Abstract
The virion incorporation of 7SL, the RNA component of the host signal recognition particle (SRP), has been shown for several simple retroviruses. Data here demonstrate that 7SL is also packaged by HIV-1, in sevenfold molar excess of genomic RNA. Viral determinants of HIV-1 genome and primer tRNA packaging were not required for 7SL incorporation, as virus-like particles with only minimal assembly components efficiently packaged 7SL. The majority of 7SL within cells resides in ribonucleoprotein complexes bound by SRP proteins, and most SRP protein exists in signal recognition particles. However, Western blot comparison of virion and cell samples revealed that there is at least 25-fold less SRP p54 protein per 7SL RNA in HIV-1 particles than in cells. Comparing 7SL:actin mRNA ratios in virions and cells revealed that 7SL RNA appears selectively enriched in virions.
Keywords: retroviral RNA packaging
Retroviruses are ribonucleoprotein complexes synthesized by metazoan cells in response to genetic information in virion RNA. The most prominent nucleic acid in virions is viral genomic RNA, a capped and polyadenylated Pol II transcript that is selectively incorporated at two copies per virion and accounts for >50% of the mass of RNA in retroviruses like HIV-1 (Berkowitz et al. 1996). The remainder of virion RNAs are host-encoded, with some more abundant on a molar basis than the viral genome (Levin and Seidman 1979; Onafuwa-Nuga et al. 2005). Viral genomic RNA is dispensable in retroviral assembly, but RNA, not necessarily of viral origin, is required (Campbell and Vogt 1995; Cimarelli et al. 2000; Muriaux et al. 2001; Wang and Aldovini 2002).
One of the first host RNAs identified in avian and murine retroviruses was a molecule with 7S sedimentation properties now known as 7SL RNA (Erikson 1969; Bishop et al. 1970; Erikson et al. 1973; Faras et al. 1973; Sawyer and Dahlberg 1973; Walker et al. 1974). The packaging of 7SL by lentiviruses has not been well studied, although one report failed to detect 7S RNA in equine infectious anemia virus (Cheevers et al. 1977). 7SL is the RNA component of signal recognition particle (SRP), the ribonucleoprotein complex that promotes cotranslational protein transport into the endoplasmic reticulum (Walter and Blobel 1982, 1983; Doudna and Batey 2004).
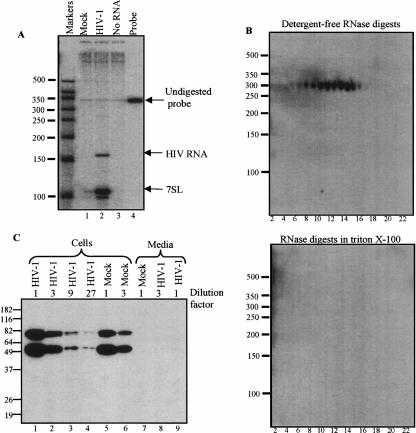
Here, we examined 7SL RNA incorporation by HIV-1. The ratio of 7SL to HIV genomic RNA was compared using an RNA probe with complementarity to both HIV-1 and 7SL sequences (Fig. 1A ▶). Results show that mock-infected cells released only trace amounts of 7SL RNA into the media, whereas virions contained about seven molecules of 7SL per HIV RNA.
FIGURE 1.
SRP RNA but not SRP protein in HIV-1. (A) RNA from virions and culture media analyzed by a ribonuclease protection assay. HIV-1BRU was propagated in CEM-SS cells obtained through the NIH AIDS Research and Reference Reagent Program. Culture media from uninfected and HIV-1BRU infected cells was concentrated by ultracentrifugation. RNA was extracted using TRIzol (Invitrogen), and it protected both a 150-nucleotide (nt) riboprobe fragment corresponding to sequences from HIV-1 and a 101-nt fragment that hybridized to 7SL RNA. The 344-nt [α-32P] rCTP-labeled riboprobe was similar to previously described chimeric viral/7SL probes (Onafuwa-Nuga et al. 2005). Protected fragments were quantified by adjusting phosphorimager values for the number of C residues in each. Markers are in nucleotides. (B) Nuclease sensitivity of virion-associated RNAs. Concentrated HIV-1 was centrifuged through 6%–18% discontinuous iodixanol gradients (Optiprep, Nycomed Pharma) using a Beckman SW41 Ti rotor at 27,500 rpm for 1 h. Micrococcal nuclease (Takara Biotech) digestion was in either the presence or absence of 0.1% Triton X-100. RNAs from digestions were separated by denaturing PAGE, electrophoretically transferred to nylon membranes (Zeta-Probe GT, BioRad), and UV cross-linked. 7SL RNA was detected with a 32P-labeled probe (5′-TGCTCCGTTTCCGACCTGGGCCGGTTCACCCCTCCTT-3′). Note that in Northern blots, 7SL migrated at ~300 nt. Hybridization to longer products was not detected (not shown), indicating that little if any of the encapsidated 7SL was incorporated as provirus read-through RNA. (C) Western blot analysis for SRP54. Cell or virion lysates prepared in RIPA buffer were cleared of particulates by centrifugation, then boiled and loaded on 12% SDS-PAGE in SDS sample buffer. After electrophoretic transfer to PDVF membranes (BioRad), proteins were detected with monoclonal mouse anti-human SRP54 (BD Biosciences Pharmingen). Antibodies were detected using biotinylated anti-mouse IgG (BRL) and HRP-Streptavidin conjugate (43–4323, Zymed), and visualized using Lumi-Light Substrate (Roche). Markers are in kDa. Note that in addition to SRP54, the monoclonal antibody detected a slower migrating protein in CEM-SS cells. This ~75-kDa species was not detectable in NIH3T3 cells using the same reagents, or by the antibody’s manufacturer in Jurkat cells. It was also not seen in blots of human proteins using less sensitive SRP54-targeted polyclonal antibodies (not shown). Despite this unidentified band, data with CEM-SS cells and the monoclonal antibody are presented here because the relatively high levels of virus production, and good antibody sensitivity with this combination best illustrated the magnitude of SRP protein deficiency in virions.
To explore the association of 7SL with HIV-1, viral particles were centrifuged through discontinuous iodixanol gradients. Virons were localized by assaying fractions for reverse transcriptase activity. To determine whether or not HIV-associated 7SL was resistant to ribonuclease, as it should be if inside particles, fractions were subsequently divided into three sets of tubes: One was left untreated and the other two were nuclease digested in the presence or absence of detergent. Virion 7SL was resistant to digestion in the absence of detergent but susceptible when detergent-exposed (Fig. 1B ▶, cf. top and bottom panels). Amounts of 7SL in undigested and detergent-free digested fractions were similar, fractions from control gradients of mock-infected cell media did not contain detectable amounts of 7SL or HIV RNA, and 7SL copurified with HIV-1 RT activity (not shown). These data suggest 7SL RNA is packaged inside virions. The possibility that non-viral vesicles contained observed 7SL cannot be ruled out by these data but seems unlikely, as such vesicles would have to be the same density as HIV and released from HIV-infected, but not mock-infected, cells.
To address whether all SRP components, and not just 7SL RNA, were associated with HIV-1, Western blots were performed using a monoclonal antibody to detect the 54-kDa protein subunit of SRP (SRP54) in cell and viral samples (Fig. 1C ▶). Amounts of protein per lane were normalized to the 7SL content of parallel samples (not shown), and a mock-infected media sample was generated by concentrating the same volume of media required to generate the virion sample. Serial dilutions of proteins were examined to compare amounts of SRP54 in cells and virions (Fig. 1C ▶).
Within cells, ~75% of all 7SL resides in SRP RNPs, and immunological detection of SRP proteins in cell fractions shows that ~85% exist in either free or membrane-associated SRPs, both of which require 7SL RNA for their structural integrity (Walter and Blobel 1983). Thus, most SRP protein in cells is associated with SRP RNA. In results presented here, no detectable SRP54 was associated with HIV-1 (Fig. 1C ▶, lanes 8,9), while even cell sample dilutions containing 27-fold less 7SL RNA than the undiluted virion sample yielded easily detectable SRP54 signal (Fig. 1C ▶, lane 4).
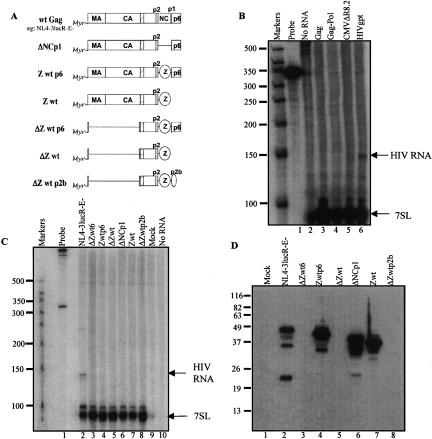
Previously described non-infectious virus-like particles that lack specific HIV components were used to address which viral determinants were required for 7SL incorporation (Fig. 2A ▶ and not shown). Because SRP is required for glycoprotein synthesis, and Env is the only HIV-1 glycoprotein, 7SL encapsidation by Env-defective particles was examined by a nuclease protection assay (Fig. 2B ▶, lane 6 and not shown). The results showed that particles that lacked Env still contained 7SL RNA.
FIGURE 2.
Viral requirements for 7SL packaging. (A) Schematic illustration of the utilized Gag variants. (Myr) myristate, (boxes) HIV-derived domains, (hatched box within CA) Major Homology Region, (lines) deleted regions, (ovals) heterologous protein domains: Z is a yeast-derived GCN4 zipper domain, and p2b contains Rous sarcoma virus late domain sequences (Accola et al. 2000). Schematic is based on information in (Accola et al. 2000); variants’ construction and other details are in (Accola et al. 2000). Minimal gag constructs were obtained from Dr. H. Gottlinger. (B) Env, genomic RNA, and Pol-deficient virus-like particle RNAs analyzed by a ribonuclease protection assay. All virus-like particles were produced by calcium phosphate transient transfection of 293T cells. Medium was harvested 48 h post-transfection, filtered, and concentrated by centrifugation through 20% sucrose. HIV-gpt (an HIV proviral clone with gpt in place of env) and pNL4–3.Luc.R–E– (HIV with frameshifts in env and vpr and luciferase replacing nef) were from the NIH AIDS Research and Reference Reagent Program. pCMVΔR8.2 (Naldini et al. 1996) encodes all HIV-1 proteins except Env. Rev-independent HIV Gag or Gag-pol encoding vectors (Huang et al. 2001) were supplied by Dr. Y. Huang. Ribonuclease protection was performed as in Figure 1 ▶. (C) Gag deletion variant virus-like particle RNAs analyzed by ribonuclease protection. Approaches are as above; variants’ structures are in panel A. All variants in panel B contain wild-type Gag. (D) Western blot of Gag proteins. Performed using anti-HIV-1 monoclonal anti-p24 (AS-55–11, Microbix Biosystems), a monoclonal anitibody that recognizes a CA N-terminal epitope. Expected protein sizes: ΔZwt6, 22 kD; Zwtp6, 50 kD; ΔZwt, 14 kD; ΔNCp1, 46 kD; Zwt, 44 kD; and ΔZwtp2b, 16 kD. Note that proteins missing the N-terminal half of CA are not visualized using this antibody.
To test if 7SL:genome RNA–RNA interactions or if reverse transcriptase, which contributes to tRNA primer packaging (Sawyer and Hanafusa 1979), contributed to 7SL packaging, particles that lacked genomic RNA and/or reverse transcriptase were examined (Fig. 2B ▶, lanes 3–5) and shown to still contain 7SL.
Many parts of the Gag polyprotein are dispensable for particle production (Accola et al. 2000). Thus, 7SL packaging was examined for deletion derivatives that included only minimal Gag assembly determinants (Fig. 2A ▶). 7SL in these virus-like particles was assayed by nuclease protection (Fig. 2C ▶), and Gag proteins were visualized by Western blotting (Fig. 2D ▶). Even gross deletion derivatives missing almost all of Matrix and undetectable with anti-CA antibody because they lacked portions of Capsid (CA) (Fig. 2D ▶, lanes 3,5,8) still released pelletable 7SL RNA (Fig. 2C ▶, lanes 3,5,8).
Because HIV nucleocapsid protein, NC, forms specific protein–RNA interactions necessary for genomic RNA encapsidation (Berkowitz et al. 1996; Zhang and Barklis 1997), 7SL packaging by NC deletion or replacement variants (Accola et al. 2000) was also examined (Fig. 2C ▶, lanes 4,6,7). Neither these nor Gag late domain mutants (Fig. 2C ▶, lanes 5,7,8) displayed reduced 7SL packaging. None of the tested Gag deletion derivatives separated the 7SL packaging phenotype from the ability to produce virus particles. Because 7SL encapsidation appears to be broadly conserved across a wide range of retroviral genera (this work and Bishop et al. 1970; Faras et al. 1973; Walker et al. 1974), 7SL is not secreted by uninfected cells (Fig. 1A ▶ and not shown), and 7SL release revealed ongoing particle production even for HIV-1 derivatives that could not be assayed by RT activity or with commercially available anti-CA antibodies (Fig. 2C,D ▶; Accola et al. 2000), the presence of extracellular 7SL may, in some instances, serve as a useful surrogate marker for retroviral particle production.
7SL is an abundant cytoplasmic RNA and thus could be passively incorporated into HIV. Alternatively, 7SL could be selectively recruited. To monitor recruitment, the amount of 7SL in HIV particles was compared with that of actin mRNA. Low levels of mRNAs have been detected in retroviruses (Adkins and Hunter 1981), but like many of the host proteins observed in virions (Cantin et al. 2005), their encapsidation appears non-selective. If actin mRNA, which served here as a surrogate for randomly encapsidated RNA, and 7SL were both incorporated non-selectively, their ratios should be the same within cells and in virions.
To address 7SL enrichment, RNA was extracted from cells or virus, and actin and 7SL were quantified by real-time PCR. The results demonstrated that the amount of 7SL RNA was twofold higher in HIV-infected than uninfected cells, while the amount of actin mRNA decreased twofold in HIV-infected cells, resulting in a fourfold increase in the intracellular ratio of 7SL to actin mRNA upon HIV infection (Table 1 ▶). The observation that HIV-1 infection leads to a fourfold increase in pol III-driven 7SL RNA expression compared with actin mRNA levels may be consistent with findings for several other viruses that are known to affect pol III-driven gene expression (Panning and Smiley 1995; Russanova et al. 1995; Gottesfeld et al. 1996; Piras et al. 1996).
TABLE 1.
Real-time reverse transcriptase-PCR quantification of virus and cell RNAsa
| RNA source | 7SLb | Actinb | 7SL/actinc |
| CEM-SS cells | 5.2 ± 0.2 × 107 | 7.2 ± 0.1 × 106 | 7.2 ± 0.9 × 103 |
| Infected CEM-SS cells | 1.3 ± 0.1 × 108 | 4.2 ± 0.9 × 106 | 3.1 ± 0.7 × 101 |
| HIV-1BRU virions | 6.2 ± 0.3 × 107 | 7.8 ± 1.9 × 102 | 8.0 ± 2.0 × 103 |
aDNA was removed from samples using DNase I (Ambion). First-strand cDNA was synthesized using random primers and SuperScript II (Invitrogen). After RNaseH treatment, samples were diluted in herring DNA and PCR was performed by TaqMan using an ABI 7700 system (Perkin Elmer). Primers for 7SL: 5′-GGGCTGTAGTGCGCTATGC-3′ and 5′-CCCGGGAGGT CACCATATT-3′; probe: 6FAM-CGGGTGTCCGCACTAAGTTCGGCTAMRA. Primers for beta-actin: 5′-TCACCCACACTGTGCCCATCTACGA-3′, and 5′-CAGCGGAACCGCTCATT GCCAATGG-3′; probe: 6FAM-ATGCCCTCCCCCATGCCATCCTGCGT-TAMRA. Standard curves were generated using dilutions of plasmid target. Calculations were based on the linear response of the threshold cycle when PCR products were first detected versus log input template copy number. Linear response ranged from 10 to 106 template copies, achieving correlation coefficients of >0.99.
bAmounts of 7SL or actin sequences present in dilutions of cDNAs produced from cellular or viral RNA by the addition of random primers plus reverse transcriptase, as determined by quantitative PCR. Standard errors are indicated with ±.
cMolar ratios of 7SL to actin were calculated from presented 7SL and actin data. Values from controls in which reverse transcriptase was not included were subtracted from presented values; these -RT values were 12 ± 5.0 template copies for CEM-SS cells’ 7SL, 6.7 for CEM-SS’ actin, 160 ± 21 for infected cells’ 7SL, 24 ± 3.5 for infected cells’ actin, 1.5 ± 0.3 for virions’ 7SL, and 9.1 ± 4.1 for virions’ actin.
Comparing the 7SL:actin mRNA ratio in virions to the same ratio in infected cells (Table 1 ▶, 7SL/actin column) suggested a 250-fold greater relative abundance of 7SL RNA within virions than actin mRNA. These data are consistent with the selective recruitment of 7SL into HIV particles and/or the active exclusion of actin mRNA.
In summary, this study demonstrated that the incorporation of host 7SL/SRP RNA, at a copy number that exceeds that of viral genomic RNA, is a conserved property of retro-viruses. The determinants of 7SL RNA packaging were shown to differ from those for viral genomic RNA and for primer tRNA. Furthermore, although most 7SL RNA exists intracellularly in signal recognition particles comprised of one molecule each of 7SL RNA and six different SRP proteins (Walter and Blobel 1983; Doudna and Batey 2004), the largest of these proteins, SRP54, was absent from virions, demonstrating that HIV-associated 7SL RNA is not encapsidated in its predominant intracellular form.
Acknowledgments
We thank Heinrich Gottlinger for providing minimal gag expression constructs, Jennifer Masterson for technical assistance, support from NIH #GM64479 to A.T., and an American Heart Association pre-doctoral fellowship to A.O.-N.
Article published ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2306306.
REFERENCES
- Accola, M.A., Strack, B., and Gottlinger, H.G. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74: 5395–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins, B. and Hunter, T. 1981. Identification of a packaged cellular mRNA in virions of rous sarcoma virus. J. Virol. 39: 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz, R., Fisher, J., and Goff, S.P. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214: 177–218. [DOI] [PubMed] [Google Scholar]
- Bishop, J.M., Levinson, W.E., Sullivan, D., Fanshier, L., Quintrell, N., and Jackson, J. 1970. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology 42: 927–937. [DOI] [PubMed] [Google Scholar]
- Campbell, S. and Vogt, V.M. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69: 6487–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin, R., Methot, S., and Tremblay, M.J. 2005. Plunder and stowaways: Incorporation of cellular proteins by enveloped viruses. J. Virol. 79: 6577–6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheevers, W.P., Archer, B.G., and Crawford, T.B. 1977. Characterization of RNA from equine infectious anemia virus. J. Virol. 24: 489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli, A., Sandin, S., Hoglund, S., and Luban, J. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74: 3046–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J.A. and Batey, R.T. 2004. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 73: 539–557. [DOI] [PubMed] [Google Scholar]
- Erikson, R.L. 1969. Studies on the RNA from avian myeloblastosis virus. Virology 37: 124–131. [DOI] [PubMed] [Google Scholar]
- Erikson, E., Erikson, R.L., Henry, B., and Pace, N.R. 1973. Comparison of oligonucleotides produced by RNase T1 digestion of 7 S RNA from avian and murine oncornaviruses and from uninfected cells. Virology 53: 40–46. [DOI] [PubMed] [Google Scholar]
- Faras, A.J., Garapin, A.C., Levinson, W.E., Bishop, J.M., and Goodman, H.M. 1973. Characterization of the low-molecular-weight RNAs associated with the 70S RNA of Rous sarcoma virus. J. Virol. 12: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld, J.M., Johnson, D.L., and Nyborg, J.K. 1996. Transcriptional activation of RNA polymerase III-dependent genes by the human T-cell leukemia virus type 1 tax protein. Mol. Cell. Biol. 16: 1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Kong, W.P., and Nabel, G.J. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75: 4947–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, J.G. and Seidman, J.G. 1979. Selective packaging of host tRNAs by murine leukemia virus particles does not require genomic RNA. J. Virol. 29: 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux, D., Mirro, J., Harvin, D., and Rein, A. 2001. RNA is a structural element in retrovirus particles. Proc. Nat. Acad. Sci. 98: 5246–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini, L., Blomer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F.H., Verma, I.M., and Trono, D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272: 263–267. [DOI] [PubMed] [Google Scholar]
- Onafuwa-Nuga, A.A., King, S.R., and Telesnitsky, A. 2005. Nonrandom packaging of host RNAs in moloney murine leukemia virus. J. Virol. 79: 13528–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panning, B. and Smiley, J.R. 1995. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J. Mol. Biol. 248: 513–524. [DOI] [PubMed] [Google Scholar]
- Piras, G., Dittmer, J., Radonovich, M.F., and Brady, J.N. 1996. Human T-cell leukemia virus type I Tax protein transactivates RNA polymerase III promoter in vitro and in vivo. J. Biol. Chem. 271: 20501–20506. [DOI] [PubMed] [Google Scholar]
- Russanova, V.R., Driscoll, C.T., and Howard, B.H. 1995. Adenovirus type 2 preferentially stimulates polymerase III transcription of Alu elements by relieving repression: A potential role for chromatin. Mol. Cell. Biol. 15: 4282–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, R.C. and Dahlberg, J.E. 1973. Small RNAs of Rous sarcoma virus: Characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J. Virol. 12: 1226–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, R.C. and Hanafusa, H. 1979. Comparison of the small RNAs of polymerase-deficient and polymerase-positive Rous sarcoma virus and another species of avian retrovirus. J. Virol. 29: 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, T.A., Pace, N.R., Erikson, R.L., Erikson, E., and Behr, F. 1974. The 7S RNA common to oncornaviruses and normal cells is associated with polyribosomes. Proc. Nat. Acad. Sci. 71: 3390–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P. and Blobel, G. 1982. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 299: 691–698. [DOI] [PubMed] [Google Scholar]
- ———. 1983. Subcellular distribution of signal recognition particle and 7SL–RNA determined with polypeptide-specific antibodies and complementary DNA probe. J. Cell Biol. 97: 1693–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.W. and Aldovini, A. 2002. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 76: 11853–11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. and Barklis, E. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71: 6765–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]