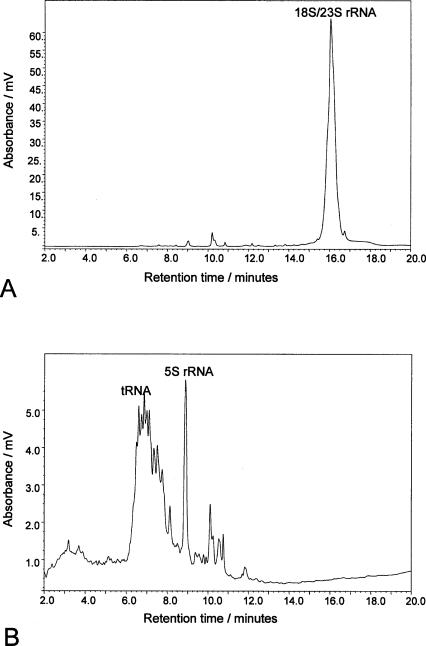
We have previously shown that using ion pair reverse phase chromatography in conjunction with alkylated poly(styrene-divinylbenzene) particles enables high-resolution separations of RNA and the rapid analysis of total RNA populations. We have used the same ion pair methodology to extract total RNA, and in particular, to specifically enrich the small RNAs from total RNA. Solid phase extraction columns were prepared using 8 μm alkylated poly(styrene-divinylbenzene) particles; the total RNA extracted from HeLa cells was adsorbed to the stationary phase in the presence of 0.2 M TEAA in 10% acetonitrile (pH 7). Under these conditions only the 18S and 23S rRNAs are adsorbed: The smaller RNA species are not retained. A stronger ion pair reagent (tetrabutylammonium bromide) is subsequently added to the unbound fraction (small RNA species) and passed down a second solid phase extraction column. Under these ion pair conditions the small RNAs are retained by the stationary phase. In both cases the extraction columns are washed in the presence of the appropriate ion pair reagent and acetonitrile to remove salts and contaminants. The RNA species are subsequently eluted from the column by the addition of 50% acetonitrile. The results of the solid phase extraction procedures are shown in Figure 2 ▶. Figure 2A ▶ shows the RNA species retained on the initial solid phase extraction column. The results demonstrate that the rRNA species are retained while the small RNA species pass directly through the column. Figure 2B ▶ shows the eluted fraction following addition of the stronger ion pair reagent (tetrabutylammonium bromide) to the unbound species. The results show that the small RNA species are now retained on the stationary phase and can be subsequently eluted with acetonitrile. Furthermore, the enriched small RNAs are completely free of any contaminating rRNA (see Fig. 2B ▶). These results demonstrate that alkylated poly(styrene-divinylbenzene) particles can be used successfully for the fractionation and purification of small RNAs from total RNA preparations, using ion pair methodology under stabilizing conditions.
FIGURE 2.
Solid phase enrichment and fractionation of RNA using alkylated poly(styrene-divinylbenzene) particles. A shows the IP RP HPLC chromatogram of the enriched and fractionated large RNA species from total RNA extracts. B shows the IP RP HPLC chromatogram of the enriched small RNA species in the absence of larger rRNA species.
RNA analysis using capillary HPLC
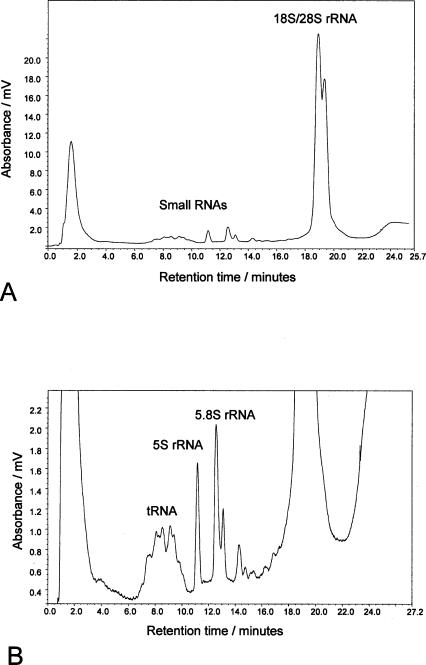
Capillary chromatography using ion pair reverse phase conditions on poly(styrene-divinylbenzene) monoliths has previously been used for the separation of DNA, at very high resolution and over short analysis times with the advantage of increased sensitivity compared to traditional separations using conventional analytical columns (Oberacher and Huber 2002; Walcher et al. 2002). We have used 200 μm i.d. poly(styrene-divinylbenzene) monoliths in the analysis of RNA under denaturing conditions. The analysis of RNA using capillary chromatography was also extended to the analysis of total RNA extracted from HeLa cells: The results are shown in Figure 3 ▶. A similar elution profile of the RNA species is obtained as previously, with the early eluting small RNA followed by the coeluting rRNA species. The analysis using the 200 μm i.d. monoliths further illustrates the increase in sensitivity that can be achieved in the analysis of RNA. Nine nanograms (9 ng) of total RNA was analyzed in comparison to the microgram quantities of RNA required using conventional analytical columns to visualize the lower abundance small RNA and rRNAs. Analysis of the abundant rRNA species alone requires sub-nanogram quantities. In addition, fractionation of the RNA species during the chromatography enables the isolation of the RNA species in low volumes due to the low flow rates (3 μL/min) used during the chromatography. Therefore, no further sample manipulation and downstream processing of the sample is required, thereby significantly reducing potential RNase contamination.
FIGURE 3.
IP RP μHPLC chromatogram of eukaryotic total RNA. The chromatogram shows the analysis of 9.2 ng total RNA extracted from Hela cells using gradient 4 (see Materials and Methods). A shows the typical profile of the early eluting small RNA species and the later coeluting 18S and 23S rRNA. B shows an enhanced view of the smaller RNA species. The tRNA, 5.8S, and 5S rRNA are highlighted.
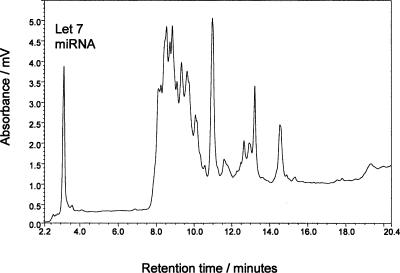
To demonstrate the ability to enrich and fractionate miRNAs from total RNA populations, a total RNA fraction was spiked with a synthetic miRNA corresponding to the let-7 miRNA sequence (see Materials and Methods). The small RNAs were subsequently enriched using solid phase extraction as previously described, and the enriched small RNAs were analyzed using IP RP μHPLC (see Fig. 4 ▶). These results demonstrate the ability to efficiently enrich the miRNAs and subsequently separate the miRNAs from the smaller RNA species.
FIGURE 4.
IP RP μHPLC chromatogram of the enriched small RNAs from total RNA spiked with synthetic let-7 miRNA. The chromatogram shows the analysis of the small RNA fraction obtained following the solid phase enrichment of a total RNA (20 μg) spiked with synthetic miRNA (let-7 150 ng) using gradient 5 (see Materials and Methods). The let-7 miRNA species is highlighted.
CONCLUSIONS
Ion pair reverse phase chromatography of RNA under denaturing conditions enables the high-resolution separation of RNA in relatively short analysis times. We have shown that both small RNAs (tRNAs, 5S rRNA) and larger rRNA species can be selectively separated during chromatography. Such approaches could be extended to the detection of microorganisms on the basis of their 16S rRNA gene sequences. The retention time is likely to be affected by both the size and the sequence of the rRNA and may allow the discrimination of closely related 16S rRNA genes.
Recently the focus of much molecular biology research has been the roles played by a group of small RNA species referred to as small nuclear RNAs (including siRNAs and miRNAs). The increasing importance of the identification of such miRNAs requires the development of technology to overcome many of the problems associated with such methods, including the enrichment of small RNAs from total RNA extracts. Here we have demonstrated the enrichment of small RNAs (tRNA, 5S rRNA) from the larger, more abundant rRNA species using solid phase extraction centered around ion pair methodology. Using such methodology also allows the enrichment of siRNA and miRNAs from total RNA extracts and was demonstrated using synthetic RNA corresponding to the let-7 miRNA sequence. Furthermore the use of solid phase extraction methodology will allow the scale up of large quantities of RNA and control over the elution parameters to selectively elute the RNAs of interest. In addition, capillary chromatography in conjunction with ion pair methodology has enabled very sensitive approaches to be developed in the analysis of RNA, requiring only sub-low nanogram quantities of RNA. Using such techniques allows the detection of very low levels of RNA and therefore will prove particularly useful in the analysis/fractionation of miRNA/siRNA and from procedures where only very low amounts of RNA can be extracted from tissues/cells or micro-organisms.
MATERIALS AND METHODS
IP RP HPLC
All samples were analyzed by IP RP HPLC on an Agilent 1100 HPLC using a DNAsep column 4.6 × 50 mm i.d. (Transgenomic). The stationary phase of the column consists of a nonporous, alkylated poly(styrene-divinylbenzene) matrix. Chromatograms, analyzed using UV detection, were recorded at a wavelength of 260 nm.
The IP RP HPLC analysis was performed using the following conditions:
Gradient 1: Buffer A, 0.1 M TEAA (pH 7.0) (Fluka); buffer B, 0.1 M TEAA (pH 7.0) containing 25% acetonitrile (Sigma). Starting at 30% buffer B the gradient was extended to 32% buffer B in 2 min, followed by an extension to 55% buffer B over 15 min at a flow rate of 1.0 mL/min.
Gradient 2: Buffer A, 0.1 M TEAA (pH 7.0) (Fluka); buffer B, 0.1 M TEAA (pH 7.0) containing 25% acetonitrile. Starting at 20% buffer B the gradient was extended to 22% buffer B in 2 min, followed by an extension to 52% buffer B over 15 min, followed by an extension to 65% buffer B over 2.5 min at a flow rate of 1.0 mL/min.
IP RP μHPLC
All samples were analyzed by IP RP μHPLC on an Ultimate capillary HPLC system (LC Packings) using a poly(styrene-divinylbenzene) monolith column 60 × 0.2 mm i.d. (LC Packings). Chromatograms, analyzed using UV detection, were recorded at a wavelength of 260 nm using a 45 nL flow cell (LC Packings).
The IP RP μHPLC analysis was performed using the following conditions:
Gradient 3: Buffer A, 0.1 M TEAA (pH 7.0) (Fluka); buffer B, 0.1 M TEAA (pH 7.0) containing 25% acetonitrile. Starting at 25% buffer B the gradient was extended to 30% buffer B in 2 min, followed by an extension to 50% buffer B over 15 min at a flow rate of 3.0 μL/min.
Gradient 4: Buffer A, 0.1 M TEAA (pH 7.0) (Fluka); buffer B, 0.1 M TEAA (pH 7.0) containing 25% acetonitrile. Starting at 25% buffer B the gradient was extended to 30% buffer B in 2 min, followed by an extension to 55% buffer B over 15 min, followed by an extension to 60% buffer B over 2 min at a flow rate of 3.0 μL/min.
Gradient 5: Buffer A, 0.1 M TEAA (pH 7.0) (Fluka); buffer B, 0.1 M TEAA (pH 7.0) containing 25% acetonitrile. Starting at 15% buffer B the gradient was extended to 20% buffer B in 2 min, followed by an extension to 45% buffer B over 15 min, followed by an extension to 60% buffer B over 2 min at a flow rate of 3.0 μL/min.
Solid phase extraction and enrichment of small RNAs
Total RNA was extracted from 1 × 106 Hela cells (CILBIOTECH) using standard phenol/chloroform extraction procedures. Following extraction the total RNA was resuspended in 0.1 M TEAA (Fluka) (pH 7.4), 10% acetonitrile (HPLC grade, Sigma). Solid phase extraction columns were prepared using 8 μm alkylated poly(styrene-divinylbenzene) (Transgenomic) particles suspended in 50% acetonitrile, 0.2 M TEAA using 5 mL columns (Pierce) The columns were equilibrated with two column volumes of 0.2 M TEAA in 10% acetonitrile. The total RNA fraction was passed down the solid phase extraction column and the unbound fraction retained. Subsequently tetrabutylammonium bromide (TBAB) (Fluka) was added to the unbound fraction to a final concentration of 5 mM. This fraction was then added to a second solid phase extraction column that had previously been equilibrated with two column volumes of 5 mM TBAB. Both columns were washed with five column volumes of the appropriate ion pair solution. Elution of the RNA from the columns was performed by the addition of 1–2 column volumes of 50% acetonitrile. Let-7 miRNA (5′-UGAGGUAGUAGGUUGUAUUAGUU-3′) was obtained from Sigma Proligo. Twenty micrograms (20 μg) of total RNA was spiked with 150 ng of let-7 miRNA. Subsequently the small RNAs were enriched using solid phase extraction as previously described.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2278606.
REFERENCES
- Ambros, V. 2004. The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- Azarani, A. and Hecker, K.H. 2001. RNA analysis by ion-pair reversed-phase high performance liquid chromatography. Nucleic Acids Res. 29: e7. http://nar.oupjournals.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.M. and Davidson, N. 1976. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal. Biochem. 70: 75–85. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Boom, R., Sol, C.J., Salimans, M.M., Jansen, C.L., Wertheim-van Dillen, P.M., and van der Noordaa, J. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28: 495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Floyd, R.W., Stone, M.P., and Joklik, W.K. 1974. Separation of single-stranded ribonucleic acids by acrylamide-agarose-urea gel electrophoresis. Anal. Biochem. 59: 599–609. [DOI] [PubMed] [Google Scholar]
- Gjerde, D.T., Hanna, C.P., and Hornby, D.P. 2002. DNA chromatography. Wiley-VCH, Weinheim, Germany.
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., and Altman, S. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35: 849–857. [DOI] [PubMed] [Google Scholar]
- Huber, C.G. 1998. Micropellicular stationary phases for high-performance liquid chromatography of double-stranded DNA. J. Chromatogr. A 806: 3–30. [DOI] [PubMed] [Google Scholar]
- Huber, C.G., Oefner, P.J., Preuss, E., and Bonn, G.K. 1993. High-resolution liquid chromatography of DNA fragments on non-porous poly(styrene-divinylbenzene) particles. Nucleic Acids Res. 21: 1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger, K., Grabowski, P.J., Zaug, A.J., Sands, J., Gottschling, D.E., and Cech, T.R. 1982. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31: 147–157. [DOI] [PubMed] [Google Scholar]
- Lehrach, H., Diamond, D., Wozney, J.M., and Boedtker, H. 1977. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical re-examination. Biochemistry 16: 4743–4751. [DOI] [PubMed] [Google Scholar]
- McMaster, G.K. and Carmichael, G.G. 1977. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc. Natl. Acad. Sci. 74: 4835–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberacher, H. and Huber, C.G. 2002. Capillary monoliths for the analysis of nucleic acids by high performance liquid chromatography-electrospray ionization mass spectrometry. Trends Anal. Chem. 21: 166–174. [DOI] [PubMed] [Google Scholar]
- Pinder, J.C., Staynov, D.Z., and Gratzer, W.B. 1974. Electrophoresis of RNA in formamide. Biochemistry 13: 5373–5378. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2d ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sebo, T.J. and Schmit, J.C. 1982. Analytical gel electrophoresis of high-molecular-weight RNA in acrylamide-agarose gels containing methylmercuric hydroxide. Anal. Biochem. 120: 136–145. [DOI] [PubMed] [Google Scholar]
- Tsang, S.S., Yin, X., Guzzo-Arkuran, C., Jones, V.S., and Davison, A.J. 1993. Loss of resolution in gel electrophoresis of RNA: A problem associated with the presence of formaldehyde gradients. Biotechniques 14: 380–381. [PubMed] [Google Scholar]
- Walcher, W., Oberacher, H., Troiani, S., Holzl, G., Oefner, P., Zolla, L., and Huber, C.G. 2002. Monolithic capillary columns for liquid chromatography-electrospray ionization mass spectrometry in proteomic and genomic research. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 782: 111–125. [DOI] [PubMed] [Google Scholar]